Abstract
The ordered assembly of a functional preinitiation complex (PIC), composed of general transcription factors (GTFs), is a prerequisite for the transcription of protein-coding genes by RNA polymerase II. TFIID, comprised of the TATA binding protein (TBP) and 13 TBP-associated factors (TAFs), is the GTF that is thought to recognize the promoter sequences allowing site-specific PIC assembly. Transcriptional cofactors, such as SAGA, are also necessary for tightly regulated transcription initiation. The contribution of the two TAF10-containing complexes (TFIID, SAGA) to erythropoiesis remains elusive. By ablating TAF10 specifically in erythroid cells in vivo, we observed a differentiation block accompanied by deregulated GATA1 target genes, including Gata1 itself, suggesting functional cross talk between GATA1 and TAF10. Additionally, we analyzed by mass spectrometry the composition of TFIID and SAGA complexes in mouse and human cells and found that their global integrity is maintained, with minor changes, during erythroid cell differentiation and development. In agreement with our functional data, we show that TAF10 interacts directly with GATA1 and that TAF10 is enriched on the GATA1 locus in human fetal erythroid cells. Thus, our findings demonstrate a cross talk between canonical TFIID and SAGA complexes and cell-specific transcription activators during development and differentiation.
INTRODUCTION
Initiation of RNA polymerase II (RNA pol II) transcription in eukaryotes is a process involving the stepwise recruitment and assembly of the preinitiation complex (PIC) at the core promoter of a transcriptional unit. Transcription factor TFIID, comprised of the TATA binding protein (TBP) and a series of TBP-associated factors (TAFs), is the general transcription factor (GTF) that, by recognizing the promoter sequences and surrounding chromatin marks, allows the site-specific assembly of the PIC (see reference 1 and references therein). Binding of the TFIID complex is aided by TFIIA and is followed by the remainder of the general transcription machinery, including TFIIB, RNA pol II/TFIIF, TFIIE, and TFIIH complexes. Additional cofactors, including the Mediator complex, histone modifiers, and chromatin remodelers, facilitate the communication between gene-specific transcription factors and the general transcription machinery.
The TFIID complex binds not only to TATA box-containing promoters but also to TATA-less promoters, and this led to the idea that TAFs could provide TFIID with additional functional features (2, 3). Indeed, 9 out of 13 TAFs contain a histone fold domain (HFD) (4) favoring the formation of TAF heterodimers. For instance, the TAF6-TAF9 heterodimer has been found to bind promoter elements downstream of the TATA box (5–7) and is a direct target of transcriptional activators (8, 9). Moreover, it has been shown that TAF knockouts (KOs) and in vitro TAF-knockdown experiments result in both the down- and upregulated expression of subsets of genes (10, 11). All these results together suggest that TFIID is a highly flexible regulator of transcription, functioning both in gene activation and in repression.
Additionally, coactivator complexes with histone acetyltransferase (HAT) activity, responsible for gene activation-associated interactions, including the ATAC (Ada-two-A-containing) and SAGA (Spt-Ada-Gcn5-acetyltransferase) complexes, appear to have distinct functional roles by targeting either promoters or enhancers, or both (see reference 12 and references therein).
TAF10 is a subunit of both the TFIID and the SAGA coactivator HAT complexes (13). The role of TAF10 is indispensable for early embryonic transcription and mouse development, as TAF10-KO embryos die early in gestation (between embryonic day 3.5 [E3.5] and E5.5), at about the stage when the supply of maternal protein becomes insufficient (14). However, when analyzing TFIID stability and transcription, it was noted that not all cells and tissues were equally affected by the loss of TAF10. For example, ablation of TAF10 in keratinocytes impaired skin barrier formation and deregulated a subset of related genes when inactivated during the fetal stage but resulted in no detectable effect in adult keratinocytes (15). Moreover, studies in which TAF10 was conditionally ablated in fetal or adult liver demonstrated the essential role of TAF10 in liver development, revealing the necessity of TAF10 for TFIID stability to repress specific genes in the liver in postnatal life (10). These findings together confirm that TAF10, probably as a subunit of TFIID and/or SAGA, is essential during mouse development and suggest that TAF10 plays an important role during embryonic development and homeostasis in a tissue-dependent manner. Understanding the interplay of TAF10-containing TFIID and SAGA complexes with developmentally important and tissue-specific transcription factors is crucial to obtain a more comprehensive view of cell differentiation throughout development.
Erythropoiesis is the process by which red blood cells are formed (16). There are two waves of erythropoiesis in mammals, primitive and definitive. Definitive erythropoiesis starts in the fetal liver and later during gestation moves to the spleen and bone marrow, which in mice remain the sites of erythropoiesis during adulthood. The fetal and adult stages of definitive erythropoiesis differ at the transcriptional level, exemplified in humans by the type of beta-hemoglobin chain expressed. Many tissue-specific transcription factors have been studied in order to provide mechanistic clues to this process of developmental stage-specific hemoglobin expression (17). GATA1 is one of them; it is expressed in lineage-committed cells (erythroid, megakaryocytic, eosinophilic, mast, and dendritic cells) and plays an important role in the regulation of differentiation and survival of these lineages (18, 19). Embryos lacking Gata1 die at approximately embryonic day 11.5 due to the maturation arrest of primitive erythroid cells (20), while conditional knockout of Gata1 in adults leads to red blood cell aplasia and severe thrombocytopenia (21). However, the composition of general transcription complexes, such as TFIID and SAGA, and the role of TAFs during developmental erythropoiesis have not been investigated yet.
Alternative TFIID and other GTF complexes have been implicated in providing alternative pathways leading to gene regulation during differentiation (22, 23). To gain insight into the role of GTF in mouse erythropoiesis, we carried out the specific inactivation of TAF10, a cornerstone subunit of the TFIID and SAGA complexes (10), in the erythroid cell compartment by crossing TAF10lox mice (14) with EpoR-Cre mice (24). We found that TAF10 ablation results in a block of erythropoiesis, leading to severe anemia, which is lethal at E13.5. Several GATA1 target genes, including Gata1 itself, were deregulated when TAF10 was ablated. We also analyzed by mass spectrometry (MS) the composition and stoichiometry of the TAF10-containing transcription complexes TFIID and SAGA in proliferating erythroid cell precursors (proerythroblasts) and synchronously differentiated erythroid cells of mouse and human origin. Interestingly, we found that TAF10 interacts physically with the master regulator of erythroid cell differentiation, the GATA1 transcription factor, and we observed enrichment of TAF10 binding to the GATA1 locus in human erythroid cells. Collectively, these data suggest that the interaction of TAF10 with GATA1 is important to facilitate the recruitment of TFIID and/or SAGA to GATA1-responsive promoters and that the autologous control of GATA1 expression (25) requires the presence of TAF10 in these complexes.
MATERIALS AND METHODS
Mice.
The TAF10lox/KO mice (14) and EpoR-Cre mice (24) used in this study have been described previously. TAF10lox/KO:EpoR-Cre+/− mice (here called TAF10KOcEry mice) and mice with single genetic modifications were maintained in a C57BL/6 mouse background. All experiments described in this article have been approved and conducted according to the guidelines of the Animal Welfare Committee of Erasmus MC, Rotterdam, The Netherlands.
Human material.
Samples of fetal liver and buffy coats from peripheral blood were provided by the clinic and the Sanquin Blood Bank in The Netherlands, in compliance with the guidelines of the Erasmus MC Ethical Guidelines Committee.
Flow cytometry.
Fetal livers were dissected from embryos at E11.5, E12.5, and E13.5 and resuspended in phosphate-buffered saline (PBS) supplemented with 1% bovine serum albumin (BSA) as single-cell suspensions. Cells were stained on ice for 20 min and washed twice with PBS–1% BSA before acquisition on a FACSAria II instrument (Becton Dickinson [BD]). Antibodies used for staining included TER119-peridinin chlorophyll protein-Cy5.5 (BD), KIT (CD117)-phycoerythrin (PE)-Cy7 (BD), and CD71-PE (BD). Since the EpoR-Cre allele includes a green fluorescent protein (GFP) reporter (24), EpoR-driven Cre expression was followed by GFP expression in the fluorescein isothiocyanate channel. Dead cells were excluded by Hoechst staining (Invitrogen), and analysis was performed with FlowJo software (Tree Star, Inc.).
Extract preparation and IP assays.
Nuclear extracts were prepared as described previously (10, 26). TAF10 immunoprecipitations (IPs) were performed as previously described for mouse cells (10) and human cells (27). Mouse monoclonal antibodies 23TA 1H8 (28) and 6TA 2B11 (14) were used for human (29) and mouse TAF10 IPs, respectively, and a mix of monoclonal antibodies 6TA 2B11 and 6TA 4G2 (27) (dilution, 1/1,000) against the TAF10 protein was used for the assessment of IP efficiency by Western blotting. Mock IP was performed using a glutathione S-transferase (GST) antibody (sc-80004 antibody; Santa Cruz) according to the manufacturer's recommendations.
Mass spectrometry was performed as described before (30) on an LTQ-Orbitrap mass spectrometer (Thermo).
Purification of proteins.
GST-GATA1 and GST proteins (pGEX expression vectors) were expressed in Escherichia coli BL21 upon IPTG (isopropyl-β-d-thiogalactopyranoside; 0.4 mM) induction for 3 h at 30°C. Bacteria were lysed (50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 5% glycerol, 1 mM glutathione, 2.5 mM phenylmethylsulfonyl fluoride, 50 μg/ml DNase, 1 mM MgCl2, complete protease inhibitor cocktail [Roche]) and passed through a pressure plunger three or four times. After centrifugation at 20,000 rpm for 20 min at 4°C, the extract was incubated with preequilibrated (with wash buffer without glutathione) glutathione beads (glutathione-Sepharose 4B; GE Healthcare) for 1 h at room temperature. After three washes (50 mM Tris-HCl, pH 8.0, 1 M NaCl, 5% glycerol, 1 mM glutathione, complete protease inhibitor cocktail), the proteins were eluted (50 mM glutathione, 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, complete protease inhibitor cocktail).
