Abstract
We investigated perchlorate (ClO4−) and chlorate (ClO3−) (collectively (per)chlorate) in comparison with nitrate as potential inhibitors of sulfide (H2S) production by mesophilic sulfate-reducing microorganisms (SRMs). We demonstrate the specificity and potency of (per)chlorate as direct SRM inhibitors in both pure cultures and undefined sulfidogenic communities. We demonstrate that (per)chlorate and nitrate are antagonistic inhibitors and resistance is cross-inducible implying that these compounds share at least one common mechanism of resistance. Using tagged-transposon pools we identified genes responsible for sensitivity and resistance in Desulfovibrio alaskensis G20. We found that mutants in Dde_2702 (Rex), a repressor of the central sulfate-reduction pathway were resistant to both (per)chlorate and nitrate. In general, Rex derepresses its regulon in response to increasing intracellular NADH:NAD+ ratios. In cells in which respiratory sulfate reduction is inhibited, NADH:NAD+ ratios should increase leading to derepression of the sulfate-reduction pathway. In support of this, in (per)chlorate or nitrate-stressed wild-type G20 we observed higher NADH:NAD+ ratios, increased transcripts and increased peptide counts for genes in the core Rex regulon. We conclude that one mode of (per)chlorate and nitrate toxicity is as direct inhibitors of the central sulfate-reduction pathway. Our results demonstrate that (per)chlorate are more potent inhibitors than nitrate in both pure cultures and communities, implying that they represent an attractive alternative for controlling sulfidogenesis in industrial ecosystems. Of these, perchlorate offers better application logistics because of its inhibitory potency, solubility, relative chemical stability, low affinity for mineral cations and high mobility in environmental systems.
Introduction
Owing to its toxic, explosive and corrosive nature, inadvertent hydrogen sulfide (H2S) production by sulfate-reducing microorganisms (SRMs) poses significant health and operational risks to a broad diversity of industries (WHO 2000). Anthropogenic H2S sources are dominated by the oil industry where microbially produced H2S in reservoir gases and fluids (denoted as souring) has an associated annual cost on the order of $90 billion globally. Identifying inhibitors of SRM that are potent, cost-effective and environmentally benign is essential for providing safe and sustainable industrial practices. For over 60 years, researchers have studied the inhibition of SRM by sulfate analogs, biocides and other compounds (Postgate 1952; Greene et al., 2006; Gieg et al., 2011), and both pure cultures and microcosm studies have yielded a wide range of possible treatments (Gieg et al., 2011). In the case of oil reservoir souring, nitrate injection is the primary strategy to control SRM activity and inhibit sulfidogenesis (Youssef et al., 2008). Although the exact mechanism is still uncertain, its effectiveness is thought to be due to a combination of factors (Hubert 2010; Gieg et al., 2011) that can be classified as direct or indirect. These involve putative inhibition of the ATP sulfurylase enzyme that catalyzes the first step of sulfate reduction as previously shown for some eukaryotic proteins (Farley et al., 1976); thermodynamic preference of nitrate respiration over sulfate respiration; sulfide reoxidation by nitrate-reducing microorganisms; and inhibition of SRM by biogenic nitrite or nitric oxide toxicity (Sorensen et al., 1980; Greene et al., 2003; Hubert 2010; Gieg et al., 2011).
However, at lower concentrations (<10 mM) nitrate is not directly inhibitory to SRM and many SRM can alternatively respire nitrate as a suitable electron acceptor, allowing for the establishment of robust populations that are poised for active sulfate reduction once nitrate is depleted. In addition, a broad phylogenetic diversity of SRM express an Nrf nitrite reductase and are insensitive to nitrite toxicity (Greene et al., 2003). Furthermore nitrite and nitric oxide intermediates are chemically and biologically labile, have a limited half-life in a reduced reservoir matrix, and may be reacted out before they have a significant impact on the SRM population. Finally, many SRM have well-characterized mechanisms for coping with reactive nitrogen species, including evasion through chemotactic responses and dedicated nitric oxide detoxification systems (Fischer and Cypionka 2006; Zhou et al., 2011; Yurkiw et al., 2012).
Perchlorate and chlorate, collectively (per)chlorate, represent an attractive alternative to nitrate as inhibitors of sulfide production (Engelbrektson et al., 2014; Gregoire et al., 2014). As with nitrate, both direct and indirect inhibition mechanisms are possible. Over 60 years ago, Postgate (1952) evaluated the effect of perchlorate on hydrogen consumption by a Desulfovibrio and hypothesized that it could be an inhibitor of sulfate respiration. In eukaryotic systems, chlorate is well known as an inhibitor of sulfation (Baeuerle and Huttner 1986; Hoogewerf et al., 1991), whereas kinetic and structural studies with purified ATP sulfurylase suggest that chlorate functions as both a competitive and allosteric inhibitor of sulfate binding and activation (Ullrich et al., 2001; Hanna et al., 2002). Alternatively, indirect inhibition of sulfidogenesis by (per)chlorate can proceed by thermodynamic preference of these compounds as respiratory electron acceptors (Coates and Achenbach 2004; Engelbrektson et al., 2014) and inhibition of SRM by biogenic reactive chlorine and oxygen species. Furthermore, all dissimilatory (per)chlorate-reducing microorganisms tested innately coupled H2S oxidation to (per)chlorate reduction (Bruce et al., 1999), producing elemental sulfur as an inert primary end product (Gregoire et al., 2014) thus removing the principle cause of souring.
In this paper, we present evidence of the specificity and potency of (per)chlorate as inhibitors of mesophilic respiratory sulfate reduction in both pure culture models and undefined sulfidogenic communities. We demonstrate that nitrate is antagonistic to (per)chlorate inhibition in batch systems and resistance is cross-inducible implying that these compounds share at least one common target or mechanism of resistance. In combination with our previous studies (Engelbrektson et al., 2014; Gregoire et al., 2014) these studies indicate the great potential for (per)chlorate as an attractive alternative technology for controlling sulfidogenesis in industrial ecosystems. Because of its unique physical and chemical properties relative to chlorate (Motzer 2001; Urbansky 2002; Coates and Achenbach 2004; Engelbrektson et al., 2014), perchlorate is seen as a more practical solution for industrial application.
Materials and methods
Media and cultivation conditions
Desulfovibrio species were cultivated in basal tris-buffered lactate/sulfate media. The media contained 8 mM MgCl2, 20 mM NH4Cl, 0.6 mM CaCl2, 2 mM KH2PO4, 0.06 mM FeCl2 and 30 mM Tris–HCl. A quantity of 60 mM sodium lactate and 30 mM sodium sulfate was added. Trace elements and vitamins were added from stocks according to a recipe in Mukhopadhyay et al. (2006); Price et al. (2013) and the media was brought to a pH of 7.4 with 0.5 M HCl. The media was degassed with N2 and either sterile filtered in an anaerobic chamber for microplates or dispensed into anoxic vials. The incubation temperature for all growth experiments was 30 °C. Desulfovibrio cultures were always recovered from 1 ml freezer stocks in 10 ml anoxic basal media in sealed Hungate tubes with 1 g l−1 yeast extract and 1 mM sodium sulfide. Active cultures were washed in basal media to remove residual yeast extract prior to inoculation of microplates or tubes for growth experiments.
