Abstract
Planktonic bacterial lineages with streamlined genomes are prevalent in the ocean. The base composition of their DNA is often highly biased towards low G+C content, a possible source of systematic error in phylogenetic reconstruction. A total of 228 orthologous protein families were sampled that are shared among major lineages of Alphaproteobacteria, including the marine free-living SAR11 clade and the obligate endosymbiotic Rickettsiales. These two ecologically distinct lineages share genome sizes of <1.5 Mbp and genomic G+C content of <30%. Statistical analyses showed that only 28 protein families are composition-homogeneous, whereas the other 200 families significantly violate the composition-homogeneous assumption included in most phylogenetic methods. RAxML analysis based on the concatenation of 24 ribosomal proteins that fall into the heterogeneous protein category clustered the SAR11 and Rickettsiales lineages at the base of the Alphaproteobacteria tree, whereas that based on the concatenation of 28 homogeneous proteins (including 19 ribosomal proteins) disassociated the lineages and placed SAR11 at the base of the non-endosymbiotic lineages. When the two data sets were concatenated, only a model that accounted for compositional bias yielded a tree identical to the tree built with composition-homogeneous proteins. Ancestral genome analysis suggests that the first evolved SAR11 cell had a small genome streamlined from its ancestor by a factor of two and coinciding with an ecological transition, followed by further gradual streamlining towards the extant SAR11 populations.
Introduction
Planktonic bacterial lineages with streamlined genomes are widespread in the ocean (Swan et al., 2013; Giovannoni et al., 2014). Prominent examples are alphaproteobacterial SAR11 (Giovannoni et al., 2005), gammaproteobacterial SAR86 (Dupont et al., 2012), cyanobacterial Prochlorococcus (Dufresne et al., 2003; Rocap et al., 2003) and betaproteobacterial OM43 (Giovannoni et al., 2008). Members of these lineages are either uncultivated or difficult to propagate when cultures are available. It is generally assumed that these streamlined bacteria evolved from lineages with larger genomes through genome reduction processes. To address this hypothesis, the evolutionary relationships of the streamlined lineages and their non-streamlined relatives need to be resolved. For instance, ancestral reconstruction based on a robust phylogeny in which Prochlorococcus evolved from their larger Synechococcus relatives supported the genome streamlining hypothesis for Prochlorococcus (Luo et al., 2011).
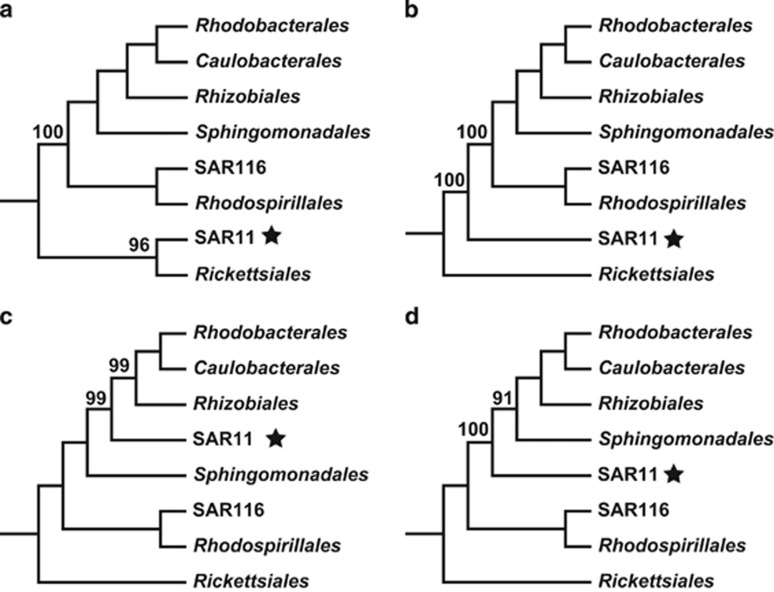
In the case of SAR11, however, several alternate evolutionary positions have been proposed in the Alphaproteobacteria tree, all of which have strong statistical support in the evolutionary model underlying the analysis (Thrash et al., 2011; Rodríguez-Ezpeleta and Embley, 2012; Viklund et al., 2012; Luo et al., 2013) (Figure 1). Although the first evolved SAR11 cell is consistently predicted to have a streamlined genome, the genome size of its immediate ancestor varies considerably depending on where SAR11 is located in the Alphaproteobacteria tree (Luo et al., 2013). If SAR11 and Rickettsiales form a monophyletic clade at the basal node of the tree (Thrash et al., 2011;Figure 1a), it is predicted that the immediate ancestor had a similar genome size as the first SAR11 cell, and thus genome streamlining following the divergence of the SAR11 lineage is not well supported. If SAR11 does not cluster with Rickettsiales but is basal to other Alphaproteobacteria lineages (Luo et al., 2013; Figure 1b), the first SAR11 cell is predicted to be a descendant of an intermediate-size ancestor. If SAR11 is positioned at the middle of the non-endosymbiotic lineages (Viklund et al., 2012; Luo et al., 2013; Figures 1c and d), the first SAR11 is predicted to have evolved from an ancestor with a large genome size, and the genome streamlining hypothesis is most strongly supported. Therefore, collecting additional evidence to help resolve the evolutionary position of SAR11 was the primary goal of the present study.
Figure 1.
Four alternate evolutionary positions of the SAR11 clade in the Alphaproteobacteria phylogeny. The statistical support values were obtained from previous publications (Thrash et al., 2011; Viklund et al., 2012; Luo et al., 2013).
