Highlight
TaFAR5, a fatty acyl-coenzyme A reductase, catalyses the synthesis of primary alcohols in wheat cuticle and is linked to response to abiotic stress in an ABA-dependent manner.
Key words: Abiotic stress, cuticular wax, endoplasmic reticulum, fatty acid, fatty acyl-CoA reductase, leaf blade, primary alcohol, wheat.
Abstract
A waxy cuticle that serves as a protective barrier against non-stomatal water loss and environmental damage coats the aerial surfaces of land plants. It comprises a cutin polymer matrix and waxes. Cuticular waxes are complex mixtures of very long chain fatty acids (VLCFAs) and their derivatives. Results show that primary alcohols are the major components of bread wheat (Triticum aestivum L.) leaf blade cuticular waxes. Here, the characterization of TaFAR5 from wheat cv Xinong 2718, which is allelic to TAA1b, an anther-specific gene, is reported. Evidence is presented for a new function for TaFAR5 in the biosynthesis of primary alcohols of leaf blade cuticular wax in wheat. Expression of TaFAR5 cDNA in yeast (Saccharomyces cerevisiae) led to production of C22:0 primary alcohol. The transgenic expression of TaFAR5 in tomato (Solanum lycopersicum) cv MicroTom leaves resulted in the accumulation of C26:0, C28:0, and C30:0 primary alcohols. TaFAR5 encodes an alcohol-forming fatty acyl-coenzyme A reductase (FAR). Expression analysis revealed that TaFAR5 was expressed at high levels in the leaf blades, anthers, pistils, and seeds. Fully functional green fluorescent protein-tagged TaFAR5 protein was localized to the endoplasmic reticulum (ER), the site of primary alcohol biosynthesis. SDS–PAGE analysis indicated that the TaFAR5 protein possessed a molecular mass of 58.4kDa, and it was also shown that TaFAR5 transcript levels were regulated in response to drought, cold, and abscisic acid (ABA). Overall, these data suggest that TaFAR5 plays an important role in the synthesis of primary alcohols in wheat leaf blade.
Introduction
All of the primary aerial organs of land plants are covered with a cuticle that is essential for their protection and interaction with the environment. The cuticle comprises two main types of lipids: cutin, the core structural polymer composed of a three-dimensional polymer of mostly C16 and C18 hydroxy fatty acids cross-linked by ester bonds (Beisson et al., 2012); and cuticular waxes, which are the complex mixtures of very long chain fatty acids (VLCFAs) and their derivatives, including primary and secondary alcohols, aldehydes, alkanes, ketones, esters, triterpenes, sterols, and flavonoids (Bianchi et al., 1990; Kunst and Samuels, 2009), with chain lengths of >20 carbons. Wax composition varies according to species, organ, and developmental state (Samuels et al., 2008). Cuticular wax is believed to play important roles in limiting non-stomatal water loss, a feature contributing to drought tolerance (Hall and Jones, 1961; Premchandra et al., 1992; Kerstiens, 2006; Kosma and Jenks, 2007). It also protects the plant from UV radiation (Yeats and Rose, 2013), defends against bacterial and fungal pathogens (Jenks et al., 1994), and prevents inappropriate organ fusion during development (Sieber et al., 2000). The cuticular waxes of most plants are often present in the form of a film or microcrystals which are mainly classified into platelets, tubules, and rod-shaped structures and give the plant surface a glaucous or grey appearance (Post-Beittenmiller, 1996; Jenks and Ashworth, 1999; Barthlott et al., 1998; Jeffree, 2006).
The first step in wax biosynthesis is the elongation of C18:0 fatty acid produced in the plastid to generate VLCFA wax precursors by joining the C2 building blocks of acetyl-coenzyme A into chains of up to 34 carbons in length (Baker, 1982; Post-Beittenmiller, 1996; Samuels et al., 2008). VLCFAs are formed by a microsomal fatty acid elongation (FAE) system involving four consecutive enzymatic reactions that are catalysed in turn by a β-ketoacyl-CoA synthase (KCS), a β-ketoacyl-CoA reductase (KCR), a β-hydroxyacyl-CoA dehydratase (HCD), and an enoyl-CoA reductase (ECR). Subsequently, VLCFAs are converted through several biosynthetic pathways to the remaining wax components. In Arabidopsis, two principal wax biosynthetic pathways are found: (i) the acyl reduction pathway, which is initiated by the reduction of VLCFAs to aldehydes, followed by a further reduction of aldehydes by an aldehyde reductase to produce even-chained primary alcohols (Kolattukudy, 1971); and (ii) the decarbonylation pathway, leading to aldehydes, alkanes, secondary alcohols, and ketones, which account for 70–80% of total wax in Arabidopsis (Kolattukudy, 1971; Kunst and Samuels, 2003; Samuels et al., 2008).
Long-chain primary alcohols synthesized by the acyl reduction pathway are found throughout the biological world, and are widespread on the surfaces of plants and animals (Metz et al., 2000). In diverse plant species and organs, the important compounds are primary alcohols with a chain length preference of C26 or C28 and, in some systems, C30 or C32 (Baker, 1982). The biochemistry of primary alcohol formation has been examined in a variety of plants (Kolattukudy, 1971; Vioque and Kolattukudy, 1997; Metz et al., 2000). The partial purification of reducing activities from Brassica oleracea initially led to the proposal that primary alcohol production is a two-step process carried out by two separate enzymes, an NADH-dependent acyl-CoA reductase required to reduce VLCFAs to aldehydes and an NADPH-dependent aldehyde reductase required to reduce aldehydes further to primary alcohols (Kolattukudy, 1971; Kunst and Samuels, 2003). Biochemical support for the two-step process leading to alcohol formation comes from experiments in B. oleracea, in which an aldehyde intermediate was isolated (Kolattukudy, 1971). Subsequent studies have demonstrated that primary alcohols are not formed in two steps. Initially, the green alga Euglena gracilis was found to generate alcohols from acyl-CoA precursors without releasing aldehyde intermediates (Kolattukudy, 1970). Subsequent biochemical studies revealed that VLCFA precursors are reduced to primary alcohols by a single fatty acyl-coenzyme A reductase (FAR) in jojoba (Simmondsia chinensis) embryos (Pollard et al., 1979) and pea (Pisum sativum) leaves (Vioque and Kolattukudy, 1997) (Fig. 1). Furthermore, functional expression of FARs from jojoba (Metz et al., 2000), silkmoth (Bombyx mori) (Moto et al., 2003), mouse (Mus musculus), and human (Homo sapiens) cells (Cheng and Russell, 2004) in heterologous systems led to the formation of primary alcohols from fatty acid precursors. To date, eight FAR genes have been identified in Arabidopsis. MS2 (FAR2), a gene previously implicated in exine production, catalyses the conversion of fatty acids to fatty alcohols when expressed in Escherichia coli (Aarts et al., 1997; Doan et al., 2009; Dobritsa et al., 2009). CER4 (FAR3) is specifically involved in the production of C24 to C28 very long chain primary alcohols (Rowland et al., 2006). FAR1, FAR4, and FAR5 can generate the fatty alcohols found in root, seed coat, and wound-induced leaf tissue (Domergue et al., 2010). Ectopic expression of TAA1a from wheat resulted in the production of fatty alcohols in tobacco seeds and in E. coli (Wang et al., 2002).
Fig. 1.
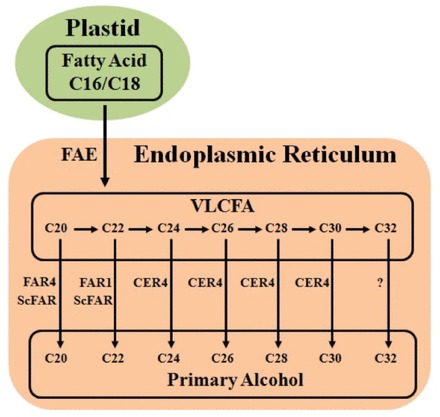
Proposed biosynthetic pathway of very long chain primary alcohol in plants (cuticle and seed coat). FAE, FAR1, FAR4, and CER4 are the wax biosynthetic enzymes from Arabidopsis. ScFAR is the wax biosynthetic enzyme from jojoba. (This figure is available in colour at JXB online.)
