Abstract
Genome-wide association studies (GWAS) of colorectal cancer (CRC) have identified 23 susceptibility loci thus far. Analyses of previously conducted GWAS indicate additional risk loci are yet to be discovered. To identify novel CRC susceptibility loci, we conducted a new GWAS and performed a meta-analysis with five published GWAS (totalling 7,577 cases and 9,979 controls of European ancestry), imputing genotypes utilising the 1000 Genomes Project. The combined analysis identified new, significant associations with CRC at 1p36.2 marked by rs72647484 (minor allele frequency [MAF] = 0.09) near CDC42 and WNT4 (P = 1.21 × 10−8, odds ratio [OR] = 1.21 ) and at 16q24.1 marked by rs16941835 (MAF = 0.21, P = 5.06 × 10−8; OR = 1.15) within the long non-coding RNA (lncRNA) RP11-58A18.1 and ~500 kb from the nearest coding gene FOXL1. Additionally we identified a promising association at 10p13 with rs10904849 intronic to CUBN (MAF = 0.32, P = 7.01 × 10-8; OR = 1.14). These findings provide further insights into the genetic and biological basis of inherited genetic susceptibility to CRC. Additionally, our analysis further demonstrates that imputation can be used to exploit GWAS data to identify novel disease-causing variants.
Twin studies indicate that heritable factors account for 35% of the variation in risk of developing colorectal cancer (CRC)1. However, only 5% of CRC can be attributed to the inheritance of high-penetrance mutations in the known genes2,3. Genome-wide association studies (GWAS) conducted primarily in European4,5,6,7,8,9,10,11,12 but also Asian13,14,15,16 populations have vindicated the long-held belief that part of the heritable risk of CRC is attributable to common, low-risk variants. These GWAS have provided insights into the biological basis of CRC, highlighting the role of genes within the bone morphogenetic protein signalling pathway (BMP2, BMP4, GREM1 and SMAD7)4,11 and some candidate genes (e.g. CDH1/CDH3), as well as genes not previously implicated in CRC (e.g. POLD3, TERC, CDKN1A and SHROOM2)5,6.
Despite the success of GWAS the risk SNPs so far identified in European populations account for only 8% of the familial CRC risk (Supplementary Table 1). Together with the over-representation of association signals in GWAS strongly suggests that additional risk SNPs remain to be discovered. The statistical power of individual GWAS is limited by the modest effect sizes of the genetic variants and the requirement for a stringent threshold to establish statistical significance in order to avoid type 1 errors. Meta-analysis of GWAS data therefore offers the opportunity to identify new CRC risk loci and provide further insights into tumour biology. Furthermore, imputation of untyped variants in GWAS data using publicly available reference datasets increases the number of variants that can be tested for an association with CRC risk.
To identify new CRC susceptibility loci, we conducted an independent primary scan of CRC using patient samples from the COIN trial and performed a genome-wide meta-analysis with five previously published GWAS. To recover untyped genotypes, thereby maximising the prospects of identifying risk variants, we imputed over 10 million SNPs in the six GWAS datasets, using data from the 1000 Genomes Project17 as reference (see Materials & Methods for details).
Methods
Primary GWAS
The COIN GWAS was based on 2,244 CRC cases (64% male, mean age 61 years, SD = 10) ascertained through two independent Medical Research Council clinical trials of advanced/metastatic CRC; COIN and COIN-B18. Cases were genotyped using Affymetrix Axiom Arrays according to the manufacturer’s recommendations (Affymetrix, Santa Clara, CA 95051, USA), using duplicate samples and sequencing of significantly associated SNPs in a subset of samples to confirm genotyping accuracy. For all SNPs >99% concordant results were obtained. For controls, we made use of Wellcome Trust Case Control Consortium 2 (WTCCC2) Affymetrix 6.0 array data on 2,674 individuals from the UK Blood Service Control Group. Individuals were excluded with: <95% successfully genotyped SNPs (n = 122), discordant sex information (n = 8), classed as out of bounds by Affymetrix (n = 30), duplication or cryptic relatedness (identity by descent >0.185, n = 4), evidence of non-white European ancestry using PCA in conjunction with HapMap samples (n = 130; cut-off based on the minimum and maximum values of the top two principal components of the controls; Supplementary Figure 2). The details of all sample exclusions are provided in Supplementary Figure 3. We excluded SNPs from the analysis with: call rate <95%; different missing genotype rate between cases and controls at P < 10−5; MAF < 0.01; departure from Hardy–Weinberg equilibrium in controls at P < 10−5. The adequacy of the case–control matching and the possibility of differential genotyping of cases and controls were assessed using quantile-quantile (Q–Q) plots of test statistics.
