Abstract
Setd8 is the sole histone methyltransferase in mammals capable of monomethylating histone H4 lysine 20 (H4K20me1). Setd8 is expressed at significantly higher levels in erythroid cells than any other cell or tissue type, suggesting that Setd8 has an erythroid-cell-specific function. To test this hypothesis, stable Setd8 knockdown was established in extensively self-renewing erythroblasts (ESREs), a well-characterized, nontransformed model of erythroid maturation. Knockdown of Setd8 resulted in impaired erythroid maturation characterized by a delay in hemoglobin accumulation, larger mean cell area, persistent ckit expression, incomplete nuclear condensation, and lower rates of enucleation. Setd8 knockdown did not alter ESRE proliferation or viability or result in accumulation of DNA damage. Global gene expression analyses following Setd8 knockdown demonstrated that in erythroid cells, Setd8 functions primarily as a repressor. Most notably, Gata2 expression was significantly higher in knockdown cells than in control cells and Gata2 knockdown rescued some of the maturation impairments associated with Setd8 disruption. Setd8 occupies critical regulatory elements in the Gata2 locus, and knockdown of Setd8 resulted in loss of H4K20me1 and gain of H4 acetylation at the Gata2 1S promoter. These results suggest that Setd8 is an important regulator of erythroid maturation that works in part through repression of Gata2 expression.
INTRODUCTION
The production of mature red blood cells from committed erythroid progenitors is a complex and incompletely understood process that involves significant changes in gene expression during a time of rapid cell division and nuclear condensation. At the molecular level, erythroid maturation is driven by the complex interaction of transcription factors and chromatin-modifying enzymes that act in concert to drive the expression of erythroid-cell-specific genes, while silencing most other genes in preparation for nuclear condensation and enucleation. The core unit of chromatin is the nucleosome, which is composed of 146 bp of DNA wound around a protein octamer composed of two copies each of histones H2A, H2B, H3, and H4. Posttranslational modification of the N-terminal “tail” of histone proteins is an important determinant of cell-type- and developmental-stage-specific gene expression and higher-order chromatin structure (1–6). Methylation (either mono-, di-, or trimethylation) of the lysine residues on the N-terminal tail of histone proteins is one of the most common posttranslational modifications and can result in either gene activation or repression depending on the lysine residue methylated (7). The consequences of histone H3 lysine methylation on gene expression are fairly well understood (e.g., methylation of histone H3 lysine 4 is associated with transcriptional activation, and methylation of histone H3 lysine 9 or 27 is generally associated with transcriptional repression) (7). In addition, the function of histone H3 lysine methylation has been extensively studied during erythropoiesis, with histone H3 methyltransferases and demethylases interacting with lineage-restricted transcription factors such as Gata1 to mediate changes in gene expression (8–11).
Setd8 is the sole histone methyltransferase in mammals capable of monomethylating histone H4 lysine 20 (H4K20me1) (12). In contrast to histone H3 lysine methylation, the functional consequences of histone H4 lysine methylation are not well understood. Knockout of Setd8 and subsequent loss of H4K20me1 are embryonic lethal preimplantation at approximately the 4- to 8-cell stages due to abnormalities in cell cycle progression and impaired nuclear condensation (12). Monomethylation of histone H4 lysine 20 has been further implicated in a number of biologic processes central to erythropoiesis, including DNA replication (13), chromatin compaction (12), cell cycle progression (12, 14), and DNA repair (15, 16), but little is known about its function in erythroid cells. Setd8 and H4K20me1 are also thought to be transcriptional regulators (7, 15, 17–25); however, in contrast to some of the better-characterized histone modifications (e.g., histone H3 lysine 4 methylation), the influence of this epigenetic pathway on gene expression has not been clearly defined. Disruption of the Setd8/H4K20me1 pathway has been associated with a number of human cancers, including leukemia (26–30), highlighting the relevance of this pathway to human health and disease.
Previous studies examining the role of Setd8 and H4K20me1 in transcriptional regulation have yielded conflicting results (7, 15, 17–25), suggesting that the impact of Setd8 on gene expression may be dependent on genomic and/or cellular context. Initial studies, which focused on the E2F family of genes, demonstrated that H4K20me1 occupancy over promoter regions was associated with gene repression (24, 25). Further supporting a role for Setd8 and H4K20me1 in transcriptional repression, knockdown of Setd8 in HeLa cells resulted in increased expression of H4K20me1 target genes and regions of DNA bound by H4K20me1 functioned as repressors in luciferase reporter assays (18). In contrast, other studies have associated the Setd8/H4K20me1 pathway with transcriptional activation, with knockdown of Setd8 significantly impairing activation of Wnt target genes (31) and genome-wide studies of H4K20me1 occupancy demonstrating localization of H4K20me1 downstream of the transcription start site of highly expressed genes in multiple cell types, including lymphocytes, HeLa, and erythroleukemia (K562) cells (15). To date, few studies have utilized unbiased methods to determine the net effect of Setd8 disruption on gene expression.
Setd8 is expressed at significantly higher levels in CD71+ erythroid precursors than in any other cell type (32), suggesting that it may have erythroid-cell-specific functions. Setd8 is expressed in all three murine erythroid lineages (primitive, fetal definitive, and adult definitive) (33) and throughout the erythroid differentiation of human CD34+ hematopoietic stem and progenitor cells (34); however, little is known about how Setd8 interacts with the complex network of transcription factors and chromatin modifiers that govern erythroid differentiation. To date, the only published study examining the function of Setd8 in erythroid cells was done using the G1E-GATA1ER cell line, and it suggested that Setd8 is important for Gata1-mediated gene repression (20). Taken together, these preliminary data imply that Setd8 and H4K20me1play an important role during erythropoiesis, but the function of this epigenetic pathway in erythroid cells is poorly understood.
To delineate the role of the Setd8/HK20me1 pathway in erythroid cells, we established stable knockdown of Setd8 expression in extensively self-renewing erythroblasts (ESREs), a unique model system for studying erythroid maturation. ESREs are nontransformed, cytokine-dependent cells, cultured from murine yolk sac or fetal liver, that proliferate extensively at the proerythroblast phase while retaining the ability to appropriately mature and enucleate in approximately 3 or 4 cell divisions (35, 36). Setd8 is robustly expressed in ESREs, and its expression does not change significantly during maturation (36). Knockdown of Setd8 in ESREs was associated with significant impairments in maturation, as demonstrated by morphology, benzidine staining, and imaging flow-cytometric analyses. Unbiased transcriptome analyses following Setd8 knockdown demonstrated that Setd8 functions as a transcriptional repressor in erythroid cells. Interestingly, Gata2 was expressed at significantly higher levels in Setd8 knockdown cells than in controls. In hematopoietic stem and progenitor cells, Gata2 is a critical regulator of the balance between self-renewal and differentiation, with downregulation of Gata2 expression necessary for terminal erythroid differentiation to occur (37–40). Chromatin immunoprecipitation (ChIP) analyses demonstrated occupancy of Setd8 at several key regulatory regions of the Gata2 locus and alteration of chromatin structure within the Gata2 locus following Setd8 knockdown. Gata2 knockdown was able to rescue some of the maturation impairments associated with Setd8 disruption. Taken together, these results suggest that Setd8 is an important regulator of erythroid maturation that works in part through repression of Gata2.
MATERIALS AND METHODS
Mice and tissues.
The University of Rochester's Committee on Animal Resources approved all experiments utilizing mice. C57BL/6 (Jackson Laboratories) mice were bred overnight and vaginal plugs checked after 12 h (embryonic day 0.5 [E0.5]). At E14.5, the mice were killed by CO2 narcosis, the embryos were harvested by dissection in PB2 (Dulbecco phosphate-buffered saline [Invitrogen] supplemented with 0.1% glucose [Invitrogen] and 0.3% bovine serum albumin [Fisher Scientific]), and the fetal liver was isolated.
Erythroid expansion culture.
Extensively self-renewing erythroblasts were derived as previously described (35, 36). They were maintained at a concentration of 1 × 106/ml in expansion medium composed of StemSpan serum-free expansion medium (SFEM; Stem Cell Technologies) supplemented with human recombinant erythropoietin (2 U/ml), stem cell factor (SCF; 100 ng/ml; Pepro Tech), dexamethasone (10−6 M; Sigma), insulin-like growth factor-1 (40 ng/ml; Pepro Tech), penicillin-streptomycin (Invitrogen), and cholesterol mixed to a final concentration of 0.4% (Sigma). Partial medium changes were done daily.
Erythroid cell maturation.
Maturation of ESREs was initiated as previously described (35, 36). Briefly, the cells were washed in PB2 and suspended in maturation medium, composed of Iscove's modified Dulbecco medium (IMDM; Gibco), supplemented by 2 U/ml human recombinant erythropoietin, 100 ng/ml SCF, 10% serum replacement (Invitrogen), 5% plasma-derived serum (PDS; Animal Technologies), and glutamine.
