Abstract
Rhythmic activation and repression of clock gene expression is essential for the eukaryotic circadian clock functions. In the Neurospora circadian oscillator, the transcription of the frequency (frq) gene is periodically activated by the White Collar (WC) complex and suppressed by the FRQ-FRH complex. We previously showed that there is WC-independent frq transcription and its repression is required for circadian gene expression. How WC-independent frq transcription is regulated is not known. We show here that elevated protein kinase A (PKA) activity results in WC-independent frq transcription and the loss of clock function. We identified RCM-1 as the protein partner of RCO-1 and an essential component of the clock through its role in suppressing WC-independent frq transcription. RCM-1 is a phosphoprotein and is a substrate of PKA in vivo and in vitro. Mutation of the PKA-dependent phosphorylation sites on RCM-1 results in WC-independent transcription of frq and impaired clock function. Furthermore, we showed that RCM-1 is associated with the chromatin at the frq locus, a process that is inhibited by PKA. Together, our results demonstrate that PKA regulates frq transcription by inhibiting RCM-1 activity through RCM-1 phosphorylation.
INTRODUCTION
Circadian clocks allow prokaryotic and eukaryotic organisms to regulate their daily molecular, cellular, behavioral, and physiological activities. The eukaryotic circadian clocks are composed of autoregulatory negative-feedback loops, including positive and negative elements that form the core circadian oscillators (1–6). Rhythmic transcriptional activation of the negative elements by the positive elements and rhythmic repression of positive elements by the negative elements are thought to be the main molecular basis for the generation of endogenous rhythmicity in eukaryotic clock systems. In addition to regulation at the transcriptional level, posttranslational modification of clock proteins by phosphorylation plays essential roles in clock functions (7–10).
In the filamentous fungus Neurospora crassa, the core circadian negative feedback loop is composed of the positive elements, White Collar 1 (WC-1) and White Collar 2 (WC-2), and the negative elements, FREQUENCY (FRQ) and its partner FRQ-interacting RNA helicase (FRH) (2, 3, 11, 12). WC-1 and WC-2, two Per-Arnt-Sim (PAS) domain-containing transcription factors, form the WC heterodimeric complex that binds to the Clock (C)-box of frq promoter to activate frq transcription (13–18). On the other hand, FRQ and FRH form the FFC complex to inhibit frq transcription by suppressing the activity of the WC complex by promoting WC phosphorylation (19–24). FRQ is progressively phosphorylated over time before its degradation through the ubiquitin–proteasome pathway mediated by FWD-1 (21, 25–28). Genetic analyses show that the FRQ-dependent phosphorylation of WC-1 and WC-2 that suppresses White Collar complex (WCC) activity is mostly mediated by CKI and CKII (21).
WC complex was long thought to be the only transcriptional activator of frq transcription. However, we recently discovered that frq transcription can occur in a WC-independent manner, and the suppression of the WC-independent frq transcription by RCO-1 is essential for circadian clock function (29). The homolog of RCO-1 in Saccharomyces cerevisiae is Tup1, a component of the Tup1-Ssn6 complex that represses gene expression (30, 31). Even though our previous results suggest that RCO-1 regulates the chromatin status at the frq locus, RCO-1 does not appear to be associated with the frq locus. Therefore, the mechanism of how RCO-1 represses WC-independent frq transcription and how it affects the chromatin status are not clear.
The cyclic AMP (cAMP)-dependent protein kinase A (PKA) was previously shown to act as a FRQ-independent WC kinase and serve as a priming kinase for the casein kinase-dependent WC phosphorylation, a process that inhibits WC complex activity and stabilize WC proteins (32). However, this interpretation cannot fully explain the molecular phenotype of the mcb mutant, in which PKA activity is high as a result of the mutated PKA regulatory subunit. Despite the dramatically reduced WC complex binding activity at the frq promoter in the mcb mutant, FRQ expression levels are at an intermediate level in constant darkness (DD) (32). These results suggest that PKA may have another role in regulating FRQ expression.
We show here that elevated PKA activity in Neurospora induces WC-independent frq transcription. Protein purification of RCO-1 identified RCM-1/Ssn6p as the protein partner of RCO-1 that is essential for clock function and is also required for suppressing WC-independent frq transcription. In addition, we demonstrated that RCM-1 is a substrate of PKA and the PKA-dependent phosphorylation of RCM-1 regulates the transcription repression activity of RCM-1 on frq expression. Furthermore, RCM-1 is associated with DNA at the frq locus, suggesting a mechanism for how the RCM-1/RCO-1 complex functions to suppress frq transcription.
MATERIALS AND METHODS
Strains and culture conditions.
The 87-3 (bd, a) and 301-6 (bd, his-3, A) strains were used as the wild-type strains in the present study. The bd ku70RIP strain generated previously (21) was used as the host strain for creating the pkac-1 or pde-2 knockout mutant. The bd pkac-1KO or pde-2KO strain was created by deleting the entire PKAC-1 or PDE-2 open reading frame (ORF) through homologous recombination using a protocol described previously (33). An mcb strain (FGSC 7094) was obtained from the Fungal Genetic Stock Center and crossed with a bd strain to create the bd mcb strain. The rcm-1RIP strains were created by the repeat-induced point mutation (RIP) method (34). A positive transformant (bd, A) with a plasmid containing qa-2 promoter driven the RCM-1 ORF and its 3′ untranslated region (3′-UTR) at its his-3 locus was crossed with a wild-type strain (bd, a) to get the rcm-1RIP strain. The rco-1KO, wc-1KO, wc-2KO, and frq9 strains generated previously (14, 18, 29, 35) were also used in the present study. The newly created mcb wc-1KO, mcb wc-2KO, pde-2KO wc-2KO, rcm-15E wc-2KO, rcm-1RIP wc-1KO, rcm-1RIP wc-2KO, rcm-1RIP frq9, and rco-1KO frq9 double mutants were obtained by crossing.
The medium for race tube assays contained 1× Vogel's salts, 0.1% glucose, 0.17% arginine, 50 ng of biotin/ml, and 1.5% agar with or without quinic acid (QA). Liquid culture conditions were as described previously (36, 37).
Luciferase reporter assay.
The luciferase reporter assay was performed as reported previously (29, 38, 39). The 87-3 (bd, a), frq-luc strain was used as the control strain. The rcm-1RIP strain was crossed with the 87-3 (bd, a), frq-luc strain to obtain the rcm-1RIP, frq-luc strain.
Creation of the rcm-1 knock-in strains.
To create the rcm-1KI-WT strain, a wild-type knock-in cassette with a hygromycin resistance gene (hph) was inserted into downstream of the rcm-1 3′-UTR (see Fig. 7A). The cassette was then transformed into the bd ku70RIP strain to select for hph-resistant transformants. The homokaryotic strains were obtained by microconidial purification and confirmed by DNA sequencing (21). Using this method, we created four rcm-1 knock-in strains at the endogenous locus by homologous recombination: the rcm-1KI-WT, rcm-12A, rcm-15A, and rcm-15E strains. In the rcm-12A strain, S718 and S733 were mutated to alanine residues, whereas five identified RCM-1 phosphorylation sites (S718, S733, S630, S682, and T536) were mutated to alanine residues in the rcm-15A strain or to glutamic acids to mimic phosphorylation at these residues in the rcm-15E strain.
FIG 7.
Phosphorylation of RCM-1 results in WC-independent frq transcription. (A) RCM-1 contains 10 TPR (tetratricopeptide repeat) motifs in the N-terminal region and different phosphorylation sites in the C-terminal region. The asterisks in the RCM-1 ORF indicate the location of the mutation sites. The endogenous rcm-1 gene was replaced with a knock-in cassette containing the mutated rcm-1 gene by homologous recombination and hph-resistant transformants were selected. (B) Coimmunoprecipitation (Co-IP) experiments showing that RCM-1 interacts with RCO-1 and the interactions are not affected in the rcm-1 knock-in strains. (C) Race tube assays showing the circadian conidiation rhythm in wild-type, rcm-1KI-WT, rcm-15E, and rcm-15A strains. (D) Western blot analysis of FRQ protein in wild-type, rcm-1KI-WT and rcm-15E strains grown in the dark for the indicated times. The asterisks indicate nonspecific bands. (E) Northern blot analysis of ccg-1 mRNA in wild-type, rcm-1KI-WT, and rcm-15E strains. (F) Western blot analysis of FRQ in wild-type, wc-2KO, and rcm-15E wc-2KO strains. The asterisks indicate nonspecific bands. (G) Northern blot analysis of frq mRNA in wild-type, wc-2KO, and rcm-15E wc-2KO strains.
