Abstract
Within hydrothermal plumes, chemosynthetic processes and microbe–mineral interactions drive primary productivity in deep-ocean food webs and may influence transport of elements such as iron. However, the source of microorganisms in plumes and the factors governing how these communities assemble are poorly understood, in part due to lack of data from early stages of plume formation. In this study, we examined microbial community composition of rising hydrothermal plumes from five vent fields along the Eastern Lau Spreading Center. Seafloor and plume microbial communities were significantly dissimilar and shared few phylotypes. Plume communities were highly similar to each other with significant differences in community membership only between Kilo Moana and Mariner, two vents that are separated by extremes in depth, latitude and geochemistry. Systematic sampling of waters surrounding the vents revealed that species richness and phylogenetic diversity was typically highest near the vent orifice, implying mixing of microbial communities from the surrounding habitats. Above-plume background communities were primarily dominated by SAR11, SAR324 and MG-I Archaea, while SUP05, Sulfurovum, Sulfurimonas, SAR324 and Alteromonas were abundant in plume and near-bottom background communities. These results show that the ubiquitous water-column microorganisms populate plume communities, and that the composition of background seawater exerts primary influence on plume community composition, with secondary influence from geochemical and/or physical properties of vents. Many of these pervasive deep-ocean organisms are capable of lithotrophy, suggesting that they are poised to use inorganic electron donors encountered in hydrothermal plumes.
Introduction
Hydrothermal vents are important conduits for the transfer of elements between the lithosphere and the oceans (Wu et al., 2011; Holden et al., 2012), and dispersal of these elements occurs primarily via hydrothermal plumes (Elderfield and Schultz, 1996). Hydrothermal plumes develop as hot, subsurface fluids rich in electron donors and exit the seafloor, rapidly mix with cold background seawater and disperse vertically and laterally from the vent field. The geochemistry of a plume is dictated by the composition of the host rock, fluid temperature and subsurface mixing (German and Von Damm, 2004), and thus can vary in metal content, pH, temperature and dissolved gases (Holden et al., 2012) between spatially close hydrothermal vents (German et al., 2010). It has long been recognized that plume geochemistry, which is replete with electron donors, drives microbial metabolism (Jannasch and Wirsen, 1979; Karl et al., 1980; Winn et al., 1986), and in turn influences the speciation and transport of metals such as manganese and iron (Jannasch and Mottl, 1985; Cowen et al., 1986; Dick et al., 2009, Li et al., 2014b). Thus, hydrothermal plumes support a diverse community of chemolithoautotrophic microorganisms that fuel carbon fixation through oxidation of reduced substrates, that is, H2, H2S, CH4, NH4 and Fe (McCollom, 2000; Dick et al., 2013), and are significant sources of labile carbon in the deep ocean (de Angelis et al., 1993; Lam et al., 2004). Therefore, the diversity, concentration and available energy from the oxidation of electron donors present in hydrothermal vent fluids will greatly influence the metabolisms of chemosynthetic microorganisms, and in turn ecosystem productivity (Amend et al., 2011).
Using a cultivation-independent approach, Sunamura et al. (2004) presented the first whole community-based molecular survey of microorganisms that inhabit plumes. These plumes were highly enriched with two phylotypes, SUP05 Gammaproteobacteria and SUP01 Epsilonproteobacteria. Similarly, Dick and Tebo (2010) found that Guaymas Basin plumes were dominated by SUP05 and methylotrophic Gammaproteobacteria, SAR324 Deltaproteobacteria, SAR11 Alphaproteobacteria, Thaumarchaeota (MG-I) and Marine Group II Archaea, and also resembled communities from deep waters of an adjacent basin without hydrothermal activity. Subsequent metagenomic and metatranscriptomic analysis of Guaymas plumes confirmed the clone library results (Lesniewski et al., 2012) and highlighted the physiological importance of several key microbial groups, MG-I Archaea (Baker et al., 2012), SUP05 (Anantharaman et al., 2013), methylotrophs (Li et al., 2014a) and SAR324 (Sheik et al., 2014). Interestingly, many of the groups identified at Guaymas Basin are ubiquitous in the world's oceans, including SAR11 (Morris et al., 2002), SAR324 (Brown and Donachie, 2007) and MG-I (Karner et al., 2001), while groups such as certain methylotrophs and SUP05 are restricted to specific ocean habitats such as methane cold seeps (Tavormina et al., 2010) or oxygen minimum zones (Walsh et al., 2009). Conversely, descending hydrothermal plume particles collected with sediment traps analyzed during a time-series study of two vents at 9°50'N on the East Pacific Rise revealed populations dominated by Epsilonproteobacteria likely derived from seafloor vents (Sylvan et al., 2012). While the contribution of seafloor communities to plume microbial communities is in question, the apparent functional and taxonomic linkages between plume communities and portions of the greater ocean indicate that hydrothermal plumes may have a role in facilitating the proliferation and dispersion of these important microorganisms (Dick et al., 2013). This potential is underscored by the vast expanse of ocean ridge axis and the expanding catalog of discovered vent fields (Beaulieu et al., 2013).
Given the limited number of cultivation-independent studies of plume microbiology and the complexity of this environment, several first-order questions remain concerning (i) the primary source of plume microorganisms (e.g. water column, seafloor or subseafloor); (ii) the extent to which vent fluid chemistry influences plume microbial communities and their distribution; and (iii) the extent to which plume microbial communities influence plume chemistry. Here we present a biogeographic survey of microbial communities sampled systematically from rising hydrothermal plumes at five vent fields within the Lau Basin in the western Pacific Ocean. This is the first time that plume communities were sampled and filtered in situ in a systematic way, including samples from both near-bottom (NBB) and above-plume background (APB), as well as several heights within rising plumes (RPs) from the vent orifice to as high as 200 m above the vent. We find high degrees of similarity between microbial communities across the five sites and at all points of the RPs sampled. These results indicate that the deep background water column is the primary source of plume microorganisms, with a substantial but smaller contribution from seafloor environments.
Materials and methods
Sampling location and description
The Eastern Lau Spreading Center (ELSC) is situated on a back-arc basin spreading center in the southern portion of the Lau Basin (Supplementary Figure 1) (Martinez and Taylor, 2002; Ferrini et al., 2008). The five vents sampled for this study, Kilo Moana, Tahi Moana, ABE, Tui Malila and Mariner, are demarcated by north to south gradients of depth (∼2500–1900 m below surface) and geochemistry (low to high metal concentrations) (Kamenetsky et al., 1997; Baker et al., 2005; German et al., 2006; Mottl et al., 2011) (Figure 1). In general, higher concentrations of iron and manganese have been observed at Mariner vents relative to Kilo Moana, Tahi Moana, ABE and Tui Malila, whereas temperature, pH, sulfide and methane are similar among all vent fields (Figure 1, inset table) (Mottl et al., 2011; Flores et al., 2012).
Figure 1.
Approximate depth, latitudinal location and general characteristics of hydrothermal vents sampled along the spreading ridge in Lau Basin. Arrows give location of vents with respect to spreading center. Figure inset gives distance in kilometers of each vent relative to one another. The table describes location and previously reported geochemistry of end-member fluids associated with each vent. Bathymetry was generated using GeoMapApp (http://www.geomapapp.org) and plotted in R.