Materials and methods
Taxon sampling
A total of 66 alphaproteobacterial and 8 gammaproteobacterial and betaproteobacterial outgroup genome sequences were obtained from GenBank. The alphaproteobacterial genomes include 10 associated with the marine Roseobacter clade, 1 with Parvularculales, 3 with Hyphomonadaceae, 5 with Caulobacterales, 14 with Rhizobiales, 6 with Sphingomonadales, 5 with the marine SAR116 clade, 7 with Rhodospirillales, 8 with the marine SAR11 clade and 7 with Rickettsiales. Among the 10 Roseobacter clade members, genomes of five strains are closed and the remaining were estimated to be complete or nearly so (Luo et al., 2013). Among the eight SAR11 genomes, all are closed except that strain HIMB114 consists of scaffold with one contig (Grote et al., 2012). Among the five SAR116 genomes, one (HIMB100) has 10 contigs (Grote et al., 2011) and three are uncultivated single cell genomes (SCGC AAA015-N04, SCGC AAA536-K22, SCGC AAA536-G10) with a variable success in recovering the genomic DNA (69%–91%) (Swan et al., 2013). Genomes of all other lineages are closed. In the subsequent analyses, all of the 66 alphaproteobacterial genomes were used in phylogenetic tree reconstructions, whereas the three single-cell genomes and HIMB100 were not included in ancestral genome reconstruction because of their relatively low recovery of genome content. Taxon sampling was carried out to maximize the phylogenetic diversity by sampling the major taxonomic units (Family/Genus) in each well-accepted Order of Alphaproteobacteria, and to minimize the total number of taxa so that the computation for phylogenomic reconstruction could be completed with a reasonable amount of time. Under this principle, the strains used for phylogenomic analyses were chosen randomly.
Ortholog identification, character selection and phylogenomic tree reconstruction
Orthologous gene families among the above 74 genomes were identified using the OrthoMCL software (Li et al., 2003). Inparalog, copies in a gene family were discarded. A total of 228 gene families, including 43 ribosomal protein families, were chosen for phylogenetic analyses, each of which contains at least 6 gene members affiliated with the Roseobacter clade, 3 with Caulobacterales, 8 with Rhizobiales, 4 with Sphingomonadales, 4 with marine SAR116 clade, 4 with Rhodospirillales, 5 with the marine SAR11 clade and 5 with Rickettsiales, and 4 with outgroup. This relatively small number of shared genes is presumably influenced by the inclusion of the free-living marine SAR11 clade and the endosymbiotic Rickettsiales, two streamlined lineages with their genomic content shaped by their distinct environments, and by the presence of partial genomes of three single cells.
To obtain a more reliable alignment, seven independent alignment programs were used to align the orthologous amino acid sequences for each of the selected gene families. These programs are ClustalW (Larkin et al., 2007), MAFFT (Katoh et al., 2005), MUSCLE (Edgar, 2004), T-coffee (Notredame et al., 2000), DIALIGN (Morgenstern, 2004), Kalign (Lassmann and Sonnhammer, 2005) and OPAL (Wheeler and Kececioglu, 2007). Unreliable regions of the alignment were trimmed using the trimAl software (Capella-Gutiérrez et al., 2009) using the parameters ‘-automated1 -resoverlap 0.55 -seqoverlap 60'. Some short partial sequences were automatically discarded using the above parameter setting; this is important in the analysis because of the many missing nucleotides in parts of the single cell genomes. The seven trimmed alignments for each gene family were then compared using trimAl and the one with the largest fraction of sites showing consistency with other alignments was selected. The selected alignments were subject to a ProtTest (Abascal et al., 2005) analysis, which determines the best-fit amino acid substitution matrix and whether the among-site rate heterogeneity model is applicable.
As there is a substantial variation of G+C content among lineages, which is known to result in compositional heterogeneity among lineages at the amino acid sequence level (Gu et al., 1998; Foster and Hickey, 1999; Singer and Hickey, 2000), the validity of the stationarity (compositional homogeneity) assumption for each of the 228 families was specifically tested using the posterior predictive simulation implemented in the P4 Bayesian phylogenetic software package (Foster, 2004).
For phylogenomic analyses, three data sets were compiled: the concatenation of the 28 composition-homogenous proteins (including 19 ribosomal proteins), of the 24 composition-heterogeneous ribosomal proteins and of the combined 52 proteins. The standard maximum likelihood method implemented in the MPI version of RAxML v7.3.0 software (Stamatakis, 2014) was used to analyze the three data sets separately. To account for the possibility that different proteins may have undergone distinct patterns of amino acid replacement, a data partition model was applied so that proteins are grouped into categories and proteins within each category have similar substitution patterns. The optimal partitioning scheme for each of the three data sets was determined separately by the PartitionFinder software (Lanfear et al., 2012) according to Bayesian information criterion score. The RAxML tree was constructed using the ‘PROTGAMMALG' model, which assumes amino acid substitution rates among sites follow a gamma distribution. The concatenated super-alignment was partitioned according to the optimal partitioning scheme. To obtain statistical confidence of internal branches, 100 pseudoreplicates were generated using the ‘rapid bootstrap' method in RAxML.