Lentivirus-mediated shRNA.
Lentiviral short hairpin RNA (shRNA) constructs targeting Setd8 (Sigma) or nontargeting scramble control constructs (Sigma) were transduced into ESREs by spin inoculation. Briefly, 200 μl of viral solution was mixed with 0.25 μl of Polybrene and 50 μl of ESREs at a density of 1 million cells per ml. The cells were then centrifuged at room temperature for 90 min at 1,600 × g. Following centrifugation, 250 μl of expansion medium was added and the cells were incubated overnight. Following the overnight incubation, the cells were spun at 300 × g for 7 min and then resuspended in expansion medium containing 1 μg/μl of puromycin.
siRNA knockdown of Gata2.
SMARTpool siGENOME Gata2 small interfering RNA (siRNA; number M-062114-01) or nontargeting control (Dharmacon, GE Healthcare) was transfected into Setd8 knockdown and scramble control cultures using RNAiMax Lipofectamine (Life technologies) diluted in Dharmafect cell culture reagent (Dharmacon, GE Healthcare) according to the manufacturer's instructions. Following transfection, cells were cultured in antibiotic-free ESRE expansion medium for 48 to 72 h prior to maturation studies. All reactions were performed in triplicate.
Imaging flow cytometry.
ESREs were stained with CD71-fluorescein isothiocyanate (CD71-FITC; eBioscience), c-Kit–phycoerythrin (eBioscience), 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich), and Draq5 (eBioscience) and run on ImageStream (Amnis). The data were analyzed with IDEAS software (Amnis) as previously published (36, 41).
Quantitative ChIP.
ChIP assays were performed as previously described (6) with approximately 10 million cells used for each assay. Briefly, DNA was cross-linked to DNA binding proteins using 1% formaldehyde. The cells were lysed, and the DNA was isolated and sonicated into ∼200-bp fragments using a Diagenode Bioruptor. The DNA-protein complexes were immunoprecipitated with an antibody to Setd8 (Abcam), H4K20me1 (Abcam), or H4 acetyl (Millipore). DNA-protein complexes were recovered with protein G magnetic beads (Invitrogen). Immunoprecipitated DNA was subjected to quantitative PCR, and enrichment was calculated relative to total input control as previously described (6).
Analysis of gene expression using qPCR.
RNA was isolated using TRIzol, and cDNA was generated using the SuperScript III First Strand kit (Invitrogen). Quantitative reverse transcription-PCR (qPCR) using Sybr green was used to assess gene expression (36). Data were normalized to either 18S or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression. P values were calculated using Student's t test. Primers are listed in Table S1 in the supplemental material.
RNA-seq and bioinformatic analyses.
RNA was isolated from Setd8 knockdown and scramble control cultures during self-renewal and following 6 h of maturation using Qiagen's RNeasy kit. RNA was assessed with an Agilent Bioanalyzer (Santa Clara, CA) prior to poly(A) selection and library preparation according to Illumina instructions. Libraries were subjected to 65-bp single-end sequencing on the Illumina HiSeq2500 instrument (Illumina, San Diego, CA). Sequence reads were aligned to the mouse reference genome (UCSC assembly mm9, NCBI build 37) using SHRiMP2 (42). Differential gene expression was assessed using the Cuffdiff suite of software (43).
Nucleotide sequence accession number.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (44) and are accessible through GEO Series accession number GSE62647.
RESULTS
Knockdown of Setd8 impairs erythroid maturation.
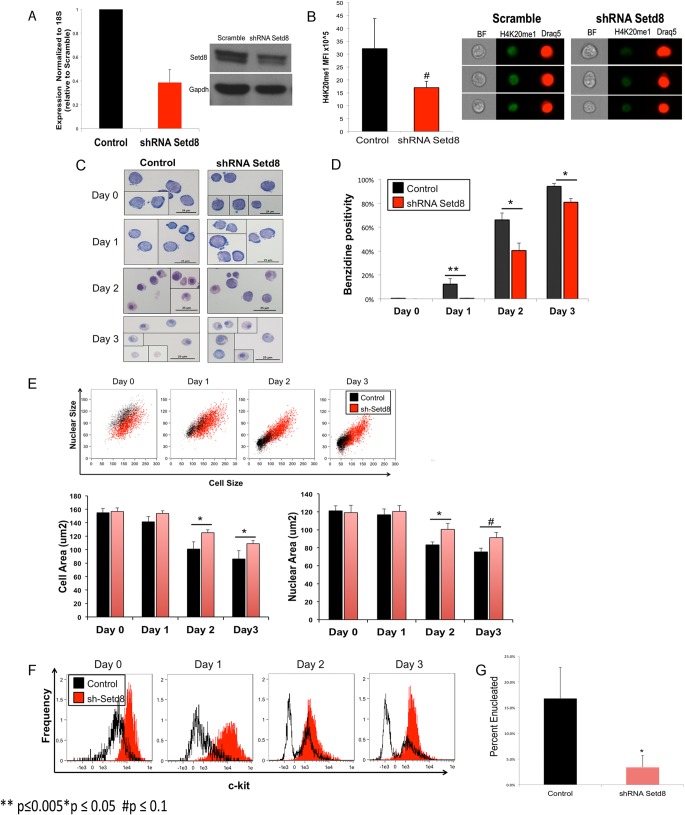
To study the role of Setd8 in erythroid maturation, we utilized lentivirus-mediated shRNA to establish stable knockdown of Setd8 in extensively self-renewing erythroblasts (ESREs), with approximately 75% knockdown at the RNA level and approximately 50% knockdown at the protein level (Fig. 1A). As expected, imaging flow-cytometric analyses demonstrated a decrease in H4K20me1 levels in knockdown cultures compared to scramble control (Fig. 1B).
FIG 1.
Knockdown of Setd8 impairs erythroid maturation. (A) shRNA targeting Setd8 reduces Setd8 expression at both the mRNA and protein levels. (B) Imaging flow-cytometric analyses using an antibody specific to H4K20me1 conjugated to Alexa Fluor 488–goat anti-rabbit IgG (Life Technologies) demonstrate lower H4K20me1 mean fluorescence intensity (MFI) in Setd8 knockdown cultures than in scramble control. Data represent the means and standard errors of the means (SEM) from three independent experiments. (C) Representative images from Setd8 knockdown and scramble control cultures. Setd8 knockdown and scramble control cells are morphologically similar while self-renewing (day 0), resembling proerythroblasts. After 3 days of maturation, the control culture consists primarily of late-stage erythroid precursors, enucleated cells, and pyrenocytes. In contrast, the Setd8 knockdown cultures are more heterogeneous, with persistence of cells resembling earlier erythroid precursors and a paucity of enucleated cells and pyrenocytes. (D) Setd8 knockdown results in delayed hemoglobin accumulation, as determined by benzidine staining. (E) Imaging flow-cytometric analyses demonstrate that during maturation, Setd8 knockdown cells have larger mean cell and nuclear areas than do scramble control cells. (F) Setd8 knockdown cells fail to downregulate ckit during maturation. (G) Imaging flow-cytometric analyses, comparing cell area and Draq intensity, demonstrate that Setd8 knockdown is associated with a significant deficit in enucleation. *, P < 0.05; #, P < 0.1.
Erythroid maturation is characterized by hemoglobin accumulation, changes in cell surface marker expression, and a progressive decrease in cell and nuclear size that culminates in enucleation (45). ESREs recapitulate this process within 3 days of placement in maturation medium (35, 36). Setd8 knockdown did not alter the proliferation or morphology of self-renewing cells but significantly impaired multiple facets of erythroid maturation. Following 3 days of maturation, the control culture was comprised mainly of late-stage erythroid precursors, enucleated reticulocytes, and pyrenocytes. In contrast, the Setd8 cultures were heterogeneous in appearance, containing a spectrum of erythroblasts at various stages of maturation, with almost no enucleated cells present (Fig. 1C). Knockdown of Setd8 also resulted in slower hemoglobin accumulation (Fig. 1D) and a higher level of ckit expression that was not appropriately downregulated with maturation (Fig. 1F).
Differences in mean cell area, mean nuclear area, and rates of enucleation following Setd8 disruption were quantified using imaging flow-cytometric analyses. Prior to maturation, knockdown and control cultures had similar mean cell and nuclear areas; however, by day 2 of maturation, the knockdown cells had larger mean cell and nuclear areas than did control cells (Fig. 1E). Enucleation is a hallmark of terminal erythroid differentiation. Rates of enucleation were determined by comparing Draq5 staining to nuclear area, with Setd8 knockdown cultures demonstrating a significant enucleation deficit (Fig. 1G). The culture conditions do not effectively support maturation past 3 days, making it difficult to determine if the Setd8 knockdown cells have the capacity to complete terminal erythroid maturation if given a longer time frame.