Generation of antiserum against PKAC-1 or RCM-1.
The glutathione S-transferase (GST)/PKAC-1 fusion protein (containing PKAC-1 amino acids 1 to 318) or GST/RCM-1 (containing RCM-1 amino acids 480 to 883) was expressed in BL21 cells, and the recombinant proteins were purified and used as the antigens to generate rabbit polyclonal antisera as described previously (36, 37). RCO-1 antiserum was generated previously (29).
Protein and RNA analyses.
Protein extraction, quantification, Western blot analysis, and immunoprecipitation assays were performed as previously described (40, 41). Nuclear and cytoplasmic protein extracts were prepared as previously described (23). For Western blot analyses, equal amounts of total protein (40 μg) were loaded in each protein lane. After electrophoresis, proteins were transferred onto polyvinylidene difluoride membrane, and Western blot analysis was performed. To monitor PKA phosphorylation in vivo, substrate proteins were precipitated under denaturing conditions, with protease and phosphatase inhibitors present at all steps. The level of PKA phosphorylation was then assessed by Western blotting with an anti-PKA substrate antibody (Cell Signaling, catalog no. 9621). RNA was extracted as described previously and then analyzed by Northern blotting (29). For Northern blot analysis, equal amounts of total RNA (20 μg) were loaded onto agarose gels for electrophoresis, and the gels were blotted and probed with RNA probes specific for frq or ccg-1. For Western and Northern blot analyses, each experiment was independently performed at least three times.
In vitro phosphorylation.
For the in vitro phosphorylation assay, GST/RCM-1 fusion protein were purified from Escherichia coli BL21 cells and incubated with bovine PKA (Promega) or partially purified Myc/His/PKAC-1 from Neurospora. Standard reaction mixtures (total 30 μl) were set up containing 5 μg of substrate and 0.5 μg of kinase in the reaction buffer containing 20 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 25 μM ATP, and 3 μCi of [γ-32P]ATP for 30 min at 30°C with or without cAMP (1 μM). The proteins were separated by SDS-PAGE, and the level of phosphorylation was assessed by autoradiography. Each experiment was independently performed at least three times.
Purification of RCM-1 from N. crassa, followed by MS/MS.
RCM-1 was immunoprecipitated using RCM-1 antibody from wild-type strain in the presence of protease and phosphatase inhibitors in the extraction buffer. The precipitates were analyzed by SDS-PAGE (4 to 20%), which was subsequently stained with colloidal blue. The colloidal blue-stained RCM-1 protein bands were excised from SDS gels and subjected to in-gel digestion with trypsin. The phosphopeptide sequences and sites of phosphorylation were identified by nano-electrospray tandem mass spectrometry (MS/MS).
ChIP analyses.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously, and 1 ml of protein (2 mg/ml) was used for each immunoprecipitation reaction (29). The ChIPs were carried out with 2 μl of WC-2 antibody or RCM-1 antibody. Immunoprecipitated DNA was quantified using real-time PCR. The primer sets used are listed in a previously published study (29). Occupancies were normalized to a sample of input DNA (10 μl), and data are presented as the percentage of input DNA. Each experiment was independently performed at least three times.
RESULTS
MCB, the regulatory subunit of PKA, is required for suppressing WC-independent frq transcription.
To identify new components that are involved in the regulation of the WC-independent frq transcription, we generated double mutants by crossing the wcKO single mutants with some of the known Neurospora clock mutants. mcb encodes the regulatory subunit of PKA that inhibits PKA activity. A previously isolated mutant, mcb, had high PKA activity due to a small deletion in the mcb regulatory region that results in low mcb expression levels (32). In this mcb mutant, the circadian oscillations of FRQ protein levels and FRQ phosphorylation patterns were completely abolished and WC binding to the frq promoter in DD was dramatically reduced, an observation consistent with a role for PKA in phosphorylating WCC to inhibit its activity. However, the levels of FRQ expression were constant at an intermediate level in the mcb strain (32), suggesting another unknown role of PKA in regulating frq expression. To test this hypothesis, we created mcb wc-1KO and mcb wc-2KO double mutants and determined the expression levels of frq and FRQ in these strains that were cultured in constant light (LL). Western blot analyses showed that no WC-1 or WC-2 signals and decreased levels of MCB proteins were detected in the mcb wc-1KO or mcb wc-2KO double mutants, respectively, confirming their genetic identity (Fig. 1A). In contrast to the almost undetectable FRQ expression levels in the wc-1KO or wc-2KO single mutant, the mcb wcKO double mutants expressed significant amounts of FRQ protein. For cultures grown in DD (Fig. 1B), constant FRQ expression at an intermediate levels were observed at different time points in the mcb wcKO double mutants. Similar results were also seen in the Northern blot analyses, which showed intermediate levels of frq mRNA in the mcb wcKO double mutants in DD that are comparable to those in the mcb single mutant in DD (Fig. 1C and D), suggesting that FRQ expression in the mcb single mutant is mostly WC independent in DD. Together, these results demonstrate that MCB is required for the suppression of WC-independent frq transcription.
FIG 1.
MCB is required for suppressing WC-independent frq transcription. (A) Western blot analyses were performed with antibodies against FRQ, WC-1, WC-2, or MCB in wild-type, mcb, wc-1KO, wc-2KO, mcb wc-1KO, and mcb wc-2KO strains. Three different mcb wc-1KO and mcb wc-2KO strains were used. (B) Western blot analysis of FRQ in wild-type, mcb, wc-1KO, wc-2KO, mcb wc-1KO, and mcb wc-2KO strains. Asterisks indicate nonspecific bands. (C) Northern blot analysis of frq mRNA in wild-type, mcb, wc-1KO, wc-2KO, mcb wc-1KO, and mcb wc-2KO strains in LL and DD8. (D) Northern blot analysis of frq mRNA in wild-type, mcb, wc-1KO, wc-2KO, mcb wc-1KO, and mcb wc-2KO strains at different time points in DD.
Elevated PKA activity results in WC-independent frq transcription.
In Neurospora, pkac-1 (NCU00682) and pkac-2 (NCU06240) encode the PKA catalytic subunits, and PKAC-1 is the major catalytic subunit (32, 42). To determine whether the WC-independent frq transcription in mcb mutant is due to the increase of PKA activity but not due to a role of MCB in another process, we introduced a QA-inducible Myc-His-tagged PKAC-1 construct into the wild type or a pkac-1KO, wc-1KO, or wc-2KO single mutant strain. Race tube assay showed that the induction of Myc/His/PKAC-1 expression in the pkac-1KO strain by QA (10−2 M) was able to rescue the growth and circadian conidiation defects of the pkac-1KO strain (Fig. 2A), indicating that Myc-His-tagged PKAC-1 was functional. Interestingly, race tube assay showed that the induction of Myc/His/PKAC-1 expression by QA (10−2 M) in the wild-type strain significantly reduces the conidiation rhythm (Fig. 2B), a phenotype that is similar to but not as severe as the mcb mutant (Fig. 2E). As shown in Fig. 2C and D, the induction of Myc-His-PKAC-1 in the wc-1KO or wc-2KO strain results in FRQ and frq expression in the wc-1KO or wc-2KO strain in LL and DD. It should be noted that the qa-2 promoter is not a strong promoter that can only express Myc/His/PKAC-1 to a level that is similar to the endogenous PKAC-1 in these wcKO mutants (Fig. 2C). This may explain the modest levels of frq and FRQ expression in the wcKO mutants after induction.
FIG 2.