ChIP.
Preparation of chromatin from human erythroid progenitor cell (HEP) cultures was previously described (10). Fetal liver and adult HEPs were cultured (31) and used for chromatin immunoprecipitation (ChIP) reactions, which were performed as described previously (32) with the 23TA 1H8 antibody clone against the human TAF10 protein, the GATA1 antibody (ab11852), and a CD71 antibody (347510; BD Biosciences) as a negative control. Quantitative PCR (qPCR) was performed on the input and immunoprecipitated samples using primers for the HS2 binding site (a palindromic GATA1 binding site) or a more proximal GATA1 binding site at the GATA1 promoter. The relative fold enrichment was calculated using the ΔΔCT threshold cycle (CT) method (33), setting the relative fold enrichment of CD71 background binding equal to 1. The primers used were Hs GATA1 palindromic binding site forward (Fw) primer 5′-AGACTTATCTGCTGCCCCAG-3′, Hs GATA1 palindromic binding site reverse (Rev) primer 5′-CCAGGCTAAGCCTGCAGGC-3′ or Rev primer 5′-TAGAGCCTGTGGGATACCTTG-3′, Hs GATA1 binding site at kb −3 Fw primer 5′-GGGATGAGGGAATAGTGGTG-3′, and Hs GATA1 binding site at kb −3 Rev primer 5′-GCTCTTTGTCTCTGTGTCTCTGTC-3′.
Gene expression (RNA-seq and quantitative reverse transcription-PCR [qRT-PCR]).
The transcriptome sequencing (RNA-seq) library was generated according to the Illumina protocol using a TruSeq RNA sample preparation kit (version 2). Five hundred nanograms of total RNA was initially extracted with the TRIzol reagent (Invitrogen) from the livers of TAF10KOcEry and wild-type (WT) mouse embryos at E12.5. The quality of the RNA was checked with a NanoDrop analyzer and on an Agilent Technologies Bioanalyzer 2100 apparatus. Sequencing was performed on an Illumina HiSeq 2000 instrument. The reads were aligned by use of the Tophat (version 2.0.8) program (34) on a UCSC mm10 reference genome using the Ensembl (version 75) gene models (35).
qRT-PCR.
cDNA synthesis was performed from 1 μg of RNA with a DyNamo cDNA synthesis kit (F-470; Finnzymes), cDNA was diluted to a 10-ng/μl concentration, and 10 ng of cDNA was used per reaction mixture with a 20-μl total volume. The conditions of the qRT-PCR and the primer sequences used have already been described (10), and reactions were performed in Applied Biosystems thermal real-time PCR instruments (ABI 7900).
RNA-seq analysis.
Raw counts were measured with the htseq-count (version 0.6.0) tool using −m union −s no −a 20 as settings (36). The counted data were normalized by the size factor of the libraries, and subsequently, the number of fragments per kilobase of exon per million fragments mapped (FPKM) was calculated (GEO accession number GSE68083). The differentially expressed genes were called using a generalized linear negative binomial model that controlled for the effect of the RNA sample preparation date. The calculations were performed by the DESeq2 R package (37). The false discovery rate (FDR) was calculated by the method of Hochberg and Benjamini (38), and the threshold value was set equal to 0.01. Gene ontology (GO) gene enrichment analyses were carried out with the GO-stat package (39) using a conditional hypergeometrical test for overrepresented Ensembl gene identifiers using a threshold P value of 1 × 10−6. Principal component analysis (PCA) was carried out using Pearson's correlation matrix after blind variance stabilizing transformation of the normalized counts (37).The RNA-seq data were joined with the GATA1 ChIP sequencing (ChIP-seq) data (40) and plotted in a Venn diagram. The statistical package R was used for calculations and plotting of the data (41).
Culture of erythroid cells.
Erythroid cell culture conditions have been described for mouse (42) and human erythroid progenitor cells (31). Cells were kept under proliferation or differentiation conditions, and they were followed by cell density and size monitoring with a Casy instrument (Innovatis, Roche Diagnostics GmbH).
Proteomics data accession number.
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD000729 (http://www.ebi.ac.uk/pride/archive/projects/PXD000729).
RESULTS
Erythroid cell-specific knockout of TAF10 shows its requirement for erythropoiesis.
To better understand the role of TFIID and SAGA in erythroid cell differentiation, we generated mice in which TAF10 was specifically ablated in erythroid cells. TAF10+/− mice are normal (14); therefore, we generated mice bearing a TAF10-knockout allele and a TAF10 LoxP-flanked allele in order to reduce the recombination requirements to one allele when generating conditional knockout mice. In order to study the TAF10 loss of function specifically in erythroid cells, we generated TAF10lox/KO animals bearing the erythroid cell-specific EpoR-Cre allele (24), and we refer to them as TAF10KOcEry mice. The expression of EpoR starts at E8.0 in yolk sac erythroid progenitor cells (43) and increases during definitive erythropoiesis in BFU-E (burst forming unit- erythroid) progenitors, while reaching maximal levels in CFU-E (colony forming unit-erythroid) progenitors.
Definitive erythropoiesis starts at E10.5 in the fetal liver, but definitive erythroid cells are detected in the circulation after E11.5 and reach an approximate 1:1 ratio with still circulating primitive erythroid cells derived from the yolk sac only at E13.5 (44, 45). In order to analyze this developmental transition period, we performed an analysis of TAF10KOcEry and control embryos at E11.5, E12.5, and E13.5. From E12.5 onwards, TAF10KOcEry embryos were paler than control or heterozygous littermates, and all TAF10KOcEry embryos were dead by E13.5. A representative picture of E12.5 embryos is shown in Fig. 1A. Gross morphological analysis revealed that the fetal liver size was considerably reduced at E12.5 in TAF10KOcEry embryos compared to the size of the livers of control littermates (Fig. 1A). The fetal liver size and total blood cell counts were significantly reduced at E13.5 (Fig. 1B). Flow cytometry analysis of fetal liver cells showed a decrease in live cells at E13.5 (Fig. 1C and Table 1), in concordance with the apoptotic phenotype that has previously been shown in other cell types lacking TAF10 (10, 14). This decrease started to be noticeable at E12.5 and was accompanied by reduced differentiation, as measured by flow cytometry (see below). In summary, the erythroid cell-specific loss of TAF10 dramatically affected the erythroid cell compartment between E12.5 and E13.5.
FIG 1.
Developmental analysis of TAF10KOcEry. (A) Representative pictures of control and TAF10KOcEry embryos at E12.5. TAF10KOcEry embryos are pale, and the fetal livers of TAF10KOcEry embryos are much smaller than those of the control embryos. (B) Cell counts at E13.5 from fetal liver and embryonic blood of control and TAF10KOcEry embryos. (C) Flow cytometry analysis of single-cell suspensions of fetal liver from control and TAF10KOcEry embryos at different developmental stages (E11.5, E12.5, and E13.5). Numbers on the right indicate the percentages of live cells (Hoechst negative).
TABLE 1.
Flow cytometry analysis of fetal livers
| Parameter | Embryonic day | Result for: |
P valuea | |
|---|---|---|---|---|
| Control mice | Taf10KOcEry mice | |||
| % live cells | E11.5 | 39.55 ± 7.51 | 38.20 ± 22.49 | NS |
| E12.5 | 63.39 ± 5.94 | 52.53 ± 4.16 | <0.005 | |
| E13.5 | 60.69 ± 3.21 | 6.55 ± 6.13 | <0.001 | |
| % c-Kit-positive cells | E11.5 | 22.45 ± 5.99 | 35.15 ± 4.60 | NS |
| E12.5 | 5.61 ± 0.98 | 11.47 ± 1.24 | <0.001 | |
| E13.5 | 3.85 ± 0.43 | 0.35 ± 0.40 | <0.001 | |
| % c-Kit- and CD71-positive cells | E11.5 | 37.78 ± 6.96 | 20.95 ± 15.20 | NS |
| E12.5 | 16.30 ± 2.84 | 26.43 ± 5.67 | <0.05 | |
| E13.5 | 8.03 ± 1.10 | 0.05 ± 0.03 | <0.001 | |
| % CD71-positive cells | E11.5 | 17.10 ± 5.85 | 7.79 ± 6.38 | NS |
| E12.5 | 69.48 ± 3.19 | 50.95 ± 6.16 | <0.01 | |
| E13.5 | 82.92 ± 2.09 | 14.50 ± 6.85 | <0.001 | |
| % CD71- and Ter119-positive cells | E11.5 | 29.00 ± 9.29 | 14.95 ± 10.54 | NS |
| E12.5 | 78.15 ± 1.99 | 55.93 ± 7.21 | <0.01 | |
| E13.5 | 89.99 ± 1.64 | 14.57 ± 8.79 | <0.001 | |
| c-Kit MFI | E11.5 | 11,121.63 ± 1,768.39 | 10,149.00 ± 2,604.98 | NS |
| E12.5 | 2,645.51 ± 126.30 | 3,331.04 ± 618.45 | NS | |
| E13.5 | 1,440.50 ± 331.33 | 221.98 ± 188.65 | <0.001 | |
| CD71 MFI | E11.5 | 41,095.75 ± 5,407.63 | 19,293.50 ± 12,581.55 | NS |
| E12.5 | 36,305.74 ± 2,721.79 | 31,207.59 ± 1,445.95 | <0.001 | |
| E13.5 | 43,745.10 ± 2,266.78 | 3,034.50 ± 1,274.59 | <0.001 | |
| Ter119 MFI | E11.5 | 1,312.75 ± 250.17 | 647.50 ± 7.78 | <0.001 |
| E12.5 | 3,175.30 ± 215.79 | 2,194.38 ± 323.44 | <0.005 | |
| E13.5 | 3,734.50 ± 339.00 | 359.75 ± 92.00 | <0.001 | |
Statistical analysis derived from the flow cytometry analysis of fetal livers of Taf10KOcEry and control embryos at E11.5, E12.5, and E13.5. At least 3 embryos were analyzed per genotype per stage. NS, no significant difference.