Published GWAS
Statistical and bioinformatic analysis
Analyses were undertaken using R(v3.02)24 and PLINK25 software. The association between each SNP and the risk of CRC was assessed by the Cochran–Armitage trend test. ORs and associated 95% CIs were calculated by unconditional logistic regression. Phasing of GWAS SNP genotypes was performed using SHAPEIT(v2.644)26. Prediction of the untyped SNPs was carried out using IMPUTE(v2.3.0)27 based on the data from the 1000 Genomes Project (Phase 1 integrated variant set, v3.20101123)28 as reference. Imputed data were analyzed using SNPTEST(v2.4.1)29. Association meta-analyses only included markers with info scores >0.4, imputed call rates/SNP > 0.9 and MAFs > 0.01. The fidelity of imputation, as assessed by the concordance between imputed and sequenced SNPs, was examined in a subset of 200 UK cases. Meta-analyses were carried out using META(v2.4-1)30, under an inverse-weighted fixed-effects model using the genotype probabilities from IMPUTE, where a SNP was not directly typed. We calculated Cochran’s Q statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation that was caused by heterogeneity –I2 values ≥75% are considered characteristic of large heterogeneity31. Associations by sex, age and clinico-pathological phenotypes were examined by logistic regression in case-only analyses. The familial relative risk of CRC attributable to each variant was calculated as detailed by Pharoah et al.32 assuming the overall familial risk of CRC, as shown in epidemiological studies, is 2.233.
To explore epigenetic profiles of association signals, we used ChromHMM34. States were inferred from ENCODE Histone Modification data on the CRC cell line HCT116 (DNAse, H3K4me3, H3K4me1, H3K27ac, Pol2 and CTCF)35 binarized using a multivariate Hidden Markov Model.
To examine whether any of the SNPs or their proxies (i.e. r2 > 0.8 in 1000genomes CEU reference panel) annotate putative transcription factor binding/enhancer elements we used the CADD (combined annotation dependent depletion) web-server36. We assessed sequence conservation using: PhastCons (<0.3 indicative of conservation), Genomic Evolutionary Rate Profiling37 (GERP) (−12 to 6, with 6 being indicative of complete conservation) and CADD (>10.0 deemed to be deleterious).
Analysis of TCGA data
To examine for a relationship between SNP genotype and mRNA expression we made use of Tumor Cancer Genome Atlas (TCGA)38 RNA-seq expression and Affymetrix 6.0 SNP data (dbGaP accession number: phs000178.v7.p6) on 223 colorectal adenocarcinoma (COAD) and 75 rectal adenocarcinoma samples using a best proxy where SNPs were not represented directly. Association between normalised RNA counts per-gene and SNP genotype was quantified using the Kruskal-Wallis trend test. The frequency of somatic mutations in CRC was obtained using the CBioPortal for Cancer Genomics39,40 and TumorPortal web servers41.
Pathway analysis
To determine whether any genes mapping to the three newly identified regions act in pathways already over-represented in GWAS regions we utilized the NCI pathway interaction database42. All genes within the LD block containing each tagSNP, or linked to the SNP through functional experiments (MYC) were submitted as a Batch query using the NCI-Nature curated data source.
Assignment of microsatellite instability (MSI), KRAS, NRAS and BRAF status in cancers
Tumour MSI status in CRCs was determined using the mononucleotide microsatellite loci BAT25 and BAT26, which are highly sensitive MSI markers. Samples showing more than or equal to five novel alleles, when compared with normal DNA, at either or both markers were assigned as MSI-H (corresponding to MSI-high)43.
Tumours from the COIN study were screened for mutations in KRAS codons 12, 13, and 61 and BRAF codon 600 by pyrosequencing18. Additionally, KRAS (all three codons), BRAF (codons 594 and 600), and NRAS (codons 12 and 61) were screened for mutations by MALDI-TOF mass array (Sequenom, San Diego, CA, USA)44.
Results
In the primary scan, 2,244 advanced (stage IV) CRC cases ascertained through the Medical Research Council (MRC) trials COIN18 and COIN-B45 were analysed with control data on 2,674 individuals from the WTCCC2 UK National Blood Service Control Group. After applying strict quality control criteria (Materials and Methods), we analysed 234,675 autosomal SNPs for association with CRC risk in 1,950 cases and 2,162 controls. A Q–Q plot of observed versus expected χ2-test statistics showed little evidence for an inflation of test statistics, thereby excluding the possibility of substantive hidden population substructure, cryptic relatedness among subjects or differential genotype calling (inflation factor λ = 1.05; Supplementary Figure 1).
We performed a meta-analysis of our primary scan data with five non-overlapping GWAS case-control series of Northern European ancestry, which have been previously reported (Supplementary Table 2). The adequacy of the case-control matching and possibility of differential genotyping of cases and controls was assessed using Q-Q plots of test statistics. λGC values46 for the UK1, Scotland1, VQ58, CCFR1 and CCFR2 studies were 1.02, 1.01, 1.01, 1.02 and 1.03 respectively (Supplementary Figure 1). Any ethnic outliers or individuals identified as related were excluded (Supplementary Figure 2).