Sample collection
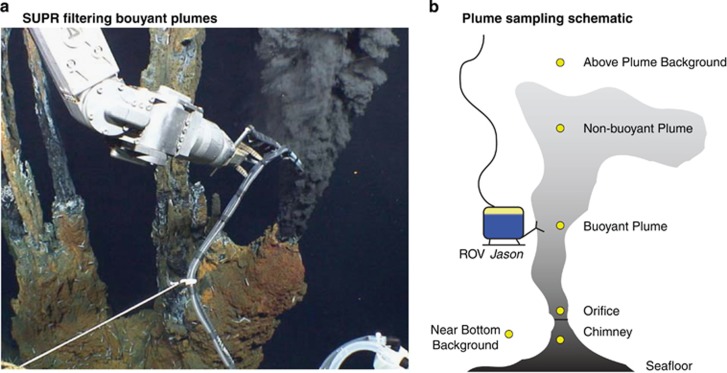
Samples were collected from five actively venting hydrothermal sites (Figure 1) located on the ELSC on two consecutive cruises aboard the R/V Thomas G Thompson in May–July 2009. Suspended mineral particles and microorganisms associated with the particles (Toner et al., 2009; Breier et al., 2012) were filtered from the water column in situ using a Suspended Particulate Rosette (SUPR) filtration device (Breier et al., 2009b) mounted to the remotely operated vehicle Jason II. When operated in this configuration, a collection hose is attached to the SUPR sampler and extended away from the vehicle into waters to be sampled using the remotely operated vehicle manipulator arm (Figure 2a). Water with suspended particles is then drawn directly into the SUPR sampler. The exceptions were the J2.424–426 dives where the inlet hose was positioned at the corner of the basket. Sampling locations, especially within the plume, were visually identified by the SUPR operator with the suite of cameras onboard Jason II and the presence of suspended particles, as determined by a beam transmissometer deployed during the J2.424–426 dives. The SUPR rosette is capable of filtering up to 24 discrete samples, whereby water is filtered through sequential, single, isolated 37 mm filters. For microbial analysis, 0.8 μm SUPOR polyethersulfone membrane filters (Pall, Port Washington, NY, USA) were used. Because plumes are highly enriched in particles including minerals, microorganisms (planktonic and particle associated) and organic aggregates, a filter pore size of 0.8 μm was selected so that sufficient material could be collected within the operational time allotted on each dive. It should be noted that some small microorganisms may pass through a 0.8 μm pore size filters, potentially influencing the results presented here. However, direct comparisons of 0.8 μm to 0.22 μm filters (see Supplementary Figure 5, dives 445.08 and 445.20, respectively) revealed highly similar community structure, suggesting minimal influence of pore size on observed community structure. Upon recovery, filters were removed from the SUPR sampler, placed in conical vials, flooded with RNAlater (Ambion, Austin, TX, USA) and frozen at −80 °C. Samples were kept frozen for shipment back to the University of Michigan and immediately placed at −80 °C until the time of DNA extraction. For geochemical analysis of filtered particles, 0.8 μm Isopore polycarbonate membrane filters (Millipore, Port Washington, NY, USA) were used. Upon recovery, filters were removed from the SUPR sampler in a nitrogen-purged glovebox, rinsed with distilled/deionized water neutralized with trace metal grade ammonia to remove sea salts, transferred to vacuum containers (Sample-Storr, Ted Pella, Inc., Redding, CA, USA) and frozen until analysis.
Figure 2.
Sampling methodology overview. (a) Operator view of Jason II sampling actively venting fluids with the SUPR sampler. Because no high-resolution pictures of SUPR from ELSC were available, the image shown here is from the Mid Cayman Rise. Jason imagery courtesy of the National Science Foundation, Woods Hole Oceanographic Institution and CR German. (b) Sampling locations relative to the vent orifice are highlighted by yellow circles.
Vents were sampled in a systematic manner (Figure 2b), which included: above plume background (APB), near bottom background (NBB), near orifice (orifice) and rising plume (RP). However, the location and number of samples depended on the dive mission and time available (i.e., the extent to which plume sampling logistics complimented other dive objectives). Sampling consisted of maneuvering Jason II to the vent orifice and sampling at <1 m from the orifice, within the actively venting waters. Plumes were tracked by temperature anomaly near the vent, and then visually or by particle density along a vertical transect. Samples were collected at semiregular intervals depending on the magnitude of plume fluid flux. Two replicate samples were collected at each sample location: one for microbial analysis and one for geochemical analysis. Along with each plume profile, samples were also collected for NBB and APB controls. NBBs were collected as Jason II approached the active vent ∼5–10 m above the seafloor; APBs were collected as Jason II returned to the surface and generally incorporated particles from the ∼2000–1000 m depth interval. Internal control filters, for both microbiology and geochemistry, were also included with every dive and served as a contamination control. These filters were handled, preserved and analyzed as all other filter membranes but were not used for sample filtration.
Geochemistry
Filters were split into ¼ segments, which were completely digested in 30 ml acid-cleaned perfluoroalkoxy vials (Savillex, Eden Prairie, MN, USA) according to methods in Bowie et al. (2010). All acids were trace metal grade (Optima; Fisher Scientific, Pittsburgh, PA, USA). Vials were heated in a temperature-controlled hot plate (Qblock; Questron Technologies, Mississauga, ON, Canada). Aliquots of sample digest, at a 1:10 dilution, were analyzed for Al, Fe and Mn by inductively coupled plasma optical emission spectrometry on a Varian 730-ES axial spectrometer (Agilent Technologies, Santa Clara, CA, USA). For samples containing Al, Fe and Mn concentrations below optical emission spectrometry detection limits, additional aliquots of sample digest, at dilutions ranging from 1:10 to 1:100000, were subsequently analyzed by inductively coupled plasma mass spectrometry on a Thermo-Finnigan Element2 (Thermo-Scientific, Waltham, MA, USA). Sample digestion and dilution were carried out at Woods Hole Oceanographic Institution (Woods Hole, MA, USA). Instrumental analysis was carried out by Activation Laboratories Ltd (Ancaster, ON, Canada). External reference standards were used for instrument calibration. Digestion and analysis were monitored by processing and comparing filter and acid blanks, geostandards (BHVO) and internal sulfide reference materials by these same methods (Raczek et al., 2001; Breier et al., 2009a).