We next aimed to determine the differentiation stage at which TAF10 is essential during the fetal erythroid cell differentiation process. Thus, we analyzed E11.5, E12.5, and E13.5 fetal livers by flow cytometry. Despite the fact that gross morphological analysis of embryos at E11.5 revealed no significant differences, the distribution of the early erythroid cell markers KIT and CD71 was altered in TAF10KOcEry fetal livers compared to that in the livers of the control littermates and expression of Ter119 was significantly lower (Table 1). At this stage, the frequency of KIT-positive (KIT+) early progenitors was higher and the frequency of KIT+ CD71+ committed erythroid progenitor cells as well as that of the more mature CD71+ cell population was reduced (Fig. 2A), but the difference was not as significant as that observed at E12.5 (Table 1). By E12.5, both KIT+ early erythroid progenitor cells and KIT+ CD71+ committed erythroid progenitor cells had accumulated at the detriment of the more mature KIT-negative CD71+ cells. By E13.5 there was an almost complete loss of KIT+ CD71+ cells. These data demonstrate a block in the differentiation of the erythroid progenitor cells in the fetal liver throughout development. In addition, by E12.5 the KIT mean fluorescence intensity (MFI) was higher in TAF10KOcEry fetal livers than in the livers of the control littermates, although the difference not statistically significant (Fig. 2C and Table 1). The CD71 MFI was reduced at E11.5 and was significantly lower at E12.5 and E13.5, further supporting the notion of a differentiation defect. The maturation delay and the reduction of mature cells (the percentage of KIT+, CD71+, and Ter119-positive [Ter119+] cells; Table 1) by E12.5 coincided with the maximum expression of EpoR-Cre, as measured by determination of the expression of the GFP reporter of EpoR-Cre mice by flow cytometry (Fig. 3) (24). Furthermore, analysis of CD71 and Ter119 expression as markers of maturation in erythroid cells revealed a significant reduction in the fraction of mature CD71+ Ter119+ cells by E12.5 and a dramatic decrease by E13.5 (Fig. 2B). The MFI of Ter119 was reduced throughout development, as measured from E11.5 to E13.5 (Fig. 2C and Table 1). These data strongly suggest that erythroid cell development requires TAF10 and that erythroid cells are blocked in differentiation upon TAF10 ablation. Since by E13.5 >80% of the fetal liver cells are mature erythroid cells (CD71+ Ter119+) in control embryos (Table 1), it is not surprising that TAF10KOcEry embryos do not survive beyond this stage.
FIG 2.
Flow cytometry of TAF10KOcEry fetal liver cells during gestation. Representative flow cytometry analysis of single-cell suspensions from the livers of control and TAF10KOcEry embryos at E11.5, E12.5, and E13.5 following the differentiation of erythroid cells. (A) Staining with KIT and CD71 markers (numbers indicate the percentages of gated live cells); (B) staining with CD71 and Ter119 markers (numbers indicate the percentages of gated live cells); (C) MFIs of KIT, CD71, and Ter119 populations expressed during development of erythroid cells of control and TAF10KOcEry fetal liver cells.
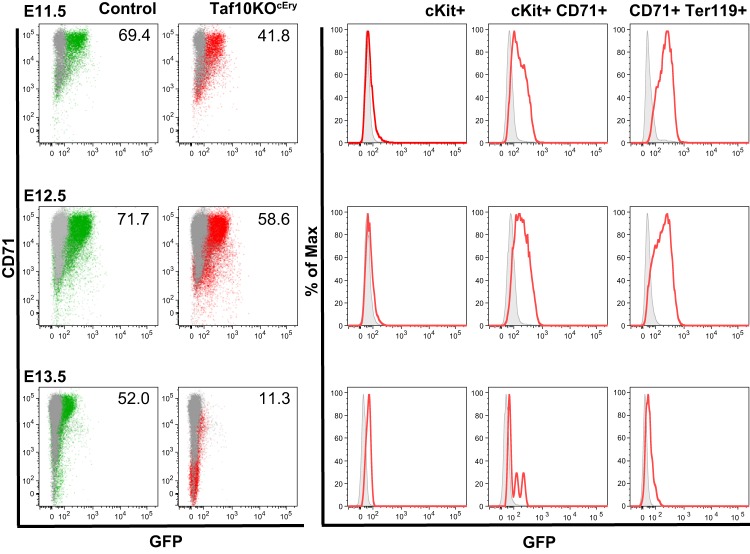
FIG 3.
Analysis of EpoR-Cre transgenic expression. Fetal liver cells were analyzed by flow cytometry following the expression of GFP in frame with Cre recombinase in EpoR-Cre mice. Expression of GFP confined to the erythroid cell compartment was detected initially in KIT+ CD71+ cells at E11.5 (green population, controls; red population, TAF10KOcEry erythroid cells). Erythroid cells were affected in TAF10KOcEry embryos, and that is why the percentage (the numbers shown in the graphs) of CD71+ GFP+ cells is lower than that of control cells at E12.5. By E13.5 most of the cells were dead.
RNA-seq analysis of E12.5 TAF10KOcEry fetal liver cells shows deregulation of several GATA1 target genes, including Gata1 itself.
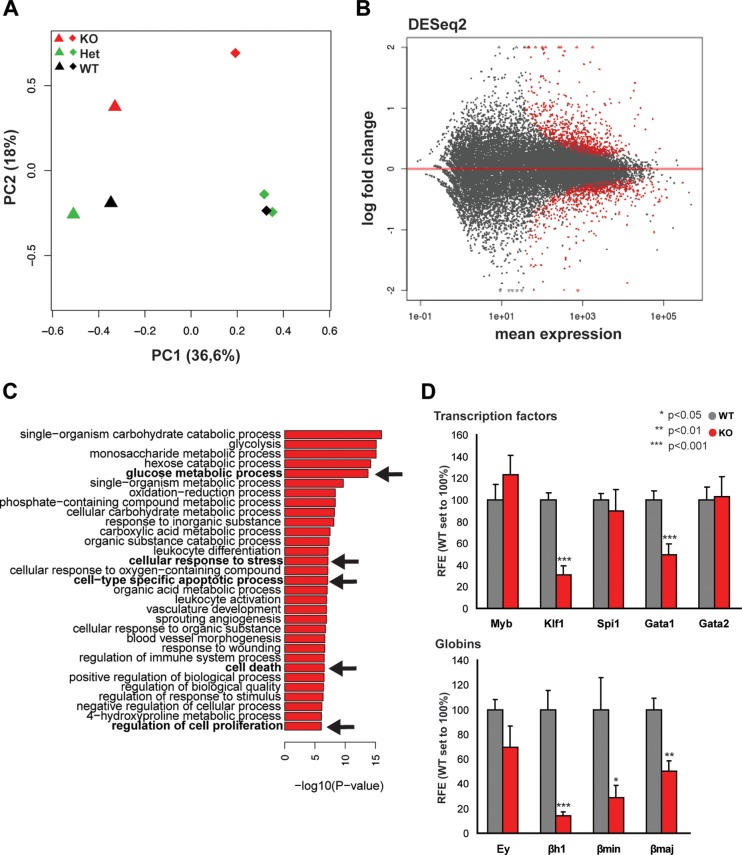
To better characterize the TAF10-regulated genes in erythroid cells, we analyzed global gene expression levels by sequencing of mRNA from E12.5 fetal liver cells from two homozygous TAF10KOcEry, two heterozygous TAF10KOcEry, and three WT embryos. By using a principal component analysis (PCA), we observed a separate clustering of the homozygous TAF10KOcEry fetal liver samples from the fetal liver samples from the heterozygous TAF10KOcEry and WT controls at E12.5 (Fig. 4A). At this stage, when live erythroid cells are still present in the fetal liver, the gene expression analysis revealed about 300 deregulated genes, including the Taf10 gene itself, with a minimum of a 1.5-fold change (Fig. 4B and D). TAF10 levels, as expected, were reduced by more than 50% in TAF10KOcEry fetal livers compared to those in the controls, which corresponds approximately to the percentage of erythroid cells (approximately 50%) beyond the KIT+ CD71+ committed erythroid progenitor cell stage in the KO fetal livers (Fig. 2A and Table 1), when EpoR expression (and, therefore, Cre expression) reaches its maximum levels (as shown in Fig. 3). Gene ontology (GO) analysis revealed that major metabolic pathways, the cellular response to stress, cell-type-specific apoptosis, and cell death-related processes were among the most affected ones (Fig. 4C). The expression levels of TFIID subunits were not significantly changed, consistent with the findings of a previous analysis of TAF10-KO trophoblast and mouse fetal liver (10, 14), suggesting that the composition of the TFIID core complex containing five TAFs (TAFs 4, 5, 6, 9, and 12) (46) does not change upon TAF10 loss. Similarly, the expression of the majority of the SAGA subunits was not affected, with the exception of the expression of the Trrap and Atxn7l1 genes, which showed modest upregulation.
FIG 4.