After quality control procedures, the six GWAS provided data on 7,577 CRC cases and 9,979 controls. To maximise the prospects of identifying novel risk variants, we imputed over 10 million variants using 1000 Genomes Project Pilot data as a reference panel. Q-Q plots for all variants post-imputation did not show evidence of substantive over-dispersion introduced by imputation (Supplementary Figure 1).
Meta-analysis
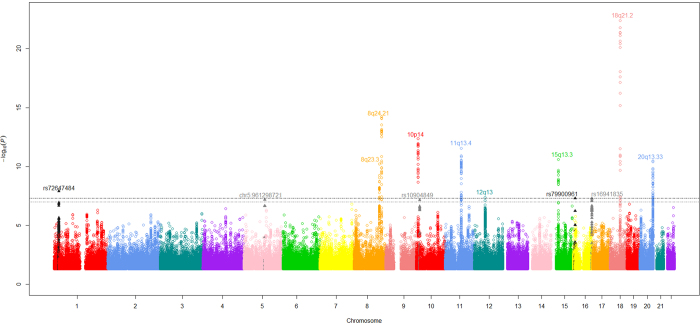
Associations for all 23 established European CRC risk SNPs showed a direction of effect consistent with previously reported studies, with eight of the loci having a P-value of <5.0 × 10−8 (Supplementary Table 3; Fig. 1). Additionally six SNPs previously identified in GWAS in Asian populations as determinants of CRC risk showed evidence for an association in this meta-analysis; albeit at varying degrees of significance (P-values ranging from 3.64 × 10−2 to 1.71 × 10−3; Supplementary Table 3); thereby providing support for trans-ethnic effects.
Figure 1. Genome-wide P-values (–log10P, y-axis) plotted against their respective chromosomal positions (x-axis).
Known regions attaining genome-wide significance (i.e. P = 5.0 × 10−8) are labelled with their chromosomal location. Variants in grey lie in novel regions that reach the significance threshold level (P = 1.0 × 10−7) required for variants to be analysed further in this study. Variants in black lie in novel regions attaining genome-wide significance.
Excluding SNPs (including those correlated with r2 > 0.8) mapping to the risk loci, five variants in distinct regions of linkage disequilibrium (LD) were associated with CRC at P < 1.0 × 10−7 (Table 1; Fig. 1).
Table 1. Summary statistics for variants showing an association with CRC risk at P < 1.0 × 10 −7 .
|
Individual study
P-values |
Meta-analysis |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Nearest gene(s) | SNP | Position (bps) | Alleles | RAF | INFO | UK1 | Scotland1 | VQ58 | CFR1 | CFR2 | COIN | OR (95% CI) | P | Phet |
| 1p36.12 | WNT4/CDC42 | rs72647484 | 22,587,728 | TC | 0.91 | 0.94 (0.85-0.99) | 3.25 ×10−2 | 3.32 ×10−5 | 4.99 ×10−2 | 4.08 ×10−2 | 4.58 ×10−2 | 3.47 ×10−2 | 1.24 (1.15-1.33) | 1.21 ×10-8 | 0.33 |
| 5q15 | ERAP1 | rs202110856 | 96,129,872 | GGC | 0.99 | 0.79 (0.66-0.92) | 2.97 ×10−1 | 5.96 ×10-8 | 2.81 ×10−2 | 4.43 ×10−1 | 3.35 ×10−1 | 3.67 ×10−1 | 1.51 (1.23-1.86) | 6.67 ×10-8 | 0.13 |
| 10p13 | CUBN | rs10904849 | 16,997,266 | GT | 0.68 | 0.98 (0.97-1.00) | 2.90 ×10−2 | 3.39 ×10−1 | 2.36 ×10−2 | 8.68 ×10−3 | 7.73 ×10−2 | 1.29 ×10−3 | 1.13 (1.08-1.19) | 7.01×10-8 | 0.83 |
| 16p13.2 | C16orf72 | rs79900961 | 9,297,812 | GA | 0.98 | 0.70 (0.61-0.74) | 2.21 ×10−1 | 8.68 ×10−2 | 1.04 ×10−3 | 2.54 ×10−2 | 2.41 ×10−1 | 1.02 ×10−3 | 1.49 (1.26-1.76) | 4.93 ×10-8 | 0.76 |
| 16q24.1 | FOXL1 | rs16941835 | 86,695,720 | GC | 0.21 | 0.97 (0.92-0.99) | 1.04 ×10−1 | 1.17 ×10−1 | 1.57 ×10−4 | 3.74 ×10−3 | 1.25 ×10−2 | 3.65 ×10−1 | 1.16 (1.09-1.22) | 5.06 ×10-8 | 0.40 |
For each variant shown along with meta-analysis test statistics are the P-values from the six individual studies and imputation Information scores. Risk alleles are given in bold.
INFO, imputation Information score; P-het, P-value of heterogeneity between studies; RAF, risk allele frequency.