Marine enrichment cultures were passaged planktonic communities from continuous flow reactor columns inoculated from marine sediments collected from San Francisco Bay (Engelbrektson et al., 2014). Yeast extract (2 g l−1) was added to autoclaved seawater or Instant Ocean (Thermo Fisher Scientific, Waltham, MA, USA) marine mix (35 g l−1) to make seawater media. Enrichments were frozen in −80 °C glycerol stocks and frozen stocks were recovered in seawater media before inoculation of cultures for all experiments. Concentrations of (per)chlorate or nitrate that inhibit 50% (IC50 values) of growth and sulfidogenesis were determined for cells pre-grown in sealed anoxic Hungate tubes that were centrifuged, resuspended in autoclaved seawater and added at 2 × dilutions to microplates containing compounds diluted in autoclaved seawater media at an initial optical density (OD) 600 of 0.02. Desulfovibrio and marine enrichment cultures were cultivated both in sealed anaerobic glass culture tubes (Hungate tubes, Bellco, Vineland, NJ, USA) and polystyrene 96-well microplates (Costar, Thermo Fisher Scientific) and 384-well microplates (Nunc, Thermo Fisher Scientific) with plate seals (Thermo Fisher Scientific) (Supplementary Methods for more details).
Data analysis for inhibition experiments was carried out in GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA, USA) and curves were fit to a standard inhibition log dose-response curve to generate IC50 values. A total of 95% confidence intervals are reported. IC50 values are the mean of at least three biological replicates. Synergy was assessed using the equation for Fractional Inhibitory Concentration Index (Supplementary Methods; European Committee for Antimicrobial Susceptibility Testing of the European Society of Clinical M, Infectious D (2000)).
Nitrate, nitrite, perchlorate and chlorate were sodium salts (Sigma-Aldrich, St Louis, MO, USA). DETANONOate (Cayman Chemical, Ann Arbor, MI, USA) is a nitric oxide donor with a half-life of 56 h at 22–25 °C and pH 7.4, but is stable in 0.1 M NaOH. Stocks in 0.1 M NaOH were added to plates or Hungate tubes and serial dilutions made immediately prior to inoculation. Nitrate, perchlorate and chlorate were measured in culture media at the beginning and end of growth curves using ion chromatography (Dionex ICS-2100, Dionex, Sunnyvale, CA, USA).
16S rRNA gene amplicon sequencing of marine enrichment cultures
For 16S rRNA gene amplicon sequencing, marine enrichment cultures were grown in 96-well plates in the presence of twofold serial dilutions of nitrate or perchlorate (gradient plates). The gradient plate cultures were inoculated at an initial OD 600 of 0.02 in a volume of 200 μl. After 48 h (OD 600∼0.3–0.4), cultures were harvested by centrifugation, 180 μl supernatant was removed, and genomic DNA was extracted from the remaining pellet and the V3V4 region of the 16S rRNA gene was amplified using unique dual-indexed primers with attached Illumina adaptors, similar to previously published primers (Kozich et al., 2013; Fadrosh et al., 2014), and sequenced using the 600 bp MiSEQ V3 kit (Illumina, San Diego, CA, USA). Reads were analyzed by a combination of custom scripts, PEAR (Zhang et al., 2014) and the QIIME pipeline (Caporaso et al., 2010; Supplementary Methods and https://github.com/polyatail/arkin).
Quantitative PCR assay for quantifying dsrA
DNA was pooled from four replicate 96-well gradient plates (∼800 μl of culture) and Taqman (Life Technologies, Grand Island, NY, USA) quantitative PCR was used to quantify dsrA gene abundance using previous methods with some modifications (Leloup et al., 2007; Bourne et al., 2011; Supplementary Methods).
Tagged-transposon pools
1 ml −80 °C frozen aliquots of Desulfovibrio alaskensis G20 tagged-transposon pools (Kuehl et al., 2014) were recovered in basal media in 10 ml Hungate tubes with 1 g l−1 yeast extract and 1 mM sodium sulfide, centrifuged and washed to remove residual yeast extract and resuspended at an initial OD of 0.02 in 10 ml of fresh media containing 1 mM sodium sulfide and various stressor compounds in sealed Hungate tubes. Growth was monitored by OD 600 (Spectronic 20d spectrophotometer, Thermo Fisher Scientific). When pools reached an OD 600∼0.8 (between five and six doublings), 1 ml aliquots were collected by centrifugation and stored at −20 °C until genomic DNA extraction.
Genomic DNA was extracted with a Qiagen (Redwood City, CA, USA) DNAeasy kit following the protocol for extraction of genomic DNA from Gram-negative bacteria. The optional RNAse treatment step was included. DNA barcodes were then PCR amplified and hybridized to a microarray as previously described (Kuehl et al., 2014).
Strain fitness was calculated as previously described (Kuehl et al., 2014) as the log2 ratio of the abundance after growth versus at the start of the experiment. Gene fitness is the average of the strain fitness values. Gene fitness values were further normalized by subtracting gene fitness values for no stress controls from gene fitness values for stress experiments. Thus, reported fitness values in Supplementary Dataset S1 are log2(stress/no stress control). Genes with fitness >1 were considered beneficial mutations and those with fitness<−1 were considered detrimental mutations. For comparison of nitrate, perchlorate and chlorate mutations, only genes for which beneficial or detrimental mutations were observed in two independent experiments were included.
Gene expression and data analysis
Cells for microarray analysis were pelleted anaerobically at 4000 r.c.f. from single replicate 50 ml mid-log phase (OD 0.3–0.5) cultures containing stressors at indicated concentrations (Figure 4). The supernatant was decanted and cells were immediately frozen at −80 °C. We followed previously published protocols for microarray experiments and data analysis (Meyer et al., 2013). Briefly, pellets were resuspended in lysis buffer and the Qiagen RNAeasy kit was used to extract RNA and the SuperScript III indirect labeling system (Life Technologies) was used to make labeled cDNA. RNA quality was checked with a bioanalyzer and labeled cDNA hybridized to custom 12-plex microarrays following the Nimblegen protocols. Microarray signals were quantile normalized and Loess normalized using R scripts and the JMP kernel smoother.
Proteomics and data analysis
Cells for proteomics were harvested from triplicate 50 ml mid-log phase cultures (OD 0.3–0.5) containing stressors at indicated concentrations (Figure 4). Cells were pelleted anaerobically at 4000 r.c.f., supernatant was decanted and cells were resuspended in 100 mM ammonium bicarbonate, pH 7.4, sonicated and digested with trypsin prior to analysis by liquid chromatography-mass spectroscopy/mass spectroscopy (Supplementary Methods).