Analysis of gene expression in TAF10KOcEry fetal livers at E12.5. (A) PCA plot of the first two components of the fetal liver samples (from TAF10KOcEry mice [KO], heterozygous TAF10KO mice [Het], and WT mice) analyzed by RNA-seq. The variance explained by each component is depicted in parentheses on the axes. (B) MA plot of the mean normalized gene count versus the log2 fold changes in gene expression in TAF10KOcEry mice compared with that in WT and heterozygous TAF10KO mice. Genes are plotted as black circles. Genes with an adjusted P value (FDR) of <0.01 are colored red. Genes that fall out of the window boundaries of −2 or 2 log2 fold change are plotted as open triangles. (C) GO analysis of the upregulated genes in the TAF10KOcEry fetal livers. Metabolic pathways, cell-type-specific apoptotic and cell death-related processes, and proliferation were among the most affected and are indicated in boldface and with arrows. (D) qRT-PCR of total mRNA of fetal liver cells at E12.5. The expression levels of transcription factors and globin genes are depicted. Bars represent standard errors of the means (SEMs). RFE, relative fold enrichment.
Initially, the expression levels of the Gata1, Gata2, Myb, Klf1, and Spi1 genes were measured by qRT-PCR (Fig. 4D), and only Gata1 and Klf1 displayed a significant change in their expression levels. Subsequently, in the RNA-seq analysis, we found that many of the differentially expressed genes were transcription factors and other erythroid cell-specific genes which have recently been identified to be direct GATA1 targets by chromatin immunoprecipitation followed by sequencing (ChIP-seq) using E12.5 mouse fetal liver cells (40). We observed the downregulation of genes encoding relevant transcription factors during erythropoiesis, such as Gata1, Klf1, Nfe2, Zptba7, Bcl2l1, and other metabolically crucial erythroid cell-specific genes, including Slc4a1, Gypa, Alas2, and Car2 (Fig. 5A and C). The genes encoding the TFDP22 coregulator, recently reported to be an essential factor during terminal erythropoiesis (47), and the E2F2 transcription factor, together with two E2F2 target genes (Dhfr and Ccna2), were significantly downregulated in TAF10KOcEry fetal livers. We also found a subset of upregulated genes (Spi1, Gata2, Runx1, Cux1, Car1, Rb1, Cbp/p300, and Myc), most of which are erythroid cell-related genes (Fig. 5B and C). Of note, Ddit3 and Trib3, which were recently reported to be coinduced during erythroid cell differentiation (47), and the Ern1 gene, which has been linked to endoplasmic reticulum (ER) stress and induced apoptosis, were found to be significantly upregulated in the TAF10-KO embryos (48–50). Genes important for erythroid cell differentiation, such as Myb, Tal1, Cdk6, and the recently described Exosc2 gene, which is part of the exosome complex (51), did not show any change in their expression levels (q value, >0.4). Most of the aforementioned genes were found to be bound by GATA1 in their regulatory sequences but in many cases were bound at different developmental stages (40, 47, 51, 52). Specifically for Myb, Cdk6, and Exosc2, the GATA1 peaks were found to present at levels below the cutoff level in the E12.5 fetal liver (40).
FIG 5.
Analysis of expression of transcription factor, erythroid cell-related, and globin genes. (A) Expression levels (in number of FPKM), determined by RNA-seq, of downregulated erythroid cell-related genes with q values of <0.05 and GATA1 binding peaks found within 10 kb of their TSSs. CTRL, control. (B) Expression levels (in number of FPKM), determined by RNA-seq, of upregulated genes with q values of <0.05 and GATA1 binding peaks found within 10 kb of their TSSs. (C) Deregulated genes with q values of >0.05 and GATA1 binding peaks found within 10 kb of their TSSs. (D) Venn diagram of deregulated genes. Upregulated, genes upregulated in TAF10KOcEry fetal liver cells identified by RNA-seq analysis (q value, <0.05); Downregulated, genes downregulated in TAF10KOcEry fetal liver cells identified by RNA-seq analysis (q value, <0.05); Gata1, GATA1 target genes described previously (40). Half of the deregulated genes have at least one GATA1 binding peak at locations within 10 kb of their TSSs.
The expression of globin genes (Hbb-bh1, Hbb-y, Hbb-bt, Hbb-bs, Hba-a2, and Hba-x) was also lower in TAF10KOcEry fetal liver cells, and especially, the Hbb-y, Hba-a2, and Hba-x genes were found to be downregulated with a high statistical power (Fig. 5A and C). Among the affected globin genes, the Hbb-bh1 gene was the only one with no peaks for GATA1 binding at E12.5.
Interestingly, the expression of eight out of nine genes coding for the DNA-binding proteins (KRAB zinc finger proteins [KRAB-ZFPs]) shown earlier (53) to be expressed in CD71+ TER119+ and/or TER119+ erythroid cells (Zfp689, Zfp13, Zfp661, Zfp92, Zfp641, Zfp551, Zfp583, and Zfp872) appeared to be unaffected (q value, >0.55). The ninth of these genes, Zfp667 (q value, 0.02), was expressed only in TER119+ cells, no GATA1 peak was identified for this gene at E12.5, and it was downregulated (Fig. 5A). In addition, the expression of TRIM28, which was recently reported to lead to a block in erythroid cell differentiation (53) when deleted, did not change, suggesting that it is not implicated in the phenotype of the embryos. Most of these genes had no peak or, exceptionally, one peak (i.e., Zfp689) for GATA1 binding in their regulatory sequences (locations at a ±10-kb range around the transcription start site [TSS]) at E12.5.
These results indicate that TAF10 ablation does not affect global expression, as observed before in hepatocyte-specific TAF10-KO cells (10). In addition, about half of the deregulated genes in the TAF10KOcEry embryos are potential GATA1 target genes, with many of them being known to play a role during erythropoiesis (Fig. 5D). In contrast, the expression of genes that have a role in erythroid cell differentiation but that are not bound by GATA1 at this developmental stage (Zfp689, Zfp13, Myb, and Exosc8) is not affected. Thus, GATA1 targets are among the primary affected genes due to the loss of TAF10 in the TAF10KOcEry embryos in vivo, suggesting an important requirement for cross talk or an interaction between GATA1 and TAF10.
The composition of TAF10-containing complexes during erythroid cell differentiation and development.
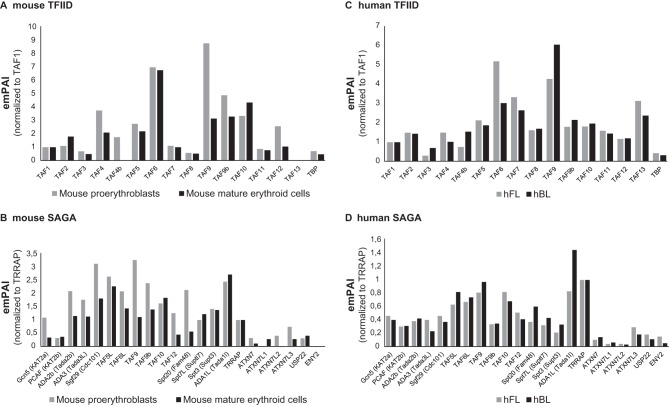
As several recent studies suggested that the composition of general transcription factor or coactivator complexes may change during differentiation and development (22, 23), we sought to analyze the composition of the TAF10-containing TFIID and SAGA complexes during erythroid cell differentiation. To this end, protein extracts were prepared from mouse fetal liver cell lines (mFLcl) at the proerythroblast stage (immature). These cells are able to differentiate into mature erythroblasts in a synchronous manner upon an increase in the dose of erythropoietin. TAF10-containing complexes were isolated by anti-TAF10 IPs from immature and differentiating (mature) but still nucleated erythroid cells and analyzed by MS. The relative abundance (exponentially modified protein abundance index [emPAI]) values of the different subunits in the isolated complexes were first normalized by comparing all abundance values of the subunits of TFIID to those of TAF1 or all the emPAI values of the subunits of SAGA to those of TRRAP, as TAF1 and TRRAP are the largest subunits in each complex (Fig. 6A and B). Interestingly, when comparing the composition of the TFIID complexes between proerythroblasts and mature erythroblasts, we noticed that TAF4b completely disappeared from the TFIID complex of mature cells and also that less TAF4 was associated with the TFIID complex in mature erythroid cells. In agreement with the observation that TAF12 is the histone fold partner of TAF4 (54, 55), the TAF4/TAF4b decrease in mature erythroid cell TFIID was accompanied by a reduction of TAF12. Note that TAF9, but not TAF9b, was also significantly decreased in TAF10 IPs from mature erythroid cell extracts. When comparing the composition of the SAGA complex at these two differentiation stages after normalizing the abundance to that of TRRAP, we observed a slight reduction in several subunits of the SAGA complex in TAF10 IPs from mature erythroid cell extracts (Fig. 6B). Out of the two homologous SAGA HATs (GCN5 and PCAF), which have been reported to be mutually exclusive in the corresponding SAGA complexes (56), the abundance of GCN5 was reduced about three times in mature erythroid cells, while that of PCAF was not reduced. Moreover, in the deubiquitination (DUB) module of SAGA, ATXN7 and its orthologue, ATXN7L2, appeared to be replaced by the orthologous ATXN7L1 at the mature stage. We note that the absence of TAF13 from TFIID and of ENY2 from SAGA in these analyses may be due to the very small sizes of these proteins.
FIG 6.
Mass spectrometry of TFIID and SAGA complexes in mouse and human erythroid cells. (A and B) TAF10-containing TFIID (A) and SAGA (B) complexes were isolated by TAF10 immunoprecipitation from protein extracts prepared from immature and differentiating (mature) mouse erythroid cells and analyzed by mass spectrometry. The relative abundance (emPAI) values of the different subunits in the isolated complexes were first normalized by comparing the abundance values of all the subunits of TFIID to those for TAF1 or the abundance values of all the subunits of SAGA to those of TRRAP. TAF1 and TRRAP are the largest subunits in each complex. (C and D) Similarly, TAF10-containing TFIID (C) and SAGA (D) complexes were isolated by TAF10 immunoprecipitation from protein extracts prepared from hFL and hPB cells and analyzed by mass spectrometry. The emPAI values presented were normalized as described for the mouse samples.