Phylogenomic tree reconstruction using a Bayesian nonstationary model
Reduced alphabets were used to overcome the computational inefficiency issue of P4 (Foster, 2004) and alleviate the compositional bias by recoding the amino acid sequences (with 20 character states) into the following six Dayhoff groups that correspond to most amino acid substitution matrices (Hrdy et al., 2004): (cysteine), (alanine, serine, threonine, proline, glycine), (asparagine, aspartic acid, glutamic acid, glutamine), (histidine, arginine, lysine), (methionine, isoleucine, leucine, valine), (phenylalanine, tyrosine, tryptophan). This recoding scheme has been used to improve topological estimation in the presence of compositional heterogeneity in a number of phylogenomic studies (Cox et al., 2008; Foster et al., 2009; Nesnidal et al., 2010), including a recent study of the evolutionary placement of the SAR11 clade (Rodríguez-Ezpeleta and Embley, 2012). The Dayhoff-recoded concatenated datasets of the 52 protein sequences (the 28 homogeneous and the 24 heterogeneous ribosomal proteins) were analyzed using multiple configurations of the NDCH (node-discrete composition heterogeneity) and NDRH (node-discrete rate matrix heterogeneity) models, general time reversible (GTR) substitution matrix plus four Gamma-distributed rate categories, and employing the polytomy prior (Lewis et al., 2005). Ten replicate runs were performed for each configuration. In each replicate run, one cold and three heated MCMC chains were run for a total of 1 500 000 generations with trees sampled every 1000 generations. The first 500 000 generations were discarded as ‘burn-in'. The model adequacy with respect to composition was assessed using the χ2 homogeneity test on posterior distributed samples which were generated by posterior predictive simulation in P4. This test rejected the stationary model (1 composition vector plus 1 GTR rate matrix across the tree) (P<0.05), while it suggested that a composition-heterogeneous model (two composition vector plus two GTR rate matrix across the tree) was adequate (P>0.05). The phylogenomic trees were reconstructed using both the stationary and nonstationary models. The average standard deviation of split support was <0.01 suggesting convergence was reached for all phylogenetic reconstructions. A majority-rule consensus tree was constructed from the post-burn-in trees.
Phylogenomic tree reconstruction using a Bayesian mixture model
Among-site compositional heterogeneity was accounted for by the CAT Bayesian mixture model (Lartillot and Philippe, 2004) implemented in the PhyloBayes MPI software package (Lartillot et al., 2013). The Bayesian MCMC analyses were run with CAT-GTR model with a Gamma distribution of rates among sites using the concatenated datasets of the 52 protein sequences. Two independent MCMC runs were performed, each with >100 000 cycles. The first 20% of all runs were discarded as ‘burn-in'. Convergence was reached with the maxdiff statistic of 0.08 and an effective sample size >400.
Computational time of the phylogenomic analyses
Phylogenomic analyses using the PhyloBayes MPI software and the P4 software are computationally expensive. All analyses were performed using a Linux cluster consisting of multiple 8-core or 12-core nodes (Intel-Xeon processors with different model numbers, including E5410, E5504, E5530 and X5650) varying in their clock speeds from 2.00 to 2.67 GHz. It took 56 days with 75 cores to complete each of the two independent PhyloBayes runs of the concatenated 52 protein sequences (74 taxa and 12 987 sites) employing the CAT-GTR model, with a maximum virtual memory used by all MPI processes of ∼20 GB. The P4 software is not coded for parallel computing, and thus only one CPU core can be assigned to run the jobs. It took on average 25 days to complete each of the 10 replicate runs of the Dayhoff recoded data set of the 52 proteins for each configuration of the NDCH and NDRH models, with a maximum virtual memory use of ∼2 GB. This computational burden imposed a constraint in the number of taxa that could be used in these phylogenomic analyses, considering that computational time rapidly increases as the number of taxa increases.
Reconstruction of ancestral genomes
For ancestral genome reconstruction using a maximum likelihood birth-and-death model implemented in the COUNT software (Csűrös and Miklós, 2009; Csűrös, 2010), the phyletic pattern (gene family presence/absence and gene copy number) of the 62 complete or nearly complete Alphaproteobacteria genomes was mapped to a rooted and compositionally unbiased Alphaproteobacteria phylogeny. The orthologous gene family table of these 62 genomes was obtained by clustering all of the predicted protein sequences from these genomes using OrthoMCL (Li et al., 2003). The procedure was repeated with 100 bootstrap data sets generated by randomly sampling the gene families (with repetitions). The number of genes in the ancestral lineages was predicted through regression analysis between the number of gene families and the number of genes at the leaf nodes. The details of the procedure follows a recent publication (Luo et al., 2013).
Results
Identification of composition-homogeneous protein families
A total of 228 orthologous gene families (Supplementary Table S1), including 43 encoding for ribosomal proteins, were selected for phylogenetic analyses at the amino acid level. These families occur across major lineages of Alphaproteobacteria. Although a majority of the included lineages are represented by members with high genomic G+C content (50–70%), the marine SAR11 clade, the Rickettsiales and a lineage in the marine SAR116 clade represented by three single cells (SCGC AAA015-N04, SCGC AAA536-K22, SCGC AAA536-G10) have low genomic G+C content (30% and below). Posterior predictive simulation generated posterior distributed samples for each of the families, and χ2 homogeneity test on the posterior samples showed that the composition-homogeneous model is adequate (P>0.05) in only 28 functionally conserved families (Supplementary Table S1), of which 19 encode for ribosomal proteins (Table 1). Among the remaining 200 composition-heterogeneous families, 24 encode for ribosomal proteins (Table 1).