We assessed the fidelity of imputation in 200 UK cases by comparing imputed genotypes with those obtained by sequencing. For the three common variants (MAF > 0.05), rs72647484, rs16941835 and rs10904849 which each had imputation info scores >0.9 there was high correlation between imputed and directly typed genotype (r2 = 0.98, 1.00 and 0.99, respectively). For the rare variant rs79900961 (MAF = 0.016), the correlation was poor (r2 = 0.60). The call rate for the rare Indel on chromosome 5q15 (rs202110856) in the sequencing data was only 71% and both imputed heterozygotes were sequenced as homozygous reference. Therefore, only the three common variants at 1p36.12, 10p13 and 16q24.1 were subject to further analyses.
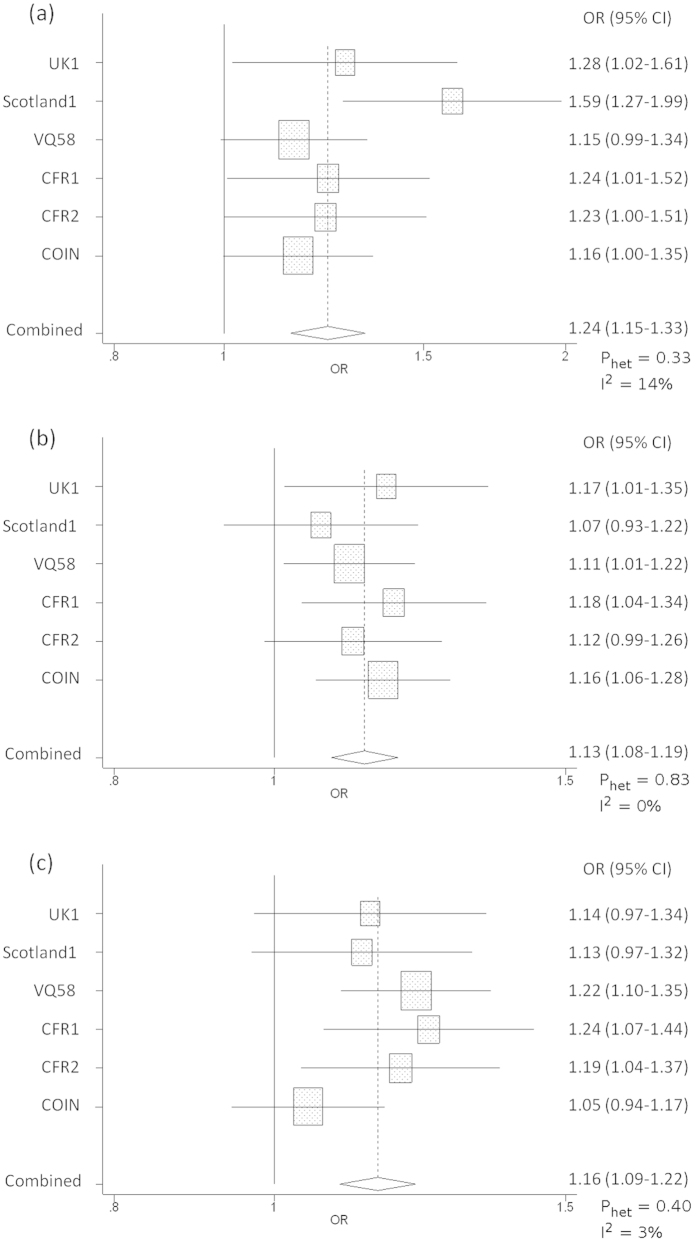
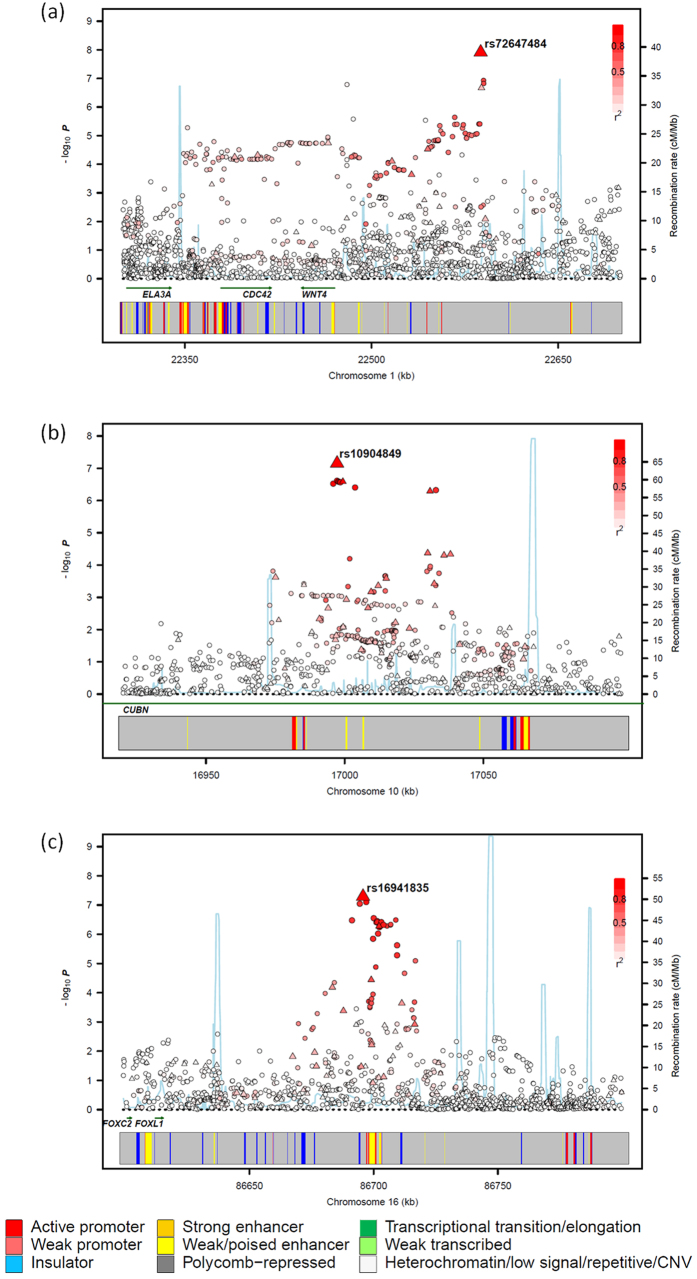
In the combined analysis of the six GWAS datasets, rs72647484, which maps to chromosome 1p36.12 (22,587,728 bps; NCBI build 37), showed the strongest evidence for association with CRC (P = 1.21 × 10−8; Phet = 0.33, I2 = 14%; Fig. 2a). rs72647484 maps within a 300 kb block of LD encompassing WNT4 (wingless-type mmtv integration site family, member 4; MIM 603490) and CDC42 (cell division cycle 42, MIM 116952; Fig. 3a). The second strongest association was provided by rs16941835 (P = 5.06 × 10−8; Phet = 0.40, I2 = 3%; Fig. 2c) which localises to the long non-coding RNA (lncRNA) RP11-58A18.1 at chromosome 16q24.1 (86,659,720 bps; NCBI build 37) within a 65 kb region of LD (Fig. 3c). The nearest coding gene, ~500 kb away, is the transcription factor FOXL1. The third strongest association was provided by rs10904849 (P = 7.01 × 10−8; Phet = 0.83, I2 = 0%; Fig. 2b) which localises to chromosome 10p13 (16,997,266 bps; NCBI build 37) within intron 31 of the gene encoding cubulin (CUBN; alias intrinsic factor-cobalamin receptor [IFCR], MIM 602997; Fig. 3b).
Figure 2. Forest plot of the odds ratios for the association between rs72647484, rs16941835, rs10904849 and CRC.