Extraction and measurement of intracellular NADH and NAD+
NADH and NAD+ were extracted from Desulfovibrio alaskensis G20 according to a protocol modified from Sporty et al. (2008) and analyzed by high-performance liquid chromatography (Supplementary Methods).
Results
Direct and specific inhibition of SRM by (per)chlorate and nitrate
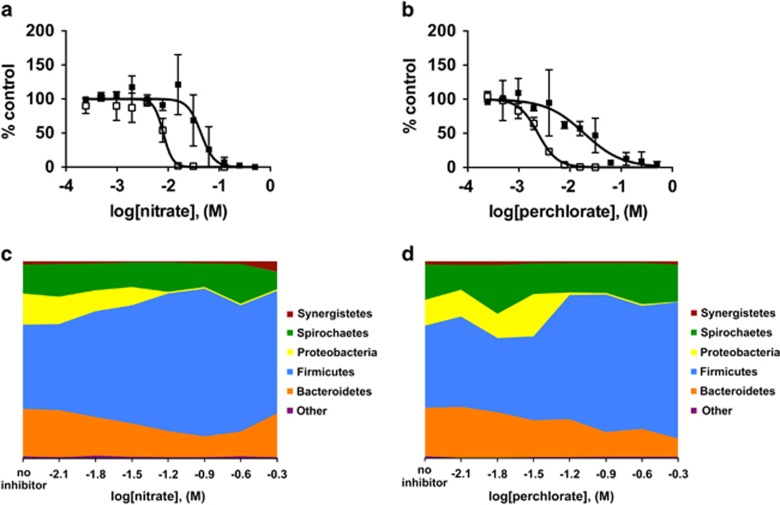
To evaluate the specificity of (per)chlorate for inhibition of sulfate reduction in complex communities, we established an active model sulfidogenic community from marine samples collected from our previous reactor studies (Engelbrektson et al., 2014). This culture was amended with a multifarious labile carbon source (yeast extract, 2 g l−1) to ensure maintenance of a phenotypically and phylogenetically diverse community membership with sulfate as the sole electron acceptor. We determined the IC50 values for (per)chlorate in comparison with nitrate against both growth and sulfide production (Figures 1a and b, Table 1). Our results indicated that although both (per)chlorate and nitrate specifically inhibited sulfidogenesis at lower concentrations than they inhibited overall growth, (per)chlorate were significantly (3.5–5 fold) more potent inhibitors than nitrate (Table 1; analysis of variance, P<0.05). 16S rRNA gene amplicon sequencing of the marine enrichment cultures revealed a relatively simple community with Desulfovibrionales as the only representative Proteobacteria and known SRM present (Figures 1c and d, Supplementary Dataset S1). When this community was grown in the presence of varying concentrations of (per)chlorate and nitrate, specific inhibition of the growth of Desulfovibrionales was observed at lower concentrations than that of other phyla in the enrichment (Figures 1c and d, Table 1; analysis of variance, P<0.05). This finding was confirmed by quantitative PCR analysis of the dissimilatory sulfite reductase (dsrA) gene, which indicated a decrease of several orders of magnitude of the community dsrA copy number in the presence of the inhibitors (Table 1, Supplementary Figure S1). In fact, the IC50 values against sulfide production, Desulfovibrionales abundance and dsrA copy number were identical (Table 1, Supplementary Figure S1). This result indicates the loss of sulfidogenic function from the community rather than adaptation of resident SRM to alternative metabolisms (for example, fermentation or syntrophism). Furthermore, no measurable consumption of nitrate or (per)chlorate (Supplementary Table S1) was observed during incubation, indicating that the loss of the Desulfovibrionales was likely not community adaptation to the utilization of these preferential alternative electron acceptors over sulfate with a resultant outgrowth of nitrate-reducing microorganisms or dissimilatory (per)chlorate-reducing microorganisms.
Figure 1.
Nitrate and perchlorate are specific inhibitors of sulfate reduction in marine enrichment cultures. (a–b) Growth (closed symbols) and sulfide (open symbols) after 48 h in marine enrichment growth cultures in the presence of varying concentrations of (a) nitrate and (b) perchlorate as a percent of uninhibited control cultures. Concentrations are presented on a log10 scale (c–d) Bacterial phyla observed by 16S amplicon sequencing of marine enrichment cultures after 48 h in the presence of varying concentrations of (c) nitrate and (d) perchlorate versus a no inhibitor control. Concentrations are presented on a log10 scale. The sole Proteobacterial genera observed was Desulfovibrionales (Supplementary Dataset S1).
Table 1. Inhibitory effect of nitrate and (per)chlorate against sulfate-reducing bacteria, IC50 (mM) (95% confidance interval)a.
| NO3− | ClO4− | ClO3− | NO2− | ClO2− | |
|---|---|---|---|---|---|
| (A) Marine enrichment culture | |||||
| Growth | 46 (34–62) | 21 (14–31) | 44 (29–60) | 5.5 (3.3–9.3) | 2.8 (1–7.5) |
| Sulfide | 8.0 (7.0–9.0) | 2.3 (2.0–2.6) | 1.6 (1.4–1.8) | 0.12 (0.1–0.4) | 1.17 (0.8–1.7) |
| Desulfovibrionales | 6.1 (3.5–10) | 1.7 (1.3–2.2) | NM | NM | NM |
| dsrA copy # | 9.5 (7.8–12) | 1.9 (1.3–2.7) | NM | NM | NM |
| (B) Desulfovibrio alaskensis G20 | |||||
| Wild type | 51 (40–65) | 24 (20–32) | 6.3 (4.9–8) | 0.42 (0.32–0.56) | 4.7 (0.5–41) |
| tn5::rex (Rex mutant) | 250 (90–300) | 51 (37–70) | 25 (20–31) | 0.1 (0.09–1.2) | 9.6 (6.7–13) |
Abbreviations: ANOVA, analysis of variance; NM, not mentioned.
(A) IC50 against growth, sulfide production, dsrA copy number and Desulfovibrionales 16S abundance in marine enrichment cultures and (B) growth of Desulfovibrio alaskensis G20 and tn5::rex. Values for the 95% confidence intervals are shown in parentheses. Italicized values for sulfide or tn5::rex growth represent significant differences between growth and wild-type IC50 values respectively, ANOVA, P<0.05. Underlined values for sulfide or wild-type G20 growth represent significant differences between IC50 values relative to nitrate, ANOVA, P<0.05. IC50 values against Desulfovibrionales 16S abundance and dsrA copy number were not significantly different from IC50 values against sulfide production, ANOVA, P>0.05.