Elevated PKA activity results in WC-independent frq transcription. (A) Circadian conidiation rhythm of the wild-type and pkac-1KO strains and the pkac-1KO, qa-Myc-His-PKAC-1 transformants in the race tube in the presence of QA (10−2 M). (B) Race tube assays of wild-type and wild-type, qa-Myc-His-PKAC-1 strains in the presence of QA in DD. (C) Western blot analyses were performed using antibodies against FRQ or PKAC-1 in wild-type, pkac-1KO, wc-1KO, and wc-2KO strains and in wc-1KO, qa-Myc-His-PKAC-1, and wc-2KO, qa-Myc-His-PKAC-1 transformants in the presence of QA (10−2 M). (D) Northern blot analysis of frq mRNA in wild-type, wc-1KO, and wc-2KO strains and in wc-1KO, qa-Myc-His-PKAC-1, and wc-2KO, qa-Myc-His-PKAC-1 transformants in the presence of QA (10−2 M). (E) Race tube assays of wild-type, mcb, and pde-2KO strains in DD. (F) Western blot analyses were performed using antibodies against FRQ or WC-2 in wild-type, pde-2KO, wc-2KO, and pde-2KO wc-2KO strains. Three independent pde-2KO wc-2KO strains were used.
pde-1 (NCU00237) and pde-2 (NCU00478) encode cAMP phosphodiesterase in Neurospora, and PDE-2 is the major high-affinity enzyme that can inhibit PKA activity (43, 44). To further confirm our conclusion, we created a pde-2KO strain. As shown by race tube assay, the circadian conidiation rhythm was initially dampened and later entirely abolished in the pde-2KO strain, which is similar to that of the mcb mutant, suggesting that the deletion of pde-2 resulted in elevated PKA activity (Fig. 2E). To examine whether the WC-independent frq transcription in the pde-2KO strain is also impaired, we generated pde-2KO wc-2KO double mutants. As shown in Fig. 2F, nearly wild-type levels of FRQ expression were seen in the pde-2KO wc-2KO double mutants in LL. Taken together, these results demonstrate that PKA promotes WC-independent frq transcription.
RCM-1 is the partner of RCO-1 and is required for suppressing WC-independent frq transcription.
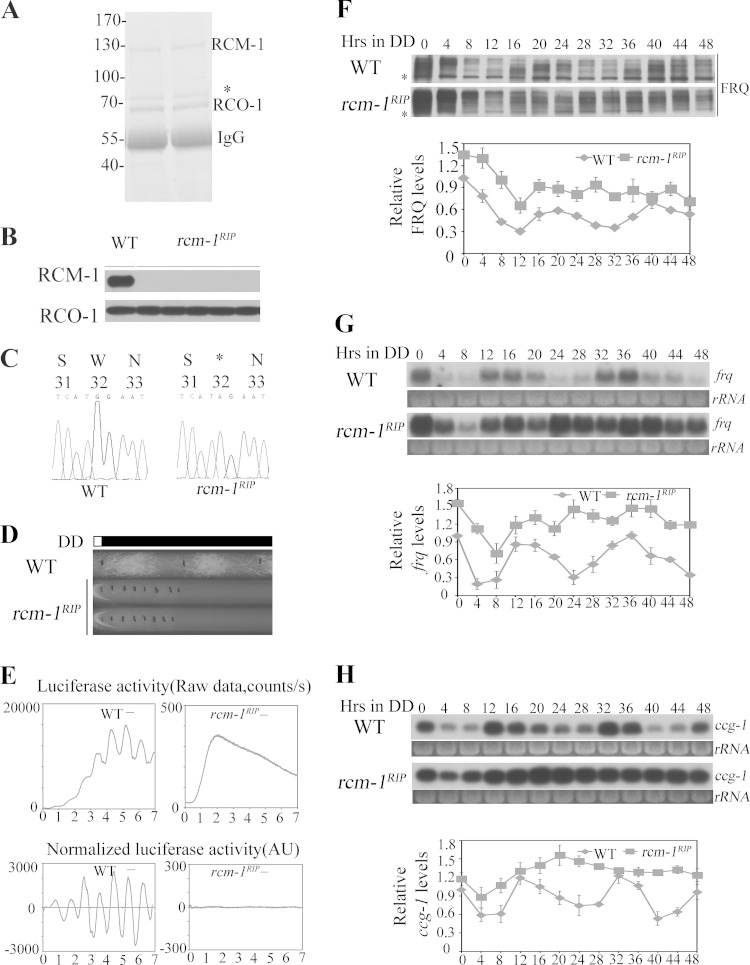
In S. cerevisiae, the homolog of RCO-1, Tup1, forms a complex with Ssn6 to repress gene expression (30, 31). To examine whether the Neurospora Ssn6 homolog RCM-1 (NCU06842) has a similar role, we generated a RCM-1 antibody and used it to purify RCM-1 from Neurospora by immunoprecipitation. As shown by colloidal blue-stained SDS-PAGE gel in Fig. 3A, RCM-1 coprecipitated with another protein, which was identified as RCO-1 by mass spectrometry analysis. These results confirm that RCO-1 and RCM-1 form a complex, as previously shown by Sancar et al. and as suggested by Olmedo et al. (45, 46).
FIG 3.
RCM-1 is required for rhythmic frq transcription. (A) The colloidal blue-stained SDS-PAGE gel showing the purification of the RCM-1/RCO-1 complex from the wild-type strain. The asterisk indicates a nonspecific band. (B) Western blot analysis showing that there is no RCM-1 signal in rcm-1RIP strains, while the levels of RCO-1 protein are not affected by mutation of rcm-1. (C) DNA sequencing showing that the rcm-1RIP strain contains many G-C to A-T point mutations in the rcm-1 allele, including premature stop codon (a replacement of a TGG codon by a TAG stop codon at nucleotide +32) in the RCM-1 ORF of one rcm-1 allele. (D) Race tube assays of wild-type and rcm-1RIP strains in DD. (E) Luciferase reporter assay showing the frq promoter activity in WT (left) and rcm-1RIP (right) strains grown in DD for several days. Raw data were normalized to subtract the baseline calculated by LumiCycle analysis software (38, 39). (F) Western blot analysis of FRQ protein in wild-type and rcm-1RIP strains grown in the dark for the indicated times. The asterisks indicate nonspecific bands. (G) Northern blot analysis of frq mRNA in wild-type and rcm-1RIP strains. (H) Northern blot analysis of ccg-1 mRNA in wild-type and rcm-1RIP strains.
To examine the role of RCM-1 in the clock, we generated rcm-1 disruption mutants by repeat-induced point mutation (RIP) (34). As shown by Western blotting in Fig. 3B, the expression of RCM-1 was abolished in the rcm-1RIP strains. DNA sequencing revealed that they contained many G-C to A-T point mutations in the rcm-1 gene, including several premature stop codons in the RCM-1 ORF (Fig. 3C). Race tube assays showed that the rcm-1RIP strains exhibited a slow-growth phenotype and lacked a conidiation rhythm (Fig. 3D), which are similar to those of the rco-1KO mutants (29). To confirm the lack of circadian rhythm at the molecular level, we introduced a frq promoter driven luciferase reporter into the rcm-1RIP strain (38). As shown in Fig. 3E, the robust circadian rhythm of luciferase activity seen in a wild-type strain was abolished in the rcm-1RIP strain.
Western blot of FRQ in DD showed that, after the initial light/dark transition, FRQ levels stayed high after DD16 and there was no obvious rhythm of FRQ phosphorylation profile (Fig. 3F). Northern blot analysis of frq and ccg-1 mRNA showed that the circadian rhythms of frq and ccg-1 mRNA were also abolished in the rcm-1RIP strain and the levels of frq mRNA were constantly high (Fig. 3G and H), a finding consistent with a role for RCM-1 in suppressing frq expression. Together, these results indicate that RCM-1, like RCO-1, is required for a functional clock.