Here the identification of TaFAR5 encoding an alcohol-forming FAR from wheat leaf blades, which is identical to TAA1b, previously reported as an anther-specific gene (Wang et al., 2002), is reported. Evidence is also presented for a new function for TaFAR5 in the primary alcohol biosynthesis of leaf blade cuticular wax in wheat. The expression pattern of TaFAR5 coincides with known sites of primary alcohol deposition. The TaFAR5 protein is localized to the endoplasmic reticulum (ER), the site of wax synthesis. Furthermore, TaFAR5 is regulated by drought and cold stresses in an abscisic acid (ABA)-independent manner.
Materials and methods
Plant materials and stress treatments
The hexaploid wheat cultivar Xinong 2718 was used for cDNA cloning, genomic PCR, quantitative real-time PCR (qRT-PCR), and cuticular waxes analysis. Chinese Spring nullisomic–tetrasomic lines were provided by Dr L.W. Shan and used to determine the chromosomal position of TaFAR5, and were grown at the experimental farm of Northwest A&F University in Yangling during the 2011 and 2012 wheat-growing seasons. Simultaneously, seeds of Xinong 2718 were also soil-grown in a glasshouse at 22 °C with 8h dark/16h light, after which 4-week-old seedlings were subjected to various stress treatments. For drought treatment, seedlings were dehydrated on filter paper and harvested at various time points. For polyethylene glycol (PEG) 6000 and ABA treatments, seedlings were transferred to solutions containing 20% (w/v) PEG 6000 and 100 μM ABA, respectively. For cold stress treatment, seedlings were maintained at 4 °C.
Wax extraction and chemical characterization
Wax load was determined on seedlings and flowering plants. Leaf blades were harvested and immediately immersed in chloroform for 1min at room temperature to extract cuticular waxes (Leide et al., 2007; Wang et al., 2011; Zhang et al., 2013). After extraction, n-tetracosane (C24 alkane) was added as an internal standard, and the solvent was completely evaporated under a stream of nitrogen. Samples were transferred to a GC autosampler vial, dried under nitrogen, and derivatized with 100 μl of bis-N, N-(trimethylsilyl)trifluoroacetamide (Sigma) and 100 μl of pyridine (Fluka) for 60min at 70 °C. Solvent was again evaporated under nitrogen gas and the waxes were resuspended in 500 μl of chloroform for gas chromatography–mass spectroscopy (GC-MS) analysis. The qualitative composition of the wax was analysed using a capillary gas chromatograph equipped with a Rxi-5ms column (length 30 m, id 0.25mm, film thickness 0.25 μm) and attached to a mass spectrometer (GCMS-QP2010, Shimadzu, Japan) using helium as the carrier gas. The initial temperature of 50 °C was held for 2min, increased at 20 °C min–1 to 200 °C, held for 2min at 200 °C, increased again at 2 °C min–1 to 320 °C, and held for 15min at 320 °C. Injector and detector temperatures were set at 250 °C. The molecular identities of individual wax compounds were identified by comparing their mass spectra with those of authentic standards and literature data. The compounds were quantified by comparing the peak areas with that of the internal standard. The total amount of cuticular wax was expressed per unit of leaf surface area. The leaf areas were determined using ImageJ software (http://rsb.info.nih.gov/ij/) based on digital images of the leaves and multiplying by 2.
Scanning electron microscopy (SEM) analysis
For epidermal wax crystal examination, leaf blades of wheat cv Xinong 2718 were collected from seedlings and flowering plants. The samples were air-dried for 2 d in a desiccator at room temperature and carefully dissected. Dried leaf blade pieces were mounted on specimen stubs using double-sided copper tape and sputter-coated with gold particles using 90 s bursts from a sputter coater. Coated surfaces were viewed using a Hitachi S4800 scanning electron microscope at an accelerating voltage of 10kV and at a working distance of 12mm.
Cloning of wheat TaFAR5 full-length cDNA
Total RNA was extracted from durum wheat cv Xinong 2718 leaves using Trizol reagent (Invitrogen). To remove contaminating genomic DNA, total RNA was treated with DNase I (Promega). First-strand cDNA was synthesized using PrimeScript™ reverse transcriptase (TaKaRa) according to the manufacturer’s directions for use as a template for PCR amplification. A pair of gene-specific primers were designed based on the sequence of TaFAR5 (TaFAR5F 5′-ATGGTGGGCACGCTGGATGAGG-3′ and TaFAR5R 5′-TCACTTGTGGACGTACTTCATG-3′) and used to amplify the coding region. The cDNA was amplified using the following protocol: denaturation for 5min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 55 °C, and 2min at 72 °C, with a final extension for 10min at 72 °C. The resulting PCR product was separated by agarose gel electrophoresis (1% agarose) and extracted using a Gel Extraction Kit (TIANGEN). A 1.6kb DNA band was recovered, cloned into the pMD™ 18-T vector (TaKaRa) using T4 DNA ligase (TaKaRa), and subsequently transformed into DH5α cells. Successful isolation of TaFAR5 cDNA was confirmed by sequencing. The genomic sequence of TaFAR5 was deposited in the GenBank database under the accession number KJ725345.
Heterologous expression in yeast
The coding sequence of TaFAR5 was amplified from wheat cv Xinong 2718 cDNA using a primer labelled as TaFAR5-YS (Supplementary Table S1 available at JXB online). The corresponding PCR fragment was cloned into the yeast expression vector pYES2 (Invitrogen) to yield pYES-TaFAR5 under the control of the GAL1 promoter. The construct pYES-TaFAR5 was transformed into DH5α cells. Sequencing of an individual clone confirmed that there were no errors in TaFAR5. The pYES-TaFAR5 vector and an empty vector were transformed into the mutant yeast strain INVSc1 according to Gietz and Woods (2002). Transgenic yeast cells were grown on synthetic complete (SC) selection medium without uracil. Three individual cell lines were selected from each transgenic strain. After galactose induction and 24h of incubation in 0.1M potassium phosphate-containing glucose and haemin, cells were collected, refluxed for 5min in 20% (w/v) KOH/50% (v/v) ethanol, and extracted twice with hexane. Following phase separation, the chloroform phase was transferred to a fresh tube. The samples were evaporated to dryness under a stream of nitrogen, and then derivatized with 100 μl of bis-N, N-(trimethylsilyl)trifluoroacetamide plus 100 μl of pyridine at 70 °C for 60min, and analysed by GC-MS as described above for the plant wax analysis.
Agrobacterium tumefaciens-mediated genetic transformation of tomato
The TaFAR5 coding region was amplified from wheat cv Xinong 2718 cDNA using the LA Taq DNA polymerase (TaKaRa). The corresponding PCR fragment was cloned into the binary vector pCXSN which had been digested with XcmI (NEB) restriction enzyme under the control of the 35S promoter. Positive clones were identified by colony PCR and sequencing. Construct pCXSN-TaFAR5 and the empty vector were individually introduced into Agrobacterium tumefaciens strain GV3101, and verified by colony PCR and restriction digestion. Tomato cv MicroTom plants were transformed as described by Dan et al. (2006) with minor modifications. The transgenic plants were screened via hygromycin selection and confirmed by PCR.
Protein alignment and phylogenetic analysis
Multiple sequences were aligned with the ClustalW 1.83 program (Thompson et al., 1997) using default parameters (http://www.ebi.ac.uk/clustalw). The aligned sequences were imported into BioEdit for manual editing (http://www.mbio.ncsu.edu/BioEdit/bioedit.html; Hall, 1999). A Neighbor–Joining tree was constructed using MEGA software (version 3.1; http://www.megasoftware.net/index; Kumar et al., 2004) with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates, random seed).