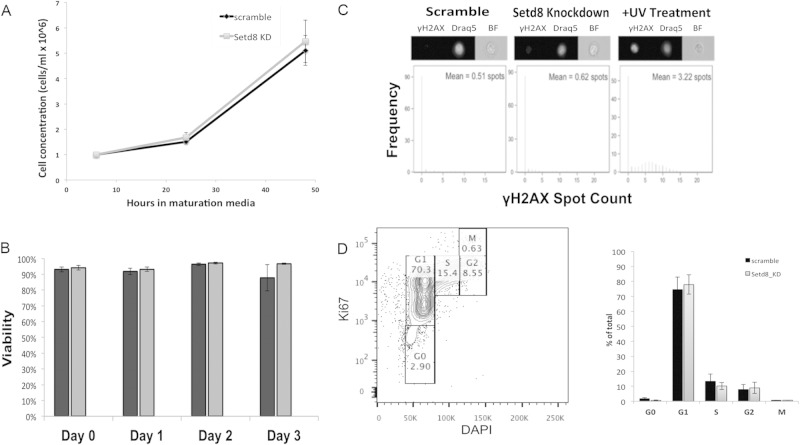
In some cell types, loss of Setd8 results in cell cycle arrest and accumulation of DNA damage (12, 16, 46); however, alterations in viability and accumulation of DNA damage do not appear to underlie the abnormalities observed in erythroid maturation. Cell counts and trypan blue staining demonstrated that knockdown of Setd8 did not affect ESRE proliferation or viability, consistent with a previous study in erythroid cells (20) (Fig. 2A and B). In addition, flow-cytometric analyses utilizing an antibody specific for γ-H2Ax, a well-established marker of DNA double-strand breaks (47), did not detect significant accumulation of DNA damage in either scramble control or Setd8 knockdown cultures (Fig. 2C). The normal rates of proliferation in Setd8 knockdown cells make a cell cycle defect unlikely, and flow-cytometric analyses utilizing an antibody specific for Ki-67 confirmed that there was no abnormality in cell cycle progression following Setd8 disruption (Fig. 2D).
FIG 2.
Setd8 knockdown in ESREs is not associated with abnormal proliferation, decreased viability, or accumulation of DNA damage. (A) During maturation, cell counts are similar in control and knockdown cultures. Data represent the means and SEM from a minimum of three independent experiments. (B) Trypan blue staining demonstrates similar viability in control and knockdown cultures. Data represent the means and SEM from a minimum of three independent experiments. (C) Image stream analyses using an antibody specific to γ-H2Ax did not detect significant numbers of γ-H2AX foci in knockdown or control cultures. A UV-treated sample is included as a positive control. (D) Setd8 knockdown is not associated with altered cell cycle progression in erythroid cells. Flow-cytometric analyses utilizing an antibody specific for Ki-67 demonstrate that the scramble control and knockdown cultures have similar proportions of cells in each phase of the cell cycle. The left panel is an example of the gating strategy. The bar graph demonstrates the percentage of cells in each phase of the cell cycle for the Setd8 knockdown and control cultures. The means and SEM from three independent experiments are shown.
Setd8 functions as a transcriptional repressor in erythroid cells.
The role of Setd8 in transcriptional regulation is unclear, with Setd8 acting as an activator (15, 21, 31) in some cellular and genomic contexts and a repressor in others (15, 18, 24, 25). To determine the influence of Setd8 on gene expression in erythroid cells, global gene expression was analyzed by transcriptome sequencing (RNA-seq) in Setd8 knockdown and scramble control samples prior to maturation. To gain insights into the molecular mechanisms underlying the impaired maturation associated with Setd8 disruption, gene expression was also assessed following 6 h in maturation medium, a time point at which phenotypic differences between the control and Setd8 knockdown cultures are not yet evident.
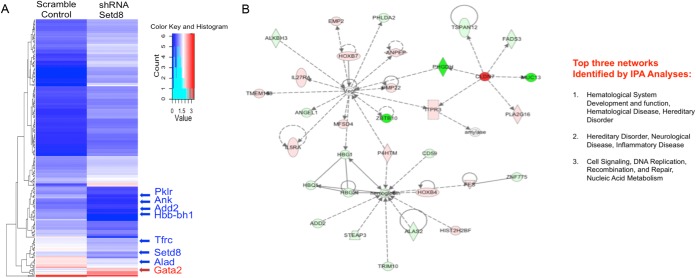
In ESREs, prior to maturation, 1,048 genes were differentially expressed (P < 10−3; false discovery rate, <0.01) (see Table S2 in the supplemental material) between Setd8 knockdown cells and scramble controls. Setd8 acts primarily as a repressor in erythroid cells, with 679 genes upregulated and 369 genes downregulated following Setd8 disruption. Ingenuity pathway analyses (IPA) of the differentially expressed genes identified “cell morphology, cellular function and maintenance, and cellular growth and proliferation” as the top network (score, 44; focus molecules, 31) (see Fig. S1 in the supplemental material). The differentially expressed genes were further analyzed by the upstream regulator function of IPA, which uses the well-curated IPA database to predict which regulatory molecules are upstream of observed changes in gene expression (48). Kit (P < 5.74 × 10−10), Gata1 (P = 4.06 × 10−9), and Stat3 (P = 1.24 × 10−6) were predicted as upstream regulators of the gene expression changes that occur following Setd8 knockdown.
Following 6 h of maturation, 1,149 genes were differentially expressed (P <10−3; false discovery rate, <0.01) (see Table S3 in the supplemental material) between Setd8 knockdown and control cells. Consistent with a repressor function for Setd8 in erythroid cells, 780 genes were upregulated and 369 genes were downregulated (Fig. 3A). IPA of the differentially expressed genes identified “hematologic development and disease” (score, 43; focus molecules, 32) as the top network (Fig. 3B). The top disease or functions annotation identified by IPA was “function of red blood cells” (P = 6.27 × 10−17). In addition, both Gata1 (P = 9.28 × 10−10) and Kit (P = 1.89 × 10−9) were predicted as upstream regulators. The prediction that well-characterized erythroid transcription factors are upstream of the gene expression changes observed following Setd8 knockdown supports the hypothesis that Setd8 participates in the intricate network of chromatin modifiers and transcriptional regulators that govern the terminal erythroid maturation program.
FIG 3.
Setd8 promotes transcriptional repression in erythroid cells. (A) Heat map of differentially expressed genes in maturing Setd8 knockdown and scramble control cultures. (B) Ingenuity pathway analyses of differentially expressed genes. The top network identified (hematologic system, development, and function) is shown.
Setd8 disruption alters the expression of multiple transcriptional regulators, most notably Gata2.
Setd8 has been shown to directly regulate the expression level of key transcriptional regulators during cellular differentiation (22, 46) and can act in a cell-type- and developmental-stage-specific manner (14, 22). We carefully examined the list of genes differentially expressed upon Setd8 knockdown and determined that knockdown of Setd8 in erythroid cells alters the expression of a number of chromatin modifiers and transcription factors known to regulate terminal erythroid maturation. In self-renewing ESREs, prior to maturation, Setd8 knockdown decreased the expression of Fog1 (fold change, −3.97; P < 5.7 × 10−6) (see Table S2 in the supplemental material) and the Wnt effector Tcf7l2 (fold change, −4.0; P < 0.0003) (see Table S2 in the supplemental material); however, these genes were not differentially expressed in maturing cells (see Table S3 in the supplemental material). The expression of several other transcriptional regulators was altered following Setd8 knockdown in both self-renewing and maturing cells, including Sox6 (for self-renewing cells, fold change was −4.3, P < 0.0002; for maturing cells, fold change was −3.1, P < 0.001), Hes1, a downstream effector of the Notch signaling pathway (for self-renewing cells, fold change was 9.1, P < 5 × 10−9; for maturing cells, fold change was 16.0, P < 5 × 10−14), and Smad1, a downstream effector of the BMP pathway (for self-renewing cells, fold change was 4.8, P < 0.0008; for maturing cells, fold change was 6.3, P < 8.8 × 10−7).