Table 1. List of 28 composition-homogeneous and 24 composition-heterogeneous ribosomal proteins.
|
Composition-homogeneous |
Composition-heterogeneous |
||
|---|---|---|---|
| COG | Protein | COG | Protein |
| COG0522 | Ribosomal protein S4 | COG0261 | Ribosomal protein L21 |
| COG0096 | Ribosomal protein S8 | COG0203 | Ribosomal protein L17 |
| COG0100 | Ribosomal protein S11 | COG0359 | Ribosomal protein L9 |
| COG0238 | Ribosomal protein S18 | COG0200 | Ribosomal protein L15 |
| COG0199 | Ribosomal protein S14 | COG0098 | Ribosomal protein S5 |
| COG0093 | Ribosomal protein L14 | COG0256 | Ribosomal protein L18 |
| COG0186 | Ribosomal protein S17 | COG0097 | Ribosomal protein L6P/L9E |
| COG0197 | Ribosomal protein L16/L10E | COG0094 | Ribosomal protein L5 |
| COG0091 | Ribosomal protein L22 | COG0092 | Ribosomal protein S3 |
| COG0185 | Ribosomal protein S19 | COG0089 | Ribosomal protein L23 |
| COG0090 | Ribosomal protein L2 | COG0088 | Ribosomal protein L4 |
| COG0051 | Ribosomal protein S10 | COG0049 | Ribosomal protein S7 |
| COG0222 | Ribosomal protein L7/L12 | COG0081 | Ribosomal protein L1 |
| COG0080 | Ribosomal protein L11 | COG0539 | Ribosomal protein S1 |
| COG0211 | Ribosomal protein L27 | COG0268 | Ribosomal protein S20 |
| COG0099 | Ribosomal protein S13 | COG0103 | Ribosomal protein S9 |
| COG0102 | Ribosomal protein L13 | COG0360 | Ribosomal protein S6 |
| COG0184 | Ribosomal protein S15P/S13E | COG0087 | Ribosomal protein L3 |
| COG0048 | Ribosomal protein S12 | COG0244 | Ribosomal protein L10 |
| COG1278 | Cold shock proteins | COG0198 | Ribosomal protein L24 |
| COG0050 | GTPases—translation elongation factors | COG0227 | Ribosomal protein L28 |
| COG0361 | Translation initiation factor 1 | COG0335 | Ribosomal protein L19 |
| COG1158 | Transcription termination factor | COG1825 | Ribosomal protein L25 |
| COG0055 | F0F1-type ATP synthase, beta subunit | COG0228 | Ribosomal protein S16 |
| COG3118 | Thioredoxin domain-containing protein | ||
| COG0740 | ATP-dependent Clp proteases | ||
| COG0468 | RecA/RadA recombinase | ||
| COG0443 | Molecular chaperone | ||
The COG (Cluster of Orthologous Groups) annotations of these protein families are provided.
Phylogenetic position of SAR11 using composition-homogeneous and -heterogeneous data
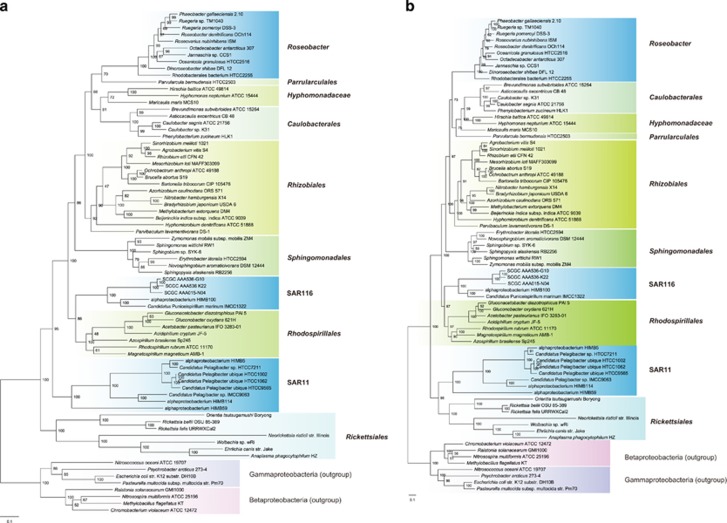
To investigate the effect of character sampling on phylogenetic reconstruction of genomes displaying striking compositional variation, two independent data sets of amino acid sequences were compiled: the concatenated 28 composition-homogeneous proteins and the concatenated 24 composition-heterogeneous ribosomal proteins. Intriguingly, the maximum likelihood RAxML software (Stamatakis, 2014) places the SAR11 bacteria in different evolutionary positions depending on which data set is used, whereas the branching order of other alphaproteobacterial lineages remains identical. The composition-homogeneous protein set places Rickettsiales at the base of Alphaproteobacteria phylogeny and SAR11 at the base of the remaining lineages (Figure 2a), whereas the composition-heterogeneous protein set clusters SAR11 and Rickettsiales at the base of the tree in a monophyletic group (Figure 2b). The different branching patterns in these analyses suggest that the clustering of SAR11 with Rickettsiales, as has been reported previously (Thrash et al., 2011), is an artifact due to the attraction of sequences with compositional similarity.
Figure 2.
Maximum likelihood phylogeny of Alphaproteobacteria using the RAxML v7.3.0 software. (a) Tree based on a concatenation of the 28 composition-homogeneous proteins, in which 19 are ribosomal proteins. (b) Tree based on a concatenation of the 24 composition-heterogeneous ribosomal proteins. A data partition model was employed to allow subsets of component proteins to evolve independently in amino acid replacement processes, which was determined using the PartitionFinder software. Values at the nodes show the number of times the clade defined by that node appeared in the 100 bootstrapped datasets. Trees are rooted using species from Betaproteobacteria and Gammaproteobacteria.