Quantitative RT-PCR
RNA samples from various organs, including leaf, leaf sheath, root, internode, young panicle, anther, awn, glume, filling seed, and seedlings, under stress treatments were extracted using the Trizol reagent (Invitrogen) according to the manufacturer’s protocol. An additional DNase I (Promega) treatment was included to eliminate any contaminating DNA. The RNA was quantified spectrophotometrically. Then, total RNA was used to synthesize the oligo(dT)18-primed first-strand cDNA using PrimeScript™ reverse transcriptase (TaKaRa) following standard protocols. Quantitative real-time PCR (RT-PCR) was performed in a 25 μl volume using a SYBR® Premix Ex Taq™ Kit (TaKaRa) on a CFX96 real-time PCR detection system (Bio-Rad) according to the manufacturer’s instructions. The data were analysed using Opticon monitor software (Bio-Rad). Three replicates were performed for each sample. Wheat TaActin gene was used as an internal control.
Subcellular localization of TaFAR5
To determine the localization of TaFAR5 protein in plant cells, the coding region of TaFAR5 without a termination codon was cloned into the pA7-GFP vector, yielding a TaFAR5–green fluorescent protein (GFP) fusion construct. The fusion construct and ER marker mCherry–HDEL (Nelson et al., 2007) were transiently co-expressed in rice leaf protoplasts by PEG according to the previously described protocol (Chen et al., 2006). The fluorescence was observed with a confocal laser scanning microscope (Leica TCS-SP4).
Results
Chemical composition and chain-length distribution of the cuticular wax in wheat leaf blade
The chemical composition of the wheat leaf surface was analysed by GC-MS. Three or four leaf blades were obtained from the different seedling and flowering plants of wheat cv Xinong 2718, which is derived from Shaanxi Province, China. During the seedling stage (~80 d old), the leaf blades exhibited 594.9±21.0 μg dm–2 of total wax load. Primary alcohols were the major components of the wax extract, accounting for 78.4±0.5% of the total wax load, followed by alkanes (11.3±0.7%), fatty acids (1.6±0.2%), aldehydes (1.2±0.2%), and esters (0.9±0.1%) (Table 1). The chain lengths of primary alcohols ranged from C20 to C32, of which C28 was the most dominant. Chain length distributions of alkanes varied from C23 to C33, with C27 being the dominant chain length. The major fatty acid constituent was C26, and lesser amounts of C20, C22, C24, and C28 fatty acids were found (Fig. 2A).
Table 1.
Amount and percentage of cuticular wax in leaf blades of seedling and flowering wheat cv Xinong 2718Mean values (μg dm–2) of total wax loads and coverage of individual compound classes are given with the SD (n=3).
| Wax composition | Measurement | Seedling | Flowering |
|---|---|---|---|
| Fatty acids | Amount | 9.8±1.1 | 17.8±1.4 |
| Percentage | 1.6±0.2 | 2.2±0.1 | |
| Aldehydes | Amount | 7.5±0.8 | 9.7±1.7 |
| Percentage | 1.2±0.2 | 1.2±0.1 | |
| Alkanes | Amount | 66.9±3.6 | 145.4±9.8 |
| Percentage | 11.3±0.7 | 18.3±0.8 | |
| β-Disketone | Amount | ND | 260.7±25.8 |
| Percentage | ND | 32.8±2.6 | |
| OH-β-Disketone | Amount | ND | 5.7±1.3 |
| Percentage | ND | 0.7±0.2 | |
| Primary Alcohols | Amount | 466.1±17.2 | 277.7±13.9 |
| Percentage | 78.4±0.5 | 35.0±1.5 | |
| Esters | Amount | 5.5±0.5 | 6.2±0.2 |
| Percentage | 0.9±0.1 | 0.8±0.1 | |
| Unidentified | Amount | 42.4±7.3 | 39.2±5.5 |
| Percentage | 7.1±1.0 | 4.9±0.6 | |
| Total Load | Amount | 594.9±21.0 | 794.8±38.6 |
ND, not detected.
Fig. 2.
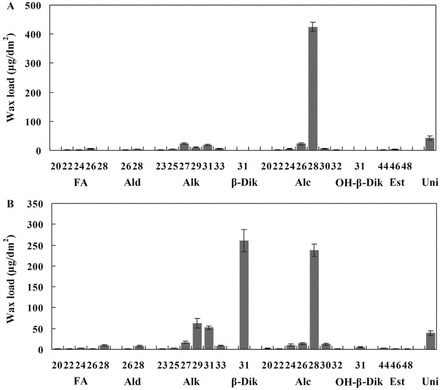
Cuticular wax composition in the leaf blade of wheat cv Xinong 2718. Wax coverage is expressed as μg dm–2 of leaf blade surface area. (A) Composition of the leaf cuticular wax of seedling plants. (B) Composition of the leaf cuticular wax of flowering plants. Each wax constituent is designated by carbon chain length, and is labelled by chemical class along the x-axis. Each value is the mean of three independent measurements of individual plants. FA, fatty acids; Ald, aldehydes; Alk, alkanes; β-Dik, β-diketone; Alc, primary alcohols; OH-β-Dik, hydroxy-β-diketone; Est, esters; Uni, unindentified compounds. Error bars indicate the SD.
After 150 d, the total wax load on leaf blades of flowering Xinong 2718 increased to 794.8±238.6 μg dm–2. It was mainly composed of primary alcohols (35.0±1.5%) and β-diketone (32.8±2.6%), followed by alkanes (18.3±0.8%), fatty acids (2.2±0.1%), aldehydes (1.2±0.1%), and esters (0.8±0.1%) (Table 1). It is interesting to note that although primary alcohols remained the major component of the cuticular waxes, the relative proportions of primary alcohols decreased 43.4% compared with seedling primary alcohols. The most drastic changes in wax composition occurred during the flowering stage: the percentages of β-diketone and OH-β-diketone increased significantly from 0 to 32.8±2.6% and from 0 to 0.7±0.2%, respectively (Table 1). The percentages of alkanes and fatty acids also increased relatively. The flowering leaf blades exhibited a similar chain length distribution of primary alcohols to that observed in the seedling leaf blades; even-numbered chain lengths ranging from C20 to C32 were observed, and C28 chains predominated (Fig. 2B). Furthermore, the chain length distribution of alkanes ranged from C23 to C33, and a shift in the dominant chain length towards C29 could be observed. This trend also occurred for fatty acids, the chain lengths of which ranged from C20 to C28, with a shift in the dominant chain length from C26 to C28. A single carbon chain length, C31, was detected for β-diketone and OH-β-diketone (Fig. 2B). The developmental changes in composition and chain length distribution confirm that wax biosynthesis occurs largely during leaf surface expansion.
Cuticular wax morphology on the leaf blades of wheat
To gain insight into the micromorphology of cuticular wax, the wax crystallites deposited on the adaxial and abaxial surfaces of seedling and flowering flag leaf blades were examined by SEM. The cuticular wax structure and density were similar on both the adaxial and abaxial sides of seedling leaf surfaces, which all formed platelets and were connected to a dense network (Fig. 3A–F). At the seedling stage, SEM showed no differences in wax morphology between the adaxial and abaxial leaf surfaces of the leaf blades. Strikingly, a clearly different morphology of wax deposition was observed between the adaxial and abaxial surfaces of flowering flag leaf blades. The adaxial cuticular wax layer was characterized by a covering of platelet-shaped wax crystals with irregular margins, some of which were connected to their neighbouring crystals. In contrast, the rod-shaped wax crystallites deposited on the abaxial surface densely covered the guard cells (Fig. 3G–L). Tulloch et al. (1980) detected a different morphology of wax deposition between adaxial and abaxial surfaces of the flag leaf in bread wheat. Wax morphology is determined by wax composition (Zhang et al., 2013). Since the amount of β-diketone significantly increased from 0 μg dm–2 at the seedling stage to 277.7 μg dm–2 at anthesis, it is inferred that the wax composition of rod-shaped wax crystallites may be comprised of β-diketone. These results indicate that the cuticular wax crystals on the leaf blades change with developmental stages in wheat.