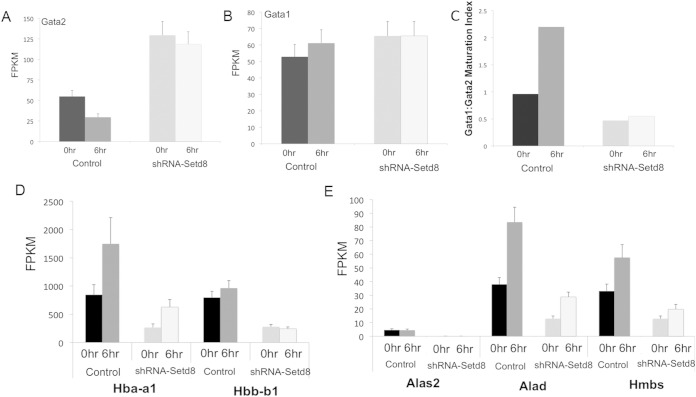
Gata2 is a critical regulator of the balance between maturation and self-renewal in hematopoietic stem and progenitor cells, and Gata2 overexpression is sufficient to impair erythroid maturation (37, 39). Gata2 is robustly expressed in proliferating ESREs but is rapidly silenced during maturation (36). One of the most intriguing findings following Setd8 disruption was the elevation of Gata2 expression in both self-renewing cells (fold change, 2.3; P < 0.0009) (Fig. 4A; see also Table S2 in the supplemental material) and maturing cells (fold change, 4; P < 5 × 10−5) (Fig. 4A; see also Table S3 in the supplemental material). We validated this increase in Gata2 expression by both quantitative PCR and Western blotting (see Fig. S2A in the supplemental material). Interestingly, Setd8 knockdown did not significantly alter the expression of other major erythroid transcription factors, including Gata1, Klf1, and Tal1 as determined by CuffDiff analyses or by qPCR validation (Fig. 4B; see also Fig. S2B and D and Tables S2 and S3 in the supplemental material).
FIG 4.
Setd8 knockdown alters the expression of Gata2, the globins, and several heme synthesis enzymes. Data represent the fragments per kilobase of exon per million fragments mapped (FPKM) attained from RNA-seq studies. Black bars represent self-renewing control samples. Dark gray bars represent maturing control samples. Light gray bars represent self-renewing Setd8 knockdown samples. The lightest bars represent maturing Setd8 knockdown samples. Error bars represent SEM estimated from Cuffdiff analyses. (A) Gata2 is differentially expressed in Setd8 knockdown cells compared to control in both self-renewing (fold change, 2.3; P = 0.0009) and maturing (fold change, 4; P = 5 × 10−5) cells. (B) Setd8 knockdown does not significantly alter Gata1 expression in either self-renewing (fold change, 1.16; P = 0.59) or maturing (fold change, 1.0; P = 0.99) cells. (C) The Gata1/Gata2 ratio is altered in Setd8 knockdown cells compared to scramble control cells. (D) The expression of Hba-a1 (for self-renewing cells, fold change = −3.17, P = 0.00042; for maturing cells, fold change = −2.8, P = 0.0004) and that of Hbb-b1 (for self-renewing cells, fold change = −2.8, P = 0.00023; for maturing cells, fold change = −3.9, P = 9.04 ×10−7) are significantly altered following Setd8 knockdown. (E) The expression levels of Alas2 (for self-renewing cells, fold change = −50, P < 1 × 10−10; for maturing cells, fold change = −65, P < 1 × 10−10), Alad (for self-renewing cells, fold change = −2.9, P = 0.00055; for maturing cells, fold change = −2.9, P = 0.00017), and Hmbs (for self-renewing cells, fold change = −2.6, P = 0.0029; for maturing cells, fold change = −2.9, P = 0.0018) are also significantly altered in Setd8 knockdown cells compared to control cells.
During erythroid maturation, the expression of Gata1 increases and the expression of Gata2 is silenced (49). As the Gata1/Gata2 ratio changes, a “Gata switch” occurs, with Gata1 replacing Gata2 at numerous regulatory regions genome-wide, driving the terminal differentiation program (50, 51). Setd8 knockdown alters the temporal expression pattern of Gata2 in maturing ESREs. Following 6 h of maturation, Gata2 levels decrease by approximately 50% in the control cultures but remain unchanged in the Setd8 knockdown samples (Fig. 4A; see also Fig. S2A in the supplemental material). The elevated Gata2 level in Setd8 knockdown cells results in an altered Gata1/Gata2 ratio (Fig. 4C; see also Fig. S2C in the supplemental material). Consistent with this change, a number of “Gata switch” targets are differentially expressed following Setd8 knockdown, most notably the globins and heme synthesis enzymes (Fig. 4D and E, respectively). These genes are initially expressed at significantly lower levels in the knockdown cultures and fail to gain the robust induction of expression seen in the control samples.
We next sought to determine how many of the genes differentially expressed following Setd8 knockdown were associated with Gata2/Gata1 switch sites. Gata2/Gata1 switch sites were identified using published genome-wide profiles of Gata2 and Gata1 occupancy, done in G1E and induced Gata1-G1E-ER4 cells, respectively (51). G1E cells are derived from murine Gata1 null embryonic stem cells and are arrested at approximately the proerythroblast stage of maturation. Gata1-GIE-ER4 cells carry an inducible Gata1 allele and when treated with estradiol have gene expression changes consistent with erythroid maturation (52). The G1E system has been used extensively to study how Gata factor interplay regulates erythroid gene expression (51–55). ChIP-seq (chromatin immunoprecipitation coupled with high-throughput sequencing) identified 4,905 sites of Gata2 occupancy in G1E cells and 11,491 sites of Gata1 occupancy in induced Gata1-G1E-ER4 cells (51). We used the Galaxy platform (56) to identify Gata2/Gata1 switch sites, defined as regions that bound Gata2 in G1E cells and Gata1 in induced Gata1-G1E-ER4 cells. In total, 2,165 Gata1/Gata2 switch sites were identified. The Homer program (57) was used to identify the nearest gene to each Gata2/Gata1 switch site, and the list of genes associated with Gata2/Gata1 switch sites was compared to the list of genes differentially regulated following Setd8 knockdown. Approximately 12% (141/1,149) of differentially expressed genes following Setd8 knockdown were associated with a Gata2/Gata1 switch site.
The only previously published study examining the role of Setd8 in erythroid cells suggested that Setd8 plays an important role in Gata1-mediated transcriptional repression (20). Consistent with that study, the majority of differentially expressed genes associated with Gata2/Gata1 switch sites are upregulated upon Setd8 knockdown (93/141; 66%). Taken together, these data suggest that for a subset of genes elevated Gata2 expression and perturbed Gata2/Gata1 switching may contribute to the altered gene expression observed following Setd8 knockdown.
Setd8 occupies critical regulatory elements in the Gata2 locus in both primary fetal liver-derived erythroblasts and ESREs.
The Gata2 locus has several well-characterized regulatory elements, including both general (1G) and hematopoiesis-specific (1S) promoters (58, 59). In addition, genomic and functional studies have identified three enhancers immediately upstream of the Gata2 gene at kb −3.9, −2.8, and −1.8 (54, 60–62) as well as an enhancer located at kb +9.5 from the transcription start site that controls its spatial and temporal expression (63, 64). Knockout of Gata2 or deletion of the +9.5 enhancer is embryonic lethal due to severe anemia (63, 65). The enhancers upstream of the transcription start site (the −3.8, −2.8, and −1.8 enhancers) modulate Gata2 levels but are not essential for Gata2 expression (42–45).
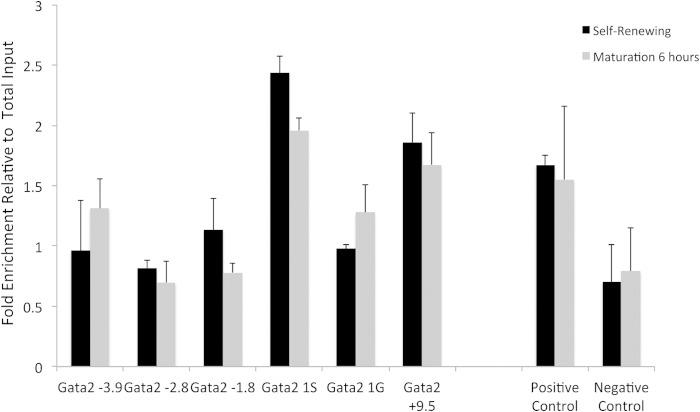
To determine if Gata2 is a direct target of Setd8, we interrogated key regulatory regions within the Gata2 locus for Setd8 occupancy using quantitative chromatin immunoprecipitation (ChIP). To avoid the possibility of cell culture artifacts, quantitative ChIP studies were done in primary uncultured fetal liver erythroblasts obtained from E14.5 embryos. In primary fetal liver-derived erythroblasts, Setd8 occupancy occurs in multiple regulatory regions within the Gata2 locus, including both the general (1G) and hematopoiesis-specific (1S) promoters, as well as the −3.9 and +9.5 enhancers (Fig. 5), suggesting that Setd8 may directly regulate Gata2 expression.
FIG 5.
Chromatin immunoprecipitation demonstrates that Setd8 occupies key regulatory elements in the Gata2 locus in uncultured fetal liver erythroblasts, including the −3.8 and +9.5 enhancers and the 1S and 1G promoters. The positive control is beta actin intron, and the negative control is a region upstream of the Gata2 locus with no known function. Data are presented as fold enrichment relative to total input control and represent the means and SEM from three independent experiments.