With this composition-unbiased data set of 28 concatenated homogeneous protein sequences, testing was carried out in the alternate SAR11 evolutionary positions that have been reported (Figure 1) using the approximately unbiased test (Shimodaira, 2002) and the more conservative Shimodaira-Hasegawa test (Shimodaira and Hasegawa, 1999), both allowing for statistical comparison of tree topologies. These methods strongly support the tree in Figure 2a (as outlined in Figure 1b), lend weak support (P=0.051) to the tree in Figure 2b (as outlined in Figure 1a) and strongly reject (P<0.001) other placements of SAR11 (Figures 1c and d).
Phylogenetic position of SAR11 using different models
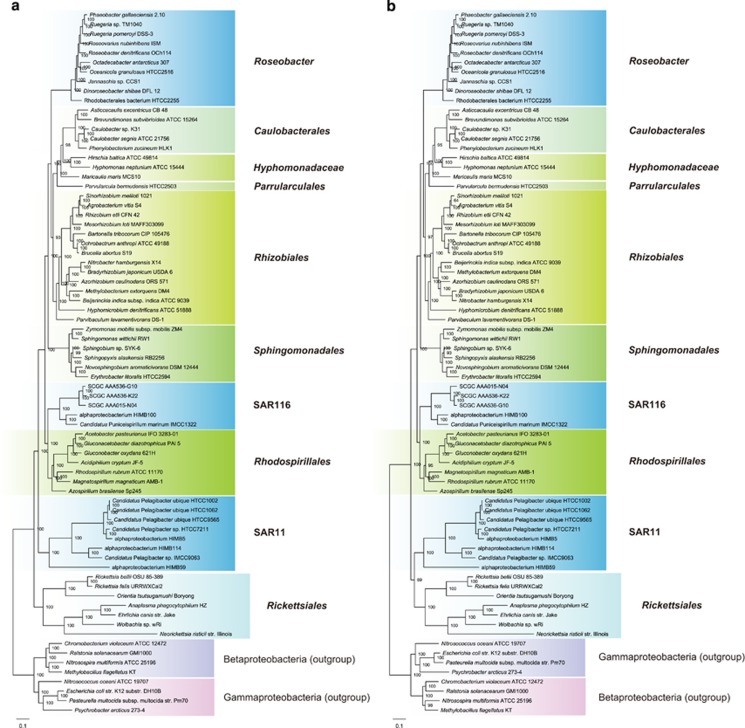
Most phylogenomic analyses do not separate proteins into homogeneous and heterogeneous classes in regard to composition, and often combined data sets with both homogeneous and heterogeneous sequences are used. The ability of phylogenetic models to accommodate heterogeneity was thus tested using the concatenation of the 28 homogeneous and 24 heterogeneous ribosomal proteins. As expected, the RAxML software (Stamatakis, 2014) clustered SAR11 and Rickettsiales at the base of the tree (Supplementary Figure S1), but this clustering had less statistical support (Supplementary Figure S1) compared with that of the RAxML tree based solely on the 24 heterogeneous proteins (Figure 2b), as a result of conflicting phylogenetic signals contained in the two protein subsets. Intriguingly, the CAT model (Lartillot and Philippe, 2004) in the PhyloBayes MPI software (Lartillot et al., 2013) yielded a phylogeny (Supplementary Figure S2) displaying an identical topology to the RAxML tree (Supplementary Figure S1), which is at odds with the previous PhyloBayes analyses that were based on concatenated data sets that, although distinct from this 52-protein data set, also consist of both composition-homogeneous and heterogeneous protein sequences (Viklund et al., 2012; Luo et al., 2013; Viklund et al., 2013); these previous analyses placed SAR11 in the middle of the non-endosymbiotic lineages (Figures 1c and d).
The P4 Bayesian software offers the NDCH model that allows the amino acid composition to vary across lineages (Foster, 2004). This NDCH model generated a P4 phylogeny (Figure 3a) with a branching order identical to the RAxML tree based on the 28 homogeneous proteins (Figure 2a). When this model was not invoked, the resulting P4 tree (Figure 3b) displayed a topology identical to the RAxML tree based on the 24 heterogeneous proteins (Figure 2b). The robustness of this NDCH model is further confirmed by the posterior predictive simulation, followed by the χ2-test showing that this 52-protein data set can be adequately modeled (P>0.05) only when the NDCH model is invoked. These analyses strongly support the disassociation of SAR11 and Rickettsiales as outlined in Figure 1b.
Figure 3.
Bayesian phylogeny of Alphaproteobacteria using the P4 software. (a) Tree employing a composition-heterogeneous model that is adequate to the data. (b) Tree employing a composition-homogeneous model that significantly violates the data. Both trees are based on a Dayhoff-recoded sequence with a concatenation of 52 proteins, in which 28 are composition-homogeneous while the remaining 24 are composition-heterogeneous. The value near each internal branch is the posterior probability for that branch. Trees are rooted using species from Betaproteobacteria and Gammaproteobacteria.