DNA extraction, amplification and sequencing
Total DNA was extracted from ¼ of a filter using previously described chemical and physical lysis methods (Dick and Tebo, 2010). The 16S ribosomal gene was PCR amplified with the 515-For and 806-Rev primers containing 454 sequencing adapters and barcodes (Fierer et al., 2008) that are capable of amplifying both Bacteria and Archaea (Bates et al., 2011). PCRs were performed in triplicate 25 μl reactions for each sample with 12.5 μl 5PRIME HotMasterMix (5PRIME, Gaithersburg, MD, USA), 9.5 μl PCR grade water (Ambion, Life Technologies, Grand Island, NY, USA), 1 μl each of forward and reverse primer (15 μM) and 1 μl DNA. PCR was performed with the following conditions: 94 °C for 4 min followed by 30 rounds of 94 °C for 30 s, 50 °C for 1 min, 72 °C for 1 min and a final extension step of 72 °C for 10 min. Triplicate PCR reactions were pooled, cleaned with the MoBio UltraClean PCR Clean-Up Kit (MoBio, Carlsbad, CA, USA) and quantified by PicoGreen (Invitrogen, Life Technologies). Pooled PCR-DNA was combined with other pooled samples at near equivalent concentrations. Reads were sequenced from the B-adapter in the Laboratory of Dr Vince Young at the University of Michigan (Ann Arbor, MI, USA) with 454 Titanium chemistry (454 Life Sciences-a Roche company, Branford, CT, USA). Pyrosequencing reads can be obtained through NCBI's short read archive SRR1138581.
454-Read processing and statistical analysis
Pyrosequencing reads were extracted from flowgram files and fastq files generated using Mothur v.1.28 (Schloss et al., 2009). Sequence reads were quality checked and truncated to a uniform length using the Uparse OTU (operational taxonomic unit) pipeline (Edgar, 2013) with the following flags: fastq_maxee=0.5 and fastq_trunclen=250. Trunclen length was optimized for length for our pyrosequencing run to maximize the phylogenetic signal (a function of read length) while simultaneously minimizing the number of reads rejected due to poor quality at the read's 3′ end. OTUs were clustered at 97% similarity, checked for chimeras and abundance calculated with Uparse. OTUs were reverse complemented using an in-house python script. Representative OTU sequences were classified to the Silva v.111 taxonomic database (Pruesse et al., 2007) with Mothur using the naive Bayesian algorithm (Wang et al., 2007). In Mothur, rare OTUs were removed (<3 sequences per sample) and subsampled to a uniform depth of 1500 sequences. A phylogenetic tree was generated with FastTree (Price et al., 2009) (parameters: −gtr, −nt, −gamma) using OTUs aligned to a representative subset of the Silva database. The phylogenetic tree and subsampled OTU table were used to estimate the phylogenetic diversity (Faith et al., 2009), standard effect size of mean pairwise distances (SES.mpd) (−1*Net Relatedness Index) and mean nearest taxon distance (SES.mntd) (−1*Nearest Taxon Index) in Picante (Kembel et al., 2010) implemented in R (R Core Team, 2013). Non-metric multidimensional scaling ordination plots were generated with Jaccard distance metrics in vegan (Oksanen et al., 2013). Differences in community structure were quantified using the adonis, anosim, betadisper and permutest functions in the vegan package. Tukey's pairwise comparisons were calculated with the Tukey's HSD (honest significant difference) base function in R.
Investigation of top OTUs present across all Lau sequences revealed five OTUs (97% similarity) that were suspected as shipboard contaminants. OTUs were confirmed as contaminants by cloning and sequencing amplified 16S rRNA genes from DNA extracted from control filters included on each dive. These putative contaminant OTUs represented 10–50% of the total community in three sequential Jason dives at two differing vent sites and were present in several other samples but at very low abundance (<0.1%). These OTUs were classified taxonomically as Psuedomonadales and Sphingomonads similar to sequences derived from surface-ocean and terrestrial clones. Burkholderia were also identified but represented few sequences and were present in a small subset of filters. After removal of these OTUs from the entire data set, samples that were heavily contaminated did not meet subsampling sequence thresholds and were subsequently removed from downstream analyses. A total of five samples were removed from the downstream analysis: three plume and two APB samples.
To identify linkages between the seafloor sulfide-hosted communities and RPs, 454-reads from this study were clustered at 97% similarity with 16S rRNA sequences from a previous study of Lau Basin seafloor hydrothermal vent communities (Flores et al., 2012). Because the seafloor data was generated with bacterial primer sets 560/803 (forward and reverse), reverse complemented reads generated in this study (515/806 sequenced from the B′ primer) and seafloor reads were aligned to the Silva database and columns with no overlap were removed using the filter.seqs command in Mothur. OTUs were then clustered with the Uparse pipeline and postprocessing methods described above.
To quantify the effect hydrothermal plumes have on defining niche space and the subsequent structure of microbial communities, we calculated the SES.mpd and SES.mntd. These metrics are equivalent to −1 times the net relatedness index and nearest taxon index. Null communities were generated with the independent swap method, which randomizes species co-occurrences but maintains sample richness and species frequencies and is robust at detecting niche processes (Kembel, 2009).
Results
Dilution effects of rising hydrothermal plumes
Plume dilution, as quantified by the particulate elemental ratio of Al to the sum of Al, Fe and Mn (e.g., Al/(Mn+Fe+Al)) (Boström and Peterson, 1969), was positively correlated with height above seafloor owing to vent fluids being greatly enriched in Fe and Mn relative to Al. Al in plume particles is predominantly sourced from marine detrital material. Mariner plume samples were highly enriched in Fe and Mn and had particulate Al/(Mn+Fe+Al) ratios as low as 0.02 near the vent orifice. In general, the ratio increased with vertical distance above all vents, reaching values as high as 0.78 in background samples (Supplementary Table 1). These Al/(Mn+Fe+Al) ratios provide geochemical evidence of hydrothermal plume origin and the relative degree to which these samples were influenced by hydrothermal material. NBB suspended material had intermediate Al/(Mn+Fe+Al) ratios, indicating that they too contain material of hydrothermal origin.
Microbial community diversity patterns and seafloor linkages across the ELSC
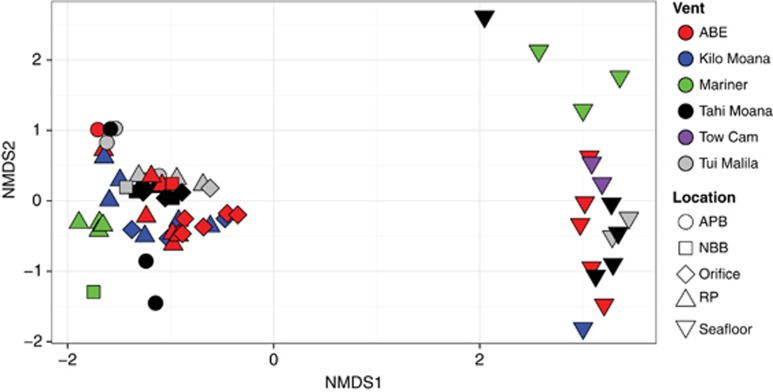
The microbial communities of the 48 ELSC plume and background samples analyzed here were compared with each other and with those of seafloor hydrothermal deposits from the same five ELSC vent fields (Flores et al., 2012). A clear difference between water-column and seafloor sulfide communities was apparent (Figure 3). Differences between water-column microbial communities were smaller but showed some clustering consistent with vent field and sample type (Figure 4a). All of the Mariner samples clustered together. Most of the Kilo Moana and ABE samples formed their own clusters, and another cluster included plume samples from Tui Malila, ABE and Tahi Moana. APB waters were generally more similar to each other than plume-associated communities. Four samples did not adhere to these patterns: an ABE RP clustered with APB; a Kilo Moana RP was similar to ABE RP; and two APB samples from Tahi Moana, while quite distinct, were most similar to ABE and Kilo Moana plume samples (Figure 4a). Interestingly, samples taken on the same leg of the cruise clustered together (Figure 4a).