We further utilized quantitative ChIP to determine the occupancy of Setd8 within the Gata2 locus of ESREs. Similar to the data obtained from primary fetal liver cells, in ESREs Setd8 occupies both the Gata2 1S promoter and the essential +9.5 enhancer (Fig. 6 and 7A). Knockdown of Setd8 resulted in loss of Setd8 occupancy at these regulatory elements (Fig. 7A). ESREs are a uniform population of cells that morphologically appear to be at the proerythroblast stage, while fetal liver-derived erythroid cells represent a more heterogeneous population of erythroid cells at various stages of maturation. This heterogeneity in cell types and maturation stages may underlie the differences in Setd8 occupancy observed between ESREs and primary fetal liver-derived erythroblasts.
FIG 6.
Chromatin immunoprecipitation for Setd8 in self-renewing and maturing wild-type ESREs. There is no significant change in Setd8 occupancy at key regulatory elements of the Gata2 locus following 6 h of maturation. The positive control is beta actin intron, and the negative control is a region upstream of the Gata2 locus with no known function. Data are presented as fold enrichment relative to total input control and represent the means and SEM from three independent experiments.
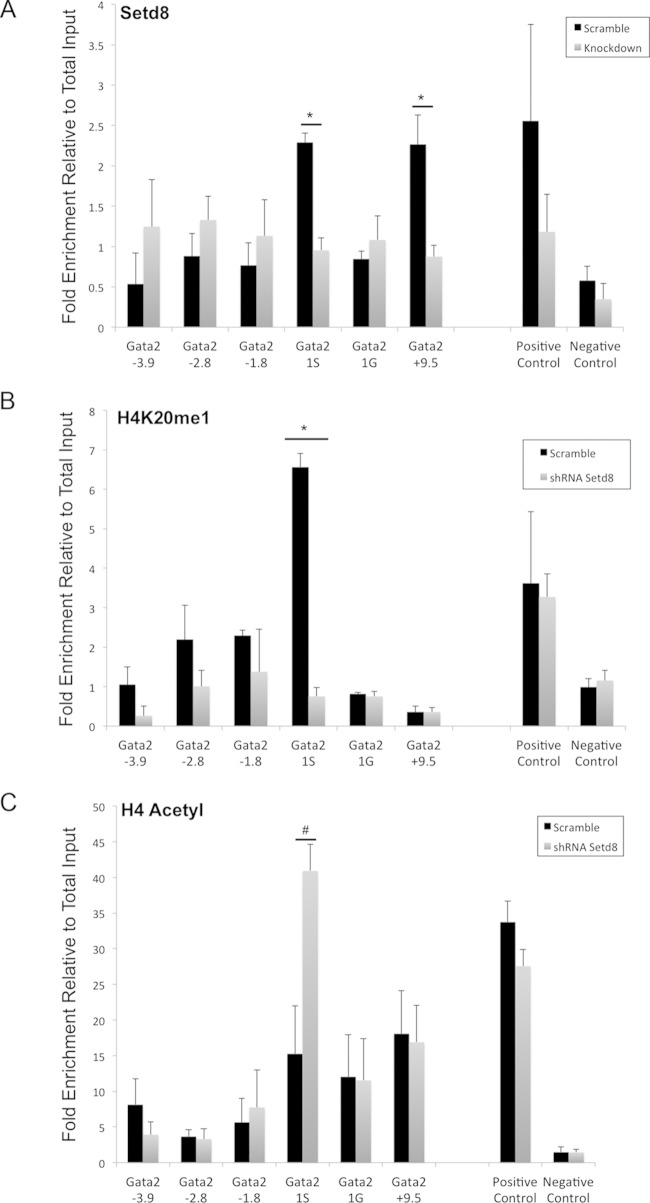
FIG 7.
Setd8 knockdown results in significant changes in chromatin architecture in the Gata2 locus. ChIP assays were performed in self-renewing Setd8 knockdown and scramble control ESREs. Data are presented as fold enrichment relative to total input control and represent the means and SEM from a minimum of three independent experiments. (A) Setd8 occupies the Gata2 1S promoter and +9.5 enhancer in scramble control ESREs. Setd8 knockdown is associated with loss of Setd8 occupancy in the Gata2 locus. The positive control is the beta actin intron, and the negative control is a region upstream of the Gata2 locus with no known function. (B) There is occupancy of H4K20me1 at the Gata2 1S promoter that is lost with Setd8 knockdown. The positive control is beta actin intron, and the negative control is a region upstream of the Gata2 locus with no known function. (C) Setd8 knockdown results in increased histone H4 acetylation at the Gata2 1S promoter. The positive control is the beta actin promoter. The negative control is the necdin gene, which is not expressed in erythroid cells.
We next sought to determine whether Setd8 occupancy at the Gata2 locus changed during ESRE maturation. We used quantitative ChIP to profile Setd8 occupancy at key regulatory regions in the Gata2 locus following 6 h of maturation, a time point when Gata2 expression has decreased by approximately 50% in control cells (Fig. 4A; see also Table S2 in the supplemental material). Occupancy of Setd8 did not vary significantly between the two time points (Fig. 6), consistent with the literature demonstrating that Gata2 repression during terminal erythroid maturation is mediated by a number of different transcription factors and chromatin modifiers (54, 66).
Knockdown of Setd8 results in loss of H4K20me1 and gain of H4 acetylation in the Gata2 locus.
Deposition of H4K20me1 at promoters is associated with gene repression that can be mediated by multiple mechanisms. In some cell types, H4K20me1 is able to recruit proteins that mediate transcriptional repression, such as L3MBTL1 [lethal(3) malignant brain tumor-like protein] (67, 68), a polycomb group protein expressed in fetal brain but not expressed at significant levels in maturing erythroid cells (32, 36). To date, H4K20me1-associated repressors have not been described in erythroid cells. Alternatively, Setd8 and H4K20me1 can regulate transcription by altering the chromatin landscape to promote transcriptional repression. For example, H4K20me1 occupancy may interfere with histone H4 acetylation (69, 70), which facilitates active transcription (17, 18).
To test this possibility, we used quantitative ChIP to compare the levels of histone H4 modifications (H4K20me1 and H4 acetylation) within the Gata2 locus in Setd8 knockdown and scramble control cells. We observed that knockdown of Setd8 and loss of Setd8 occupancy within the Gata2 locus were accompanied by significant changes in the epigenetic landscape. In control cultures, there was significant occupancy of H4K20me1 at the Gata2 1S promoter, which was abolished by Setd8 knockdown (Fig. 7B). Consistent with the elevated expression of Gata2 observed in knockdown cultures, the loss of H4K20me1 occupancy was accompanied by a corresponding increase in H4 acetylation at the Gata2 1S promoter (Fig. 7C). These observations support the hypothesis that Setd8 promotes transcriptional repression in erythroid cells by altering the chromatin landscape.
Knockdown of Gata2 rescues some of the maturation impairment associated with Setd8 disruption.
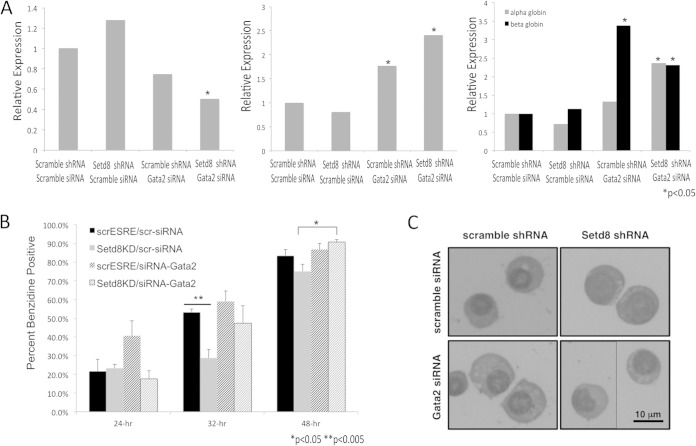
We next sought to determine whether knockdown of Gata2 could rescue some of the maturation impairments associated with Setd8 knockdown. Setd8 knockdown and scramble control cultures were transfected with either siRNA targeting Gata2 or nontargeting (scramble) control siRNA and then cultured in ESRE expansion medium for 48 to 72 h prior to the induction of maturation. Treatment of the Setd8 knockdown cultures with siRNA targeting Gata2 (shRNA Setd8, siRNA Gata2) resulted in significant Gata2 knockdown (Fig. 8A, left panel). Consistent with its role in regulating erythroid maturation, knockdown of Gata2 resulted in a significant increase in Gata1 and beta globin expression in both the scramble control and Setd8 knockdown cultures prior to placement in maturation medium (Fig. 8A, middle and right panels). In addition, scramble control cultures treated with siRNA targeting Gata2 had a rate of benzidine accumulation higher than those observed under all other conditions. In contrast, Setd8 knockdown cells treated with nontargeting control siRNA demonstrated a significant lag in benzidine accumulation (P < 0.005 compared to shRNA scramble control cultures treated with nontargeting siRNA) (Fig. 8B). Setd8 knockdown cells treated with siRNA targeting Gata2 demonstrated significant improvements in benzidine staining (P < 0.017 at 48 h compared to Setd8 knockdown cells treated with nontargeting siRNA) (Fig. 8B) and morphology (Fig. 8C). Other parameters of terminal erythroid maturation, such as enucleation, were not assessed due to the limited cell number in small-scale siRNA experiments. These data further suggest that altered Gata2 expression may contribute to the maturation abnormalities associated with Setd8 knockdown.