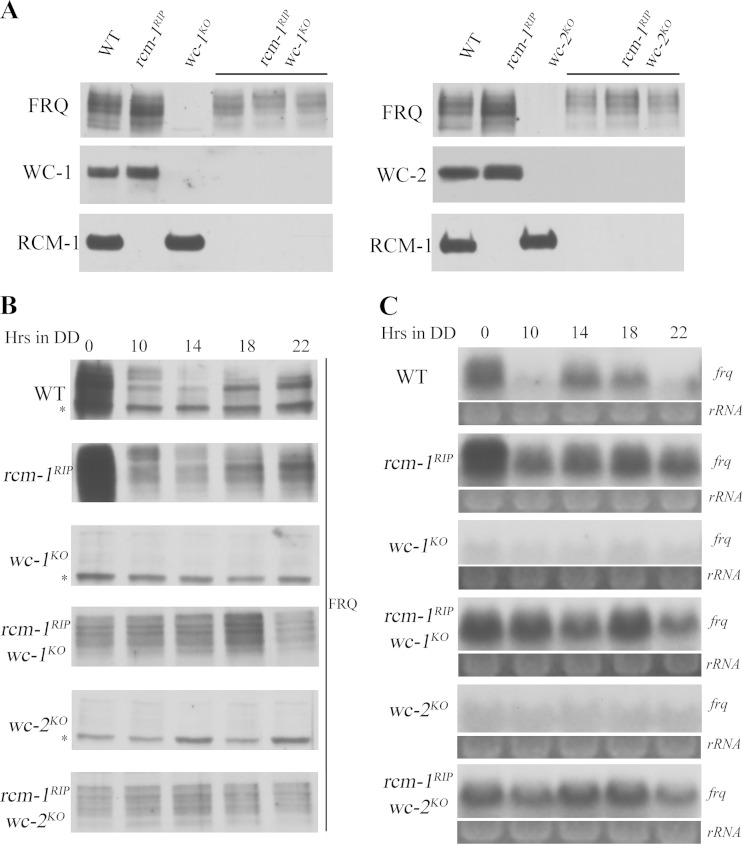
WC proteins are required to function as a complex for their clock and light responses in Neurospora (15). To examine the role of RCM-1 in suppressing WC-independent frq transcription, we generated rcm-1RIP wc-1KO or rcm-1RIP wc-2KO double mutants. Western blot analyses using the WC and RCM-1 antibodies confirmed the molecular identifies of these mutants (Fig. 4A). As expected, nearly normal levels of FRQ expression were observed in both rcm-1RIP wc-1KO and rcm-1RIP wc-2KO double mutants in LL and DD (Fig. 4B). Similarly, high levels of frq mRNA were also observed in these double mutants (Fig. 4C), indicating that RCM-1 is required for suppressing WC-independent transcription of frq.
FIG 4.
RCM-1 is required for suppressing WC-independent frq transcription. (A) Western blot analyses were performed with antibodies against FRQ, WC-1, WC-2, or RCM-1 in wild-type, rcm-1RIP, wc-1KO, wc-2KO, rcm-1RIP wc-1KO, and rcm-1RIP wc-2KO strains. (B) Western blot analysis of FRQ protein in wild-type, rcm-1RIP, wc-1KO, wc-2KO, rcm-1RIP wc-1KO, and rcm-1RIP wc-2KO strains. The asterisks indicate nonspecific bands. (C) Northern blot analysis of frq mRNA in wild-type, rcm-1RIP, wc-1KO, wc-2KO, rcm-1RIP wc-1KO, and rcm-1RIP wc-2KO strains.
RCM-1 is a substrate of PKA.
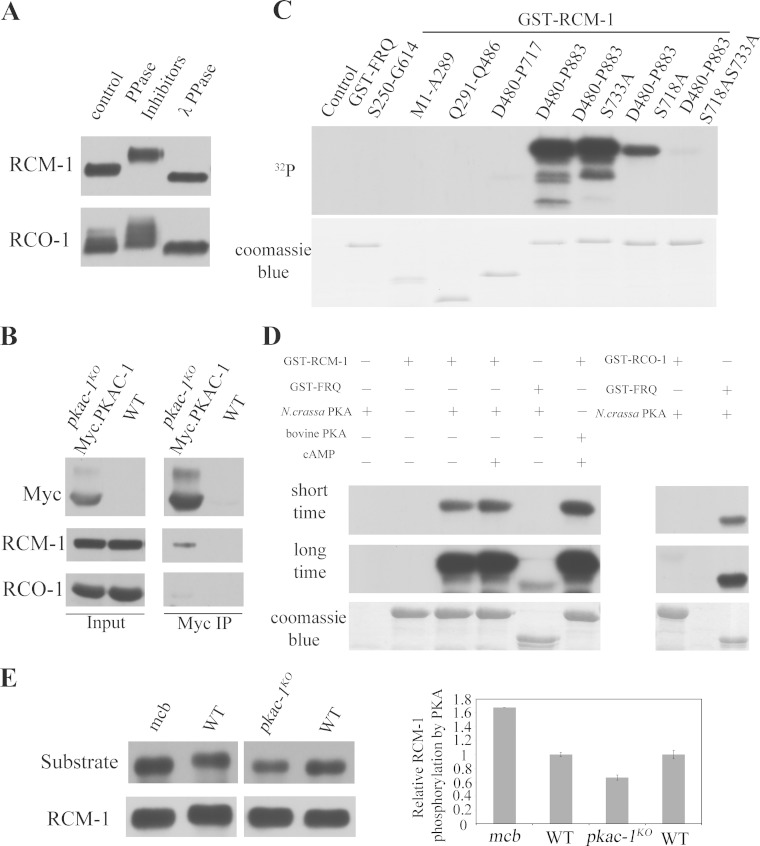
The regulation of WC-independent transcription of frq by PKA activity prompted us to examine whether PKA can exert its effect by phosphorylating the RCO-1/RCM-1 complex. Consistent with this hypothesis, we found that both RCM-1 and RCO-1 are phosphoproteins in Neurospora as indicated by protein mobility shifts in the presence of phosphatase inhibitors and after lambda phosphatase treatment (Fig. 5A). To examine whether RCO-1 and RCM-1 proteins are substrates of PKA, we performed immunoprecipitation assays to check whether PKAC-1 interacts with RCO-1 or RCM-1 in Neurospora. As shown in Fig. 5B, Myc/His/PKAC-1 expressed in the pkac-1KO strain coprecipitated with RCM-1, whereas it only interacted weakly with RCO-1, suggesting that RCM-1 but not RCO-1 interacts with PKAC-1. It is possible that PKAC-1 prefers to interact with the monomeric RCM-1 or that the the interaction between PKAC-1 and RCO-1 is transient.
FIG 5.
RCM-1 is a substrate of PKA. (A) Western blot analyses showing the phosphorylation profiles of RCM-1 and RCO-1 in the presence of phosphatase inhibitors and after treatment with λ phosphatase. (B) Immunoprecipitation (IP) assay showing that RCM-1 interacts with Myc/PKAC-1. The Myc-tagged PKAC-1 was immunoprecipitated from Neurospora extracts, and the relative levels of the associated RCM-1 and RCO-1 were assessed by Western blotting using their specific antibodies. (C) In vitro kinase assay showing that purified Neurospora PKA phosphorylates RCM-1 at both S718 and S733. Different portions of RCM-1 were fused to GST and purified from E. coli BL21 cells and GST-FRQ (S250-G614) was used as a positive control. These proteins were incubated with Neurospora PKAC-1 and [γ-32P]ATP for 30 min at 30°C. The reaction products were then separated using SDS-PAGE, and the level of PKA phosphorylation was assessed by autoradiography. (D) In vitro kinase assay showing that the C-terminal fragment of RCM-1 (D480-P883) is phosphorylated by either the bovine or Neurospora PKA, and this phosphorylation is increased after the addition of cAMP. Full-length RCO-1 is phosphorylated weakly by the Neurospora PKA compared to those of the positive control GST-FRQ (S250-G614). (E) Western blot analyses showing hyperphosphorylation of RCM-1 in the mcb strain with elevated PKA activity and hypophosphorylation of RCM-1 in the pkac-1KO strain with decreased PKA activity.
We then examined whether RCM-1 can be phosphorylated by PKA in vitro. A series of GST/RCM-1 recombinant proteins were purified from E. coli BL21 cells and were used in in vitro kinase assays (Fig. 5C). We found that the C-terminal region of RCM-1 could be efficiently phosphorylated by both the bovine PKA and the Neurospora PKAC-1 (Fig. 5C and D). In addition, RCO-1 was only weakly phosphorylated by the Neurospora PKA (Fig. 5D) compared to RCM-1 or FRQ, suggesting that RCO-1 is not an efficient substrate for PKA.
To confirm that PKA is involved in phosphorylating RCM-1 in vivo, we took advantage of a commercially available antibody (Cell Signaling Technologies) that specifically recognizes the phosphorylated forms of PKA target sites. This antibody recognizes phosphorylated serine or threonine residues that are preceded by an arginine residue at the −3 position. It is a useful tool in identifying substrates of AGC family kinases, including PKA and PKC. We immunoprecipitated RCM-1 proteins using RCM-1 antibody and then examined whether the RCM-1 proteins could be recognized by the PKA substrate-specific antibody in Western blot analysis. As shown in Fig. 5E, the immunoprecipitated wild-type RCM-1 was indeed recognized by this PKA-substrate specific antibody. More importantly, the level of the PKA phosphorylated RCM-1 decreased in the pkac-1KO mutant (note that PKAC-1 is one of the two PKA catalytic subunits in Neurospora and other AGC family kinases may also phosphorylate those sites) but increased in the mcb mutant. Together, these results strongly suggest that RCM-1 is directly phosphorylated by PKA.