Figure 3.
Non-metric multidimensional scaling (NMDS) ordination of Jaccard distances showing similarity of RP (RP), orifice (< 1 m), NBB, APB and sulfide hosted microbial communities (seafloor). Samples are colored by vent field at ELSC. NMDS stress=0.096.
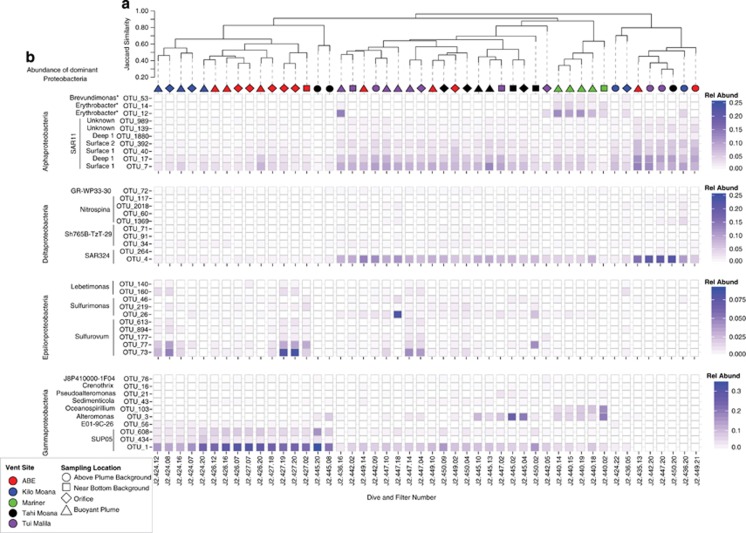
Figure 4.
Abundance of dominant Alpha-, Delta-, Epsilon- and Gammaproteobacteria OTUs across all water samples from Lau Basin. The abundance of proteobacterial OTUs97 is designated by color intensity; note that the color scale maximums change with each class to provide definition. Individual filter samples are arranged by Jaccard similarity and represent the entire community and the sampling location designated by symbol and color. Stars indicate potential contaminants.
The significance of these differences in community structure was assessed with three statistical models and pairwise analysis of beta-dispersion values. All three models detected significant differences in community structure between water-column samples (Supplementary Table 2a). Pairwise analysis of beta-dispersion indicates that much of the variance stems from Mariner communities (P-values near 0.05, Supplementary Table 2b). However, only differences between Kilo Moana and Mariner were significant (Supplementary Table 2b). Seafloor communities were all significantly different from all water-column communities (Supplementary Table 2). Observed separation by cruise leg was not significant.
Community assembly was quantified by SES.mpd and SES.mntd, which on a per sample basis calculate the mean phylogenetic distance between all community members and the mean phylogenetic distance between a member and its closest relative, respectively, and then compare these distances to those from randomly constructed communities from all samples (null). All water-column communities, including plume, orifice and background, had z-scores below zero, indicating a degree of phylogenetic similarity between individuals within the community. However, these water-column communities were not significantly underdispersed (z-score ⩽−2; Supplementary Figures 4a and b; see Supplementary Figure 2 for individual vent scores). Seafloor sulfide communities were consistently, significantly underdispersed when compared with null communities (Figures 4a and b). Kruskal–Wallis rank-sum tests detected significant sampling site differences for both SES.mpd (P<0.0001, d.f.=4, χ2=39.524) and SES.mntd (P<0.0001, d.f.=4, χ2=48.258), but pairwise analysis revealed that the differences were primarily between water-column communities and seafloor sulfide communities (Supplementary Figures 3 a and b).
Species richness and phylogenetic diversity, both components of alpha diversity, were highly variable across the ELSC (Supplementary Figure 4). Because of sampling limitations at some vents, patterns of diversity were difficult to ascertain. Thus, we chose to integrate all samples across the ELSC and focus on broad patterns of alpha diversity. When combining all data, Kruskal–Wallis rank-sum tests detected significant sampling site differences for both species richness (P=0.00013, d.f.=4, χ2=22.921) and phylogenetic diversity (P<0.0001, d.f.=4, χ2=28.42). Rank-sum tests of water-column community diversity found no significant differences; however, a peak in mean diversity was observed for vent orifice samples (Supplementary Figures 3 c and d). Mean diversity of nearly all water-column communities was significantly higher than seafloor communities, with the primary exception being NBB communities (Supplementary Figures 3 c and d).
Assessment of dominant phylotypes across the ELSC
Proteobacteria were the most frequently encountered phylum at ELSC, accounting for 50% or more of the community in most samples (Supplementary Figure 5b). Within the Proteobacteria, Gamma and Alpha classes were the most abundant and fluctuated highly between sampling location relative to the vent and vent field. NBB, plume and orifice communities consisted of four main groups: sulfur-oxidizing SUP05 Gammaproteobacteria, sulfur-oxidizing Epsilonproteobacteria, SAR324 and SAR11 (OTUs0.97; Figure 4b). The SUP05 and SAR324 phylotypes were highly similar (>97%) to genotypes recovered from Guaymas Basin hydrothermal plumes (Anantharaman et al., 2013; Sheik et al., 2014) and were distantly related (∼90%) to clones from low-temperature rock communities at Lau (Sylvan et al., 2013). Plume communities at Lau also had higher proportions of sulfur oxidizing Sulfurimonas and Sulfurovum Epsilonproteobacteria (Figure 4b and Supplementary Figure 5b). Clustering of microbial communities (Figure 4a) was driven in part by phylotypes classified as deep and shallow SAR11 ecotypes (Alphaproteobacteria), SUP05 (Gammaproteobacteria), SAR324 (Deltaproteobacteria) and MG-I (Thaumarchaeota) (Figure 4b and Supplementary Figure 5c). Mariner RP and orifice communities, which exhibited a high degree of dissimilarity to most samples, were dominated by two SAR11 phylotypes (one deep and one shallow ecotype, OTUs 17 and 7, respectively), uncharacterized Oceanospirillium (OTU_103) and Alteromonas (OTU_3) phylotypes (Gammaproteobacteria), SAR324 (OTU_4) and Erythrobacter (OTU_12, Alphaproteobacteria) (Figure 4b).