Fig. 3.
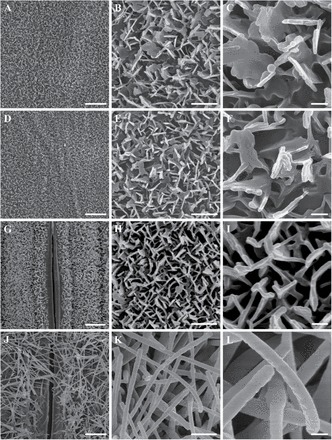
SEM analysis of the wax crystals on the leaf surface of wheat cv Xinong 2718 at the seedling stage and flowering stage. (A–C) The adaxial sides of leaf blades at the seedling stage. (D–F) The abaxial sides of leaf blades at the seedling stage. (G–I) The adaxial sides of leaf blades at the flowering stage. (J–L) The abaxial sides of leaf blades at the flowering stage. A, D, G and J were detected by SEM at ×2000 magnification. B, E, H and K were detected by SEM at ×10 000 magnification. C, F, I and L were detected by SEM at ×30 000 magnification. Scale bars=4 μm (A, D, G, and J), 1 μm (B, E, H, and K), 0.2 μm (C, F, I, and L).
Isolation of TaFAR5 from wheat
The study demonstrates that primary alcohols are the most important components of cuticular waxes on the surface of wheat leaves. In order to isolate the primary alcohol synthetic gene in wheat cuticle, a BLASTN search of the wheat GenBank database was performed using the open reading frame (ORF) nucleotide sequence of Arabidopsis CER4 (GenBank accession no. NP_567936), which encodes a FAR involved in primary alcohol formation (Rowland et al., 2006). Eight sequences in the wheat genome exhibited a significant similarity to CER4 over their entire length, one of which was TAA1b, a cDNA that has previously been found to be expressed specifically in sporophytic tapetum cells (Wang et al., 2002). However, the cuticular wax synthesis function of TAA1b remains unknown. TAA1b is referred to hereafter as TaFAR5. PCR amplification was performed with a degenerate primer designed according to identified cDNA and gDNA sequences derived from leaf blades of wheat cv Xinong 2718 (Fig. 4A), and then the products were sequenced. The full-length cDNA of TaFAR5 is 1949bp and contains a 1569bp coding region, a 73bp 5’-untranslated region, and a 308bp 3’-untranslated region. The full-length TaFAR5 spans 4237bp of genomic DNA and contains eight exons and seven introns. The exons range from 91bp to 413bp, and the introns range from 78bp to 1758bp (Fig. 4B).
Fig. 4.
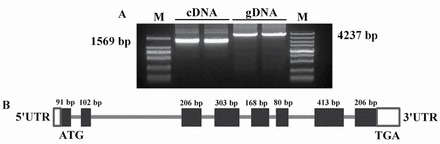
Homology cloning of TaFAR5. (A) Amplification of TaFAR5 from wheat cv Xinong 2718. (B) Schematic representation of the TaFAR5 gene structure. The thick black bars represent exons with the predicted translation start site (ATG) and stop code (TGA); the thin lines represent the introns. White boxes indicate the 5’- and 3’-untranslated regions (UTRs). gDNA, genomic DNA; M, DNA ladder DL2000 (left) and DL5000 (right).
Wheat TaFAR5 expression in yeast and transgenic tomato leaves results in primary alcohol production
To characterize the catalytic function of TaFAR5, its coding region was cloned into a yeast expression vector pYES2 under the control of the GAL1-inducible promoter. Subsequently, the recombinant plasmid was transformed into the yeast mutant strain INVSc1 (Invitrogen), which is deficient in storage lipid biosynthesis, thereby serving as a good host system to examine the involvement of TaFAR5 in wax biosynthesis. The recombinant strains containing pYES2-TaFAR5 or the empty vector as a negative control were induced with galactose for 24h, and then lipophilic compounds were extracted with hexane. An analysis of the wax indicated that the yeast cells that were transformed with TaFAR5 produced C22:0 primary alcohol (based on the GC-MS characteristics); in contrast, no primary alcohols were detected when using the empty vector control (Fig. 5A). This result confirms that TaFAR5 protein possesses a FAR activity, directly catalysing the reduction of fatty acids to primary alcohols using an endogenous C22:0 fatty acid as a substrate.
Fig. 5.
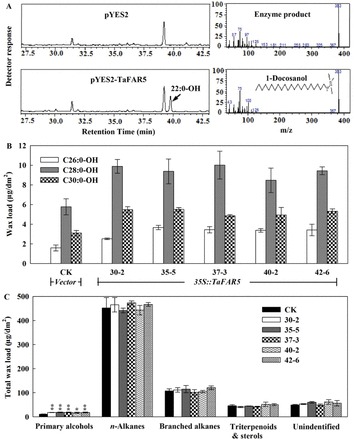
Heterologous expression of TaFAR5 in yeast and tomato. (A) GC analysis of primary alcohols in transgenic yeast. Lipids were hexane extracted from yeast cells and analysed by GC-MS. In the empty vector control, no primary alcohols were detected. In contrast, the yeast strains expressing TaFAR5 were found to contain the C22:0 primary alcohol. (B) Primary alcohol accumulation in mature leaves of transgenic T1 generation tomato cv MicroTom. (C) Quantitative analysis of total leaf wax load. CK is the empty pCXSN vector control. The other lines contain the TaFAR5 coding region under the control of the 35S promoter. Significant differences are marked with asterisks (t-test: *P<0.05; **P<0.01).
Furthermore, TaFAR5 coding region fragments were subcloned into the binary vector pCXSN (Chen et al., 2009), and transformed into tomato cv MicroTom under the transcriptional control of the Cauliflower mosaic virus (CaMV) 35S promoter via A. tumefaciens infiltration (Supplementary Fig. S1 at JXB online). The transgenic lines harbouring the empty vector were used as control. No significant morphological differences were found between T1 transgenic lines carrying the TaFAR5 gene and control lines. Subsequently, the lipidic compositions of chloroform-extractable leaf cuticular waxes derived from the transgenic lines were analysed by GC-MS. As anticipated, the total primary alcohol content per leaf area was dramatically higher in five independent T1 transgenic lines harbouring pCXSN-TaFAR5 (Fig. 5B), whereas the contents of n-alkanes, branched alkanes, and triterpenoids were only slightly affected in the transgenic leaves (Fig. 5C). No new compound was identified in the transgenic lines. The largest change in the primary alcohol deposition was observed in transgenic line 37-3, which exhibited an increase of 75.3% in the amount of total primary alcohol, with a 116.9% increase in C26:0 primary alcohol, a 73.6% increase in C28:0 primary alcohol, and a 57.0% increase in C30:0 primary alcohol. These primary alcohols are the major components of the primary alcohols present (Fig. 5B). In addition, to determine further whether the quantitative differences in the primary alcohol content in the transgenic lines were caused by an alteration of the crystal pattern, TaFAR5 transgenic lines and empty vector control plants were examined by SEM. A comparison of the leaf cuticular wax crystalline patterns showed no significant differences between the transgenic lines and the control plants, the waxes of which were deposited as a smooth film without any crystalline structures present (Supplementary Fig. S2A–D). It was inferred that the increased primary alcohol content was not sufficient to form wax crystals on the leaf surface of tomato. Similar to the results for transgenic tomato leaves, the total primary alcohols of fruits, including C28:0-OH, C30:0-OH, C32:0-OH, and C34:0-OH, showed a significant increase in three independent transgenic lines (data not shown). Taken together, these results suggest that TaFAR5 synthesizes primary alcohol of the plant cuticle.