Identification of RCM-1 phosphorylation sites in vivo.
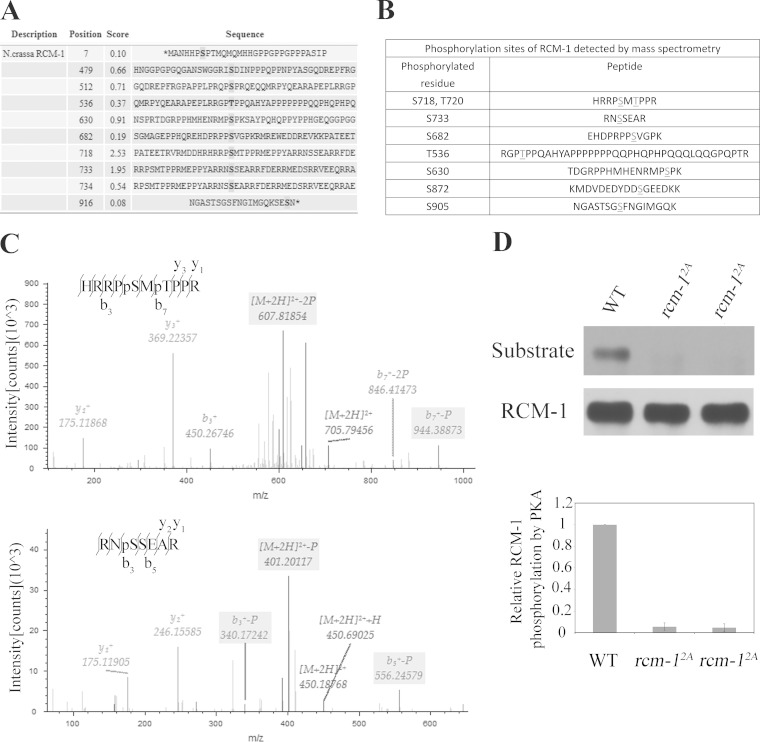
Bioinformatics analysis (http://mendel.imp.ac.at/sat/pkaPS) of RCM-1 protein revealed two PKA consensus sites (S718 and S733) and other nonconsensus sites (Fig. 6A). To identify phosphorylated residues of RCM-1 in vivo, we affinity purified RCM-1 in the presence of phosphatase inhibitors from the wild-type strain grown in LL. The colloidal blue-stained protein band corresponding to RCM-1 was excised from SDS-PAGE gels and subjected to trypsin digestion. The resulting peptides were then analyzed by nano-electrospray MS/MS. Our MS analyses (sequence coverage of 63.69%) led to identification of eight RCM-1 phosphorylation sites (Fig. 6B). Figure 6C shows two representative results of the MS/MS analysis for two of the phosphopeptides of RCM-1, which carry phosphorylated serine-718 and threonine-720 or a single phosphorylated serine-733. Among the sites identified by MS analyses, Ser-718, Ser-733, Ser-630, Thr-536, and Ser-682 are also potential PKA phosphorylation sites predicted by bioinformatics analysis. In addition, the peptides containing these eight phosphorylation sites are located in C terminus of RCM-1 (Fig. 6B), an observation consistent with our in vitro kinase assay results (Fig. 5C).
FIG 6.
Identification of RCM-1 phosphorylation sites in vivo. (A) PKA phosphorylation sites on RCM-1 are predicted using a database (http://mendel.imp.ac.at/sat/pkaPS). Among them, S718 and S733 are PKA consensus sites. (B) Phosphorylation sites of RCM-1 detected by mass spectrometry. (C) Identification of RCM-1 phosphorylation sites by mass spectrometry. The mass spectrometry of the productions resulting from collision-induced dissociation of two representative phosphorylated RCM-1 peptides are shown. The upper panel shows the results of an analysis of a trypsin fragment containing S718 and T720 of RCM-1; the lower panel shows the results of an analysis of a trypsin fragment containing S733 of RCM-1. (D) RCM-1 was recognized by an anti-PKA substrate antibody in an S718- and S733-dependent manner.
To further confirm the MS results, we created rcm-12A mutants in which S718 and S733 were mutated to alanine residues and used the PKA substrate-specific antibody to examine the phosphorylation of RCM-1. As shown in Fig. 6D, the levels of RCM-1 recognized by the PKA substrate-specific antibody were almost completely abolished in the rcm-12A mutants, indicating that S718 and S733 are the major PKA sites on RCM-1. Taken together, these results indicate that PKA is a major kinase that phosphorylates RCM-1.
Phosphorylation of RCM-1 by PKA results in WC-independent frq transcription.
To test whether the observed derepression of WC-independent frq transcription by elevated PKA activity was due to phosphorylation of RCM-1 by PKA at the identified phosphorylation sites, we created rcm-1 knock-in strains at the endogenous locus by homologous recombination, i.e., the rcm-1KI-WT, rcm-15A, and rcm-15E strains (21). In the rcm-1KI-WT strain, a wild-type knock-in cassette with a hygromycin resistance gene (hph) was inserted downstream from rcm-1 3′-UTR, while five identified RCM-1 phosphorylation sites (S718, S733, S630, S682, and T536) were mutated to alanine residues in the rcm-15A or to glutamic acids to mimic phosphorylation at these residues in the rcm-15E strains (Fig. 7A). Mutation of these phosphorylation sites in the rcm-15E strain had no effect on the interaction between RCM-1 and RCO-1 (Fig. 7B), which is consistent with previous studies that TPR (tetratricopeptide repeat) domain in the N terminus of Ssn6 is required for its interaction with Tup1 in S. cerevisiae (47, 48).
Race tube assays showed that both WT and rcm-1KI-WT strains exhibit robust conidiation rhythms (Fig. 7C), whereas the rcm-15A strain exhibited a modestly short period with low-amplitude conidiation rhythms (Fig. 7C), and the rcm-15E strain exhibited a severely dampened low-amplitude conidiation rhythm (Fig. 7C). This result suggests that the proper phosphorylation of RCM-1 at these sites is important for its role in the Neurospora circadian clock. We then investigated the circadian rhythms of these strains at the molecular levels. As shown in Fig. 7D, while robust oscillations of FRQ amounts and its phosphorylation profiles in DD were seen in the wild-type or rcm-1KI-WT strain, such oscillations were damped in the rcm-15E strain. Moreover, the circadian rhythm of ccg-1 mRNA was also severely damped in the rcm-15E strain (Fig. 7E). It should be noted that the circadian rhythms are not completely abolished in the rcm-15E strain, suggesting the existence of other functional phosphorylation sites. Taken together, these results suggest that these phosphorylation events on RCM-1 negatively regulate the function of RCM-1 in the Neurospora circadian clock.
To examine whether mimicking phosphorylation in the rcm-15E strain leads to WC-independent frq transcription, we created rcm-15E wc-2KO double mutants and compared the expression of FRQ to those in the wild type and the wc-2KO single mutant. As expected, significant levels of FRQ expression were observed in LL and DD in the rcm-15E wc-2KO strain (Fig. 7F). Similarly, near normal levels of frq mRNA were seen in the rcm-15E wc-2KO strain (Fig. 7G). These results suggest that the phosphorylation of these PKA sites on RCM-1 inhibits its function and leads to WC-independent frq transcription. Therefore, PKA regulates WC-independent frq transcription, at least in part, through its direct phosphorylation of RCM-1 protein.
RCM-1 associates with the frq locus, and its phosphorylation reduces its binding.