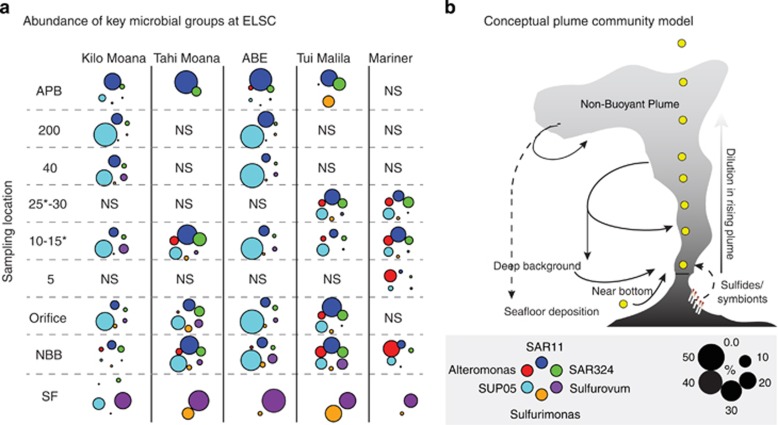
Although the abundance of Alpha, Delta, Epsilon and Gamma Proteobacterial classes was high, >75% of this abundance was attributed to just the top 10 OTUs in each class. The exception was the Epsilonproteobacteria, which showed more evenness (Supplementary Figure 6). Simper (similarity percentage) analysis, which assesses the contribution of OTUs to the total Bray dissimilarity between communities by grouping factor (here by vent field and sampling location), identified several of the dominant OTU phylotypes mentioned above. SUP05 (OTU_1) was an important driver of community structure for both sampling location relative to the vent and across vent fields (Supplementary Tables 3 a and b). A second SUP05 phylotype, OTU_608, was generally less abundant than OTU_1, and also contributed to the community structure differences observed in RPs versus orifice samples and for several vent fields. SAR324 and Alteromonas also contributed highly to the structure of nearly all vent fields and in the case of SAR324 all five sampling locations (Supplementary Tables 3 a and b). To better visualize spatial trends of these OTUs, the abundance of the major microbial groups was plotted in relation to the vent height and across vent fields (Figure 5). SUP05 was consistently abundant in plume samples, with no discernable trends with regard to the RP. SAR11 and SAR324 were consistently abundant in both backgrounds and plumes, with generally high abundance in background. Epsilonproteobacteria (Sulfurovum and Sulfurimonas) and Alteromonas were more patchily distributed. Sulfurovum and Sulfurimonas were generally more abundant in NBB and rising portions of the plume and were absent from APB with the exception of one Tui Malila sample. Although not directly targeted, Archaea communities accounted for 5–20% of the total recovered community and also displayed similar dominance patterns, whereby communities were primarily composed of MG-I, -II and -III (Supplementary Figure 5c). However, none of the archaeal phylotypes were identified as important by Simper analysis.
Figure 5.
(a) Abundance of dominant bacterial groups in NBB, orifice, RP (indicated by meters above orifice), and APB. Color, arrangement and size of the circles indicate the identity and abundance of the groups as shown in the legend. Note that in some cases samples from the same vent site were collected on different dives. NS indicates plume heights that were not sampled. Asterisks indicate height at which samples were taken at Mariner. (b) A conceptual model of potential sources of microorganisms within plumes. Solid and dashed arrows denote hypothesized strength of influence (solid arrows=stronger influence; dashed=weaker).
Discussion
Microorganisms are ecological lynchpins in deep-ocean hydrothermal ecosystems, as they fuel symbiosis (Petersen et al., 2012), dominate near-vent food webs (Bennett et al., 2013) and mediate biogeochemical processes in plumes that are of significance to the greater oceans (Kadko, 1993; Toner et al., 2009; Wu et al., 2011; Li et al., 2014b). Yet, our understanding of the ecology of hydrothermal plume microorganisms and their relationship to those of the seafloor and surrounding water column is still in its infancy (Dick et al., 2013). Thus, we used an in situ filtration approach (Breier et al., 2009b) to systematically sample and characterize microbial community composition in RPs and background waters at five hydrothermal vents of varying geochemistry along the ELSC.
The elevated values of species richness and phylogenetic diversity in samples taken near vent orifices (Supplementary Figures 3 c and d), which represent the origin of plumes, is consistent with physical mixing of several communities, that is, shallow subsurface (provided that temperature is sufficiently low), chimney, surrounding rock, symbionts, near-bottom waters and background waters entrained as the plume rises (Dick et al., 2013). Indeed, in many orifice and lower RP samples there was a high abundance of Epsilonproteobacteria (Figures 4 and 5), which are a dominant microbial component of vent fluids, chimney structures and symbiotic associations with animals (Campbell et al., 2006; Huber et al., 2007; Sievert et al., 2008; Sylvan et al., 2012). Also abundant in all plumes were SUP05, sulfur-oxidizing bacteria that often dominate hydrothermal plumes (Anantharaman et al., 2013, Mattes et al., 2013) and diffuse flow (Huber et al., 2003, Bourbonnais et al., 2012, Anderson et al., 2013) globally.
Despite the consistent occurrence of vent-associated organisms in the RP, we observed a clear difference in membership between plume and rock-hosted sulfide communities (Figure 3). Although these differences could stem in part from the different PCR primers used in the two studies, which may cause amplification bias, the universal forward primer used by Flores et al. (2012) showed sufficient coverage and few mismatches to our bacterial sequences. Furthermore, Sulfurovum and Sulfurimonas phylotypes were shared between the two studies but were quickly diluted within most RPs (Figure 5). This would suggest that differences observed between the two environments were due to the stark contrast between the two habitats rather than primers. In contrast to the cold (2–4 °C), oligotrophic background waters, the rock-hosted sulfide communities are continuously bathed in hot, chemically reduced fluids. These rock-hosted communities were significantly lower in diversity and significantly phylogenetically underdispersed (Supplementary Figure 4), indicating that they are composed of highly specialized microorganisms. Indeed, Flores et al. (2012) identified communities that were dominated primarily by thermophilic and hyperthermophilic bacteria and archaea. Our spatially resolved analysis of the RP indicates that such seafloor organisms are transferred to the water column, but that they are quickly reduced in abundance, likely due primarily to dilution with seawater (see Sulfurovum and Sulfurimonas groups in Figure 5) and the metabolic and physiological challenges faced when transitioning from high temperature, nutrient-rich conditions to cold, oligotrophic waters. Thus, early stages of the plume are substantially influenced by seafloor-derived microbes, but these populations give way to seawater and/or plume-adapted organisms (such as SUP05) as the plumes rise, cool and dilute.
As in the previous studies of vent-associated microbial communities across the ELSC (Flores et al., 2012; Sylvan et al., 2013), we found that the plume communities from the Mariner vent field were the most clearly distinct from the others (Figure 4). Indeed, the only significant difference in community composition was observed between Kilo Moana and Mariner (Supplementary Table 2), the ELSC fields that have the greatest separation in depth (∼Δ800 m) and lateral distance (∼Δ240 km), and clear distinctions in morphology and geochemistry (Ferrini et al., 2008; Mottl et al., 2011). Although all four factors may influence plume microbial community structure, Mariner geochemistry is uniquely distinct from the other vent sites owing to the influence of an actively degassing subsurface magma chamber (Mottl et al., 2011). Particularly important is the high concentration of Fe and other sulfide mineral-forming metals, which reduce the bioavailability of H2S/HS− for chemosynthetic organisms (Luther et al., 2001; Hsu-Kim et al., 2008; Yucel et al., 2011; Gartman et al., 2014). Consistent with the bioavailability of sulfur being a key factor, OTUs of sulfur-oxidizing SUP05 and Epsilonproteobacteria were absent or at low abundance at Mariner (Figures 4 and 5a). Physical factors may provide a complementary or alternative explanation for the distinct microbial communities. The vent edifices at Mariner are significantly taller than those of the other fields, up to several tens of meters, and although flux measurements are lacking from the ELSC, visual observations suggest that Mariner vents have significantly greater fluid flux. Both factors would act to increase background seawater entrainment into the plume (Jiang and Breier, 2014). However, the bulk of that entrained water is further removed from the seafloor so that the entrainment of NBB, diffuse flow and animal-associated communities, which often contain high abundances of SUP05 and Epsilonproteobacteria, could be less than at other ELSC fields. Finally, the abundance of Alteromonas at Mariner suggests that microbial iron chelation and uptake could influence iron speciation (Li et al., 2014b). Taken together, these Mariner results are in line with previous observations that geochemistry influences the microbial ecology of both surface and plume chemolithoautotrophs (Tivey et al., 2012).