Characterization of the predicted TaFAR5 protein
Sequence analysis indicated that TaFAR5 encodes a polypeptide of 522 amino acids. Protein domain searches against the Pfam database revealed that the deduced protein TaFAR5 contains an NAD_binding_4 (NADB) domain and a Sterile domain (Fig. 6A). Previous reports revealed that EgFAR (Euglena gracilis) and DPW (rice) both contain an NADB domain, and a Male Sterile 2 (MS2) domain encodes a FAR; therefore, it is possible that TaFAR5 encodes a FAR protein that is involved in the synthesis of primary alcohol. Based on TMHMM program analysis, TaFAR5 appears to be an integral membrane protein possessing a highly conserved transmembrane-spanning domain at its C-terminus (data not shown). To determine the molecular mass of TaFAR5, the coding region was cloned into the pET28a vector (Novagen) and expressed in E. coli BL21 (DE3). Recombinant strains containing pET-TaFAR5 or the empty vector as a control were induced with 0.5mM isopropyl-β-d-1-thiogalactopyranoside for 48h and shaken slowly at 20 °C to improve expression of TaFAR5. In agreement with the predicted size, the molecular mass of TaFAR5 protein was estimated to be 58.4kDa by SDS–PAGE analysis (Fig. 6B). Multiple alignments were performed for the eight plant proteins with high similarity to the TaFAR5 protein. TaFAR5 exhibits 74% identity with TAA1a and 74% identity with TAA1c, which are the most similar orthologues to TaFAR5, and ~40% identity with five Arabidopsis homologues CER4, FAR1, FAR4, FAR5, and FAR8 (Fig. 6C). Phylogenetic analysis of the 18 proteins with high similarities to TaFAR5 showed that these proteins can be grouped into two clades. TaFAR5, TAA1a, TAA1c, and two rice proteins were grouped into the first clade. Five Arabidopsis proteins (CER4, FAR1, FAR4, FAR5, and FAR8) belong to the second clade. Interestingly, the monocot proteins form one clade, and the dicot proteins form the second clade (Fig. 6D). These results suggest that TaFAR5 and its homologues play key roles in primary alcohol biosynthesis among divergent monocot and dicot plants.
Fig. 6.
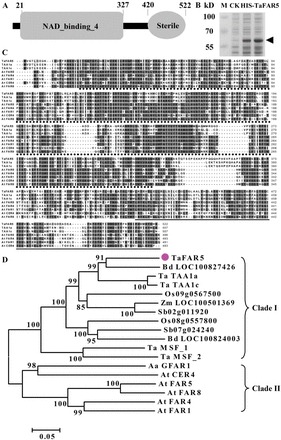
A schematic diagram illustrating the functional domains of TaFAR5, and multiple alignments and phylogenic analysis of TaFAR5-related proteins. (A) Protein structure of TaFAR5. (B) SDS–PAGE of TaFAR5 protein. Arrows indicate the HIS-TaFAR5 protein. (C) Multiple alignment of wheat TaFAR5 with seven related proteins with the ClustalX 1.83 program using default parameters. Identical and similar amino acid residues are shown on black and grey backgrounds, respectively. Gaps required for optimal alignment are indicated by dashes. NAD_binding_4 and Sterile domains are indicated by stars and grey lines under the sequences, respectively. (D) Phylogenetic tree of the TaFAR5-related proteins constructed by MEGA 3.1 with the Neighbor–Joining method. The number for each interior branch shows the percentage of the bootstrap value (1000 replicates), and only values >50% are shown. Homologous genes have the following accession numbers: Triticum aestivum Ta TAA1a (AJ459249), Ta TAA1c (AJ459253), Ta MSF_1 (CBI75514), and Ta MSF_2 (CBI75517); Brachypodium distachyon Bd LOC100827426 (XP_003578677) and Bd LOC100824003 (XP_003574925); Oryza sativa Os09g0567500 (NP_001063962) and Os08g0557800 (NP_001062488); Sorghum bicolor Sb02g011920 (XP_002459832) and Sb07g024240 (XP_002445686); Zea mays Zm LOC100501369 (NP_001183038); Arabidopsis thaliana At CER4 (NP_567936), At FAR1 (NP_197642), At FAR4 (NP_190040), At FAR5 (NP_190041), and At FAR8 (NP_190042); Artemisia annua Aa GFAR1 (ADK66305).
Expression pattern of TaFAR5 and the subcellular localization of the TaFAR5 protein
In order to gain a better understanding of the function of TaFAR5, its expression pattern was examined by quantitative RT-PCR analysis. Total RNA was isolated from various vegetative and reproductive organs of wheat cv Xinong 2718. As shown in Fig. 7A, a high level of TaFAR5 transcript was detected in leaf blades, anthers, pistils, and seeds of the wheat, and modest transcript levels were observed in internodes, leaf sheaths, young panicles, and glumes. Meanwhile, a low level of TaFAR5 transcript was detected in the roots. It is possible that TaFAR5 catalyses the production of primary alcohols in leaf blades, where primary alcohols were detected.
Fig. 7.
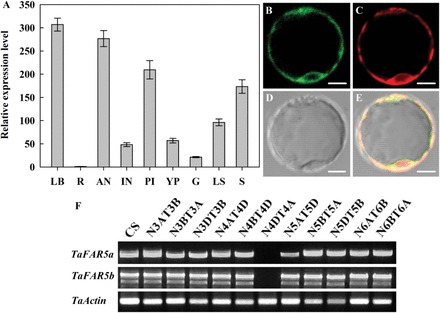
Expression analysis of TaFAR5 and subcellular localization of the TaFAR5 protein. (A) Quantitative RT-PCR analysis showing the expression of TaFAR5. LB, young leaf blades; LS, leaf sheaths; R, roots; IN, internodes; YP, young panicles; AN, anthers; PI, pistils; G, glumes; S, seeds. Wheat TaActin was used as reference. Error bars indicate the SE of three independent samples. (B–E) Subcellular localization of TaFAR5 in the rice protoplast. (B) GFP fluorescence of the protoplast cell expressing 35S:TaFAR5–GFP. (C) ER marker mCherry–HDEL. (D) Bright-field image of the protoplast cell. (E) A merged image of B, C, and D. Scale bars=5 μm. (F) Chromosomal localization of TaFAR5. RT-PCR was performed on 11 nullisomic–etrasomic (N-T) lines of wheat cultivar Chinese Spring with two pairs of primers, TaFAR5a and TaFAR5b, which specifically amplify different chromosomal assignment of TaFAR5, respectively. (This figure is available in colour at JXB online.)
Most of the known wax synthesis-related enzymes are located in the ER (Xu et al., 2002; Zheng et al., 2005; Rowland et al., 2006; Greer et al., 2007; Haslam et al., 2012; Mao et al., 2012; Rajangam et al., 2013). To investigate the subcellular localization of TaFAR5, TaFAR5 was transiently expressed in rice leaf protoplasts. The C-terminus of the TaFAR5 coding sequence was fused with GFP under the control of the CaMV 35S promoter, and the TaFAR5–GFP construct was transferred into rice leaf protoplasts by the PEG-mediated method. To mark the ER, a red fluorescent protein (RFP) mCherry–HDEL was co-transformed into the protoplasts (Nelson et al., 2007; Mao et al., 2012). As expected, overlap of the GFP signal generated by TaFAR5–GFP and the RFP signal generated by HDEL–RFP results in an orange fluorescent signal (Fig. 7B–E). Fluorescence microscopy revealed that the ER marker fluorescence and the TaFAR5–GFP fluorescence completely overlapped, indicating that TaFAR5 is located in the ER.