We also determined the IC50 values for (per)chlorate and nitrate on growth for D. alaskensis G20 (Table 1) and three other Desulfovibrio species (Supplementary Table S2). As previously observed for the enriched community, the inhibitory potency of (per)chlorate was greater than that of nitrate for the pure cultures (analysis of variance, P<0.05). If (per)chlorate were reduced in these Desulfovibrio cultures, they could be converted into reactive chlorine species (RCS) (chlorite, hypochlorite) that are non-specific and potent inhibitors of microbial growth. This mechanism of inhibition by chlorate is well documented in studies of Escherichia coli (Stewart 1988; Neidhardt et al., 1996). To test for this possibility we determined the IC50 for these RCS against growth of G20 and monitored (per)chlorate concentrations in active cultures. Our results indicated that, although these reactive intermediates were inhibitory at concentrations below 1 mM (Table 1) no consumption of (per)chlorate was detected over the course of the experiments (Supplementary Table S1). Although we cannot completely rule out the possibility that inhibition was partly due to RCS formed, some (per)chlorate consumption should be measurable in this event. Taken together, our data suggest that inhibition was primarily due to the (per)chlorate anions. Similar results were observed for nitrate and its metabolite nitrite supporting the idea that the nitrate anion is also a direct inhibitor of SRM.
Chemogenomic profiling of (per)chlorate stress in D. alaskensis G20
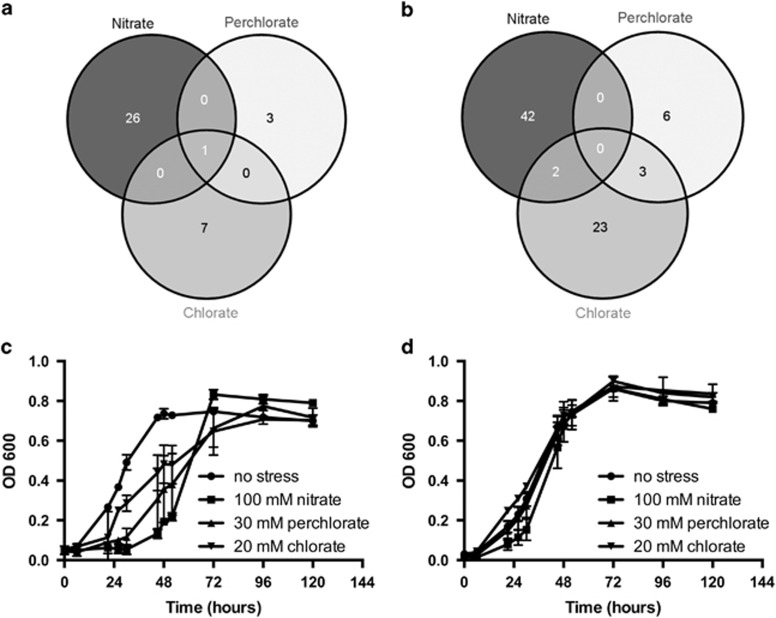
To further understand the genetic basis for direct SRM inhibition by (per)chlorate, we grew DNA-barcoded transposon pools of D. alaskensis G20 in the presence of (per)chlorate, chlorite, hypochlorite, nitrate, nitrite and nitric oxide at concentrations equal to or above the IC50. We defined genes to be important for resisting a stress if mutants in those genes were twofold reduced in abundance after growth in that stress relative to their abundance after growth in plain media. Conversely, we defined genes to be detrimental to resisting a stress if mutants were twofold enriched relative to their abundance in plain media. We also required the gene(s) to change abundance in at least two replicate experiments (Materials and methods). Comparison of the fitness profiles between inhibitors reveals overlapping and distinct genes important for tolerance or sensitivity to (per)chlorate and nitrate (Figures 2a and b, Supplementary Table S3, Supplementary Dataset S1). To confirm the results of the pool experiments, we selected a subset of genes and grew the individual transposon mutant strains alongside wild-type G20. In every case we tested, the individual mutant strains recapitulated and confirmed the phenotypes observed in the pool fitness results (beneficial versus detrimental; Supplementary Table S3).
Figure 2.
Comparison of important and detrimental genes for growth in the presence of nitrate or (per)chlorate. Genes were designated as important for resisting a stress if mutants in those genes resulted in a twofold reduction in abundance after growth in that stress relative to their abundance after growth in plain media. Conversely, genes were designated as detrimental to resisting a stress if mutants were twofold enriched relative to their abundance in plain media (Materials and methods). The complete data set is presented in Supplementary Dataset S1 and summarized in Supplementary Table S2. Only one gene, dde_2702 (rex), was observed as a common detrimental gene (beneficial mutation) between nitrate and (per)chlorate. (a) Detrimental genes (b) Important genes. Growth curves for (c) wild-type Desulfovibrio alaskensis G20 and (d) tn5::rex transposon mutant strains in the absence and presence of 100 mM nitrate, 30 mM perchlorate and 20 mM chlorate.
Transposon insertions in only one gene (dde_2702) exhibited a common phenotype in the presence of (per)chlorate and nitrate stress conferring resistance in each case (Table 1, Figures 2c and d). Dde_2702 is Rex, a transcriptional repressor that responds to the intracellular ratio of NADH:NAD+. In other bacteria, increased levels of NADH leads to Rex-mediated derepression of core respiratory enzymes (Bitoun et al., 2012; Ravcheev et al., 2012), and this is also true for G20 (Kuehl et al., 2014) and D. vulgaris Hildenborough (Geoff Christenson and Judy Wall, personal communication).
Interestingly, rex mutants were not resistant to RCS, nitrite or nitric oxide (Table 1). This finding suggests that derepression of the Rex regulon does not confer resistance to reactive chlorine or nitrogen species, which supports our earlier hypothesis that inhibition of sulfidogenesis is the result of the (per)chlorate and nitrate anions directly and not caused by inadvertent production of RCS or reactive nitrogen species (RNS) (Table 1).
Because the rex transposon mutant fitness phenotypes suggested a direct inhibitory effect of (per)chlorate and nitrate on one or more components of the respiratory sulfate-reduction pathway, we looked for fitness phenotypes of known electron transfer complexes. As the tagged-transposon pools were generated under sulfate-reducing conditions, and most of the core Rex-regulated genes are essential for growth, there were no mutant strains for these genes in the pools (Supplementary Dataset S1). However, other electron transfer complexes were represented in the pools and did display unique phenotypic responses. For example, mutant strains of the high-molecular weight cytochrome complex and the uncharacterized Hdr/Flox-1 complex (putatively involved in electron bifurcation to generate more reduced ferredoxin from NADH (Price et al., 2014)) were sensitive to both chlorate and perchlorate but not nitrate (Supplementary Table S3).