Previous studies found that the composition of microbial communities from both low- and high-temperature seafloor vent deposits was correlated with geochemistry across the ELSC (Flores et al., 2012; Sylvan et al., 2013). In contrast, we found that the differences in community composition in water-column samples were not correlated with geochemistry, as evident by the lack of significant clustering of microbial community structure (Figure 3). This similarity between water-column communities extended to sites that were distinct in terms of geography, depth and geochemistry, suggesting that these factors are not primary determinants of ELSC plume community composition. The consistent microbial community structure observed across all ELSC water-column samples studied here likely reflects a large contribution from background deep waters owing to extensive physical mixing and dilution of buoyant hydrothermal plumes. Initial plume dilution rates are high, typically 10 000:1 (seawater:vent fluid) (Lupton et al., 1985), thus the abundance in plumes of microbial groups such as SAR11 and SAR324 that are ubiquitous in the deep oceans (Ghiglione et al., 2012; Thrash et al., 2014) is to be expected. The persistent abundance of vent-associated organisms such as SUP05 in the upper portions of plumes (Figure 5) suggests that either (i) they must grow rapidly to keep pace with dilution by background seawater or (ii) they are entrained from waters surrounding the plume, where they may be abundant owing to plume influence. Given the rapid dilution rates, scenario (ii) seems more likely. The possibility that hydrothermal plumes may influence the abundance of microbial community members in surrounding seawater has been suggested but not yet rigorously tested (Dick et al., 2013). Such regional influences could feedback into the plume and reinforce lithotrophic signals in plume microbial communities (Figure 5). Our data also suggest that dispersal of plumes throughout the Lau Basin is likely extensive because of weak stratification, diapycnal mixing over rough topography, deep-ocean currents and recirculation of detached eddies (Speer et al., 2002; Speer and Thurnherr, 2012). Such mesoscale effects may also contribute to the temporal effect observed in our data, wherein microbial community composition clusters by cruise leg (Figure 4a). Taken together, our data suggest that hydrothermal plume communities at ELSC, and potentially other hydrothermal vent fields, are sourced from a combination of oligotrophic background waters, near-vent minerals and animal symbionts, plume particles and aged plumes (see conceptual model in Figure 5b).
Conclusions
To date, studies of hydrothermal plumes and their associated microbiome have focused primarily on the neutrally buoyant portions of the plume, due in part to the difficulty in precisely sampling the rising portions of the plume. Using an in situ filtering approach we were able to sample, with high spatial resolution, RP microbial communities. Our results suggest that deep-ocean microbes are important plume community members, and that plume dilution and mixing has a large role in combining microbial communities reliant on hydrothermal fluids with background deep-sea microbial communities. Although much work is needed to understand the function and dispersal of these microbial communities, our results highlight putative microbial linkages between plumes and the deep ocean. Given that many of these deep-ocean microorganisms possess the ability to fix carbon, understanding their role in plumes is critical to accurately quantifying deep-ocean carbon budgets.
Acknowledgments
We thank the Capitan and crew of the R/V Thomas G Thompson as well as the crew of the remotely operated vehicle Jason II. We also thank Drs AL Reysenbach, M Tivey, C Fisher, P Girguis, G Luther and the Eastern Lau Spreading Center 2009 scientific parties for allowing us to participate in their cruises (National Science Foundation Grants: OCE-0424953, OCE-02040985, OCE-0728391, OCE-0752469 and OCE-0751839). We are grateful to Dr AL Reysenbach for providing access to seafloor sequences and Dr Sheri White for assistance with sampling. This work was funded by National Science Foundation Grants (OCE-1038006, OCE-1029242 and OCE-1038055) and the Gordon and Betty Moore Foundation (GBMF 2609). JAB was supported by Grant OCE-1038055, and the Frank and Lisina Hock Endowed fund was provided in support of the Scientific Staff. We gratefully acknowledge the constructive comments of two anonymous reviewers, which greatly improved the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amend JP, McCollom TM, Hentscher M, Bach W. Catabolic and anabolic energy for chemolithoautotrophs in deep-sea hydrothermal systems hosted in different rock types. Geochim Cosmochim Acta. 2011;75:5736–5748. [Google Scholar]
- Anantharaman K, Breier JA, Sheik CS, Dick GJ. Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc Natl Acad Sci USA. 2013;110:330–335. doi: 10.1073/pnas.1215340110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RE, Beltrán MT, Hallam SJ, Baross JA. Microbial community structure across fluid gradients in the Juan de Fuca Ridge hydrothermal system. FEMS Microbiol Ecol. 2013;83:324–339. doi: 10.1111/j.1574-6941.2012.01478.x. [DOI] [PubMed] [Google Scholar]
- Baker B, Lesniewski R, Dick G. Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J. 2012;6:2269–2279. doi: 10.1038/ismej.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ET, Massoth GJ, Nakamura K-i, Embley RW, de Ronde CEJ, Arculus RJ. Hydrothermal activity on near-arc sections of back-arc ridges: results from the Mariana Trough and Lau Basin. Geochem Geophys Geosyst. 2005;6:Q09001. [Google Scholar]
- Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu SE, Baker ET, German CR, Maffei A. An authoritative global database for active submarine hydrothermal vent fields. Geochem Geophy Geosy. 2013;14:4892–4905. [Google Scholar]
- Bennett SA, Coleman M, Huber JA, Reddington E, Kinsey JC, McIntyre C, et al. Trophic regions of a hydrothermal plume dispersing away from an ultramafic-hosted vent-system: Von Damm vent-site, Mid-Cayman Rise. Geochem Geophy Geosy. 2013;14:317–327. [Google Scholar]
- Boström K, Peterson MNA. The origin of aluminum-poor ferromanganoan sediments in areas of high heat flow on the East Pacific Rise. Mar Geol. 1969;7:427–447. [Google Scholar]