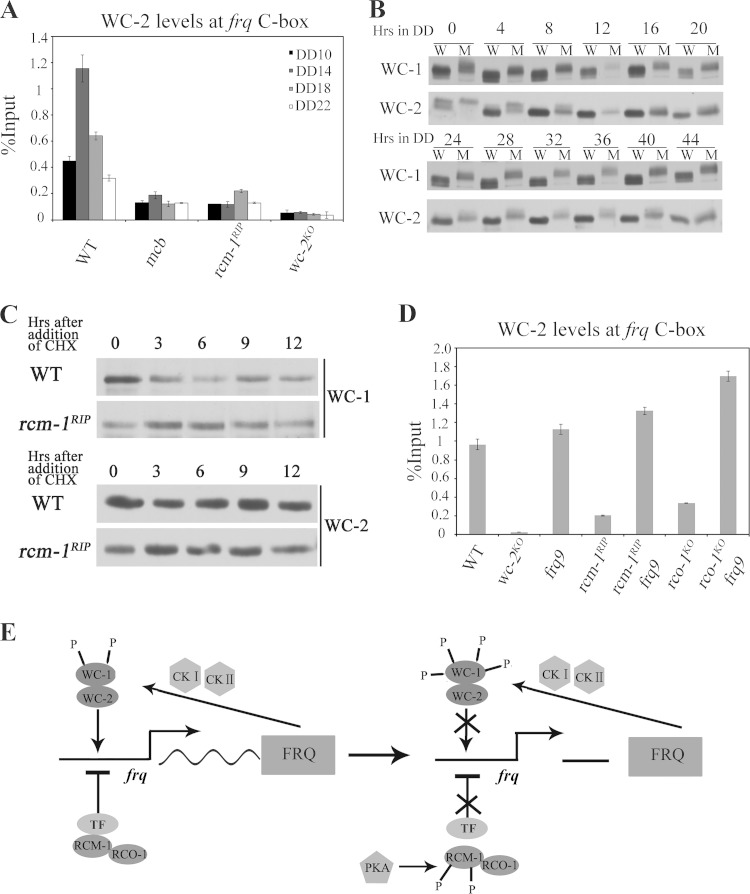
How does PKA inhibit RCM-1 activity? We previously showed that RCO-1 regulates the chromatin status at the frq locus, but how RCO-1 acts is not known since RCO-1 was not found to be associated with chromatin at the frq locus (29). The identification of RCM-1 as the partner of RCO-1 (45, 46) prompted us to examine whether the complex acts to regulate the chromatin structure at the frq locus via RCM-1. ChIP assays using the RCM-1 antibody showed that RCM-1 is enriched throughout the frq locus with a peak at the C-box of frq promoter (Fig. 8A), suggesting that the RCO-1/RCM-1 complex regulates the chromatin structure at the frq locus via the recognition of chromatin by RCM-1. Importantly, levels of RCM-1 enrichment at the frq locus were dramatically reduced in the mcb strain in different time points in DD (Fig. 8B), suggesting that elevated PKA activity reduces RCM-1 association at the frq promoter. Moreover, we examined the phosphorylation of RCM-1 in different time points using the PKA substrate-specific antibody. As shown in Fig. 8C, the levels of RCM-1 recognized by the PKA substrate-specific antibody were rhythmic at different time points in DD. Importantly, the enrichment of RCM-1 at the chromatin was low when it was hyperphosphorylated and was high when it was hypophosphorylated, suggesting that the PKA-mediated phosphorylation of RCM-1 inhibits its association at the frq promoter.
FIG 8.
The association of RCM-1 with chromatin at the frq locus is affected by PKA-mediated phosphorylation. (A) ChIP analysis showing the recruitment of RCM-1 at different regions of the frq locus in wild-type and rcm-1RIP strains at DD18. (B) ChIP analysis showing the enrichment of RCM-1 at the C-box of the frq promoter in wild-type, mcb, and rcm-1RIP strains at the indicated time points. (C) Western blot analysis showing PKA-mediated phosphorylation of RCM-1 in different time points in constant darkness (DD). (D) Western blot analysis showing that RCM-1 is stable in wild-type, mcb, and pkac-1KO strains. Protein stability was determined by measuring RCM-1 levels after the addition of CHX (10 μg/ml). (E) Western blot analysis showing the localization of RCM-1 in wild type, mcb, or pkac-1KO strains. RCM-1 is localized in the Neurospora nuclei. Total (T), nuclear (N), and cytosolic (C) protein extracts were isolated from wild type, mcb, or pkac-1KO strains, separated by SDS-PAGE, and hybridized with RCM-1-specific antibody. As controls, we used antibodies against the nuclear protein WC-1 and against cytosolic protein MCB.
Phosphorylation may also regulate protein stability and cellular localization. However, we found that the stability of RCM-1 in the mcb or pkac-1KO strain was not affected (Fig. 8D). In addition, the nuclear localization of RCM-1 was similar in wild-type and mcb strains (Fig. 8E). These results suggest that PKA does not play a significant role in regulating RCM-1 stability and cellular localization. Therefore, PKA represses RCM-1 activity mainly through inhibition of RCM-1 chromatin association activity by phosphorylation of RCM-1.
Derepression of WC-independent frq transcription results in reduced binding of WC-2 at the C-box of frq promoter.
How does the derepression of WC-independent frq transcription affect the circadian expression of frq? In wild-type strain, the circadian rhythm of frq transcription is driven by the WC complex through its rhythmic binding of the frq promoter at the C-box. ChIP assays using WC-2 antibody showed that the enrichment of WC-2 at the C-box was dramatically decreased at different time points in DD and lacked an obvious rhythm in both rcm-1RIP and mcb mutants (Fig. 9A), indicating that the transcriptional activity of WC complex is impaired. How is WC complex activity inhibited when WC-independent frq transcription is derepressed? FRQ is known to inhibit the WC complex activity by promoting its phosphorylation (20, 21, 49). Thus, we compared WC-1 and WC-2 phosphorylation profiles between wild-type and rcm-1RIP strains at different time points in DD. As shown in Fig. 9B, the phosphorylations of WC-1 and WC-2 were markedly increased in the rcm-1RIP strain at these time points. The hyperphosphorylated WC proteins in the rcm-1RIP strain suggest that constitutive WC-independent FRQ expression promotes WC phosphorylation.
FIG 9.
WC-independent frq transcription results in reduced binding of WC-2 at the C-box of frq promoter. (A) ChIP analysis showing the enrichment of WC-2 at the C-box of the frq promoter in wild-type, mcb, rcm-1RIP, and wc-2KO strains at the indicated times. (B) Western blot analyses showing the phosphorylation profiles of WC-1 and WC-2 in wild-type and rcm-1RIP strains at the indicated DD time points. W, wild type; M, rcm-1RIP mutants. (C) Western blot analyses showing that WC-1 and WC-2 are stable in wild-type and rcm-1RIP strains. Protein stability was determined by measuring WC-1 and WC-2 levels after the addition of CHX (10 μg/ml). (D) ChIP analysis showing the recruitment of WC-2 at the C-box of the frq promoter in wild-type, wc-2KO, rcm-1RIP, rco-1KO, rcm-1RIP frq9, and rco-1KO frq9 strains at DD14. (E) Model for the role of PKA in the Neurospora circadian clock by phosphorylating RCM-1. RCO-1-RCM-1 complex is recruited to the frq locus by an unknown transcription factor for suppressing WC-independent frq transcription, and disruption of rcm-1 or rco-1 results in constitutive WC-independent frq transcription. PKA directly phosphorylates RCM-1, which may disrupt the interaction between RCM-1 and the transcription repressor and relieve RCO-1/RCM-1-mediated repression of WC-independent frq transcription. The accumulation of WC-independent and WC-dependent FRQ mediates WC phosphorylation through CKI and CKII, resulting in the dissociation of WCC from frq promoter and inactivation of WC-dependent frq transcription.
WCC activity is also regulated by its stability (23). We examined WC-1 and WC-2 stability in the rcm-1RIP strain after the addition of cycloheximide (CHX) in LL and found that the stability of WC-1 and WC-2 were not affected (Fig. 9C). This result further suggested that WC-independent FRQ expression regulates WC activity mainly through phosphorylation.
To exclude the possibility that RCM-1 or RCO-1 regulates WC activity independent of FRQ, we generated rcm-1RIP frq9 and rco-1KO frq9 double mutants. In frq9 mutants, a frameshift mutation causes production of a truncated FRQ protein and the loss of circadian negative-feedback loop (35). ChIP assays with WC-2 antibody showed that even though the enrichment of WC-2 at the frq C-box was dramatically decreased in an rcm-1RIP or an rco-1KO single mutant, the WC-2 enrichment in the rcm-1RIP frq9 and rco-1KO frq9 double mutants were comparable to that of the frq9 single mutant (Fig. 9D). These results indicate that the effect of RCM-1 or RCO-1 on WC activity is dependent on FRQ. Together, our results suggest that constitutive WC-independent FRQ expression promotes WC phosphorylation, which results in reduced binding of WC-2 at the C-box of frq promoter and abolishes the rhythmic transcription of frq driven by the WC complex. Thus, the constant expression of WC-independent FRQ leads to inhibition of WC activity.