TaFAR5 is located on wheat chromosome 4D
To determine the chromosomal location of TaFAR5, a query of the wheat genome database (http://wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST) with the genomic sequence of TaFAR5 using BLAST search programs revealed that the entire 4237bp of TaFAR5 sequence completely matches the sequence of the wheat chromosome 4DS contig, without nucleotide mutation, indicating that TaFAR5 is located on wheat chromosome 4DS. In addition, a set of 11 wheat cv Chinese Spring nullisomic–tetrasomic substitution lines were used as templates for cDNA amplification using two pairs of TaFAR5-specific primers, named TaFAR5a and TaFAR5b, respectively. As shown in Fig. 7F, no PCR product was detected in N4DT4A which lacked the whole of chromosome 4A and had the whole of chromosome 4D added; in contrast, the PCR product was obtained from all of the other 10 Chinese Spring nullisomic–tetrasomic substitution lines. Consistent with the result of the BLAST search, TaFAR5 was mapped to wheat chromosome 4DS.
Transcriptional regulation of TaFAR5 under abiotic stress and ABA treatments
Previous investigations have established that Arabidopsis wax synthesis is induced by water deficit, sodium chloride, and ABA treatments (Kosma et al., 2009). To examine how the level of TaFAR5 transcript is induced, the abundance of TaFAR5 under stress conditions was analysed. Quantitative RT-PCR was performed to measure the abundance of TaFAR5 transcripts in 4-week-old chamber-grown seedlings of wheat cv Xinong 2718 subjected to various stress treatments at six different times (0, 2, 4, 6, 12, and 24h). When seedlings were exposed to the air, dehydration led to an increase of the relative abundance of TaFAR5 transcripts by 2.8-fold for up to 2h compared with 0h, and then dropped to normal levels by 4h (Fig. 8A). Consistent with the dehydration induction, TaFAR5 transcripts increased 2.6- and 2.2-fold within 4h under 20% PEG 6000 and cold treatments, respectively (Fig. 8B, C). Based on these observations, it is concluded that transcriptional control plays a major role in the regulation of TaFAR5 during drought and cold stress. ABA is an important signal in abiotic stress responses. Previous studies have shown that some wax genes are induced in ABA-dependent stress signalling. To explore this notion further, the ABA-induced expression of TaFAR5 was analysed in 4-week-old wheat seedlings exposed to 100 μM ABA treatments. As expected, TaFAR5 expression exhibited a continual rise up to at least 4h after induction by ABA, thereby demonstrating that TaFAR5 is induced by drought and cold stresses in an ABA-dependent manner (Fig. 8D).
Fig. 8.
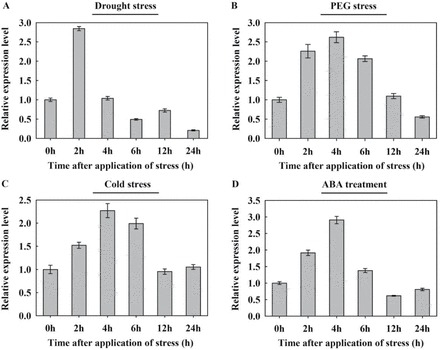
Expression analysis of TaFAR5 in wheat cv Xinong 2718 seedlings under abiotic stress and ABA treatment. Four-week-old soil-grown plants were exposed to various stress treatments. Relative transcript levels were determined by quantitative RT-PCR using the TaActin gene as an internal control. Error bars represent the SE of three independent experiments. Drought treatment, plants were removed from the soil and allowed to dry on filter paper; PEG treatment, 20% (w/v) PEG 6000; cold treatment, 4 °C; ABA treatment, 100 μM ABA.
Discussion
Analysis of primary alcohols in cuticular wax from wheat leaf blades has shown that they are all saturated with C20–C32 carbons, and C28 carbons are the most frequent species (Bianchi and Figini, 1986; Koch et al., 2006; Adamski et al., 2013; Zhang et al., 2013). Very little is known about the importance of FAR enzymes in governing the composition of the primary alcohols in wheat cuticle. The principal goal of this study was to identify and characterize a primary alcohol biosynthesis-related gene that is involved in the acyl reduction pathway. Here, a new function for TaFAR5, which is identical to TAA1b, an anther-specific gene (Wang et al., 2002), is shown, namely its involvement in the primary alcohol biosynthesis of leaf blade cuticular wax in wheat.
The present study shows that the components, amount, and micromorphology of cuticular waxes vary considerably during various stages of growth, and primary alcohols are major components of cuticular waxes in the leaf blades of wheat. These results are consistent with those of Tulloch (1973), who revealed that primary alcohols are major components of wax from leaf blades of spring wheat varieties Selkirk and Manitou. Before sheaths and flag leaf are completely developed, the major component is octacosanol. When sheath development is complete, β-diketone content is greatest. Additional studies in other species have also supported the present view. Cuticular wax of leaves in Hedera helix contained high percentages of alkanes, aldehydes, primary alcohols, and fatty acids early on, and increasing amounts of alkyl esters during seasonal development (Hauke and Schreiber, 1998). In fact, primary alcohols are always the major components of cuticular waxes on wheat leaf blades; when β-diketone is present, the primary alcohol content is correspondingly lower. Based on the observed differences in the component and micromorphology of wheat leaf blades, it is proposed that platelet-shaped wax crystals are associated with primary alcohols, and rod-shaped wax crystallites are characteristic of β-diketones. This view is supported by the findings that lobed plates of cuticular wax on Quercus robur are associated with primary alcohols (Gülz, 1994), and long thin tubes are characteristic of β-diketones on Hordeum vulgare (barley) lemma (von Wettstein-Knowles, 1995).
The TaFAR5 gene cloned from wheat leaf blade encodes a FAR with a predicted NADB domain and a Sterile domain. SDS–PAGE confirmed that TaFAR5 has a molecular mass of 58.4kDa, which is similar to the molecular masses of Arabidopis CER4 (Rowland et al., 2006), jojoba FAR (Metz et al., 2000), and E. gracilis EgFAR (Teerawanichpan and Qiu, 2010). Sequence analysis revealed that TaFAR5 shares ~40% identity with five Arabidopsis homologues, CER4, FAR1, FAR4, FAR5, and FAR8. Therefore, it is most likely that TaFAR5 enzyme has a similar function in catalysing primary alcohol synthesis to Arabidopis FARs, jojoba FAR, and E. gracilis EgFAR. Indeed, two pieces of evidence clearly indicate that TaFAR5 functions as a FAR enzyme in primary alcohol biosynthesis for the production of cuticular waxes. First, functional analysis of TaFAR5 in yeast indicated that TaFAR5 effectively converts C22:0 fatty acid to the corresponding primary alcohol. Secondly, the expression of TaFAR5 in transgenic tomato results in the accumulation of C26:0, C28:0, and C30:0 primary alcohols in leaves. Previous studies have demonstrated that expression of CER4 in yeast results in the production of C24:0-OH and C26:0-OH (Rowland et al., 2006; Domergue et al., 2010). Moreover, in yeast, recombinant FAR1, FAR4, and FAR5 are able to form alcohols using distinct but overlapping substrates with a chain length ranging from C18:0 to C24:0 (Domergue et al., 2010). Expression of E. gracilis EgFAR in yeast led to the accumulation of C14:0 and C16:0 primary alcohols (Teerawanichpan and Qiu, 2010), and expression of jojoba FAR in E. coli resulted in accumulation of C16:0-OH and C18:1-OH (Metz et al., 2000). TaFAR5 possesses a narrower substrate range than other biochemically characterized FARs and only uses saturated fatty acid with a C22 carbon chain as substrate, with the C22:0 fatty acid preferred. Interestingly, it was found that TaFAR5 has very distinct substrate specificities between yeast and tomato, with C22:0 fatty acid preferred in yeast, but C26:0, C28:0, and C30:0 fatty acids preferred in tomato. Previous studies have established that wheat TAA1a can produce C16:0 and C18:1-OH when expressed in E. coli, but that transgenic tobacco accumulates C18:1, C24:0, and C26:0 primary alcohols (Wang et al., 2002). Expression of CER4 in yeast results in the production of C24:0-OH and C26:0-OH, but molecular complementation of cer4-1 produced C24:0, C26:0, C28:0, and C30:0 primary alcohols (Rowland et al., 2006). This raises the questions of to what extent the specificity of the FAR governs the composition of the primary alcohol produced in the plant, and to what extent this specificity is governed by FAR in yeast cells.