Next, we sought to analyze the composition of human TAF10-containing complexes during development (i.e., fetal and adult stages). Although the human erythroid cells are at the proerythroblast stage at both developmentally different niches (fetal and adult) and should not be directly compared to the mouse situation, we have made similar observations regarding the slight stoichiometric changes that occur in the corresponding subunits of TFIID and SAGA. Our analyses demonstrate that all of the subunits of the two complexes are present in the corresponding complexes at these two developmental stages (Fig. 6C and D).
Importantly, these results together indicate that the canonical composition of the two analyzed TAF10-containing complexes, TFIID and SAGA, does not change dramatically during mouse and human erythroid cell differentiation and development. However, especially in mouse complexes, we often observed significant stoichiometric changes of those specific subunits that have orthologues (TAF4/TAF4b, GCN5/PCAF, TAF9/TAF9b, and ATXN7/ATXN7L1/ATXNL2), which may slightly affect the function of these transcription complexes.
TAF10 and GATA1 interact in mouse and hFL cells.
As our TAF10-KO experiments suggested a functional cross talk between GATA1 and TAF10, we next analyzed whether specific erythroid cell transcription factors, such as GATA1, would coimmunoprecipitate with TAF10 from human fetal liver (hFL) or human peripheral blood (hPB) erythroid progenitor cell cultures and mouse fetal liver cell lines. Importantly, GATA1, together with other previously reported activators and cofactors, such as LDB1 and TAL1, which are components of the so-called pentameric complex (57), was identified by MS to be a TAF10 interactor in hFL cells (Table 2).
TABLE 2.
Selected activators and cofactors found in TAF10 IPs by MSa
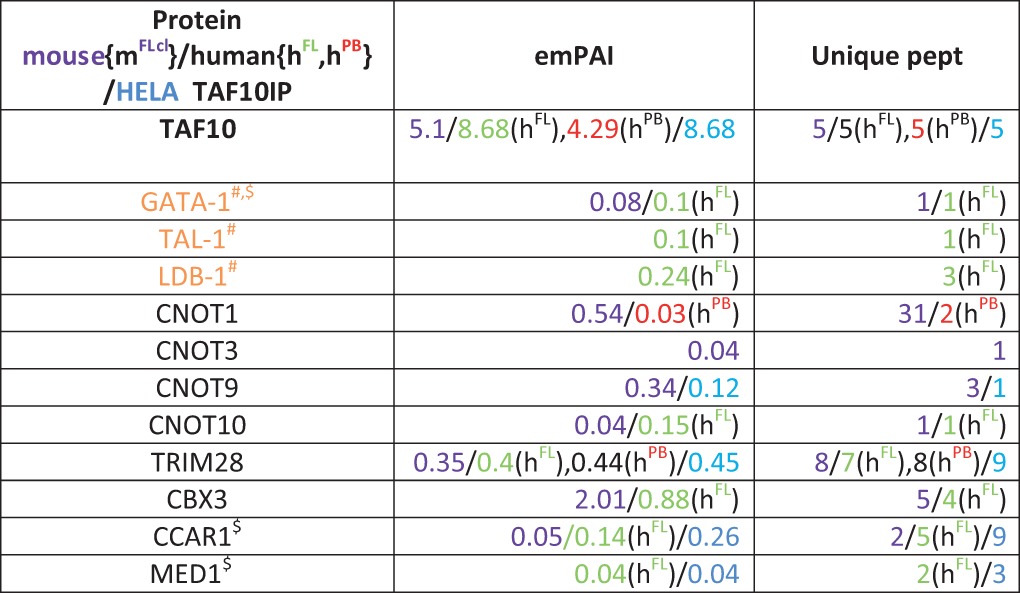
Nuclear extracts from hFL (green) and hPB (red) erythroid progenitor cells and mouse fetal liver cell lines (mFlcl [purple]) were used. Parameters of MS (emPAI and number of unique peptides [Unique pept]) for each protein in different colors indicating the species of origin and cell type are given. Symbols: #, proteins found in the same complex; $, proteins reported to interact with each other.
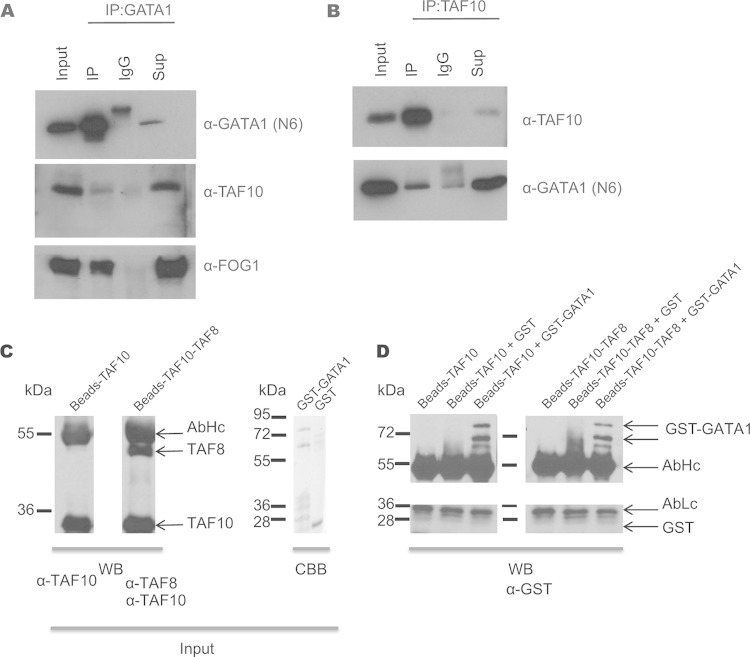
We also analyzed GATA1 interactors by carrying out an anti-GATA1 IP and subsequent MS in murine erythroleukemia (MEL) cells. Endogenous TAF10 together with other TAFs and SAGA subunits were identified in the MS analysis (Table 3). Consequently, immunoprecipitation of GATA1 from MEL cells revealed that endogenous TAF10 coimmunoprecipitated with GATA1 and the friend of GATA1 (FOG1) cofactor (Fig. 7A). In addition, the interaction of endogenous TAF10 and GATA1 was verified by immunoprecipitating TAF10 from nuclear extracts prepared from MEL cells, and the coimmunoprecipitated GATA1 was analyzed by Western blotting (Fig. 7B). Similar IP assays were performed with E12.5 fetal liver cells or MEL cells that express biotinylated GATA1 (bio-GATA1) (Fig. 8). The results of these experiments further confirmed the interaction between TAF10-containing complexes and GATA1 previously identified from the MS data.
TABLE 3.
MS of GATA1 IP from MEL cells
| TFIID or SAGA subunita | emPAI | No. of unique peptides |
|---|---|---|
| TAF1b | 0.02 | 4 |
| TAF4ab | 0.1 | 2 |
| TAF6b | 0.23 | 5 |
| TAF5Lc | 0.46 | 8 |
| TAF9d | 3.34 | 12 |
| TAF9bd | 0.94 | 6 |
| TAF10d | 0.17 | 1 |
| TBPb | 0.35 | 4 |
| PCAFc (KAT2B) | 0.12 | 5 |
| ADA3c (TADA3L) | 0.25 | 5 |
| Spt20c (Fam48) | 0.06 | 2 |
| Spt3c (Supt3) | 0.19 | 3 |
TFIID and SAGA subunits were identified by mass spectrometry after GATA1 IP (bio-GATA1) of MEL cell nuclear extracts. Subunits that were found in BirA (control) cells after streptavidin pulldown were subtracted from the list.
TFIID subunit.
SAGA subunit.
TFIID and SAGA shared subunits.
FIG 7.
Immunoprecipitation of TAF10 and GATA1 in MEL cells. (A) GATA1 immunoprecipitation in MEL cells. Anti-GATA1 antibody (N6) was used for Western blot analysis. TAF10 and FOG1 are coimmunoprecipitated. (B) TAF10 immunoprecipitation in MEL cell nuclear extracts using the 6TA 2B11 antibody clone. IgG antibody was used as a control. GATA1 is detected in the IP fraction, confirming the MS results. Sup, supernatant. (C) TAF10 alone or a TAF8-TAF10 heterodimer was immunopurified using an anti-TAF10 antibody from SFf9 cell extracts and tested by Western blotting (WB) using the indicated antibodies. The GST-GATA1 protein or GST was also purified and tested by Coomassie brilliant blue (CBB) staining. (D) The purified proteins were combined as indicated above the gel and incubated, and after several washes the bead-bound proteins were denatured, resolved on an SDS-polyacrylamide gel, and analyzed by Western blotting using an anti-GST antibody. The antibody heavy chain (AbHc) and antibody light chain (AbLc) are indicated.
FIG 8.