Reconstruction of ancestral processes giving rise to the SAR11 bacteria
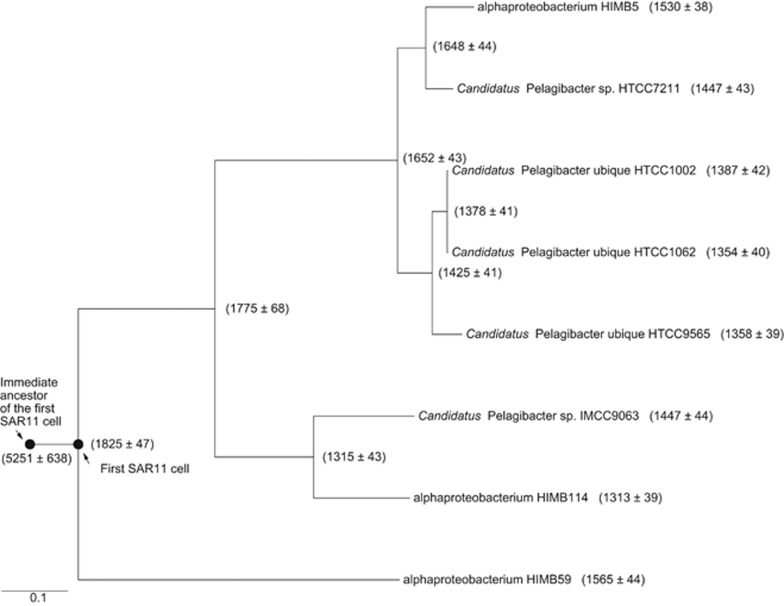
Ancestral genome content reconstruction requires a rooted species tree topology and phyletic pattern (presence/absence and copy number variation) of orthologous gene families in extant genomes. On the basis of the analyses above, the tree topology shown in Figure 2a (and Figure 3a) was selected for reconstruction. A maximum-likelihood ancestral reconstruction approach using the phylogenetic birth-and-death model (Csűrös, 2010) predicted that the first evolved SAR11 cell contained approximately 1800 genes, while its immediate ancestor had >4000 genes (Figure 4). Although over half of the genome content was lost at this early stage, genome streamlining continued until the extant lineages that contain approximately 1300–1500 genes (Figure 4). The predicted genome content of the first SAR11 (Supplementary Table S2) and its immediate ancestor (Supplementary Table S3) is significantly different according to functional categorization by Clusters of Orthologous Groups (Tatusov et al., 1997)(χ2-test; P<0.001). Using the Xipe resampling technique (Rodriguez-Brito et al., 2006), the latter genome was predicted to have been significantly enriched in transcriptional regulation, signal transduction, cell motility, and lipid transport and metabolism, which are characteristic functional categories of patch-adapted marine bacteria (Luo et al., 2013), whereas the former was significantly enriched in translation, ribosomal structure and biogenesis, amino acid transport and metabolism, nucleotide transport and metabolism, as well as coenzyme transport and metabolism, which are diagnostic categories of free-living planktonic cells (Luo et al., 2013) (P<0.01). This evidence for systematic gene loss implies that a change in ecological strategy accompanied the origin of SAR11.
Figure 4.
Reconstructed gene numbers of each ancestral node associated with the marine SAR11 clade using the COUNT software. The standard deviation was calculated based on maximum likelihood mapping of each of 100 bootstrap data sets generated by randomly sampling the gene families (with repetitions). Closed circles represent the first SAR11 cell and its immediate ancestor.
Discussion
Free-living planktonic marine bacteria with streamlined genomes have reduced the metabolic cost and increased the surface-to-volume ratio (because cell size can be correspondingly smaller) for efficient nutrient uptake, and thus streamlining has been considered an ecological advantage in inhabiting nutrient-poor ocean waters (Giovannoni et al., 2014). Study of the origin of the SAR11 lineage, the most abundant and streamlined bacterioplankton in the global oceans, requires resolving its evolutionary position in the Alphaproteobacteria tree. This is a challenge because genomes of the ecologically distinct SAR11 and Rickettsiales lineages consistently exhibit low G+C content whereas most members of the remaining alphaproteobacterial lineages contain G+C-rich genomes. Such variability in nucleotide ratios frequently results in a clustering pattern influenced by compositional similarity rather than biological relatedness in phylogenomic reconstruction (Galtier and Gouy, 1995; Jermiin et al., 2004; Collins et al., 2005; Cox et al., 2008; Foster et al., 2009; Sheffield et al., 2009; Nesnidal et al., 2010; Guy et al., 2014). Here, the evolutionary position of the SAR11 clade was shown to be better resolved in two ways, either by applying a standard phylogenetic program (for example, RAxML) to a least biased data set, or by applying a composition-heterogeneous model to a data set that contains bias. With both approaches, the G+C-poor SAR11 and Rickettsiales lineages do not emerge as a monophyletic lineage at the base of the Alphaproteobacteria tree.
Half of the ribosomal protein families were shown to have biased amino acid composition across the alphaproteobacterial lineages and their inclusion resulted in distorted phylogenetic structure. This compositional issue has not been reported in previous studies using the ribosomal proteins as phylogenetic characters. In fact, using a concatenated sequence based on a full set of ribosomal proteins has become a common approach to resolve deep evolutionary relationships in prokaryotic phylogenomics (Matte-Tailliez et al., 2002; Brochier-Armanet et al., 2008; Fournier and Gogarten, 2010; Lasek-Nesselquist and Gogarten, 2013). The major advantage of using these proteins as phylogenomic markers for prokaryotic organisms is that these genes are rarely subject to horizontal gene transfer (Ciccarelli et al., 2006; Ramulu et al., 2014), which has been generally accepted as the prevalent source of error in prokaryotic systematics (Bapteste and Boucher, 2008). Results from the current study showing different branching patterns depending on which ribosomal proteins are used caution against their indiscriminate use for systematics of Proteobacteria and perhaps other prokaryotic groups.