Studies were weighted according to the inverse of the variance of the log of the OR calculated by unconditional logistic regression. Horizontal lines: 95% confidence intervals (95% CI). Box: OR point estimate; its area is proportional to the weight of the study. Diamond (and broken line): overall summary estimate, with confidence interval given by its width. Unbroken vertical line: null value (OR = 1.0).
Figure 3.
Regional plot of association results and recombination rates for the (a) 1p36.12, (b) 10p13 and (c) 16q24.1 risk loci. Association results of both genotyped (triangles) and imputed (circles) SNPs in the GWAS samples and recombination rates within the loci at 1p36.12 (a), 10p13 (b) and 16q24 (c). For each plot, −log10 P values (y axis) of the SNPs are shown according to their chromosomal positions (x axis). The top imputed SNP in each combined analysis is shown as a large triangle and is labelled by its rsID. The colour intensity of each symbol reflects the extent of LD with the top SNP: white (r2 = 0) through to dark red (r2 = 1.0), with r2 estimated from the 1000 Genomes Phase 1 data. Genetic recombination rates (cM/Mb), are shown with a light blue line. Physical positions are based on NCBI build 37 of the human genome. Also shown are the relative positions of genes and transcripts mapping to each region of association. The lower panel shows the chromatin state segmentation track (ChromHMM).
Bioinformatic analysis of risk variants
To gain insight into the biological basis of the associations we analysed publicly available RNA-seq expression and SNP data from TCGA on 223 colonic and 75 rectal cancers using rs10904850 and rs2744753 as proxies for rs10904849 (r2 = 0.97; D’ = 1.00) and rs72647484 (r2 = 0.64; D’ = 0.89) respectively. After adjustment for multiple testing, no significant associations were seen between SNP genotype and expression of genes mapping to any of the three risk loci (Supplementary Table 4).