DISCUSSION
Suppression of the WC-independent frq transcription is required for the Neurospora circadian clock function. In the present study, we showed that RCO-1 and RCM-1 function as a complex (45, 46) to regulate WC-independent frq transcription and that the PKA signaling pathway is a key regulator of this process by phosphorylating and inhibiting RCM-1. Our results demonstrated that the activation of the PKA pathway induces frq transcription in a WC-independent manner, which interferes with the circadian negative feedback loop. Our findings identified RCM-1 as a direct target of PKA. PKA interacts and phosphorylates RCM-1, and the phosphorylation of RCM-1 relieves RCO-1/RCM-1-mediated repression of WC-independent frq transcription. Thus, PKA, in addition to its previously identified role in phosphorylating WCC directly (32), is a critical regulator of frq transcription by regulating RCO-1-RCM-1 activity.
In S. cerevisiae, Ssn6 and Tup1 are known to be phosphorylated (50, 51). However, the role of their phosphorylation on their function is not clear. In the present study, we demonstrate that PKA regulates WC-independent frq transcription by phosphorylating RCM-1. Several lines of evidence presented here indicate that RCM-1 is a direct substrate of PKA. First, immunoprecipitation assays showed that PKA and RCM-1 associate with each other in Neurospora (Fig. 5B). Second, PKA can efficiently phosphorylate the C-terminal part of RCM-1 in vitro (Fig. 5C). Third, mass spectrometry analyses of RCM-1 purified from Neurospora led to the identification of eight in vivo RCM-1 phosphorylation sites at the C terminus of RCM-1, including two PKA consensus sites S718 and S733 and other noncanonical PKA sites (Fig. 6B). Fourth, Western blot analyses using PKA substrate-specific antibody showed that the level of RCM-1 phosphorylation is altered when PKA activity is altered in Neurospora, and the mutation of S718 and S733 of RCM-1, two PKA consensus phosphorylation sites, abolishes most of RCM-1 phosphorylation (Fig. 5E and 6D). Together, these results indicate that PKA is the major kinase that phosphorylates RCM-1 in vivo.
Our results suggest that the phosphorylation of RCM-1 inhibits its activity that suppresses WC-independent frq transcription. A knock-in strain containing five mutated PKA phosphorylation sites of RCM-1 that mimic constant phosphorylation exhibited severely dampened circadian conidiation rhythms and WC-independent frq transcription (Fig. 7), indicating that the proper phosphorylation of RCM-1 is critical for circadian clock function. This is consistent with the loss of circadian rhythm and upregulation of WC-independent frq transcription in the mcb mutant (Fig. 1), which has elevated PKA activity, further indicating the role of PKA in regulating WC-independent frq transcription by phosphorylating RCM-1. Although the circadian rhythms are abolished in rcm-1RIP and mcb mutants, it should be noted that despite reduced amplitude, the clock function was not completely abolished in the rcm-15E strain. This is likely due to the existence of other unidentified functional PKA sites on RCM-1.
How RCO-1 represses frq gene transcription was previously unclear. It has been shown that there is no detectable association between RCO-1 and chromatin (29). Surprisingly, we showed here that RCM-1 associates with chromatin at the frq locus (Fig. 8A), providing a mechanism for how RCO-1-RCM-1 complex regulates the chromatin status at the frq locus. Thus, RCM-1 may recruit RCO-1 to the chromatin, which may explain the lack of detectable RCO-1/chromatin association by ChIP assays at the frq locus. However, neither RCO-1/Tup1 nor RCM-1/Ssn6 has a known DNA binding domain, and thus they may interact with DNA through another partner capable of binding to specific DNA sequences (30, 31). The identity of such a factor (indicated as TF in Fig. 9E), if it exists, is still unknown. In addition, we showed that the RCM-1/chromatin association was severely impaired in the mcb mutant (Fig. 8B). We also observed that the enrichment of RCM-1 was low when it was hyperphosphorylated, while the enrichment of RCM-1 was high when it was hypophosphorylated (Fig. 8C). These results suggest that PKA-dependent phosphorylation of RCM-1 inhibits its association with chromatin, thus affecting its role in regulating chromatin structure.
How does WC-independent frq transcription affect circadian frq transcription? A previous study suggests that PKA regulates frq expression by acting as a priming kinase for WC phosphorylation that leads to their phosphorylation by CKI and CKII (32). The increase in PKA activity in the mcb mutant drastically reduces the WCC binding to the frq C-box. Here, we showed that the mcb mutation also results in WC-independent frq transcription and increased RCM-1 phosphorylation, indicating that PKA has two distinct roles in regulating frq transcription. In the rcm-1RIP mutants, in which WC-independent frq transcription cannot be repressed, both WC-1 and WC-2 are hyperphosphorylated and WCC binding to C-box is constantly low (Fig. 9A and B). These results indicated that the WC-independent FRQ expression leads to WC phosphorylation by FRQ-dependent kinases and inhibition of WC activity. As a result, the circadian negative-feedback loop is disrupted.
cAMP is implicated in the regulation of entrainment or maintenance of clock functions and clock output in molluscs, birds, and the mammalian SCN (52, 53). In mammals, cAMP-dependent signaling oscillates in a circadian manner, which in turn regulates core circadian oscillators (54–57). In Neurospora, the level of cAMP is controlled by the circadian clock (58). The elevated intracellular levels of cAMP result in the activation of PKA (59). Thus, the daily rhythm of cAMP may regulate PKA activity rhythmically. In the present study, we showed that the phosphorylation profile of RCM-1 and the enrichment of RCM-1 at the frq C-box were rhythmic (Fig. 8B and C). Our results suggest that PKA may play a role in the on-off switch of WC-independent frq repression mediated by the RCO-1/RCM-1 complex. Low PKA activity controlled by low levels of cAMP results in hypophosphorylation of RCM-1 and helps to maintain the repressed state of WC-independent frq transcription, allowing the activation of frq transcription by the WC complex. High levels of cAMP in cells, on the other hand, lead to elevated activity of PKA and phosphorylation of RCM-1, resulting in WC-independent frq transcription. The accumulation of WC-independent and WC-dependent FRQ mediates WC phosphorylation and results in dissociation of WCC from frq promoter and inactivation of WC-dependent frq transcription (Fig. 9E). Thus, the control of RCM-1 phosphorylation by PKA should contribute to the daily rhythm of frq transcription.
ACKNOWLEDGMENTS
We thank Zhijun Wang, Zhipeng Zhou and Huijie Chen for critical reading of the manuscript, and we thank Huiqiang Lou and members of the He lab for helpful discussions and proofreading the manuscript. We also thank Xuemei Cao and Yan Gao for technical assistance.
This project is supported by grants from a project supported by the State Key Program of National Natural Science of China (31330004) and National Basic Research Program of China (973 Program) grant (2012CB947600) to Qun He and by grants from the National Institutes of Health (GM084283 and GM062591) and the Welch Foundation (I-1560) to Yi Liu.