Interestingly, genes involved in molybdopterin biosynthesis were detrimental for chlorate stress, but not perchlorate or nitrate indicating another differentiation in the respective mechanisms of these sister compounds (Supplementary Table S3). Molybdopterin guanine dinucleotide is the active site cofactor in respiratory nitrate, chlorate and (per)chlorate reductases (Thorell et al., 2003; Coates and Achenbach 2004) all of which reduce chlorate to produce RCS as their toxic end products. This result suggests that chlorate toxicity may partially be attributable to RCS biogenesis from chlorate by G20. Although ion chromatography analysis of active cultures indicated immeasurable chlorate consumption (Supplementary Table S1), the potency of any produced RCS to G20 would likely be significantly enhanced if they were generated intracellularly where they would be immune to abiotic removal through reaction with H2S. The ability of the nitrate reductase NarGHI to reduce chlorate to RCS has been widely reported (Yoshimatsu et al., 2000; Afshar et al., 2001; Bell et al., 2001) and was used to identify genes for Mo cofactor biosynthesis, molybdate transport and nitrate regulation (Stewart 1988). In these previous studies, growth inhibition by chlorate was relieved by a mutation in the molybdopterin biosynthesis pathway (Stewart and MacGregor 1982). This mode of toxicity is less likely to extend to perchlorate, which is generally not a substrate for nitrate reductases because of its large activation energy and slow rate of inadvertant reduction by metals and metalloenzymes (Urbansky 2002; Clark et al., 2013). Although it is conceivable that nitrate would similarly be reduced to nitrite as part of its inhibitory mechanism, Desulfovibrio species have several mechanisms to respond to intracellular nitrite toxicity including nitrite and nitric oxide reduction by Dsr, NrfHA and hybrid cluster proteins (Wolfe et al., 1994; Yurkiw et al., 2012). Although there are no obvious gene candidates for NrfHA in G20, Dsr is an essential electron transfer complex and as such would not be represented in the mutant pools. However, the hcp was present in the G20 mutant pools (dde_2641) and was important for tolerance to nitric oxide consistent with previous studies in D. vulgaris (Yurkiw et al., 2012). Nevertheless, it was not important for tolerance to nitrate or nitrite in our G20 pool experiments (Supplementary Dataset S1, Supplementary Table S4), suggesting a more direct mechanism of inhibition by these compounds.
The gene cluster Dde_0598-Dde_0605 was specifically detrimental for nitrate stress (Supplementary Table S3). Similar findings were recently reported by Korte et al. (2014)) in their study investigating genes involved in tolerance and sensitivity to nitrate stress in Desulfovibrio species. Pool experiments profiling the chemogenomic response to 0.25 mM nitrite and 0.125 mM DETANONOate, a slow-releasing nitric oxide donor in comparison with nitrate (Supplementary Dataset S1) revealed a few common putative tolerance genes, but no common detrimental genes (Supplementary Table S4). Genes important for tolerance to nitrate and nitrite include genes involved in iron uptake including Dde_3483, a component of anthranilate synthase, Dde_2673, a ferrous iron transporter, and Dde_2676, the ferric uptake regulator, suggesting a possible relationship between iron homeostasis and nitrogen oxide toxicity. We also observed that detrimental genes for survival on nitrate, Dde_2702 (Rex) and Dde_0598-Dde_0605, were not detrimental for nitrite or nitric oxide (Supplementary Table S2, Supplementary Table S4) and (Korte et al., 2014). Taken together, our findings suggest that nitrite and nitric oxide could partially contribute to the observed nitrate stress in G20, but that a significant component of nitrate stress is due to direct nitrate toxicity. This finding was further supported by the observation that no measurable nitrate was consumed in our cultures over the course of G20 growth curves in the presence of nitrate.
Antagonism, cross-inducibility and synergy of resistance
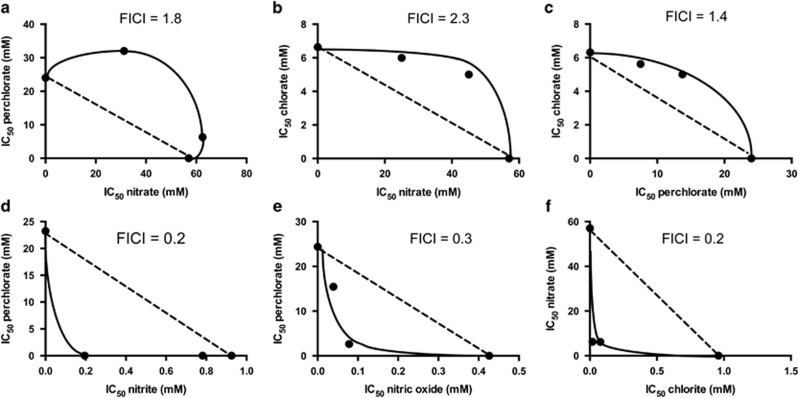
We sought to obtain further evidence that (per)chlorate and nitrate directly inhibit D. alaskensis G20 through the same mechanism. Inhibitor antagonism is frequently observed for two compounds with the same mechanism of action, whereas synergy is often observed for two compounds with different mechanisms of action. We evaluated the potential for synergy or antagonism against growth of D. alaskensis G20 for combinations of (per)chlorate and nitrate and found that inhibition was additive, but approaches antagonism, suggesting a possible common mode of action (Figure 3). In contrast, perchlorate was synergistic with nitrite or nitric oxide, and nitrate was synergistic with chlorite suggesting that these compounds likely have disparate mechanisms of inhibition (Figure 3). For example, nitrite may inhibit the dissimilatory sulfite reductase, Dsr (Wolfe et al., 1994), whereas (per)chlorate or nitrate may inhibit the ATP sulfurylase or sulfate transporters.
Figure 3.
Synergy and antagonism between nitrate, (per)chlorate and respiratory intermediates. Isobolograms are shown for Desulfovibrio alaskensis G20 inhibition by nitrate and (per)chlorate in combination with each other and respiratory intermediates nitrite, nitric oxide and chlorate. A FICI (Materials and methods) was calculated for each pair of compounds. FICI >2 implies antagonism, FICI<0.5 implies synergism. (a) Perchlorate and nitrate, (b) chlorate and nitrate, (c) chlorate and perchlorate, (d) perchlorate and nitrite, (e) perchlorate and nitric oxide, (f) nitrate and chlorite.
To determine whether similar resistance mechanisms were responsible for tolerance to nitrate and (per)chlorate, we evaluated the ability of cells pre-grown on one stressor to resist the others and observed that wild-type D. alaskensis G20 cells pre-grown on perchlorate, chlorate or nitrate were more resistant to all of the inhibitors in subsequent growth assays (Table 2). Cross-inducibility of resistance implies a common mechanism of resistance to (per)chlorate and nitrate. The only detrimental gene for (per)chlorate or nitrate stress was rex. Thus, we hypothesized that an important response to (per)chlorate and nitrate inhibition in wild-type cells is derepression of the Rex regulon in response to respiratory inhibition and NADH accumulation.
Table 2. Inhibitory effect of nitrate and (per)chlorate against Desulfovibrio alaskensis G20 pre-grown on nitrate or (per)chlorate, IC50 (mM) (95% confidance interval)a.
| NO3− | ClO4− | ClO3− | |
|---|---|---|---|
| No pre-treatment | 91 (82–100) | 15 (13–17) | 17 (16–19) |
| Pre-grown 30 mM NO3− | >150 | 33 (27–42) | 38 (31–46) |
| Pre-grown 10 mM ClO4− | >150 | 43 (39–49) | 65 (17–248) |
| Pre-grown 5 mM ClO3− | >150 | 27 (22–32) | 48 (46–50) |
IC50 against initial growth rate (0–6 h) of Desulfovibrio alaskensis G20. Values for the 95% confidence intervals are shown in parentheses. Italicized values for IC50 values represent significant differences between the no pre-treatment control and pre-grown conditions, Analysis of variance, P<0.05.