We examined whether any of the SNPs or their proxies (i.e. r2 > 0.8 in 1000 Genomes CEU reference panel) lie at putative transcription factor binding/enhancer elements and derived GERP and PhastCons scores to asses sequence conservation at these positions (Supplementary Table 5).
rs16941835 maps to a regulatory feature with histone modification suggestive of an enhancer element. rs10904852, in LD with rs10904849 (r2 = 0.95, D’ = 1.00) is conserved (GERP and PhastCons scores of 1.20 and 0.47 respectively) with CADD score of 11.53. A moderate CADD score (8.21) was associated with rs7267484 (22,590,125 bps) which is strong LD with rs72647489 (r2 = 0.93, D’ = 1.00). Six proxy SNPs in LD with rs16941835 showed some evidence of transcription factor binding (Supplementary Table 5). We made use of TCGA data to examine the frequency of somatic mutation of CDC42, WNT4, FOXL1 or CUBN in CRC. None of these genes showed evidence of significant somatic mutation. Next, we conducted pathway analysis to determine whether any genes mapping to the three newly identified regions act in pathways already over-represented in GWAS. Pathways containing three or more genes are shown in Supplementary Table 6. While this analysis identifies the BMP-signalling pathway as expected, no catalogued pathways were discernible involving genes mapping to any of the newly identified regions.
It is increasingly recognized that some genetic variants can have pleiotropic effects, influencing the risk of more than one cancer type. To explore the possibility that rs72647484, rs10904849 or rs16941835 affects the risk of other malignancies, we examined the association with lung cancer47, acute lymphoblastic leukaemia48, multiple myeloma49, glioma50 and meningioma51 using data from previously reported GWASs. However, for these cancers, there was no evidence of rs72647484, rs10904849 or rs16941835 (or correlated SNP r2 ≥ 0.8) being associated with tumour risk (i.e. P > 0.05).
Finally, the relationship between clinico-pathological variables (sex, age at diagnosis, family history of CRC, tumour stage or microsatellite instability (MSI), KRAS-mutant status and BRAF-mutant status) and genotype at rs72647484, rs10904849 and rs16941835 was assessed by case-only logistic regression (Supplementary Table 7). There was evidence of a relationship between rs72647484 and KRAS-mutant status (P = 0.03) with the T risk allele associated with KRAS-mutant CRC; however this finding was not significant after accounting for multiple testing. None of the other SNPs showed any association with any of the clinico-pathological variables examined (i.e. P > 0.05).
Discussion
We have provided evidence supporting the existence of new susceptibility loci for CRC at 1p36.12, 10p13 and 16q24.1. The 1p36.12 association implicates WNT4 and/or CDC42 as possible determinates of CRC risk. WNT4 is part of a family of structurally related genes that encode cysteine-rich secreted glycoproteins that act as extracellular signalling factors. WNT4, WNT14, and WNT16 may play redundant roles in signalling through the CTNNB1-mediated canonical Wnt-pathway52 which is known to play a central role in colorectal tumorigenesis. Additionally, WNT4 signalling appears to play a pivotal role during organogenesis, acting as an autoinducer of mesenchyme-to-epithelial transition. Inactivating germline mutations in WNT4 cause mullerian aplasia and hyperandrogenism (MIM 158330) and are responsible for the autosomal recessive SERKAL syndrome (Sex Reversal and Kidney, Adrenal, and Lung dysgenesis; MIM 611812). A priori dysfunction of either WNT4 or CDC42 could be the biological basis for the 1p36.12 association. Cdc42 is a Ras-related GTP-binding protein with roles in establishment of cell polarity, regulation of cell morphology, motility, and cell cycle progression in mammalian cells, and malignant transformation53. Notably, Cdc42 regulates the actin cytoskeleton through activation of WASP proteins and cell polarity through GSK3-beta and APC. Rho-GTPase signalling has a documented role in the development of CRC54. Activation of Rho GTPase Cdc42 promotes adhesion and invasion in CRC55 and targeting Cdc42 with AZA197 suppresses primary colon cancer growth and prolongs survival in a xenograft model through down regulation of PAK156.
Since rs10904849 is intronic to CUBN and the region of LD does not encompass any other genes or transcripts, there is a high likelihood that the functional basis of the 10p13 association is mediated through CUBN. Cubilin is the intestinal receptor for the endocytosis of intrinsic factor-vitamin B12 and a receptor in epithelial apoA-I/HDL metabolism57. Additionally cubilin is an important co-receptor in the endocytic pathway for retrieval of 25(OH)D3-DBP complexes by megalin-mediated endocytosis in the kidney58. Germline mutations in CUBN cause recessive megaloblastic anemia-1 (MGA1; MIM 261100). It is conceivable that common genetic variance in CUBN, while being insufficient to cause a “MGA type phenotype” would have physiological effects by virtue of long term effect on the cellular bioavailability of B12. Although it is entirely speculative, as epidemiological studies have yet to convincingly establish levels of B12 as a risk factor for CRC59,60, its role in DNA biosynthesis makes genetically determined variation in B12 availability a plausible candidate for a role in the development of CRC.