As anticipated, TaFAR5 expression was detected most prominently in leaf blades, consistent with a role for this gene in cuticular wax production. Surprisingly, TaFAR5 expression was not restricted to the epidermis of aerial parts. Its expression was also detected in anthers, pistils, and seeds, and this result led the authors to suggest that TaFAR5 is implicated not only in wax metabolism but also in the anther cutin biosynthesis pathway. Indeed, TaFAR5 has previously been reported as an anther-specific gene (Wang et al., 2002). Moreover, this is not the first study in which anther expression of genes associated with wax metabolism has been reported. Other genes involved in cuticle formation are also expressed in anthers, namely CER1 (Aarts et al., 1995), CER2 (Xia et al., 1996), CER3 (Hannoufa et al., 1996), WAX2 (Chen et al., 2003), WIN1 (Broun et al., 2004), and CER8 (Lü et al., 2009). Additional RNA interference experiments in wheat are required to determine whether TaFAR5 is implicated in anther cutin biosynthesis. Enzymes that catalyse the initial steps of wax synthesis, the formation of VLCFA wax precursors, are associated with the ER in all plant species investigated to date (Samuels et al., 2008). It was likewise anticipated that if TaFAR5 were involved in primary alcohol synthesis, this protein would localize to the ER. Visualization of the functional TaFAR5–GFP fusion protein in rice leaf protoplasts revealed that the wheat TaFAR5 is also localized to the ER. In Arabidopsis, the midchain hydroxylase MAH1 that catalyses the final step of the decarbonylation pathway, the formation of secondary alcohols and ketones, was also confined to the ER of stem epidermal cells (Greer et al., 2007). The diacylglycerol acyltransferase WSD1 which catalyses the formation of final alkyl esters of the acyl reduction pathway of cuticular wax biosynthesis, resides in the ER membranes (Li et al., 2008). Accordingly, it is proposed that the ER is the subcellular compartment in which most of the cuticular wax components are deposited. These wax components are then delivered from this compartment to the plasma membrane for export toward the cuticle (Samuels et al., 2008).
Cuticular waxes are known to play an important role in plant resistance to environmental stresses, such as water-limited environments (Kosma and Jenks, 2007). In Arabidopsis, cuticle composition and gene expression indicate that wax synthesis is induced by water deficit, sodium chloride, and ABA treatments (Kosma et al., 2009). This suggests that TaFAR5 plays a vital role in stress response, including drought and cold stresses, as its expression was induced under these conditions. Like TaFAR5, several genes encoding enzymes involved in wax biosynthesis are induced by osmotic stresses. CER1, with a function in alkane synthesis, was highly induced by water deficit, NaCl, and ABA (Kosma et al., 2009; Bourdenx et al., 2011). CER6, a VLCFA-condensing enzyme, was induced by osmotic stress and the presence of ABA (Hooker et al., 2002). DSO is also induced by multiple stresses, including ABA, high salt, and glucose (Alvarado et al., 2004; Panikashvili et al., 2007). Nevertheless, the molecular events implicated in these regulations have not yet been established.
In conclusion, this study presents strong evidence that TaFAR5 plays an important role in leaf surface wax synthesis in the monocot wheat. Furthermore, TaFAR5 is linked to responses to drought and cold stresses in an ABA-dependent manner.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Genetic transformation of TaFAR5 in tomato cv MicroTom driven by the CaMV 35S promoter.
Figure S2. SEM images of cuticular wax crystal patterns on tomato leaves.
Table S1. Sequences of the primers used for genomic PCR, qRT-PCR, yeast expression, and subcellular localization experiments in this study.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31271794 to ZW), start-up funding from Northwest A&F University (Z109021113 to WY), and the Natural Science Foundation of Shaanxi Province, China (2014JQ3089 to WY). We thank Dr Kunneng Zhou and Professor Jianmin Wan (Nanjing Agricultural University) for their generous help with the subcellular localization assay. We would also like to thank the anonymous reviewers for their constructive comments on improving the manuscript.
Glossary
Abbreviations:
- ER
endoplasmic reticulum
- FAR
fatty acyl-coenzyme A reductase
- GC-MS
gas chromatography–mass spectroscopy
- SEM
scanning electron microscopy
- VLCFA
very long chain fatty acid.
References
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. 1997. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. The Plant Journal 12, 615–623. [DOI] [PubMed] [Google Scholar]
- Aarts MG, Keijzer CJ, Stiekema WJ, Pereira A. 1995. Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. The Plant Cell 7, 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamski NM, Bush MS, Simmonds J, Turner AS, Mugford SG, Jones A, Findlay K, Pedentchouk N, von Wettstein-Knowles P, Uauy C. 2013. The Inhibitor of wax 1 locus (Iw1) prevents formation of β- and OH-β-diketones in wheat cuticular waxes and maps to a sub-cM interval on chromosome arm 2BS. The Plant Journal 74, 989–1002. [DOI] [PubMed] [Google Scholar]
- Alvarado MC, Zsigmond LM, Kovacs I, Cseplo A, Koncz C, Szabados LM. 2004. Gene trapping with firefly luciferase in Arabidopsis. Tagging of stress-responsive genes. Plant Physiology 134, 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EA. 1982. Chemistry and morphology of plant epicuticular waxes. In: Cutler D, Alvin KL, Price CE, eds. The plant cuticle. London: Academic Press, 139–165. [Google Scholar]
- Barthlott W, Neinhuis C, Cutler D, Ditsch F, Meusel I, Theisen I, Wilhelmi H. 1998. Classification and terminology of plant epicuticular waxes. Botanical Journal of the Linnean Society 126, 237–260. [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M. 2012. Solving the puzzles of cutin and suberin polymer biosynthesis. Current Opinion in Plant Biology 15, 329–337. [DOI] [PubMed] [Google Scholar]
- Bianchi G, Figini ML. 1986. Epicuticular waxes of glaucous and nonglaucous durum wheat lines. Journal of Agricultural and Food Chemistry 34, 429–433. [Google Scholar]
- Bianchi G, Murelli C, Ottaviano E. 1990. Maize pollen lipids. Phytochemistry 29, 739–744. [Google Scholar]
- Bourdenx B, Bernard A, Domergue F, et al. 2011. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiology 156, 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. 2004. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis . Proceedings of the National Academy of Sciences, USA 101, 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. 2006. A highly efficient transient protoplast system for analyzing defence gene expression and protein–protein interactions in rice. Molecular Plant Pathology 7, 417–427. [DOI] [PubMed] [Google Scholar]
- Chen SB, Songkumarn P, Liu JL, Wang GL. 2009. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiology 150, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA. 2003. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. The Plant Cell 15, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JB, Russell DW. 2004. Mammalian wax biosynthesis: I. Identification of two fatty acyl-coenzyme A reductases with different substrate specificities and tissue distributions. Journal of Biological Chemistry 279, 37789–33797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan Y, Yan H, Munyikwa T, Dong J, Zhang Y, Armstrong CL. 2006. MicroTom: a high-throughput model transformation system for functional genomics. Plant Cell Reports 25, 432–441. [DOI] [PubMed] [Google Scholar]
- Doan TT, Carlsson AS, Hamberg M, Bülow L, Stymne S, Olsson P. 2009. Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli . Journal of Plant Physiology 166, 787–796. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Møller BL, Preuss D. 2009. CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiology 151, 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue F, Vishwanath SJ, Joubès J, et al. 2010. Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiology 153, 1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods in Enzymology 350, 87–96. [DOI] [PubMed] [Google Scholar]
- Greer S, Wen M, Bird D, Wu X, Samuels L, Kunst L, Jetter R. 2007. The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiology 145, 653–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gülz PG. 1994. Epicuticular leaf waxes in the evolution of the plant kingdom. Journal of Plant Physiology 143, 453–464. [Google Scholar]