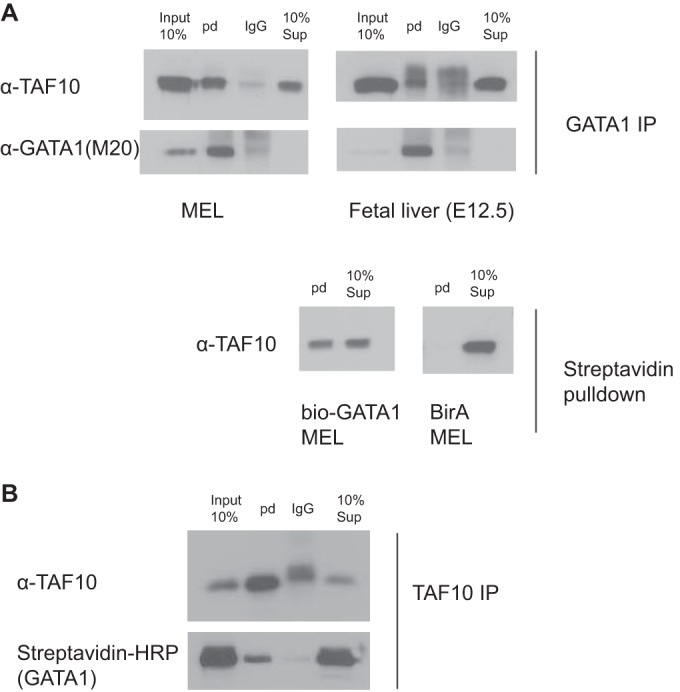
Immunoprecipitation of TAF10 and GATA1 in MEL cells and mouse fetal liver at E12.5. (A) MEL cells and mouse fetal liver nuclear cell extracts were used to immunoprecipitate GATA1 endogenous protein or bio-GATA1 overexpressed in MEL cells. GATA1 (N6) IP or streptavidin pulldown (pd) coimmunoprecipitates GATA1/bio-GATA1 and TAF10 protein, as detected by Western blotting assays using M20 (GATA1) and 6TA 2B11 (TAF10) antibodies. BirA MEL cells were used as controls. (B) TAF10 (6TA 2B11) IP in MEL cells in which bio-GATA1 is overexpressed coimmunoprecipitate GATA1 in the reverse IP. HRP, horseradish peroxidase.
We further investigated whether the GATA1-TAF10 interaction is direct by in vitro protein-protein interaction experiments. First, either TAF10 alone or the TAF8-TAF10 heterodimer was immune purified using an anti-TAF10 antibody from SFf9 cell extracts, in which the corresponding proteins were overexpressed using the baculovirus system. Next, purified GST-GATA1 protein or GST alone was added to TAF10- or TAF10-TAF8-bound beads (Fig. 7C). After several washing steps with high-salt buffer, the bead-bound proteins were denatured, resolved on an SDS-polyacrylamide gel, and analyzed by Western blotting using an anti-GST antibody. Our in vitro results indicate that GST-GATA1 bound to both TAF10- and TAF8-TAF10-containing beads but GST alone did not (Fig. 7D). These results together support a direct interaction between the key erythroid cell transcription factor GATA1 and TAF10.
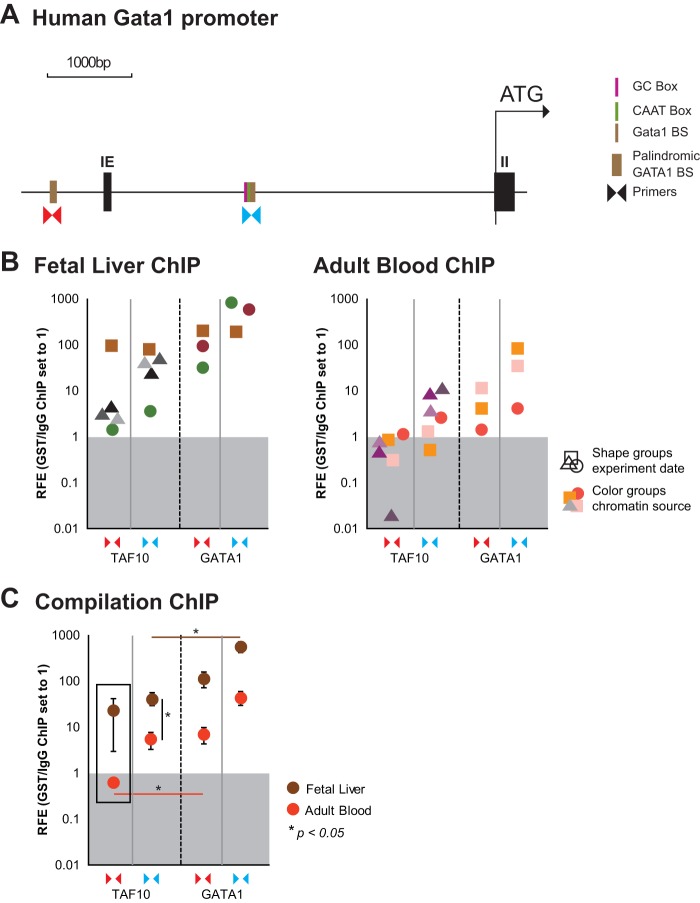
TAF10 is bound to GATA1 sites in the GATA1 locus.
In light of the downregulation of GATA1 mRNA levels, we wished to determine whether TAF10 would be present and thus possibly regulate the expression of the GATA1 gene. We looked specifically at a palindromic GATA1 binding site known to be required for normal GATA1 transcription and an additional GATA1 binding site which locates next to a TATA box at kb −3 relative to the TSS (58, 59) (Fig. 9A). We performed ChIP of TAF10 and GATA1 in hFL and hPB cells, representing cells from two distinct developmental stages, as defined by the expression of fetal and adult hemoglobin, respectively. We found that TAF10 and GATA1 binding was more enriched at both GATA1 binding sites at the human GATA1 locus in the fetal liver than in adult blood proerythroblasts (Fig. 9B and C). However, while GATA1 bound at both binding sites examined at both developmental stages, TAF10 was clearly not bound at the palindromic GATA1 binding site in the adult stage and was significantly less enriched than GATA1 at the binding site at kb −3 relative to the TSS in the fetal stage (Fig. 9C).
FIG 9.
ChIP assays of TAF10 and GATA1 in the GATA1 locus in hFL and hPB cells. A TAF10 antibody (monoclonal antibody 23TA 1H8) and a GATA1 antibody were used to immunoprecipitate the formaldehyde-cross-linked chromatin from hFL and hPB cells. (A) Primers for GATA1 binding sites of the human GATA1 locus, as indicated (the palindromic GATA1 binding site [BS] and GATA1 binding site at the kb −3 region relative to the TSS), were used to estimate the relative fold enrichment (RFE) of TAF10 and GATA1 binding by qRT-PCR. A CD71 antibody (isotype control) was used for the mock IP, and background enrichment was set equal to a value of 1. IE, exon I erythroid. (B) Enrichment for TAF10 and GATA1 at both binding sites of interest is shown in independent experiments for fetal liver and adult blood. (C) Overview of all ChIP experiments. See panel A for the positions of the primers.
GATA1 autoregulates its expression by binding to its own promoter and enhancers. We detected the TAF10-GATA1 protein-protein interaction mainly in hFL cell extracts, and ChIP results showed the selective binding of TAF10 at the palindromic GATA1 site during fetal stages. Collectively, these results support the notion that TAF10 has a role in the developmental regulation of GATA1 expression.
DISCUSSION
Erythropoiesis is a process that is controlled tightly by the regulated expression of erythroid cell-specific transcription factors and their interactions with cofactors. General transcription factors also have an active role; i.e., they have a function more cell type specific than was originally thought (60). Whether the TAF10 component of TFIID and SAGA exerts such a role by shaping the interactions with activators in erythroid cell differentiation and development was the topic of this study.
TAF10 was specifically ablated to disrupt the canonical TFIID and SAGA complexes in erythroid cells from early stages of development (E8.0) in mice by crossing TAF10lox mice with EpoR-Cre mice. This resulted in a block in erythropoiesis, leading to embryonic death at about E13.5. A progressive delay in the differentiation kinetics through development, which had already started at E11.5, was more pronounced at E12.5, with an accumulation of CD71+ TER119+ cells at the expense of mature TER119+ erythroid cells being found. Expression analysis by mRNA sequencing (RNA-seq) showed that the erythroid cell transcription factor genes Gata1, Klf1, Nfe2, Zbtb7a, and Bcl2l1 were within the downregulated group of genes and expression of Gata2, Spi1, and Myb did not change significantly, while Runx1, Rb1, Myc, and Cited2 were upregulated, following the opposite gene expression pattern that characterizes the transition from the CD71+ TER119+ toward to TER119+ mature erythroid cells (61). Along the same lines, the TFDP2 coregulator and E2F2 transcription factor were also found to be downregulated, and both genes were under the regulatory control of GATA1. Normally, these two genes are induced upon erythroid cell differentiation, and it was shown that the downregulation of Tfdp2 prevents proper erythroid cell differentiation (47). Although E2f2 was downregulated significantly, the expression level of several E2F2 target genes (Dhfr, Ccna2) was reduced, in contrast to what we would expect due to the repressive role (47) of E2F2 on its targets. Globin genes were also downregulated, which is a clear sign of the differentiation block during erythroid cell differentiation. In contrast, genes with less well defined roles in the erythroid cell lineage and no peaks for GATA1 in the ChIP-seq analysis at this developmental stage (E12.5), i.e., KRAB-ZFPs, did not change their expression levels in the TAF10KOcEry fetal livers.
Nine of these KRAB-ZFPs are known to be expressed in erythroid progenitor cells, and two of them are required for proper erythroid cell differentiation (ZFP689, ZFP13) (53). These results suggest that TAF10 is required to stabilize PIC at certain loci, explaining why the transcription of specific genes is affected upon TAF10 loss. This finding is in agreement with the notion that TAF10 loss does not affect general transcription, as would be expected for a cornerstone TAF of the TFIID (46). The specific gene expression changes in TAF10-KO erythroid cells could explain their progressive block in differentiation and subsequent apoptosis, as identified by our GO analysis in TAF10KOcEry fetal livers, since GATA1-KO erythroid cells are unable to differentiate but arrest their cell cycle and undergo apoptosis. In addition, three genes linked to ER stress and induced apoptosis (Ddit3 [48], Trib3 [62], and Ern1 [50]) were found to be significantly upregulated in the TAF10KOcEry fetal livers, supporting the notion that apoptosis is among the main causes of the phenotype observed at about E13.5.