In addition to the ongoing debate of the phylogenetic placement of the SAR11 clade, there has been disagreement in regard to the monophyly of the strain HIMB59 and other SAR11 lineages (Rodríguez-Ezpeleta and Embley, 2012; Viklund et al., 2013). These studies suggest that a monophyletic cluster of these bacteria as frequently observed in phylogenomic trees (Thrash et al., 2011; Luo et al., 2013) is a result of compositional attraction (Rodríguez-Ezpeleta and Embley, 2012; Viklund et al., 2013), and that HIMB59 is more likely to be related to the marine SAR116 clade (Rodríguez-Ezpeleta and Embley, 2012) or a broader group of Rhodospirillales that includes SAR116 (Viklund et al., 2013). Although the current study was not designed to address this question, it found no evidence to support a disassociation of HIMB59 with other SAR11 bacteria. All phylogenomic trees from the current study confidently rejected an evolutionary relatedness of HIMB59 to the SAR116 clade, even though a SAR116 lineage was included consisting of three single cells with low genomic G+C content (∼30%) that is nearly identical to that of HIMB59 (32%).
Identifying an exact evolutionary position of SAR11 requires a better sampling of major Alphaproteobacteria lineages. While eight well-accepted Orders were included here, a few under-represented but deeply branching lineages are missing in both the current and previous phylogenomic studies. A few examples are Kiloniellales, Kopriimonadales, Kordiimonadales, Sneathiellales, Rhodothalassiales and Magnetococcales (Ferla et al., 2013). Indeed, the ribosomal gene trees consistently resolved Magnetococcales as the basal Order of the Alphaproteobacteria phylogeny (Bazylinski et al., 2013; Ferla et al., 2013), and availability of the genomic sequence from Magnetococcus marinus MC-1 affiliated with this lineage allows phylogenomic validation of its basal position among the included alphaproteobacterial lineages (Supplementary Figure S3). Another consideration in future phylogenomic studies of Alphaproteobacteria and the phylogenetic placement of SAR11 is to examine the effect of taxon sampling on the resulting tree, since Ferla et al. (2013) showed that taxon selection greatly affects the branching order and monophyly of a few major lineages in the Alphaproteobacteria tree, though their analysis was based on a concatenation of easily accessible small and large subunits of rRNA genes.
Recent studies using various approaches have consistently identified statistical correlations between the ecological strategies and genome content in marine bacteria (Lauro et al., 2009; Yooseph et al., 2010; Luo et al., 2012; Luo et al., 2013). Gene functional categories involved in cell–cell interactions, such as motility, secondary metabolite synthesis and degradation, and defense mechanisms, are repeatedly found to be enriched in the genomes of marine bacteria that are associated with particles and take advantage of ephemeral patchiness of nutrients (Moran et al., 2004; Newton et al., 2010; Luo et al., 2013), while they are depleted in the genomes of marine bacteria that live as single cells in nutrient-poor bulk seawater (Giovannoni et al., 2005; Giovannoni et al., 2008). This characteristic genome content was also hypothesized in the present study, suggesting the evolutionary origin of marine SAR11 bacteria at the base of the non-endosymbiotic Alphaproteobacteria lineages may have coincided with an ecological transition from a patch-associated life-style to a free-living planktonic strategy.
Acknowledgments
I thank MA Moran for her generous support of this study and useful comments on previous versions of the manuscript, P Foster for advice on using the P4 software and the Georgia Advanced Computing Resource Center at the University of Georgia for computational resources. This research was funded in part by the Gordon and Betty Moore Foundation through grant GBMF538.01 and a Direct Grant (4930062) of the Chinese University of Hong Kong.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Bapteste E, Boucher Y. Lateral gene transfer challenges principles of microbial systematics. Trends Microbiol. 2008;16:200–207. doi: 10.1016/j.tim.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Williams TJ, Lefèvre CT, Berg RJ, Zhang CL, Bowser SS, et al. Magnetococcus marinus gen. nov., sp. nov., a marine, magnetotactic bacterium that represents a novel lineage (Magnetococcaceae fam. nov., Magnetococcales ord. nov.) at the base of the Alphaproteobacteria. Int J Syst Evol Microbiol. 2013;63:801–808. doi: 10.1099/ijs.0.038927-0. [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Micro. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Collins TM, Fedrigo O, Naylor GJP. Choosing the best genes for the job: the case for stationary genes in genome-scale phylogenetics. Syst Biol. 2005;54:493–500. doi: 10.1080/10635150590947339. [DOI] [PubMed] [Google Scholar]
- Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci USA. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csűrös M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- Csűrös M, Miklós I. Streamlining and large ancestral genomes in achaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol. 2009;26:2087–2095. doi: 10.1093/molbev/msp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, et al. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc Natl Acad Sci USA. 2003;100:10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont CL, Rusch DB, Yooseph S, Lombardo M-J, Alexander Richter R, Valas R, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferla MP, Thrash JC, Giovannoni SJ, Patrick WM. New rRNA gene-based phylogenies of the Alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability. PLoS ONE. 2013;8:e83383. doi: 10.1371/journal.pone.0083383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PG. Modeling compositional heterogeneity. Syst Biol. 2004;53:485–495. doi: 10.1080/10635150490445779. [DOI] [PubMed] [Google Scholar]
- Foster PG, Cox CJ, Embley TM. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Philos Tr Roy Soc B. 2009;364:2197–2207. doi: 10.1098/rstb.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PG, Hickey DA. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J Mol Evol. 1999;48:284–290. doi: 10.1007/pl00006471. [DOI] [PubMed] [Google Scholar]