- Hall DA, Jones RL. 1961. Physiological significance of surface wax on leaves. Nature 191, 95–96. [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98. [Google Scholar]
- Hannoufa A, Negruk V, Eisner G, Lemieux B. 1996. The CER3 gene of Arabidopsis thaliana is expressed in leaves, stems, roots, flowers and apical meristems. The Plant Journal 10, 459–467. [DOI] [PubMed] [Google Scholar]
- Haslam TM, Mañas-Fernández A, Zhao LF, Kunst L. 2012. Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiology 160, 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauke V, Schreiber L. 1998. Ontogenetic and seasonal development of wax composition and cuticular transpiration of ivy (Hedera helix L.) sun and shade leaves. Planta 207, 67–75. [Google Scholar]
- Hooker TS, Millar AA, Kunst L. 2002. Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiology 129, 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffree CE. 2006. The fine structure of the plant cuticle. In: Riederer M, Müller C, eds. Annual plant reviews 23: biology of the plant cuticle. Oxford: Blackwell, 11–125. [Google Scholar]
- Jenks MA, Ashworth EN. 1999. Plant epiculticular waxes: function, production, and genetics. Horticultural Reviews 23, 1–68. [Google Scholar]
- Jenks MA, Joly RJ, Peters PJ, Rich PJ, Axtell JD, Ashworth EA. 1994. Chemically induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) Moench. Plant Physiology 105, 1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstiens G. 2006. Water transport in plant cuticles: an update. Journal of Experimental Botany 57, 2493–2499. [DOI] [PubMed] [Google Scholar]
- Koch K, Barthlott W, Koch S, Hommes A, Wandelt K, Mamdouh W, De-Feyter S, Broekmann P. 2006. Structural analysis of wheat wax (Triticum aestivum, c.v. ‘Naturastar’ L.): from the molecular level to three dimensional crystals. Planta 223, 258–270. [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE. 1970. Reduction of fatty acids to alcohols by cell-free preparation of Euglena gracilis . Biochemistry 9, 1095–1102. [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE. 1971. Enzymatic synthesis of fatty alcohols in Brassica oleracea . Archives of Biochemistry and Biophysics 142, 701–709. [DOI] [PubMed] [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lü S, Joubès J, Jenks MA. 2009. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiology 151, 1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Jenks MA. 2007. Eco-physiological and molecular–genetic determinants of plant cuticle function in drought and salt stress tolerance. In: Jenks MA, Hasegawa PM, Jain SM, eds. Advances in molecular breeding toward drought and salt tolerant crops. Dordrecht, The Netherlands, Springer, 91–120. [Google Scholar]
- Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics 5, 150–163. [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels AL. 2003. Biosynthesis and secretion of plant cuticular wax. Progress in Lipid Research 42, 51–80. [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels L. 2009. Plant cuticles shine: advances in wax biosynthesis and export. Current Opinion in Plant Biology 12, 721–727. [DOI] [PubMed] [Google Scholar]
- Leide J, Hildebrandt U, Reussing K, Riederer M, Vogg G. 2007. The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: effects of a deficiency in a β-ketoacyl-coenzyme A synthase (LeCER6). Plant Physiology 144, 1667–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wu X, Lam P, Bird D, Zheng H, Samuels L, Jetter R, Kunst L. 2008. Identification of the wax ester synthase/acyl-coenzyme A:diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiology 148, 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S, Song T, Kosma DK, Parsons EP, Rowland O, Jenks MA. 2009. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. The Plant Journal 59, 553–564. [DOI] [PubMed] [Google Scholar]
- Mao BG, Cheng ZJ, Lei CL, et al. 2012. Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta 235, 39–52. [DOI] [PubMed] [Google Scholar]
- Metz JG, Pollard MR, Anderson L, Hayes TR, Lassner MW. 2000. Purification of a jojoba embryo fatty acyl-coenzyme A reductase and expression of its cDNA in high erucic acid rapeseed. Plant Physiology 122, 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moto K, Yoshiga T, Yamamoto M, Takahashi S, Okano K, Ando T, Nakata T, Matsumoto S. 2003. Pheromone gland-specific fatty-acyl reductase of the silkmoth, Bombyx mori . Proceedings of the National Academy of Sciences, USA 100, 9156–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, Höfer R, Schreiber L, Chory J, Aharoni A. 2007. The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiology 145, 1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, McKeon T, Gupta LM, Stumpf PK. 1979. Studies on biosynthesis of waxes by developing jojoba seed. II. The demonstration of wax biosynthesis by cell-free homogenates. Lipids 14, 651–662. [Google Scholar]
- Post-Beittenmiller D. 1996. Biochemistry and molecular biology of wax production in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 405–430. [DOI] [PubMed] [Google Scholar]
- Premchandra GS, Saneoka H, Fujita K, Ogata S. 1992. Leaf water relations, osmotic adjustment, cell membrane stability, epicuticular wax load and growth as affected by increasing water deficit in sorghum. Journal of Experimental Botany 43, 1569–1576. [Google Scholar]
- Rajangam AS, Gidda SK, Craddock C, Mullen RT, Dyer JM, Eastmond PJ. 2013. Molecular characterization of the fatty alcohol oxidation pathway for wax-ester mobilization in germinated jojoba seeds. Plant Physiology 161, 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, Kunst L. 2006. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiology 142, 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R. 2008. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annual Review of Plant Biology 59, 683–707. [DOI] [PubMed] [Google Scholar]
- Sieber P, Schorderet M, Ryser U, Buchala A, Kolattukudy P, Métraux JP, Nawrath C. 2000. Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. The Plant Cell 12, 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teerawanichpan P, Qiu X. 2010. Fatty acyl-coA reductase and wax synthase from Euglena gracilis in the biosynthesis of medium-chain wax esters. Lipids 45, 263–273. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloch AP. 1973. Composition of leaf surface waxes of ‘Triticum’ species: variation with age and tissue. Phytochemistry 12, 2225–2232. [Google Scholar]
- Tulloch AP, Baum BR, Hoffman LL. 1980. A survey of epicuticular waxes among genera of Triticeae. 2. Chemistry. Canadian Journal of Botany 58, 2602–2615. [Google Scholar]
- Vioque J, Kolattukudy PE. 1997. Resolution and purification of an aldehyde-generating and an alcohol-generating fatty acyl-CoA reductase from pea leaves (Pisum sativum L.). Archives of Biochemistry and Biophysics 340, 64–72. [DOI] [PubMed] [Google Scholar]
- von Wettstein-Knowles P. 1995. Biosynthesis and genetics of waxes. In: Hamilton RJ, ed. Waxes: chemistry, molecular biology and functions. Dundee: Oily Press, 91–129. [Google Scholar]
- Wang A, Xia Q, Xie W, Dumonceaux T, Zou J, Datla R, Selvaraj G. 2002. Male gametophyte development in bread wheat (Triticum aestivum L.): molecular, cellular, and biochemical analyses of a sporophytic contribution to pollen wall ontogeny. The Plant Journal 30, 613–623. [DOI] [PubMed] [Google Scholar]
- Wang ZH, Guhling O, Yao RN, Li FL, Yeats TH, Rose JK, Jetter R. 2011. Two oxidosqualene cyclases responsible for biosynthesis of tomato fruit cuticular triterpenoids. Plant Physiology 155, 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia YJ, Nicolau BJ, Schnable PS. 1996. Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. The Plant Cell 8, 1291–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Dietrich CR, Lessire R, Nikolau BJ, Schnable PS. 2002. The endoplasmic reticulum-associated maize GL8 protein is a component of the acyl-coenzyme A elongase involved in the production of cuticular waxes. Plant Physiology 128, 924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JK. 2013. The formation and function of plant cuticles. Plant Physiology 163, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Wang W, Li WL. 2013. Genetic interactions underlying the biosynthesis and inhibition of β-diketones in wheat and their impact on glaucousness and cuticle permeability. PLoS One 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L. 2005. Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. The Plant Cell 17, 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