In parallel with the findings of in vivo studies, we defined the composition of TAF10-containing complexes in erythroid cells in order to investigate the dynamic changes that appear to be crucial in other cell types during differentiation (63). We performed MS analysis of human and mouse cultured erythroid progenitor cells at an immature stage (proerythroblasts) and upon differentiation, which revealed that most of the TFIID and SAGA subunits are present in these complexes at all stages analyzed. Our data exclude the possibility of a total rearrangement of the TFIID or SAGA complexes in this differentiation system, in contrast to what was reported for TFIID during liver hepatocyte (64) or myoblast (65) differentiation. Nevertheless, during differentiation we observed a dynamic reorganization of some TFIID and SAGA subunits, usually affecting those that have paralogues, such as TAF4/TAF4b, TAF9/TAF9b, GCN5/PCAF, and ATXN7/ATXN7L1/ATXN7L2, without affecting the core structure of these complexes. Interestingly, Pijnappel et al. (66) demonstrated that overexpression of TAF4 with the pluripotency factors and, presumably, the incorporation of TAF4 into preexisting TFIID complexes lacking TAF4 can efficiently reprogram differentiated cells into induced pluripotent stem cells (iPSCs). The TFIID complexes that we purified from differentiated mouse erythroid cells also had reduced TAF4/TAF12 heterodimers and no TAF4b, whereas TAF4/TAF12 heterodimers and TAF4b were present at the immature stages of differentiation. This is in excellent agreement with results presented previously (66) and suggests that the low level of TAF4 and/or the lack of TAF4b is associated with a differentiated state, whereas at an immature stage cells contain a TFIID complex with stoichiometric amounts of TAF4/4b.
The idea that developmental gene regulation is dependent on protein interactions between TFIID, activators, and coactivators is also supported by our results. GATA1 and its well-known partners LDB1 and TAL1 were found to interact with endogenous TAF10 in the fetal liver cells of both mouse and human origin. This interaction was verified by reciprocal immunoprecipitation (anti-TAF10 and anti-GATA1 IPs) in MEL cells and in vitro by using purified GST-GATA1 and TAF10 (or TAF10-TAF8 heterodimer) proteins. Similar interactions between KLF1 and TAF9 for activation of the β-globin gene (67) as well as GATA1 and MED1 (68, 69) have been reported in erythroid cells.
Other transcription factors and cofactors, including subunits of the CCR4-NOT complex (e.g., CNOT3), CBX3 (a paralogue of CBX1 [HP1β]), and TRIM28, were also found to interact with TAF10 by mass spectrometry (Table 2). CNOT3, CBX1, and TRIM28 were previously reported to form a unique module involved in developmental processes (70), and TRIM28 in particular has an important role in erythropoiesis (53, 71). Of note, CNOT3 and TRIM28 do not physically interact, while there have been reports of interactions of TFIID with the CCR4-NOT complex (72). Therefore, TFIID might be acting as a scaffold protein for the assembly of the module and might play an important role in the developmental processes controlled by this module, including erythropoiesis, which would be interesting to investigate in the future.
Interestingly, our observation that TAF10 binding was enriched at the promoter of the human GATA1 locus in fetal erythroid cells compared to its level in adult erythroid cells, together with the observed TAF10-GATA1 interaction, suggests that there is a role for TAF10 in the regulation of GATA1 transcription which contributes to the phenotype observed in TAF10KOcEry embryos. This interaction preferentially occurs during the fetal stages of erythropoiesis, indicating that it is a developmental stage-specific event exerting its effect mainly, but not exclusively, on GATA1 target genes, as observed in the RNA-seq data from mouse fetal livers. We know that GATA1 expression levels do not change in human fetal and adult erythroblasts (73). However, there are dynamic changes in the occupancy of transcription factors and, consequently, in protein-protein interactions involving master regulators that could potentially activate, repress (74), or stabilize gene expression levels. When such interactions are disturbed, transcriptional deregulation is not global but depends on the transcriptional state of the gene at that developmental stage (10).
We propose that TFIID and/or SAGA contributes to this dynamic landscape of developmental stage-specific TAF10-GATA1 interaction network, thus contributing to development and differentiation of the erythroid cell lineage.
ACKNOWLEDGMENTS
We thank the PRIDE team for their help and support regarding the deposition of the mass spectrometry data. The GST and GST-GATA1 plasmids were a generous gift from K. Freson at KU Leuven (Molecular and Vascular Biology). We are also grateful to the members of T. Economou lab (Molecular Bacteriology) at KU Leuven and especially to N. Famelis and K. Tsolis for helping with the purification of the GST fusion proteins and Petros Kolovos from Erasmus MC for technical support.
This work has been partially supported by EMBO short-term fellowship ASTF 15-2010 (to P.P.), The Netherlands Organization for Scientific Research (NWO-VENI 863.09.012 to L.G.), The Netherlands Genomics Initiative (NGI Zenith 93511036), The Netherlands Proteomics Center (NPC), The Netherlands Initiative of Regenerative Medicine (NIRM), EU integrated project EuTRACC (to F.G. and L.T.), the Landsteiner Foundation for Blood Transfusion Research (LSBR; 1040 to F.G. and S.P.); ZonMw (TOP 40-00812-98-12128 and DN 82-301), EU FP7 Specific Cooperation Research Project THALAMOSS (306201 to S.P.), and ERC Advanced (Birtoaction, grant no. 340551 to L.T.).
P.P, F.G., and L.T. designed the study; P.P., L.G., J.D., D.N.P., E.K., R.V.D.L., F.P., and E.S. performed the experiments; P.P., L.G., J.D., H.J.G.V.D.W., D.H.W.D., P.V., J.S., S.P., F.G., and L.T. analyzed the data; and P.P., L.G., S.P., F.G., and L.T. wrote the paper.
REFERENCES
- 1.Muller F, Tora L. 2013. Chromatin and DNA sequences in defining promoters for transcription initiation. Biochim Biophys Acta 1893:118–126. doi: 10.1016/j.bbagrm.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Gershenzon NI, Ioshikhes IP. 2005. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics 21:1295–1300. doi: 10.1093/bioinformatics/bti172. [DOI] [PubMed] [Google Scholar]
- 3.Zhang MQ. 1998. Identification of human gene core promoters in silico. Genome Res 8:319–326. doi: 10.1101/gr.8.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gangloff YG, Romier C, Thuault S, Werten S, Davidson I. 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci 26:250–257. doi: 10.1016/S0968-0004(00)01741-2. [DOI] [PubMed] [Google Scholar]
- 5.Kaufmann J, Smale ST. 1994. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev 8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 6.Chalkley GE, Verrijzer CP. 1999. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J 18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juven-Gershon T, Kadonaga JT. 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vassallo MF, Tanese N. 2002. Isoform-specific interaction of HP1 with human TAFII130. Proc Natl Acad Sci U S A 99:5919–5924. doi: 10.1073/pnas.092025499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu WL, Coleman RA, Ma E, Grob P, Yang JL, Zhang Y, Dailey G, Nogales E, Tjian R. 2009. Structures of three distinct activator-TFIID complexes. Genes Dev 23:1510–1521. doi: 10.1101/gad.1790709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatarakis A, Margaritis T, Martinez-Jimenez CP, Kouskouti A, Mohan WS II, Haroniti A, Kafetzopoulos D, Tora L, Talianidis I. 2008. Dominant and redundant functions of TFIID involved in the regulation of hepatic genes. Mol Cell 31:531–543. doi: 10.1016/j.molcel.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Frontini M, Soutoglou E, Argentini M, Bole-Feysot C, Jost B, Scheer E, Tora L. 2005. TAF9b (formerly TAF9L) is a bona fide TAF that has unique and overlapping roles with TAF9. Mol Cell Biol 25:4638–4649. doi: 10.1128/MCB.25.11.4638-4649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spedale G, Timmers HT, Pijnappel WW. 2012. ATAC-king the complexity of SAGA during evolution. Genes Dev 26:527–541. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timmers HT, Tora L. 2005. SAGA unveiled. Trends Biochem Sci 30:7–10. doi: 10.1016/j.tibs.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Mohan WS Jr, Scheer E, Wendling O, Metzger D, Tora L. 2003. TAF10 (TAFII30) is necessary for TFIID stability and early embryogenesis in mice. Mol Cell Biol 23:4307–4318. doi: 10.1128/MCB.23.12.4307-4318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Indra AK, Mohan WS II, Frontini M, Scheer E, Messaddeq N, Metzger D, Tora L. 2005. TAF10 is required for the establishment of skin barrier function in foetal, but not in adult mouse epidermis. Dev Biol 285:28–37. doi: 10.1016/j.ydbio.2005.05.043. [DOI] [PubMed] [Google Scholar]
- 16.Wong PM, Chung SW, Eaves CJ, Chui DH. 1985. Ontogeny of the mouse hemopoietic system. Prog Clin Biol Res 193:17–28. [PubMed] [Google Scholar]
- 17.Sankaran VG, Xu J, Orkin SH. 2010. Advances in the understanding of haemoglobin switching. Br J Haematol 149:181–194. doi: 10.1111/j.1365-2141.2010.08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. 2005. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol 25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez L, Nikolic T, van Dijk TB, Hammad H, Vos N, Willart M, Grosveld F, Philipsen S, Lambrecht BN. 2007. Gata1 regulates dendritic-cell development and survival. Blood 110:1933–1941. doi: 10.1182/blood-2006-09-048322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A 93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez L, Tsukamoto S, Suzuki M, Yamamoto-Mukai H, Yamamoto M, Philipsen S, Ohneda K. 2008. Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood 111:4375–4385. doi: 10.1182/blood-2007-09-115121. [DOI] [PubMed] [Google Scholar]