FIG 8.
Knockdown of Gata2 rescues some of the maturation impairments associated with Setd8 knockdown. (A) mRNA expression of Gata2 (left panel), Gata1 (middle panel), and alpha and beta globin (right panel) in scramble control and Setd8 knockdown cultures treated with nontargeting (control) siRNA or siRNA targeting Gata2. Expression was calculated relative to Gapdh and is presented relative to the scramble shRNA, nontargeting siRNA control sample. Data represent the means and SEM from a minimum of three independent experiments. For statistical analyses, the scramble shRNA, nontargeting siRNA samples were compared to the scramble shRNA, Gata2 siRNA samples, and the Setd8 shRNA, nontargeting siRNA samples were compared to the Setd8 shRNA, siRNA Gata2 samples. (B) Percentage of cells that are benzidine positive in Setd8 knockdown or scramble control cultures treated with either siRNA targeting Gata2 or nontargeting siRNA control. scrESRE/scr-siRNA, shRNA scramble control cultures treated with nontargeting siRNA; Setd8KD/scr-siRNA, shRNA-Setd8 cultures treated with nontargeting control siRNA; scrESRE/siRNA-Gata2, shRNA scramble control cultures treated with siRNA targeting Gata2; Setd8KD/siRNA-Gata2, shRNA-Setd8 cultures, treated with siRNA targeting Gata2. Error bars represent the means and SEM from three independent reactions. One of three representative experiments is shown. (C) Cytospins demonstrating the morphology of Setd8 knockdown or scramble control cells treated with either nontargeting siRNA or siRNA targeting Gata2.
DISCUSSION
Posttranslational modifications of histone proteins and alteration of the epigenetic landscape are important determinants of cell-type- and developmental-stage-specific gene expression. Setd8 is the sole histone methyltransferase in mammals capable of generating H4K20me1 and is highly expressed in erythroblasts compared to all other cell and tissue types (32); however, prior to this study there existed few data on the function of Setd8 in erythroid cells. We have demonstrated that Setd8 is critical for terminal erythroid maturation and that Setd8 functions as a transcriptional repressor in erythroid cells. We have further shown that knockdown of Setd8 in erythroid cells alters the expression of several key transcriptional regulators, including Gata2, and that knockdown of Setd8 alters the chromatin architecture within the Gata2 locus. Lastly, knockdown of Gata2 was able to rescue some of the maturation impairment associated with Setd8 knockdown. The data presented here provide insights into the complex relationship between the chromatin modifiers and transcriptional regulators that govern erythroid maturation and suggest a novel mechanism by which histone H4 lysine methylation regulates the expression of a key transcription factor to modulate erythroid maturation.
Consistent with previous studies (69, 71), the results presented here suggest that Setd8 can influence transcription in erythroid cells by altering the chromatin landscape (i.e., increasing H4K20me1 occupancy and decreasing histone H4 acetylation) to promote transcriptional repression. In some cell types, Setd8 and H4K20me1 can also promote transcriptional repression by recruiting the repressor L3MBTL1 to target loci (68). L3MBTL1 is not expressed at significant levels in erythroid cells (33); however, it is possible that there are H4K20me1-associated repressors in erythroid cells that have yet to be described. In addition, methylation of H4K20 can influence higher-order chromatin structure by promoting chromatin compaction (72), which is also likely to result in transcriptional repression (73). Additional studies, examining Setd8 overexpression, for example, are needed to clearly delineate the mechanisms by which Setd8 and H4K20me1 regulate the expression of genes such as that encoding Gata2 in erythroid cells.
The only previously published study on the function of Setd8 in erythroid cells was done using the G1E-ER-GATA1 system. Consistent with the results presented here, microarray analyses of transient Setd8 knockdown in G1E-ER-GATA1 cells demonstrated a repressor function for Setd8 in erythroid cells and implicated Setd8 in Gata1-mediated gene repression (20). There are also some important differences between our findings and those of the previous study. Most notably, transient Setd8 disruption did not affect the maturation of G1E-ER-GATA1 cells, as determined by cell surface marker expression, and Gata2 was not identified as a differentially expressed gene. These differing results are likely attributable to differences both in the model systems and in the experimental approach.
G1E-ER-GATA1 cells have provided important insights into the transcriptional control of erythroid maturation (51–55, 64, 74); however, G1E-ER-GATA1 cells recapitulate only a narrow window of maturation and are not capable of full nuclear condensation or enucleation. In contrast to G1E-ER-GATA1 cells, ESREs are capable of completing terminal erythroid maturation, including enucleation (35, 36), and establishing stable Setd8 knockdown in ESREs allowed us to carefully assess the consequences of Setd8 knockdown on erythroid maturation. ESREs also have the advantage of doubling daily, generating an ample number of cells for high-quality functional and genomics studies that would be difficult to conduct using primary cells that are often available only in limited quantities, such as analyses of the chromatin landscape following Setd8 knockdown. Although it is possible for cell culture to introduce unwanted artifacts, Setd8 occupancy at key regulatory regions within the Gata2 locus in primary, uncultured fetal liver cells suggests that a similar occupancy demonstrated in ESREs is not spurious.
GATA2 expression is tightly controlled in a cell-type- and developmental-stage-specific manner (60, 61, 63), and dysregulated GATA2 expression underlies both inherited and acquired human genetic disease. Overexpression of GATA2 frequently occurs in acute myelogenous leukemia (49, 75–77) and is associated with a poor prognosis (77). Gata2 overexpression is also found in tumors of nonhematopoietic origin, including neuroblastoma (78) and prostate cancer (79). Conversely, mutations in the GATA2 locus that result in haploinsufficiency are associated with immunodeficiency, lymphedema, anemia, and predisposition to myelodysplasia (63, 80). Despite its importance, the molecular mechanisms regulating Gata2 expression remain incompletely understood. The data presented here, including elevated Gata2 expression following Setd8 knockdown, Setd8 occupancy at key regulatory regions in the Gata2 locus, and alterations in the chromatin structure of the Gata2 locus following Setd8 knockdown, imply that Setd8 regulates Gata2 expression in erythroid cells.
In both adipocytes and epithelial cells, Setd8 regulates lineage restricted transcription factors to modulate terminal cellular differentiation (22, 46). Our results suggest that Setd8 modulates erythroid maturation at least in part by repressing Gata2 expression. Gata2 is considered a “master regulator” of the erythroid transcription program (62), and previous studies have established a clear link between Gata2 overexpression and impaired erythroid maturation (37, 38). Dynamic interplay of Gata factor occupancy is a key driver of the terminal erythroid differentiation program, with Gata1 replacing Gata2 at numerous regulatory elements genome-wide during erythroid differentiation (51, 62). The Gata2 locus itself is a key example of Gata factor switching (64). In early erythroblasts, Gata2 occupies several important regulatory elements within the Gata2 locus, driving its own expression. As maturation proceeds, Gata1 replaces Gata2 occupancy and represses Gata2 expression (53, 54, 64). Sed8 occupies two of the well-characterized Gata2/Gata1 switch sites within the Gata2 locus (the −3.9 and +9.5 enhancers) in primary fetal liver erythroblasts. In addition, a subset of genes differentially expressed following Setd8 knockdown are associated with Gata2/Gata1 switch sites, raising the possibility that impaired Gata factor exchange contributes to the altered gene expression patterns observed following Setd8 knockdown. Future studies examining the genome-wide occupancy of Setd8 in erythroid cells will provide important insights into the role of Setd8 at Gata2/Gata1 switch sites.