- Fournier GP, Gogarten JP. Rooting the ribosomal tree of life. Mol Biol Evol. 2010;27:1792–1801. doi: 10.1093/molbev/msq057. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M. Inferring phylogenies from DNA sequences of unequal base compositions. Proc Natl Acad Sci USA. 1995;92:11317–11321. doi: 10.1073/pnas.92.24.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Cameron Thrash J, Temperton B.2014Implications of streamlining theory for microbial ecology ISME J 81553–1565.in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho J-C, et al. The small genome of an abundant coastal ocean methylotroph. Environ Microbiol. 2008;10:1771–1782. doi: 10.1111/j.1462-2920.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin K, Batista D, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Grote J, Bayindirli C, Bergauer K, Carpintero de Moraes P, Chen H, D'Ambrosio L, et al. Draft genome sequence of strain HIMB100, a cultured representative of the SAR116 clade of marine Alphaproteobacteria. Stand Genomic Sci. 2011;5:269–278. doi: 10.4056/sigs.1854551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, et al. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. MBio. 2012;3:e00252–00212. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Hewett-Emmett D, Li W-H. Directional mutational pressure affects the amino acid composition and hydrophobicity of proteins in bacteria. Genetica. 1998;102/103:383–391. [PubMed] [Google Scholar]
- Guy L, Spang A, Saw JH, Ettema TJG. 'Geoarchaeote NAG1' is a deeply rooting lineage of the archaeal order Thermoproteales rather than a new phylum. ISME J. 2014;8:1353–1357. doi: 10.1038/ismej.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdy I, Hirt RP, Dolezal P, Bardonova L, Foster PG, Tachezy J, Martin Embley T. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- Jermiin LS, Ho SYW, Ababneh F, Robinson J, Larkum AWD. The biasing effect of compositional heterogeneity on phylogenetic estimates may be underestimated. Syst Biol. 2004;53:638–643. doi: 10.1080/10635150490468648. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K-i, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E, Gogarten JP. The effects of model choice and mitigating bias on the ribosomal tree of life. Mol Phylogenet Evol. 2013;69:17–38. doi: 10.1016/j.ympev.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer E. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, et al. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA. 2009;106:15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P, Holder M, Holsinger K. Polytomies and Bayesian phylogenetic inference. Syst Biol. 2005;54:241–253. doi: 10.1080/10635150590924208. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Csűros M, Hughes AL, Moran MA. Evolution of divergent life history strategies in marine Alphaproteobacteria. MBio. 2013;4:e00373–00313. doi: 10.1128/mBio.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Friedman R, Tang J, Hughes AL. Genome reduction by deletion of paralogs in the marine cyanobacterium Prochlorococcus. Mol Biol Evol. 2011;28:2751–2760. doi: 10.1093/molbev/msr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Löytynoja A, Moran MA. Genome content of uncultivated marine Roseobacters in the surface ocean. Environ Microbiol. 2012;14:41–51. doi: 10.1111/j.1462-2920.2011.02528.x. [DOI] [PubMed] [Google Scholar]
- Matte-Tailliez O, Brochier C, Forterre P, Philippe H. Archaeal phylogeny based on ribosomal proteins. Mol Biol Evol. 2002;19:631–639. doi: 10.1093/oxfordjournals.molbev.a004122. [DOI] [PubMed] [Google Scholar]
- Moran MA, Buchan A, Gonzalez JM, Heidelberg JF, Whitman WB, Kiene RP, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- Morgenstern B. DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res. 2004;32:W33–W36. doi: 10.1093/nar/gkh373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesnidal MP, Helmkampf M, Bruchhaus I, Hausdorf B. Compositional heterogeneity and phylogenomic inference of metazoan relationships. Mol Biol Evol. 2010;27:2095–2104. doi: 10.1093/molbev/msq097. [DOI] [PubMed] [Google Scholar]
- Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010;4:784–798. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins D, Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ramulu HG, Groussin M, Talla E, Planel R, Daubin V, Brochier-Armanet C. Ribosomal proteins: Toward a next generation standard for prokaryotic systematics. Mol Phylogenet Evol. 2014;75:103–117. doi: 10.1016/j.ympev.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Rohwer F, Edwards R. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Embley TM. The SAR11 group of Alpha-Proteobacteria is not related to the origin of mitochondria. PLoS One. 2012;7:e30520. doi: 10.1371/journal.pone.0030520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield NC, Song H, Cameron SL, Whiting MF. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst Biol. 2009;58:381–394. doi: 10.1093/sysbio/syp037. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Singer GAC, Hickey DA. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol Biol Evol. 2000;17:1581–1588. doi: 10.1093/oxfordjournals.molbev.a026257. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM, et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA. 2013;110:11463–11468. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, Robbertse B, Spatafora JW, Rappe MS, Giovannoni SJ. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep. 2011;1:13. doi: 10.1038/srep00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund J, Ettema TJG, Andersson SGE. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol Biol Evol. 2012;29:599–615. doi: 10.1093/molbev/msr203. [DOI] [PubMed] [Google Scholar]
- Viklund J, Martijn J, Ettema TJG, Andersson SGE. Comparative and phylogenomic evidence that the Alphaproteobacterium HIMB59 is not a member of the oceanic SAR11 clade. PLoS One. 2013;8:e78858. doi: 10.1371/journal.pone.0078858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, Kececioglu JD. Multiple alignment by aligning alignments. Bioinformatics. 2007;23:i559–i568. doi: 10.1093/bioinformatics/btm226. [DOI] [PubMed] [Google Scholar]
- Yooseph S, Nealson KH, Rusch DB, McCrow JP, Dupont CL, Kim M, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468:60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.