Accumulation of NADH and increased expression of the Rex regulon in (per)chlorate and nitrate-stressed cultures
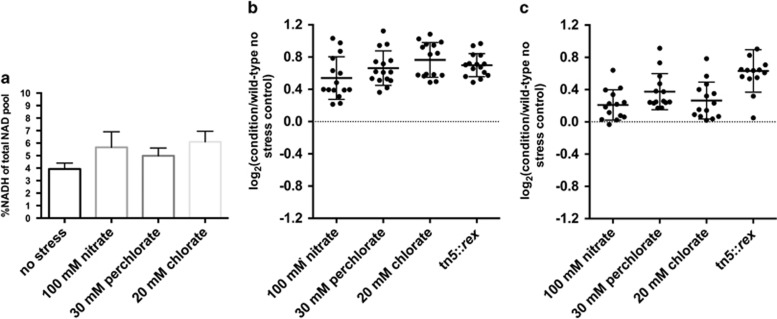
In order to determine whther wild-type G20 cells can respond to (per)chlorate and nitrate inhibition by derepression of the Rex regulon we investigated expression of the regulon components and measured intracellular NADH and NAD+ pools. Rex responds to the intracellular NADH:NAD+ ratio in other organisms and preliminary results suggest a similar response in G20 and D. vulgaris (Kuehl et al., 2014; Geoff Christenson and Judy Wall, personal communication). Inhibition of sulfate reduction should lead to excess reducing equivalents, a higher NADH:NAD+ ratio and derepression of the core Rex regulon. This, in turn, should result in increased resistance to compounds that target the sulfate reduction pathway as observed in our pre-induction experiments (Table 2) and for the Rex transposon insertion mutants (Kuehl et al., 2014).
Using a high-performance liquid chromatography-based assay, we determined the intracellular concentration of NADH and NAD+ in D. alaskensis G20. We found that the NADH:NAD+ ratio in mid-log phase cultures (OD 600=0.3–0.5) was higher (25–50%) in the presence of (per)chlorate and nitrate than in the absence (Figure 4a). For the gene targets of Rex in the presence of (per)chlorate and nitrate, we measured mRNA transcript levels using whole-genome microarrays and protein abundances as assessed by normalized peptide counts (Figures 4b and c). Previous work demonstrated that derepression in G20 rex mutants is limited to a core subset of the predicted Rex regulon, suggesting more complex regulation of some genes (Kuehl et al., 2014). This set of genes includes qmoABCD (Dde_1111:Dde_1114), sat (Dde_2265), adenylate kinase (Dde_2028), pyrophosphatase (Dde_1178), a sulfate transporter (Dde_2406), an ATP synthase, atpFFHAGD (Dde_0990:Dde_0984) and atpIIBE (Dde_2698:Dde_2701). We observed increased transcription and higher protein levels for this core set of Rex-regulated genes in response to all three stressors (perchlorate, chlorate and nitrate) and in the tn5::rex transposon mutant strain (Figures 4b and c, Supplementary Dataset S1).
Figure 4.
NADH accumulation and core Rex regulon derepression in Desulfovibrio alaskensis G20 grown to mid-log (OD 600 0.3–0.5) in the absence or presence of nitrate, perchlorate or chlorate. (a) Percentage of intracellular NAD that is reduced (NADH) for cells grown in the presence of nitrate or (per)chlorate. One-sample t-test, P<0.001 for all conditions relative to wild-type no stress control. Error bars represent s.d. values of triplicate measurements. (b) Log2(stress/wild-type no-stress control) of transcript abundances (dots represent one replicate microarray measurement) for core Rex-regulated genes for cells grown in the presence of nitrate or (per)chlorate or tn5::rex versus a wild-type no stress control. One-sample t-test, P<0.001 for all conditions relative to wild-type no stress control. Error bars represent s.d. values (c) Log2(stress/wild-type no stress control) of normalized peptide counts (dots represent average of triplicate measurements) for core Rex-regulated proteins for cells grown in the presence of nitrate or (per)chlorate or tn5::rex versus a wild-type no stress control. One-sample t-test, P<0.001 for all conditions relative to wild-type no stress control. Error bars represent s.d. values.
Taken together, our results support the hypothesis that in wild-type G20, (per)chlorate and nitrate inhibit the central pathway of sulfate reduction, leading to NADH accumulation and derepression of the core enzymes of sulfate reduction though accumulation of NADH-Rex complexes. This derepression of the core enzymes is likely a primary response mechanism to inhibition by these compounds in wild-type cells as evidenced by the increased tolerance of cells pre-grown in the presence of (per)chlorate or nitrate (Table 2) and the resistance of the Rex mutant strains (Table 1, Figure 2).
Discussion
In these studies we employed a combination of microbial community analyses and pure culture experiments to investigate the inhibition of sulfate reduction by both perchlorate and chlorate in comparison with nitrate. Our results demonstrate that (per)chlorate and nitrate are direct and specific inhibitors of the central pathway of microbial sulfate respiration and sulfide production. As nitrate and (per)chlorate are sulfate analogs and nitrate and chlorate have been shown to competitively inhibit the assimilatory sulfate-reduction enzymes in other organisms (Farley et al., 1976; Baeuerle and Huttner 1986; Hanna et al., 2002), a long-standing hypothesis has been that they inhibit the central pathway of sulfate reduction in SRM (Postgate 1952). However, evidence in support of this hypothesis has been lacking for any of these compounds. We observed that both (per)chlorate and nitrate specifically inhibit sulfide production and growth of Desulfovibrionales in marine enrichment cultures at lower concentrations than they inhibit growth of other organisms in the microbial community. In D. alaskensis G20, transposon insertion mutants in dde_2702 (rex) were more resistant to (per)chlorate and nitrate. Rex mutants expressed higher levels of central enzymes of sulfate reduction, and both (per)chlorate and nitrate induced this expression, likely through NADH accumulation and Rex derepression. Higher levels of these enzymes rendered G20 cells less sensitive to competitive inhibitors of sulfate reduction. Thus, (per)chlorate and nitrate were antagonistic inhibitors and resistance to these compounds was cross-inducible suggesting a common mode of action. However, (per)chlorate were more potent inhibitors than nitrate of sulfidogenesis in both enrichment cultures and the growth of pure cultures of SRM, suggesting that the inhibitory target(s) are universally more sensitive to (per)chlorate.