Setd8 can H4K20me1 can mediate transcriptional changes in concert with a variety of transcription factors (22, 24, 25, 46) and signaling pathways (21, 31). In addition, we were unable to determine the contribution of Gata2 overexpression to terminal erythroid maturation abnormalities, such as impaired enucleation, raising the possibility that altered expression of other factors contributes to those abnormalities. As detailed above, a number of transcriptional regulators are differentially expressed following Setd8 disruption, and it is likely that differential expressions of other transcriptional regulators and chromatin modifiers, aside from Gata2, contribute to the maturation impairments observed following Setd8 knockdown. For example, our data demonstrate that disruption of Setd8 in erythroid cells significantly increased the expression of Hes1, a downstream effector of the Notch signaling pathway. Hes1 can directly interact with Gata1 and disrupt the interaction of Gata1 with the transcriptional activator and histone acetyltransferase P300 (81). Interestingly, overexpression of Hes1 has been shown to impair erythroid differentiation (9, 81), and it is possible that Hes1 overexpression contributes to the maturation impairment noted with Setd8 knockdown. In addition, although the Setd8 knockdown and control cells appeared phenotypically similar prior to maturation, flow-cytometric analyses demonstrated higher ckit levels in the Setd8 knockdown cells and RNA-seq analyses identified 1,048 genes differentially expressed prior to maturation, and it is likely that these baseline differences contributed to the impaired maturation associated with Setd8 disruption.
Our study supports the hypothesis that Setd8 is an integral part of the network of transcriptional regulators and chromatin modifiers that regulate erythroid maturation; see also the companion article by DeVilbiss et al. for a discussion of the role of Setd8 in erythroid maturation (82). Our study does have some inherent limitations. Setd8 is highly expressed in erythroid cells, and although we screened several shRNA constructs to generate the most robust Setd8 knockdown possible, there was still significant residual Setd8 expression, as determined by qPCR and Western blotting. A more efficient Setd8 knockdown may have revealed additional phenotypes, including cell cycle defects, increased rates of apoptosis, and accumulation of DNA damage. In addition, although in vitro culture systems are excellent models, they are not always able to faithfully recapitulate the complex process of erythropoiesis. The function of Setd8 has been studied extensively in embryonic stem cells and immortalized cell lines, but in vivo studies of the Setd8 H4K20me1 pathway are lacking due to the early embryonic lethality of constitutive Setd8 deletion. The only published study to date examined the role of Setd8 in the production and maintenance of epidermis (46), and in vivo studies examining the impact of Setd8 disruption on mRNA expression and the histone landscape have yet to be done in any cell type. In vivo studies are needed to further define the role of this epigenetic pathway in modulating erythropoiesis. Those studies are also likely to provide important insights into the mechanisms by which this pathway regulates transcription and modulates terminal cellular differentiation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by K08 DK090153 from the NIDDK, the American Society of Hematology's Junior Faculty Scholar Program, and a research fellowship from the Cooley's Anemia Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01413-14.
REFERENCES
- 1.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Woodcock CL, Ghosh RP. 2010. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol 2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z, Zhang MQ. 2011. Histone modification profiles are predictive for tissue/cell-type specific expression of both protein-coding and microRNA genes. BMC Bioinformatics 12:155. doi: 10.1186/1471-2105-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steiner LA, Maksimova Y, Schulz V, Wong C, Raha D, Mahajan MC, Weissman SM, Gallagher PG. 2009. Chromatin architecture and transcription factor binding regulate expression of erythrocyte membrane protein genes. Mol Cell Biol 29:5399–5412. doi: 10.1128/MCB.00777-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. 2005. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J 24:3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross J, Mavoungou L, Bresnick EH, Milot E. 2012. GATA-1 utilizes Ikaros and polycomb repressive complex 2 to suppress Hes1 and to promote erythropoiesis. Mol Cell Biol 32:3624–3638. doi: 10.1128/MCB.00163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara T, Lee HY, Sanalkumar R, Bresnick EH. 2010. Building multifunctionality into a complex containing master regulators of hematopoiesis. Proc Natl Acad Sci U S A 107:20429–20434. doi: 10.1073/pnas.1007804107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, Bernstein BE, Fraenkel E, Cantor AB. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell 36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. 2009. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takawa M, Cho HS, Hayami S, Toyokawa G, Kogure M, Yamane Y, Iwai Y, Maejima K, Ueda K, Masuda A, Dohmae N, Field HI, Tsunoda T, Kobayashi T, Akasu T, Sugiyama M, Ohnuma S, Atomi Y, Ponder BA, Nakamura Y, Hamamoto R. 2012. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res 72:3217–3227. doi: 10.1158/0008-5472.CAN-11-3701. [DOI] [PubMed] [Google Scholar]
- 14.Yin Y, Yu VC, Zhu G, Chang DC. 2008. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle 7:1423–1432. doi: 10.4161/cc.7.10.5867. [DOI] [PubMed] [Google Scholar]
- 15.Beck DB, Oda H, Shen SS, Reinberg D. 2012. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev 26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. 2010. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell 40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karachentsev D, Sarma K, Reinberg D, Steward R. 2005. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev 19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Congdon LM, Houston SI, Veerappan CS, Spektor TM, Rice JC. 2010. PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. J Cell Biochem 110:609–619. doi: 10.1002/jcb.22570. [DOI] [PubMed] [Google Scholar]
- 19.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeVilbiss AW, Boyer ME, Bresnick EH. 2013. Establishing a hematopoietic genetic network through locus-specific integration of chromatin regulators. Proc Natl Acad Sci U S A 110:E3398–E3407. doi: 10.1073/pnas.1302771110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Sun L, Zhang Y, Wang D, Wang F, Liang J, Gui B, Shang Y. 2011. The histone modifications governing TFF1 transcription mediated by estrogen receptor. J Biol Chem 286:13925–13936. doi: 10.1074/jbc.M111.223198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wakabayashi K, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I, Kitakami J, Ihara S, Hashimoto Y, Hamakubo T, Kodama T, Aburatani H, Sakai J. 2009. The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 29:3544–3555. doi: 10.1128/MCB.01856-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talasz H, Lindner HH, Sarg B, Helliger W. 2005. Histone H4-lysine 20 monomethylation is increased in promoter and coding regions of active genes and correlates with hyperacetylation. J Biol Chem 280:38814–38822. doi: 10.1074/jbc.M505563200. [DOI] [PubMed] [Google Scholar]
- 24.Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG. 2010. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. 2010. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu N, Huangyang P, Yang X, Han X, Yan R, Jia H, Shang Y, Sun L. 2013. microRNA-7 suppresses the invasive potential of breast cancer cells and sensitizes cells to DNA damages by targeting histone methyltransferase SET8. J Biol Chem 288:19633–19642. doi: 10.1074/jbc.M113.475657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang BL, Song FJ, Zheng H, Zhang LN, Zhao YR, Chen KX. 2012. SNP rs16917496 within SET8 3′UTR is associated with the age of onset of breast cancer. Zhonghua Zhong Liu Za Zhi 34:835–837. (In Chinese.) doi: 10.3760/cma.j.issn.0253-3766.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Guo Z, Wu C, Wang X, Wang C, Zhang R, Shan B. 2012. A polymorphism at the miR-502 binding site in the 3′-untranslated region of the histone methyltransferase SET8 is associated with hepatocellular carcinoma outcome. Int J Cancer 131:1318–1322. doi: 10.1002/ijc.27352. [DOI] [PubMed] [Google Scholar]
- 29.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 30.Valente S, Lepore I, Dell'Aversana C, Tardugno M, Castellano S, Sbardella G, Tomassi S, Di Maro S, Novellino E, Di Santo R, Costi R, Altucci L, Mai A. 2012. Identification of PR-SET7 and EZH2 selective inhibitors inducing cell death in human leukemia U937 cells. Biochimie 94:2308–2313. doi: 10.1016/j.biochi.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Nie F, Wang S, Li L. 2011. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci U S A 108:3116–3123. doi: 10.1073/pnas.1009353108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW III, Su AI. 2009. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kingsley PD, Greenfest-Allen E, Frame JM, Bushnell TP, Malik J, McGrath KE, Stoeckert CJ, Palis J. 2013. Ontogeny of erythroid gene expression. Blood 121:e5–e13. doi: 10.1182/blood-2012-04-422394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An X, Schulz VP, Li J, Wu K, Liu J, Xue F, Hu J, Mohandas N, Gallagher PG. 2014. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 123:3466–3477. doi: 10.1182/blood-2014-01-548305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.England SJ, McGrath KE, Frame JM, Palis J. 2011. Immature erythroblasts with extensive ex vivo self-renewal capacity emerge from the early mammalian fetus. Blood 117:2708–2717. doi: 10.1182/blood-2010-07-299743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Getman M, England SJ, Malik J, Peterson K, Palis J, Steiner LA. 2014. Extensively self-renewing erythroblasts derived from transgenic beta-Yac mice are a novel model system for studying globin switching and erythroid maturation. Exp Hematol 42:536–46.e8. doi: 10.1016/j.exphem.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikonomi P, Rivera CE, Riordan M, Washington G, Schechter AN, Noguchi CT. 2000. Overexpression of GATA-2 inhibits erythroid and promotes megakaryocyte differentiation. Exp Hematol 28:1423–1431. doi: 10.1016/S0301-472X(00)00553-1. [DOI] [PubMed] [Google Scholar]