- Bourbonnais A, Juniper SK, Butterfield DA, Devol AH, MMM Kuypers, Lavik G, et al. Activity and abundance of denitrifying bacteria in the subsurface biosphere of diffuse hydrothermal vents of the Juan de Fuca Ridge. Biogeosciences. 2012;9:4661–4678. [Google Scholar]
- Bowie AR, Townsend AT, Lannuzel D, Remenyi TA, van der Merwe P. Modern sampling and analytical methods for the determination of trace elements in marine particulate material using magnetic sector inductively coupled plasma, Äìmass spectrometry. Anal Chim Acta. 2010;676:15–27. doi: 10.1016/j.aca.2010.07.037. [DOI] [PubMed] [Google Scholar]
- Breier JA, German CR, White SN. Mineral phase analysis of deep-sea hydrothermal particulates by a Raman spectroscopy expert algorithm: Toward autonomous in situ experimentation and exploration. Geochem Geophy Geosy. 2009;10:Q05T05. [Google Scholar]
- Breier JA, Rauch CG, McCartney K, Toner BM, Fakra SC, White SN, et al. A suspended-particle rosette multi-sampler for discrete biogeochemical sampling in low-particle-density waters. Deep-Sea Res Pt I. 2009;56:1579–1589. [Google Scholar]
- Breier JA, Toner BM, Fakra SC, Marcus MA, White SN, Thurnherr AM, et al. Sulfur, sulfides, oxides and organic matter aggregated in submarine hydrothermal plumes at 9º50 N East Pacific Rise. Geochim Cosmochim Acta. 2012;88:216–236. [Google Scholar]
- Brown MV, Donachie SP. Evidence for tropical endemicity in the Deltaproteobacteria Marine Group B/SAR324 bacterioplankton clade. Aquat Microb Ecol. 2007;46:107. [Google Scholar]
- Campbell BJ, Engel AS, Porter ML, Takai K. The versatile [epsi]-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- Cowen JP, Massoth GJ, Baker ET. Bacterial scavenging of Mn and Fe in a mid- to far-field hydrothermal particle plume. Nature. 1986;322:169–171. [Google Scholar]
- de Angelis MA, Lilley MD, Olson EJ, Baross JA. Methane oxidation in deep-sea hydrothermal plumes of the Endeavor Segment of the Juan de Fuca Ridge. Deep-Sea Res Pt I. 1993;40:1169–1186. [Google Scholar]
- Dick GJ, Clement BG, Webb SM, Fodrie FJ, Bargar JR, Tebo BM. Enzymatic microbial Mn(II) oxidation and Mn biooxide production in the Guaymas Basin deep-sea hydrothermal plume. Geochim Cosmochim Acta. 2009;73:6517–6530. [Google Scholar]
- Dick GJ, Tebo BM. Microbial diversity and biogeochemistry of the Guaymas Basin deep-sea hydrothermal plume. Environ Microbiol. 2010;12:1334–1347. doi: 10.1111/j.1462-2920.2010.02177.x. [DOI] [PubMed] [Google Scholar]
- Dick GJ, Anantharaman K, Baker BJ, Li M, Reed DC, Sheik CS. The microbiology of deep-sea hydrothermal vent plumes: ecological and biogeographic linkages to seafloor and water column habitats. Front Ext Microbiol. 2013;4:1–16. doi: 10.3389/fmicb.2013.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Elderfield H, Schultz A. Mid-ocean ridge hydrothermal fluxes and the chemical composition of the ocean. Annu Rev Earth Planet Sci. 1996;24:191–224. [Google Scholar]
- Faith DP, Lozupone CA, Nipperess D, Knight R. The cladistic basis for the phylogenetic diversity (PD) measure links evolutionary features to environmental gradients and supports broad applications of microbial ecology's ‘phylogenetic beta diversity' framework. Int J Mol Sci. 2009;10:4723–4741. doi: 10.3390/ijms10114723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrini VL, Tivey MK, Carbotte SM, Martinez F, Roman C. Variable morphologic expression of volcanic, tectonic, and hydrothermal processes at six hydrothermal vent fields in the Lau back-arc basin. Geochem Geophys Geosyst. 2008;9:Q07022. [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GE, Shakya M, Meneghin J, Yang ZK, Seewald JS, Geoff Wheat C, et al. Inter-field variability in the microbial communities of hydrothermal vent deposits from a back-arc basin. Geobiology. 2012;10:333–346. doi: 10.1111/j.1472-4669.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- Gartman A, Findlay AJ, Luther GW., III Nanoparticulate pyrite and other nanoparticles are a widespread component of hydrothermal vent black smoker emissions. Chem Geol. 2014;366:32–41. [Google Scholar]
- German C, Bowen A, Coleman M, Honig D, Huber J, Jakuba M, et al. Diverse styles of submarine venting on the ultraslow spreading Mid-Cayman Rise. Proc Natl Acad Sci USA. 2010;107:14020–14025. doi: 10.1073/pnas.1009205107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German CR, Von Damm KL.2004Hydrothemal Processesvol. 6Elsevier: Oxford, UK [Google Scholar]
- German CR, Baker ET, Connelly DP, Lupton JE, Resing J, Prien RD, et al. Hydrothermal exploration of the Fonualei Rift and Spreading Center and the Northeast Lau Spreading Center. Geochem Geophys Geosyst. 2006;7:Q11022. [Google Scholar]
- Ghiglione J-F, Galand P, Pommier T, Pedrós-Alió C, Maas E, Bakker K, et al. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc Natl Acad Sci USA. 2012;109:17633–17638. doi: 10.1073/pnas.1208160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden J, Breier J, Rogers K, Schulte M, Toner B. Biogeochemical processes at hydrothermal vents: microbes and minerals, bioenergetics, and carbon fluxes. Oceanography. 2012;25:196–208. [Google Scholar]
- Hsu-Kim H, Mullaugh K, Tsang J, Yucel M, Luther G. Formation of Zn- and Fe-sulfides near hydrothermal vents at the Eastern Lau Spreading Center: implications for sulfide bioavailability to chemoautotrophs. Geochem T. 2008;9:6. doi: 10.1186/1467-4866-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JA, Butterfield DA, Baross JA. Bacterial diversity in a subseafloor habitat following a deep-sea volcanic eruption. FEMS Microbiol Ecol. 2003;1475:1–17. doi: 10.1111/j.1574-6941.2003.tb01080.x. [DOI] [PubMed] [Google Scholar]
- Huber JA, Mark Welch D, Morrison HG, Huse SM, Neal PR, Butterfield DA. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Jannasch HW, Wirsen CO. Chemosynthetic Primary Production at East Pacific Sea Floor Spreading Centers. BioScience. 1979;29:592–598. [Google Scholar]
- Jannasch HW, Mottl MJ. Geomicrobiology of deep-sea hydrothermal vents. Science. 1985;229:717–725. doi: 10.1126/science.229.4715.717. [DOI] [PubMed] [Google Scholar]
- Jiang H, Breier JA. Physical controls on mixing and transport within rising submarine hydrothermal plumes: a numerical similation study. Deep-Sea Res Pt I. 2014;92:41–55. [Google Scholar]
- Kadko D. An assessment of the effect of chemical scavenging within submarine hydrothermal plumes upon ocean geochemistry. Earth Planet Sci Lett. 1993;120:361–374. [Google Scholar]
- Kamenetsky VS, Crawford AJ, Eggins S, Mühe R. Phenocryst and melt inclusion chemistry of near-axis seamounts, Valu Fa Ridge, Lau Basin: insight into mantle wedge melting and the addition of subduction components. Earth Planet Sci Lett. 1997;151:205–223. [Google Scholar]