The observation that the G20 Rex regulon was derepressed in the presence of (per)chlorate and nitrate suggests that wild-type G20 could overcome direct sulfate-respiration inhibition by increasing the expression of sulfate-respiratory enzymes. This ability to adapt to environmental fluxes may be beneficial for bacteria living in ecosystems in which multiple respiratory metabolisms are stratified. Some sulfate-reducing bacteria are capable of respiratory nitrate-reduction, which gives them an advantage in environments in which nitrate fluxes are present. Similarly, obligate sulfate-reducing bacteria that can tolerate higher concentrations of a competitive respiratory inhibitor (that is, perchlorate or nitrate) through Rex regulon derepression will persist and grow in the presence of higher concentrations of that inhibitor and will likely establish a wider environmental niche. Given the prevalence of Rex in diverse bacterial species and the fact that the Rex is often a repressor of electron flow pathways and redox active enzymes (Ravcheev et al., 2012), Rex regulon derepression may represent a common strategy for adapting to gradients of competitive respiratory inhibitors and fluctuating concentrations of electron donor and acceptor. However, while pure cultures may adapt in this manner, there is an energy cost to continuous overexpression of the components of the Rex regulon and in our hands adaptation of a sulfidogenic microbial community to perchlorate inhibition was never observed even after an extended treatment period of 225 days (AE and JDC, unpublished data).
The results of these studies in combination with our previous work (Engelbrektson et al., 2014; Gregoire et al., 2014) signify that perchlorate amendment is a promising new strategy for control of sulfidogenesis in industrial systems. These studies further indicate that sulfidogenesis inhibition in complex ecosystems by (per)chlorate is multifaceted. In addition to thermodynamic preference (Eo'=+797 mV for the biological couple of ClO4−/Cl−) relative to sulfate reduction (Eo'=−217 mV) and the ability of all dissimilatory (per)chlorate-reducing microorganism tested to innately oxidize H2S coupled to (per)chlorate respiration (Gregoire et al., 2014), these studies demonstrate that (per)chlorate is also directly and specifically inhibitory of SRM. As this inhibitor targets the central sulfate-respiratory pathway, which is highly conserved across all SRM regardless of phylogenetic affiliation, inhibition should be universal. Furthermore, the observed synergistic inhibition of D. alaskensis G20 in combination with nitrite, nitric oxide or chlorite suggest that combinations of (per)chlorate and nitrate may prove beneficial in ecosystems in which respiratory intermediates (nitrite, nitric oxide, chlorite) are produced by active nitrate or (per)chlorate-reducing members of the microbial community. Although chlorate is an equally effective inhibitor, it is chemically unstable in the presence of ferrous iron (Fe2+) (Engelbrektson et al., 2014) and as such may be abiotically removed. The potency and specificity of inhibition by perchlorate combined with its high-aqueous solubility and chemical stability across a broad range of environmental conditions (Urbansky 2002), suggest that it represents the most promising alternative to nitrate. Future studies are needed to investigate this possibility more thoroughly and confirm the application of perchlorate to control sulfidogenesis in industrial ecosystems.
Acknowledgments
We thank members of the Coates and Arkin groups for critical comments on this manuscript and Michi Taga and Kris Niyogi for use of their HPLC columns and instruments. We thank Mike Nold (Waters) for advice and protocols for proteomics sample preparation. Work in the laboratory of JDC on biosouring is supported by the Energy Biosciences Institute.
JDC declares that he holds IP on the application of (per)chlorate to control souring. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Afshar S, Johnson E, de Vries S, Schroder I. Properties of a thermostable nitrate reductase from the hyperthermophilic archaeon Pyrobaculum aerophilum. J Bacteriol. 2001;183:5491–5495. doi: 10.1128/JB.183.19.5491-5495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle PA, Huttner WB. Chlorate—a potent inhibitor of protein sulfation in intact cells. Biochem Biophys Res Commun. 1986;141:870–877. doi: 10.1016/s0006-291x(86)80253-4. [DOI] [PubMed] [Google Scholar]
- Bell LC, Richardson DJ, Ferguson SJ. Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. The periplasmic enzyme catalyzes the first step in aerobic denitrification. FEBS Lett. 2001;265:85–87. doi: 10.1016/0014-5793(90)80889-q. [DOI] [PubMed] [Google Scholar]
- Bitoun JP, Liao S, Yao X, Xie GG, Wen ZT. The redox-sensing regulator Rex modulates central carbon metabolism, stress tolerance response and biofilm formation by Streptococcus mutans. Plos One. 2012;7:e44766. doi: 10.1371/journal.pone.0044766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne DG, Muirhead A, Sato Y. Changes in sulfate-reducing bacterial populations during the onset of black band disease. ISME J. 2011;5:559–564. doi: 10.1038/ismej.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce RA, Achenbach LA, Coates JD. Reduction of (per)chlorate by a novel organism isolated from paper mill waste. Environ Microbiol. 1999;1:319–329. doi: 10.1046/j.1462-2920.1999.00042.x. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IC, Melnyk RA, Engelbrektson A, Coates JD. Structure and evolution of chlorate reduction composite transposons. MBio. 2013;4:pii: e00379–13. doi: 10.1128/mBio.00379-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates JD, Achenbach LA. Microbial perchlorate reduction: rocket fuelled metabolism. Nat Rev Microbiol. 2004;2:569–580. doi: 10.1038/nrmicro926. [DOI] [PubMed] [Google Scholar]
- Engelbrektson A, Hubbard CG, Tom LM, Boussina A, Jin YT, Wong H, et al. Inhibition of microbial sulfate reduction in a flow-through column system by (per)chlorate treatment. Front Microbiol. 2014;5:1–11. doi: 10.3389/fmicb.2014.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Committee for Antimicrobial Susceptibility Testing of the European Society of Clinical M, Infectious D Terminology relating to methods for the determination of susceptibility of bacteria to antimicrobial agents. Clin Microbiol Infect. 2000;6:503–508. doi: 10.1046/j.1469-0691.2000.00149.x. [DOI] [PubMed] [Google Scholar]
- Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley JR, Cryns DF, Yang YH, Segel IH. Adenosine triphosphate sulfurylase from penicillium chrysogenum. Steady state kinetics of the forward and reverse reactions. J Biol Chem. 1976;251:4389–4397. [PubMed] [Google Scholar]
- Fischer JP, Cypionka H. Analysis of aerotactic band formation by Desulfovibrio desulfuricans in a stopped-flow diffusion chamber. FEMS Microbiol Ecol. 2006;55:186–194. doi: 10.1111/j.1574-695X.2005.00024.x. [DOI] [PubMed] [Google Scholar]