- 38.Persons DA, Allay JA, Allay ER, Ashmun RA, Orlic D, Jane SM, Cunningham JM, Nienhuis AW. 1999. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood 93:488–499. [PubMed] [Google Scholar]
- 39.Briegel K, Lim KC, Plank C, Beug H, Engel JD, Zenke M. 1993. Ectopic expression of a conditional GATA-2/estrogen receptor chimera arrests erythroid differentiation in a hormone-dependent manner. Genes Dev 7:1097–1109. doi: 10.1101/gad.7.6.1097. [DOI] [PubMed] [Google Scholar]
- 40.Heyworth C, Gale K, Dexter M, May G, Enver T. 1999. A GATA-2/estrogen receptor chimera functions as a ligand-dependent negative regulator of self-renewal. Genes Dev 13:1847–1860. doi: 10.1101/gad.13.14.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGrath KE, Bushnell TP, Palis J. 2008. Multispectral imaging of hematopoietic cells: where flow meets morphology. J Immunol Methods 336:91–97. doi: 10.1016/j.jim.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.David M, Dzamba M, Lister D, Ilie L, Brudno M. 2011. SHRiMP2: sensitive yet practical SHort Read Mapping. Bioinformatics 27:1011–1012. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- 43.Roberts A, Pimentel H, Trapnell C, Pachter L. 2011. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 44.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen K, Liu J, Heck S, Chasis JA, An X, Mohandas N. 2009. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci U S A 106:17413–17418. doi: 10.1073/pnas.0909296106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Driskell I, Oda H, Blanco S, Nascimento E, Humphreys P, Frye M. 2012. The histone methyltransferase Setd8 acts in concert with c-Myc and is required to maintain skin. EMBO J 31:616–629. doi: 10.1038/emboj.2011.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma A, Singh K, Almasan A. 2012. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol Biol 920:613–626. doi: 10.1007/978-1-61779-998-3_40. [DOI] [PubMed] [Google Scholar]
- 48.Kramer A, Green J, Pollard J Jr, Tugendreich S. 2014. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vicente C, Conchillo A, Garcia-Sanchez MA, Odero MD. 2012. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol Hematol 82:1–17. doi: 10.1016/j.critrevonc.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 50.Dore LC, Chlon TM, Brown CD, White KP, Crispino JD. 2012. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood 119:3724–3733. doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, Giardine B, Schuster SC, Miller W, Chiaromonte F, Zhang Y, Blobel GA, Weiss MJ, Hardison RC. 2009. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res 19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss MJ, Yu C, Orkin SH. 1997. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol 17:1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martowicz ML, Grass JA, Boyer ME, Guend H, Bresnick EH. 2005. Dynamic GATA factor interplay at a multicomponent regulatory region of the GATA-2 locus. J Biol Chem 280:1724–1732. doi: 10.1074/jbc.M406038200. [DOI] [PubMed] [Google Scholar]
- 54.Grass JA, Jing H, Kim SI, Martowicz ML, Pal S, Blobel GA, Bresnick EH. 2006. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol 26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. 2012. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150:725–737. doi: 10.1016/j.cell.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minegishi N, Ohta J, Suwabe N, Nakauchi H, Ishihara H, Hayashi N, Yamamoto M. 1998. Alternative promoters regulate transcription of the mouse GATA-2 gene. J Biol Chem 273:3625–3634. doi: 10.1074/jbc.273.6.3625. [DOI] [PubMed] [Google Scholar]
- 59.Pan X, Minegishi N, Harigae H, Yamagiwa H, Minegishi M, Akine Y, Yamamoto M. 2000. Identification of human GATA-2 gene distal IS exon and its expression in hematopoietic stem cell fractions. J Biochem 127:105–112. doi: 10.1093/oxfordjournals.jbchem.a022570. [DOI] [PubMed] [Google Scholar]
- 60.Snow JW, Trowbridge JJ, Johnson KD, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, Bresnick EH. 2011. Context-dependent function of “GATA switch” sites in vivo. Blood 117:4769–4772. doi: 10.1182/blood-2010-10-313031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snow JW, Trowbridge JJ, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, Bresnick EH. 2010. A single cis element maintains repression of the key developmental regulator Gata2. PLoS Genet 6:e1001103. doi: 10.1371/journal.pgen.1001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. 2010. GATA switches as developmental drivers. J Biol Chem 285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson KD, Hsu AP, Ryu MJ, Wang J, Gao X, Boyer ME, Liu Y, Lee Y, Calvo KR, Keles S, Zhang J, Holland SM, Bresnick EH. 2012. Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J Clin Invest 122:3692–3704. doi: 10.1172/JCI61623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A 100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 66.Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J 24:2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, Rice JC, Nimer SD. 2008. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 27:4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trojer P, Li G, Sims RJ III, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, Wang YH, Reinberg D. 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 69.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9:1201–1213. doi: 10.1016/S1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 70.Miotto B, Struhl K. 2010. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 37:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. 2008. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol 28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, Luger K. 2008. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat Struct Mol Biol 15:1122–1124. doi: 10.1038/nsmb.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sproul D, Gilbert N, Bickmore WA. 2005. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet 6:775–781. doi: 10.1038/nrg1688. [DOI] [PubMed] [Google Scholar]
- 74.Johnson KD, Boyer ME, Kang JA, Wickrema A, Cantor AB, Bresnick EH. 2007. Friend of GATA-1-independent transcriptional repression: a novel mode of GATA-1 function. Blood 109:5230–5233. doi: 10.1182/blood-2007-02-072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimamoto T, Ohyashiki K, Ohyashiki JH, Kawakubo K, Fujimura T, Iwama H, Nakazawa S, Toyama K. 1995. The expression pattern of erythrocyte/megakaryocyte-related transcription factors GATA-1 and the stem cell leukemia gene correlates with hematopoietic differentiation and is associated with outcome of acute myeloid leukemia. Blood 86:3173–3180. [PubMed] [Google Scholar]
- 76.Ayala RM, Martinez-Lopez J, Albizua E, Diez A, Gilsanz F. 2009. Clinical significance of Gata-1, Gata-2, EKLF, and c-MPL expression in acute myeloid leukemia. Am J Hematol 84:79–86. doi: 10.1002/ajh.21332. [DOI] [PubMed] [Google Scholar]
- 77.Vicente C, Vazquez I, Conchillo A, Garcia-Sanchez MA, Marcotegui N, Fuster O, Gonzalez M, Calasanz MJ, Lahortiga I, Odero MD. 2012. Overexpression of GATA2 predicts an adverse prognosis for patients with acute myeloid leukemia and it is associated with distinct molecular abnormalities. Leukemia 26:550–554. doi: 10.1038/leu.2011.235. [DOI] [PubMed] [Google Scholar]
- 78.Hoene V, Fischer M, Ivanova A, Wallach T, Berthold F, Dame C. 2009. GATA factors in human neuroblastoma: distinctive expression patterns in clinical subtypes. Br J Cancer 101:1481–1489. doi: 10.1038/sj.bjc.6605276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bohm M, Locke WJ, Sutherland RL, Kench JG, Henshall SM. 2009. A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene 28:3847–3856. doi: 10.1038/onc.2009.243. [DOI] [PubMed] [Google Scholar]
- 80.Ostergaard P, Simpson MA, Connell FC, Steward CG, Brice G, Woollard WJ, Dafou D, Kilo T, Smithson S, Lunt P, Murday VA, Hodgson S, Keenan R, Pilz DT, Martinez-Corral I, Makinen T, Mortimer PS, Jeffery S, Trembath RC, Mansour S. 2011. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat Genet 43:929–931. doi: 10.1038/ng.923. [DOI] [PubMed] [Google Scholar]
- 81.Ishiko E, Matsumura I, Ezoe S, Gale K, Ishiko J, Satoh Y, Tanaka H, Shibayama H, Mizuki M, Era T, Enver T, Kanakura Y. 2005. Notch signals inhibit the development of erythroid/megakaryocytic cells by suppressing GATA-1 activity through the induction of HES1. J Biol Chem 280:4929–4939. doi: 10.1074/jbc.M406788200. [DOI] [PubMed] [Google Scholar]
- 82.DeVilbiss AW, Sanalkumar R, Hall BDR, Katsumura KR, de Andrade IF, Bresnick EH. 2015. Epigenetic determinants of erythropoiesis: role of the histone methyltransferase SetD8 in promoting erythroid cell maturation and survival. Mol Cell Biol 35:2073–2087. doi: 10.1128/MCB.01422-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.