- Karl DM, Wirsen CO, Jannasch HW. Deep-sea primary production at the Galapagos hydrothermal vents. Science. 1980;207:1345–1347. [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- Kembel SW. Disentangling niche and neutral influences on community assembly: assessing the performance of community phylogenetic structure tests. Ecol Lett. 2009;12:949–960. doi: 10.1111/j.1461-0248.2009.01354.x. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- Lam P, Cowen JP, Jones RD. Autotrophic ammonia oxidation in a deep-sea hydrothermal plume. FEMS Microbiol Ecol. 2004;47:191–206. doi: 10.1016/S0168-6496(03)00256-3. [DOI] [PubMed] [Google Scholar]
- Lesniewski RA, Jain S, Anantharaman K, Schloss PD, Dick GJ. The metatranscriptome of a deep-sea hydrothermal plume is dominated by water column methanotrophs and lithotrophs. ISME J. 2012;6:1–11. doi: 10.1038/ismej.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jain S, Baker BJ, Taylor C, Dick GJ. Novel hydrocarbon monooxygenase genes in the metatranscriptome of a natural deep-sea hydrocarbon plume. Environ Microbiol. 2014;16:60–71. doi: 10.1111/1462-2920.12182. [DOI] [PubMed] [Google Scholar]
- Li M, Toner BM, Baker BJ, Breier JA, Sheik CS, Dick GJ. Microbial iron uptake as a mechanism for dispersing iron from deep-sea hydrothermal vents. Nat Commun. 2014;5:3192. doi: 10.1038/ncomms4192. [DOI] [PubMed] [Google Scholar]
- Lupton JE, Delaney JR, Johnson HP, Tivey MK. Entrainment and vertical transport of deep-ocean water by buoyant hydrothermal plumes. Nature. 1985;316:621–623. [Google Scholar]
- Luther GW, Rozan TF, Taillefert M, Nuzzio DB, Di Meo C, Shank TM, et al. Chemical speciation drives hydrothermal vent ecology. Nature. 2001;410:813–816. doi: 10.1038/35071069. [DOI] [PubMed] [Google Scholar]
- Martinez F, Taylor B. Mantle wedge control on back-arc crustal accretion. Nature. 2002;416:417–420. doi: 10.1038/416417a. [DOI] [PubMed] [Google Scholar]
- Mattes TE, Nunn BL, Marshall KT, Proskurowski G, Kelley DS, Kawka OE, et al. Sulfur oxidizers dominate carbon fixation at a biogeochemical hot spot in the dark ocean. ISME J. 2013;7:2349–2360. doi: 10.1038/ismej.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom TM. Geochemical constraints on primary productivity in submarine hydrothermal vent plumes. Deep-Sea Res Pt I. 2000;47:85–101. [Google Scholar]
- Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Mottl MJ, Seewald JS, Wheat CG, Tivey MK, Michael PJ, Proskurowski G, et al. Chemistry of hot springs along the Eastern Lau Spreading Center. Geochim Cosmochim Acta. 2011;75:1013–1038. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. 2013vegan: Community Ecology PackageR package version.
- Petersen JM, Wentrup C, Verna C, Knittel K, Dubilier N. Origins and evolutionary flexibility of chemosynthetic symbionts from deep-sea animals. Bio Bul. 2012;223:123–137. doi: 10.1086/BBLv223n1p123. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria; 2013. [Google Scholar]
- Raczek I, Stoll B, Hofmann AW, Peter Jochum K. High-precision trace element data for the USGS reference materials BCR-1, BCR-2, BHVO-1, BHVO-2, AGV-1, AGV-2, DTS-1, DTS-2, GSP-1 and GSP-2 by ID-TIMS and MIC-SSMS. Geostand Newslett. 2001;25:77–86. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik CS, Jain S, Dick GJ. Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ Microbiol. 2014;16:304–317. doi: 10.1111/1462-2920.12165. [DOI] [PubMed] [Google Scholar]
- Sievert SM, Hügler M, Taylor CD, Wirsen CO.2008Sulfur Oxidation at Deep-Sea Hydrothermal Ventsvol. 238-258Springer: New York, NY, USA [Google Scholar]
- Speer K, Thurnherr AM. The Lau Basin Float Experiment (LAUB-FLEX) Oceanography. 2012;25:284–285. [Google Scholar]
- Speer KG, Maltrud ME, Thurnherr AM.2002A global view of dispersion above the mid-ocean ridgeIn: Halbach PE, Tunnicliffe V, Hein JR (eds)Energy and Mass Transfer in Marine Hydrothermal Systems Dahlem University Press: Berlin, Germany; p365 [Google Scholar]
- Sunamura M, Higashi Y, Miyako C, Ishibashi J, Maruyama A. Two Bacteria phylotypes are predominant in the Suiyo Seamount hydrothermal plume. Appl Environ Microb. 2004;70:1190–1198. doi: 10.1128/AEM.70.2.1190-1198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvan JB, Pyenson BC, Rouxel O, German CR, Edwards KJ. Time-series analysis of two hydrothermal plumes at 9°50′N East Pacific Rise reveals distinct, heterogeneous bacterial populations. Geobiology. 2012;10:178–192. doi: 10.1111/j.1472-4669.2011.00315.x. [DOI] [PubMed] [Google Scholar]
- Sylvan JB, Sia TY, Haddad AG, Briscoe LJ, Toner BM, Girguis PR, et al. Low temperature geomicrobiology follows host rock composition along a geochemical gradient in lau basin. FMICB. 2013;4:61. doi: 10.3389/fmicb.2013.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina PL, Ussler W, Joye SB, Harrison BK, Orphan VJ. Distributions of putative aerobic methanotrophs in diverse pelagic marine environments. ISME J. 2010;4:700–710. doi: 10.1038/ismej.2009.155. [DOI] [PubMed] [Google Scholar]
- Thrash CJ, Temperton B, Swan BK, Landry ZC, Woyke T, DeLong EF, et al. Single-cell enabled comparative genomics of a deep ocean SAR11 bathytype. ISME J. 2014. [DOI] [PMC free article] [PubMed]
- Tivey M, Becker E, Beinart R, Fisher C, Girguis P, Langmuir C, et al. Links from Mantle to Microbe at the Lau Integrated Study Site: insights from a Back-Arc Spreading Center. Oceanography. 2012;25:62–77. [Google Scholar]
- Toner BM, Fakra SC, Manganini SJ, Santelli CM, Marcus MA, Moffett JW, et al. Preservation of iron(II) by carbon-rich matrices in a hydrothermal plume. Nat Geosci. 2009;2:197–201. [Google Scholar]
- Walsh D, Zaikova E, Howes C, Song Y, Wright J, Tringe S, et al. Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science. 2009;326:578–582. doi: 10.1126/science.1175309. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn CD, Karl DM, Massoth GJ. Microorganisms in deep-sea hydrothermal plumes. Nature. 1986;320:744–746. [Google Scholar]
- Wu J, Wells ML, Rember R. Dissolved iron anomaly in the deep tropical, subtropical Pacific: evidence for long-range transport of hydrothermal iron. Geochim Cosmochim Acta. 2011;75:460–468. [Google Scholar]
- Yucel M, Gartman A, Chan CS, Luther GW. Hydrothermal vents as a kinetically stable source of iron-sulphide-bearing nanoparticles to the ocean. Nat Geosci. 2011;4:367–371. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.