- Gieg LM, Jack TR, Foght JM. Biological souring and mitigation in oil reservoirs. Appl Microbiol Biotechnol. 2011;92:263–282. doi: 10.1007/s00253-011-3542-6. [DOI] [PubMed] [Google Scholar]
- Greene EA, Hubert C, Nemati M, Jenneman GE, Voordouw G. Nitrite reductase activity of sulphate-reducing bacteria prevents their inhibition by nitrate-reducing, sulphide-oxidizing bacteria. Environ Microbiol. 2003;5:607–617. doi: 10.1046/j.1462-2920.2003.00446.x. [DOI] [PubMed] [Google Scholar]
- Greene EA, Brunelle V, Jenneman GE, Voordouw G. Synergistic inhibition of microbial sulfide production by combinations of the metabolic inhibitor nitrite and biocides. Appl Environ Microbiol. 2006;72:7897–7901. doi: 10.1128/AEM.01526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire P, Engelbrektson A, Hubbard CG, Metlagel Z, Csencsits R, Auer M, et al. 2014Control of sulfidogenesis through bio-oxidation of H2S coupled to (per)chlorate reduction Environ Microbiol Rep 4e-pub ahead of print 4 April 2014doi: 10.1111/1758-2229.12156 [DOI] [PubMed] [Google Scholar]
- Hanna E, MacRae IJ, Medina DC, Fisher AJ, Segel IH. ATP sulfurylase from the hyperthermophilic chemolithotroph Aquifex aeolicus. Arch Biochem Biophys. 2002;406:275–288. doi: 10.1016/s0003-9861(02)00428-9. [DOI] [PubMed] [Google Scholar]
- Hoogewerf AJ, Cisar LA, Evans DC, Bensadoun A. Effect of chlorate on the sulfation of lipoprotein lipase and heparan sulfate proteoglycans. Sulfation of heparan sulfate proteoglycans affects lipoprotein lipase degradation. J Biol Chem. 1991;266:16564–16571. [PubMed] [Google Scholar]
- Hubert C. Handbook of Hydrocarbon and Lipid Microbiology. Springer: New York City, NY, USA; 2010. Microbial ecology of oil reservoir souring and its control by nitrate injection; pp. 2753–2766. [Google Scholar]
- Korte HL, Fels SR, Christensen GA, Price MN, Kuehl JV, Zane GM, et al. Genetic basis for nitrate resistance in Desulfovibrio strains. Front Microbiol. 2014;5:153. doi: 10.3389/fmicb.2014.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl JV, Price MN, Ray J, Wetmore KM, Esquivel Z, Kazakov AE, et al. Functional genomics with a comprehensive library of transposon mutants for the sulfate-reducing bacterium Desulfovibrio alaskensis G20. MBio. 2014;5:e01041–01014. doi: 10.1128/mBio.01041-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup J, Loy A, Knab NJ, Borowski C, Wagner M, Jorgensen BB. Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ Microbiol. 2007;9:131–142. doi: 10.1111/j.1462-2920.2006.01122.x. [DOI] [PubMed] [Google Scholar]
- Meyer B, Kuehl J, Deutschbauer AM, Price MN, Arkin AP, Stahl DA. Variation among Desulfovibrio species in electron transfer systems used for syntrophic growth. J Bacteriol. 2013;195:990–1004. doi: 10.1128/JB.01959-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer WE. Perchlorate: problems, detection, and solutions. Environ Forensics. 2001;2:301–311. [Google Scholar]
- Mukhopadhyay A, He Z, Alm EJ, Arkin AP, Baidoo EE, Borglin SC, et al. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J Bacteriol. 2006;188:4068–4078. doi: 10.1128/JB.01921-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Curtiss R, Ingraham J, Lin E, Brooks Low K, Magasanik B, et al. (eds).. (1996Escherichia coli and Salmonella—Cellular and Molecular Biology ASM Press: Washington, DC [Google Scholar]
- Postgate JR. Competitive and noncompetitive inhibitors of bacterial sulphate reduction. J Gen Microbiol. 1952;6:128–142. doi: 10.1099/00221287-6-1-2-128. [DOI] [PubMed] [Google Scholar]
- Price M, Ray J, Wetmore KM, Kuehl JV, Bauer S, Deutschbauer AM, et al. 2014The genetic basis of energy conservation in the sulfate-reducing bacterium Desulfovibrio alaskensis G20 Front Microbiole-pub ahead of print 31 October 2014doi: 10.3389/fmicb.2014.00577 [DOI] [PMC free article] [PubMed]
- Price MN, Deutschbauer AM, Skerker JM, Wetmore KM, Ruths T, Mar JS, et al. Indirect and suboptimal control of gene expression is widespread in bacteria. Mol Syst Biol. 2013;9:660. doi: 10.1038/msb.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravcheev DA, Li X, Latif H, Zengler K, Leyn SA, Korostelev YD, et al. Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex. J Bacteriol. 2012;194:1145–1157. doi: 10.1128/JB.06412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen J, Tiedje JM, Firestone RB. Inhibition by sulfide of nitric and nitrous oxide reduction by denitrifying Pseudomonas fluorescens. Appl Environ Microbiol. 1980;39:105–108. doi: 10.1128/aem.39.1.105-108.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporty JL, Kabir MM, Turteltaub KW, Ognibene T, Lin SJ, Bench G. Single sample extraction protocol for the quantification of NAD and NADH redox states in Saccharomyces cerevisiae. J Sep Sci. 2008;31:3202–3211. doi: 10.1002/jssc.200800238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V, MacGregor CH. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982;151:788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988;52:190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorell HD, Stenklo K, Karlsson J, Nilsson T. A gene cluster for chlorate metabolism in Ideonella dechloratans. Appl Environ Microbiol. 2003;69:5585–5592. doi: 10.1128/AEM.69.9.5585-5592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich TC, Blaesse M, Huber R. Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation. EMBO J. 2001;20:316–329. doi: 10.1093/emboj/20.3.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbansky ET. Perchlorate as an environmental contaminant. Environ Sci Pollut Res Int. 2002;9:187–192. doi: 10.1007/BF02987487. [DOI] [PubMed] [Google Scholar]
- WHO . Hydrogen Sulfide. WHO Regional Publications, European Series: Denmark; 2000. pp. 1–7. [Google Scholar]
- Wolfe BM, Lui SM, Cowan JA. Desulfoviridin, a multimeric-dissimilatory sulfite reductase from Desulfovibrio vulgaris (Hildenborough). Purification, characterization, kinetics and EPR studies. Eur J Biochem. 1994;223:79–89. doi: 10.1111/j.1432-1033.1994.tb18968.x. [DOI] [PubMed] [Google Scholar]
- Yoshimatsu K, Sakurai T, Fujiwara T. Purification and characterization of dissimilatory nitrate reductase from a denitrifying halophilic archaeon. FEBS Lett. 2000;470:216–220. doi: 10.1016/s0014-5793(00)01321-1. [DOI] [PubMed] [Google Scholar]
- Youssef N, Elshahed MS, McInerney MJ. Microbial processes in oil fields: culprits, problems, and opportunities. Adv Appl Microbiol. 2008;66:141–251. doi: 10.1016/S0065-2164(08)00806-X. [DOI] [PubMed] [Google Scholar]
- Yurkiw MA, Voordouw J, Voordouw G. Contribution of rubredoxin:oxygen oxidoreductases and hybrid cluster proteins of Desulfovibrio vulgaris Hildenborough to survival under oxygen and nitrite stress. Environ Microbiol. 2012;14:2711–2725. doi: 10.1111/j.1462-2920.2012.02859.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, He Q, Hemme CL, Mukhopadhyay A, Hillesland K, Zhou A, et al. How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nat Rev Microbiol. 2011;9:452–466. doi: 10.1038/nrmicro2575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.