- 22.Muller F, Zaucker A, Tora L. 2010. Developmental regulation of transcription initiation: more than just changing the actors. Curr Opin Genet Dev 20:533–540. doi: 10.1016/j.gde.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Goodrich JA, Tjian R. 2010. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet 11:549–558. doi: 10.1038/ni0710-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinrich AC, Pelanda R, Klingmuller U. 2004. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood 104:659–666. doi: 10.1182/blood-2003-05-1442. [DOI] [PubMed] [Google Scholar]
- 25.Tsai SF, Strauss E, Orkin SH. 1991. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev 5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 26.Demeny MA, Soutoglou E, Nagy Z, Scheer E, Janoshazi A, Richardot M, Argentini M, Kessler P, Tora L. 2007. Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS One 2:e316. doi: 10.1371/journal.pone.0000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. 1994. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell 79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 28.Wieczorek E, Brand M, Jacq X, Tora L. 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 29.Soutoglou E, Demeny MA, Scheer E, Fienga G, Sassone-Corsi P, Tora L. 2005. The nuclear import of TAF10 is regulated by one of its three histone fold domain-containing interaction partners. Mol Cell Biol 25:4092–4104. doi: 10.1128/MCB.25.10.4092-4104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. 2010. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leberbauer C, Boulme F, Unfried G, Huber J, Beug H, Mullner EW. 2005. Different steroids co-regulate long-term expansion versus terminal differentiation in primary human progenitors. Blood 105:85–94. doi: 10.1182/blood-2004-03-1002. [DOI] [PubMed] [Google Scholar]
- 32.Follows GA, Tagoh H, Lefevre P, Hodge D, Morgan GJ, Bonifer C. 2003. Epigenetic consequences of AML1-ETO action at the human c-FMS locus. EMBO J 22:2798–2809. doi: 10.1093/emboj/cdg250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 34.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt S, Johnson N, Juettemann T, Kahari AK, Keenan S, Kulesha E, Martin FJ, Maurel T, McLaren WM, Murphy DN, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ruffier M, Sheppard D, Taylor K, Thormann A, Trevanion SJ, Vullo A, Wilder SP, Wilson M, Zadissa A, Aken BL, Birney E, Cunningham F, Harrow J, Herrero J, Hubbard TJ, Kinsella R, Muffato M, Parker A, Spudich G, Yates A, et al. 2014. Ensembl 2014. Nucleic Acids Res 42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S, Pyl PT, Huber W. 2014. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hochberg Y, Benjamini Y. 1990. More powerful procedures for multiple significance testing. Stat Med 9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 39.Falcon S, Gentleman R. 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 40.Papadopoulos GL, Karkoulia E, Tsamardinos I, Porcher C, Ragoussis J, Bungert J, Strouboulis J. 2013. GATA-1 genome-wide occupancy associates with distinct epigenetic profiles in mouse fetal liver erythropoiesis. Nucleic Acids Res 41:4938–4948. doi: 10.1093/nar/gkt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.r-project.org. [Google Scholar]
- 42.von Lindern M, Deiner EM, Dolznig H, Parren-Van Amelsvoort M, Hayman MJ, Mullner EW, Beug H. 2001. Leukemic transformation of normal murine erythroid progenitors: v- and c-ErbB act through signaling pathways activated by the EpoR and c-Kit in stress erythropoiesis. Oncogene 20:3651–3664. doi: 10.1038/sj.onc.1204494. [DOI] [PubMed] [Google Scholar]
- 43.Lee R, Kertesz N, Joseph SB, Jegalian A, Wu H. 2001. Erythropoietin (Epo) and EpoR expression and 2 waves of erythropoiesis. Blood 98:1408–1415. doi: 10.1182/blood.V98.5.1408. [DOI] [PubMed] [Google Scholar]
- 44.Isern J, Fraser ST, He Z, Baron MH. 2010. Developmental niches for embryonic erythroid cells. Blood Cells Mol Dis 44:207–208. doi: 10.1016/j.bcmd.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palis J. 2014. Primitive and definitive erythropoiesis in mammals. Front Physiol 5:3. doi: 10.3389/fphys.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I. 2013. The architecture of human general transcription factor TFIID core complex. Nature 493:699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- 47.Chen C, Lodish HF. 2014. Global analysis of induced transcription factors and cofactors identifies Tfdp2 as an essential coregulator during terminal erythropoiesis. Exp Hematol 42:464–476.e5. doi: 10.1016/j.exphem.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. 2014. Cell death. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harding HP, Zhang Y, Ron D. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 50.Brewer JW, Hendershot LM, Sherr CJ, Diehl JA. 1999. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc Natl Acad Sci U S A 96:8505–8510. doi: 10.1073/pnas.96.15.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McIver SC, Kang YA, DeVilbiss AW, O'Driscoll CA, Ouellette JN, Pope NJ, Camprecios G, Chang CJ, Yang D, Bouhassira EE, Ghaffari S, Bresnick EH. 2014. The exosome complex establishes a barricade to erythroid maturation. Blood 124:2285–2297. doi: 10.1182/blood-2014-04-571083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, Bernstein BE, Fraenkel E, Cantor AB. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell 36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, Offner S, Marquis J, Kapopoulou A, Vanicek J, Trono D. 2013. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science 340:350–353. doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gangloff YG, Werten S, Romier C, Carre L, Poch O, Moras D, Davidson I. 2000. The human TFIID components TAFII135 and TAFII20 and the yeast SAGA components ADA1 and TAFII68 heterodimerize to form histone-like pairs. Mol Cell Biol 20:340–351. doi: 10.1128/MCB.20.1.340-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leurent C, Sanders S, Ruhlmann C, Mallouh V, Weil PA, Kirschner DB, Tora L, Schultz P. 2002. Mapping histone fold TAFs within yeast TFIID. EMBO J 21:3424–3433. doi: 10.1093/emboj/cdf342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, Tora L. 2010. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell Mol Life Sci 67:611–628. doi: 10.1007/s00018-009-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van Ijcken W, Hou J, Steinhoff C, Rijkers E, Lenhard B, Grosveld F. 2010. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev 24:277–289. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicolis S, Bertini C, Ronchi A, Crotta S, Lanfranco L, Moroni E, Giglioni B, Ottolenghi S. 1991. An erythroid specific enhancer upstream to the gene encoding the cell-type specific transcription factor GATA-1. Nucleic Acids Res 19:5285–5291. doi: 10.1093/nar/19.19.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moriguchi T, Yu L, Takai J, Hayashi M, Satoh H, Suzuki M, Ohneda K, Yamamoto M. 2015. The human GATA1 gene retains a 5′ insulator that maintains chromosomal architecture and GATA1 expression levels in splenic erythroblasts. Mol Cell Biol 35:1825–1837. doi: 10.1128/MCB.00011-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naar AM, Lemon BD, Tjian R. 2001. Transcriptional coactivator complexes. Annu Rev Biochem 70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 61.Wong P, Hattangadi SM, Cheng AW, Frampton GM, Young RA, Lodish HF. 2011. Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood 118:e128–e138. doi: 10.1182/blood-2011-03-341404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. 2005. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J 24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Z, Scannell DR, Eisen MB, Tjian R. 2011. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D'Alessio JA, Ng R, Willenbring H, Tjian R. 2011. Core promoter recognition complex changes accompany liver development. Proc Natl Acad Sci U S A 108:3906–3911. doi: 10.1073/pnas.1100640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deato MD, Tjian R. 2007. Switching of the core transcription machinery during myogenesis. Genes Dev 21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pijnappel WW, Esch D, Baltissen MP, Wu G, Mischerikow N, Bergsma AJ, van der Wal E, Han DW, Bruch H, Moritz S, Lijnzaad P, Altelaar AF, Sameith K, Zaehres H, Heck AJ, Holstege FC, Scholer HR, Timmers HT. 2013. A central role for TFIID in the pluripotent transcription circuitry. Nature 495:516–519. doi: 10.1038/nature11970. [DOI] [PubMed] [Google Scholar]
- 67.Sengupta T, Cohet N, Morle F, Bieker JJ. 2009. Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc Natl Acad Sci U S A 106:4213–4218. doi: 10.1073/pnas.0808347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stumpf M, Waskow C, Krotschel M, van Essen D, Rodriguez P, Zhang X, Guyot B, Roeder RG, Borggrefe T. 2006. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc Natl Acad Sci U S A 103:18504–18509. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stumpf M, Yue X, Schmitz S, Luche H, Reddy JK, Borggrefe T. 2010. Specific erythroid-lineage defect in mice conditionally deficient for Mediator subunit Med1. Proc Natl Acad Sci U S A 107:21541–21546. doi: 10.1073/pnas.1005794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. 2009. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hosoya T, Clifford M, Losson R, Tanabe O, Engel JD. 2013. TRIM28 is essential for erythroblast differentiation in the mouse. Blood 122:3798–3807. doi: 10.1182/blood-2013-04-496166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collart MA, Timmers HT. 2004. The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol 77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 73.Xu J, Shao Z, Glass K, Bauer DE, Pinello L, Van Handel B, Hou S, Stamatoyannopoulos JA, Mikkola HK, Yuan GC, Orkin SH. 2012. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell 23:796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pimkin M, Kossenkov AV, Mishra T, Morrissey CS, Wu W, Keller CA, Blobel GA, Lee D, Beer MA, Hardison RC, Weiss MJ. 2014. Divergent functions of hematopoietic transcription factors in lineage priming and differentiation during erythro-megakaryopoiesis. Genome Res 24:1932–1944. doi: 10.1101/gr.164178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]