LncRNAs are regulators of transcription and are increasingly recognised as playing a role in cancer biology. While there is currently no evidence to implicate the RP11-58A18.1 lncRNA in CRC, lncRNAs CCAT1 and CCAT2 probably do play such roles61,62, and it is entirely plausible that the impact of variation at 16q24.1 on risk is mediated through similar long range effects.
One of the reasons for the failure to identify these CRC-loci previously is that, in addition to the issue of study power, they were not optimally tagged by SNPs featured on many commercial arrays. The power of our study to detect the major common loci conferring risks of 1.2 or greater (such as the 18q24 variant) was high. Hence, it is very unlikely there are additional CRC SNPs with similar effects for alleles with frequencies >0.2 in populations of European ancestry.
In this study, we have only considered SNPs showing evidence of an association with a stipulated P-value threshold of <1 × 10−7. There exist, however, many variants with P-values just above this threshold which may also warrant investigation in a further study (Fig. 1). Hence further efforts to expand the scale of GWAS meta-analyses, in terms of both sample size and SNP coverage, and to increase the number of SNPs taken forward to large-scale replication, may identify additional variants for CRC.
In conclusion, we have provided evidence for 3 new susceptibility loci for CRC. Our data also provide further evidence for the value of meta-analysis and the value of imputation as a means of enhancing the detection of novel risk loci thereby extending the utility of GWAS data.
Author Contributions
The study was designed and financial support was obtained by R.S.H., I.P.M.T., M.G.D. and J.P.C. The manuscript was drafted by R.S.H. with input from I.P.M.T., M.G.D. and J.P.C. All authors had access to data, analysis and had opportunity to contribute to drafting the manuscript. J.P.C. initiated and directed the GWAS of COIN and COIN-B. S.I. was responsible for COIN blood DNA extractions and quantification, R.H. for aliquoting and manifest preparation for Axiom genotyping and C.G.S. for somatic profiling of COIN tumours. N.A.A. and B.F.M. coordinated, and S.M.W. performed, the genotyping of COIN and COIN-B samples on the Axiom platform. T.S.M. was CI/co-CI of COIN and COIN-B, and R.K. provided access to linked clinico-pathological data. Cleaning of the COIN/COIN-B genotyping data and all statistical and bioinformatic analyses were conducted by N.W. and F.H., with contributions from S.D. and B.K., under the supervision of R.S.H. ICR - Sample preparation and genotyping were performed by A.L. and N.W. Oxford and local collaborators: subject recruitment and sample acquisition were done by E.B., M.G., L.M., R.K., D.K., and members of the CORGI Consortium. Sample preparation and genotyping were performed by C.P. Colon Cancer Genetics Group, Edinburgh and local collaborators: subject recruitment and sample acquisition were performed by S.M.F., C.H., H.C., I.D. and M.G.D., as well as members of SOCCS and COGS recruitment teams. Sample preparation was coordinated by S.M.F. Genotyping and analysis was performed and coordinated by S.M.F., C.H., M.G.D. and A.T. For the colon CFR datasets - D.D.B., A.K.W., J.H., M.J., F.S., G.C., S.G., N.L., P.N. and D.C. performed sample ascertainment and analysis.
Additional Information
How to cite this article: Al-Tassan, N. A. et al. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Sci. Rep. 5, 10442; doi: 10.1038/srep10442 (2015).
Supplementary Material
Acknowledgments
The COIN and COIN-B trials were funded by Cancer Research UK and the Medical Research Council and were conducted with the support of the National Institute of Health Research Cancer Research Network. COIN and COIN-B translational studies were supported by the Bobby Moore Fund from Cancer Research UK, Tenovus, the Kidani Trust, Cancer Research Wales and the National Institute for Social Care and Health Research Cancer Genetics Biomedical Research Unit (2011–2014) (all awarded to J.P.C.). We thank the patients and their families who participated in COIN and COIN-B, and gave their consent for this research, and the investigators and pathologists throughout the UK who submitted samples for assessment. N.A.A., B.F.M. and S.M.W. were funded and supported by KFSHRC. At the Institute of Cancer Research, the work was supported by Cancer Research UK (C1298/A8362 - Bobby Moore Fund for Cancer Research UK). Additional support was provided by the National Cancer Research Network and the NHS via the Biological Research Centre of the National Institute for Health Research at the Royal Marsden Hospital NHS Trust. N.W. and B.K. were in receipt of PhD studentships from the ICR. B.K additionally receives funding from the Sir John Fisher Foundation. In Edinburgh the work was supported by Programme Grant funding from Cancer Research UK (C348/A12076). In Oxford additional funding was provided by the Oxford Comprehensive Biomedical Research Centre (C.P. and I.P.M.T.) and the EU FP7 CHIBCHA grant (I.P.M.T.). Core infrastructure support to the Wellcome Trust Centre for Human Genetics, Oxford was provided by grant (090532/Z/09/Z). We are grateful to many colleagues within UK Clinical Genetics Departments (for CORGI) and to many collaborators who participated in the VICTOR and QUASAR2 trials. We also thank colleagues from the UK National Cancer Research Network (for NSCCG). Support from the European Union (FP7/207-2013) under grant 258236 and FP7 collaborative project SYSCOL and COST Action BM1206 in the UK is also acknowledged. The work of the Colon Cancer Family Registry CFR was supported by grant UM1 CA167551 from the National Cancer