REFERENCES
- 1.Dunlap JC. 1999. Molecular bases for circadian clocks. Cell 96:271–290. doi: 10.1016/S0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 2.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6:544–556. doi: 10.1038/nrg1633, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Bell-Pedersen D. 2006. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot Cell 5:1184–1193. doi: 10.1128/EC.00133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reppert SM, Weaver DR. 2001. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 5.Young MW, Kay SA. 2001. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 6.Brown SA, Kowalska E, Dallmann R. 2012. (Re)inventing the circadian feedback loop. Dev Cell 22:477–487. doi: 10.1016/j.devcel.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. 1998. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94:83–95. doi: 10.1016/S0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 8.Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. 2000. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heintzen C, Liu Y. 2007. The Neurospora crassa circadian clock. Adv Genet 58:25–66. doi: 10.1016/S0065-2660(06)58002-2. [DOI] [PubMed] [Google Scholar]
- 10.Mehra A, Baker CL, Loros JJ, Dunlap JC. 2009. Post-translational modifications in circadian rhythms. Trends Biochem Sci 34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunner M, Schafmeier T. 2006. Transcriptional and posttranscriptional regulation of the circadian clock of cyanobacteria and Neurospora. Genes Dev 20:1061–1074. doi: 10.1101/gad.1410406. [DOI] [PubMed] [Google Scholar]
- 12.Baker CL, Loros JJ, Dunlap JC. 2012. The circadian clock of Neurospora crassa. FEMS Microbiol Rev 36:95–110. doi: 10.1111/j.1574-6976.2011.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crosthwaite SK, Dunlap JC, Loros JJ. 1997. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science 276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 14.Cheng P, Yang Y, Liu Y. 2001. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc Natl Acad Sci U S A 98:7408–7413. doi: 10.1073/pnas.121170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng P, Yang Y, Gardner KH, Liu Y. 2002. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol Cell Biol 22:517–524. doi: 10.1128/MCB.22.2.517-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Froehlich AC, Loros JJ, Dunlap JC. 2003. Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc Natl Acad Sci U S A 100:5914–5919. doi: 10.1073/pnas.1030057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 18.He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y. 2002. White collar-1, a DNA binding transcription factor and a light sensor. Science 297:840–843. doi: 10.1126/science.1072795. [DOI] [PubMed] [Google Scholar]
- 19.Denault DL, Loros JJ, Dunlap JC. 2001. WC-2 mediates WC-1-FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J 20:109–117. doi: 10.1093/emboj/20.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schafmeier T, Haase A, Kaldi K, Scholz J, Fuchs M, Brunner M. 2005. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122:235–246. doi: 10.1016/j.cell.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 21.He Q, Cha J, Lee HC, Yang Y, Liu Y. 2006. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev 20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cha J, Chang SS, Huang G, Cheng P, Liu Y. 2008. Control of WHITE COLLAR localization by phosphorylation is a critical step in the circadian negative feedback process. EMBO J 27:3246–3255. doi: 10.1038/emboj.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong CI, Ruoff P, Loros JJ, Dunlap JC. 2008. Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes Dev 22:3196–3204. doi: 10.1101/gad.1706908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Q, Shu H, Cheng P, Chen S, Wang L, Liu Y. 2005. Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J Biol Chem 280:17526–17532. doi: 10.1074/jbc.M414010200. [DOI] [PubMed] [Google Scholar]
- 25.Garceau NY, Liu Y, Loros JJ, Dunlap JC. 1997. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89:469–476. doi: 10.1016/S0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- 26.He Q, Cheng P, Yang Y, Yu H, Liu Y. 2003. FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. EMBO J 22:4421–4430. doi: 10.1093/emboj/cdg425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Q, Cheng P, Liu Y. 2005. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev 19:1518–1531. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Cheng P, Liu Y. 2002. Regulation of the Neurospora circadian clock by casein kinase II. Genes Dev 16:994–1006. doi: 10.1101/gad.965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Z, Liu X, Hu Q, Zhang N, Sun G, Cha J, Wang Y, Liu Y, He Q. 2013. Suppression of WC-independent frequency transcription by RCO-1 is essential for Neurospora circadian clock. Proc Natl Acad Sci U S A 110:E4867–E4874. doi: 10.1073/pnas.1315133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith RL, Johnson AD. 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci 25:325–330. doi: 10.1016/S0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- 31.Malave TM, Dent SY. 2006. Transcriptional repression by Tup1-Ssn6. Biochem Cell Biol 84:437–443. doi: 10.1139/o06-073. [DOI] [PubMed] [Google Scholar]
- 32.Huang G, Chen S, Li S, Cha J, Long C, Li L, He Q, Liu Y. 2007. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev 21:3283–3295. doi: 10.1101/gad.1610207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cambareri EB, Jensen BC, Schabtach E, Selker EU. 1989. Repeat-induced G-C to A-T mutations in Neurospora. Science 244:1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- 35.Aronson BD, Johnson KA, Loros JJ, Dunlap JC. 1994. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Shen Y, Yang S, Wang J, Hu Q, Wang Y, He Q. 2010. Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J Biol Chem 285:4355–4365. doi: 10.1074/jbc.M109.034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H, Wang J, Hu Q, Quan Y, Chen H, Cao Y, Li C, Wang Y, He Q. 2010. DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa. PLoS Genet 6:e1001132. doi: 10.1371/journal.pgen.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, Dunlap JC. 2008. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell 7:28–37. doi: 10.1128/EC.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cha J, Zhou M, Liu Y. 2013. CATP is a critical component of the Neurospora circadian clock by regulating the nucleosome occupancy rhythm at the frequency locus. EMBO Rep 14:923–930. doi: 10.1038/embor.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Hu Q, Chen H, Zhou Z, Li W, Wang Y, Li S, He Q. 2010. Role of individual subunits of the Neurospora crassa CSN complex in regulation of deneddylation and stability of cullin proteins. PLoS Genet 6:e1001232. doi: 10.1371/journal.pgen.1001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z, Wang Y, Cai G, He Q. 2012. Neurospora COP9 signalosome integrity plays major roles for hyphal growth, conidial development, and circadian function. PLoS Genet 8:e1002712. doi: 10.1371/journal.pgen.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banno S, Ochiai N, Noguchi R, Kimura M, Yamaguchi I, Kanzaki S, Murayama T, Fujimura M. 2005. A catalytic subunit of cyclic AMP-dependent protein kinase, PKAC-1, regulates asexual differentiation in Neurospora crassa. Genes Genet Syst 80:25–34. doi: 10.1266/ggs.80.25. [DOI] [PubMed] [Google Scholar]
- 43.Ma P, Wera S, Van Dijck P, Thevelein JM. 1999. The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling. Mol Biol Cell 10:91–104. doi: 10.1091/mbc.10.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sass P, Field J, Nikawa J, Toda T, Wigler M. 1986. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 83:9303–9307. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olmedo M, Navarro-Sampedro L, Ruger-Herreros C, Kim SR, Jeong BK, Lee BU, Corrochano LM. 2010. A role in the regulation of transcription by light for RCO-1 and RCM-1, the Neurospora homologs of the yeast Tup1-Ssn6 repressor. Fungal Genet Biol 47:939–952. doi: 10.1016/j.fgb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Sancar G, Sancar C, Brugger B, Ha N, Sachsenheimer T, Gin E, Wdowik S, Lohmann I, Wieland F, Hofer T, Diernfellner A, Brunner M. 2011. A global circadian repressor controls antiphasic expression of metabolic genes in Neurospora. Mol Cell 44:687–697. doi: 10.1016/j.molcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 47.Gounalaki N, Tzamarias D, Vlassi M. 2000. Identification of residues in the TPR domain of Ssn6 responsible for interaction with the Tup1 protein. FEBS Lett 473:37–41. doi: 10.1016/S0014-5793(00)01480-0. [DOI] [PubMed] [Google Scholar]
- 48.Tzamarias D, Struhl K. 1995. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev 9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 49.Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. 2009. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell 34:354–363. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schultz J, Marshall-Carlson L, Carlson M. 1990. The N-terminal TPR region is the functional domain of SSN6, a nuclear phosphoprotein of Saccharomyces cerevisiae. Mol Cell Biol 10:4744–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redd MJ, Arnaud MB, Johnson AD. 1997. A complex composed of tup1 and ssn6 represses transcription in vitro. J Biol Chem 272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- 52.Eskin A, Takahashi JS. 1983. Adenylate cyclase activation shifts the phase of a circadian pacemaker. Science 220:82–84. doi: 10.1126/science.6298939. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi JS, Zatz M. 1982. Regulation of circadian rhythmicity. Science 217:1104–1111. doi: 10.1126/science.6287576. [DOI] [PubMed] [Google Scholar]
- 54.O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. 2008. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hastings MH, Maywood ES, O'Neill JS. 2008. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol 18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 56.O'Neill JS, Reddy AB. 2012. The essential role of cAMP/Ca2+ signalling in mammalian circadian timekeeping. Biochem Soc Trans 40:44–50. doi: 10.1042/BST20110691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, Brenner DA, Montminy M, Kay SA. 2010. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med 16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hasunuma K, Funadera K, Shinohara Y, Furukawa K, Watanabe M. 1987. Circadian oscillation and light-induced changes in the concentration of cyclic nucleotides in Neurospora. Curr Genet 12:127–133. doi: 10.1007/BF00434667. [DOI] [Google Scholar]
- 59.Toda T, Cameron S, Sass P, Zoller M, Wigler M. 1987. Three different genes in Saccharomyces cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]