Institute, National Institutes of Health and through cooperative agreements with members of the Colon CFR and Principal Investigators. Collaborating centers include the Australasian Colorectal Cancer Family Registry (U01/U24 CA097735), the USC Colorectal Cancer Family Registry (U01/U24 CA074799), Mayo Clinic Cooperative Familial Registry for Colon Cancer Studies (U01/U24 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (U01/U24 CA074783), the Seattle Colorectal Cancer Family Registry (U01/U24 CA074794), and the University of Hawaii Colorectal Cancer Family Registry (U01/U24 CA074806). The Colon CFR GWAS was supported by funding from the National Cancer Institute, National Institutes of Health (U01 CA122839 and R01 CA143237 to GC). This study made use of genotyping data from the 1958 Birth Cohort, kindly made available by the Wellcome Trust Case Control Consortium 2. A full list of the investigators who contributed to the generation of the data is available at http://www.wtccc.org.uk/. The results published here are in whole or part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions which constitute the TCGA research network can be found at http://cancergenome.nih.gov/. Finally, we would like to thank all individuals who participated in the study.
References
- Lichtenstein P. et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343, 78–85 (2000). [DOI] [PubMed] [Google Scholar]
- Aaltonen L., Johns L., Jarvinen H., Mecklin J. P. & Houlston R. Explaining the familial colorectal cancer risk associated with mismatch repair (MMR)-deficient and MMR-stable tumors. Clin. Cancer Res. 13, 356–361 (2007). [DOI] [PubMed] [Google Scholar]
- Lubbe S. J., Webb E. L., Chandler I. P. & Houlston R. S. Implications of familial colorectal cancer risk profiles and microsatellite instability status. J. Clin. Oncol. 27, 2238–2244 (2009). [DOI] [PubMed] [Google Scholar]
- Broderick P. et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet. 39, 1315–1317 (2007). [DOI] [PubMed] [Google Scholar]
- Dunlop M. G. et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat. Genet. 44, 770–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston R. S. et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet. 42, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters U. et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology 144, 799–807 e24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa A. et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 40, 631–637 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson I. et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 39, 984–988 (2007). [DOI] [PubMed] [Google Scholar]
- Tomlinson I. P. et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet. 40, 623–630 (2008). [DOI] [PubMed] [Google Scholar]
- Tomlinson I. P. et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 7, e1002105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiffin N. et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum. Mol. Genet. 23, 4729–4737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R. et al. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut 60, 799–805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W. H. et al. Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat. Genet. 45, 191–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat. Commun. 5, 4613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat. Genet. 46, 533–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan T. S. et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet 377, 2103–2114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midgley R. S. et al. Phase III randomized trial assessing rofecoxib in the adjuvant setting of colorectal cancer: final results of the VICTOR trial. J. Clin. Oncol. 28, 4575–4580 (2010). [DOI] [PubMed] [Google Scholar]
- Power C. & Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study). Int. J. Epidemiol. 35, 34–41 (2006). [DOI] [PubMed] [Google Scholar]
- Newcomb P. A. et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol. Biomarkers Prev. 16, 2331–2343 (2007). [DOI] [PubMed] [Google Scholar]
- Hunter D. J. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 39, 870–874 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M. et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 39, 645–649 (2007). [DOI] [PubMed] [Google Scholar]
- R Core Team 2013. R: A language and environment for statistical computing. . R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/ (Date of access 01/12/2014); (Accessed 01/12/2014).
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O., Marchini J. & Zagury J. F. A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2012). [DOI] [PubMed] [Google Scholar]
- Howie B. N., Donnelly P. & Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes. http://www.1000genomes.org/(Accessed 01/12/2014).
- Marchini J., Howie B., Myers S., McVean G. & Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet 39, 906–913 (2007). [DOI] [PubMed] [Google Scholar]
- Liu J. Z. et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 42, 436–440 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. P. & Thompson S. G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 (2002). [DOI] [PubMed] [Google Scholar]
- Pharoah P. D., Antoniou A. C., Easton D. F. & Ponder B. A. Polygenes, risk prediction, and targeted prevention of breast cancer. N. Engl. J. Med. 358, 2796–2803 (2008). [DOI] [PubMed] [Google Scholar]
- Johns L. E. & Houlston R. S. A systematic review and meta-analysis of familial colorectal cancer risk. Am. J. Gastroenterol. 96, 2992–3003 (2001). [DOI] [PubMed] [Google Scholar]
- Ernst J. & Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project: ENCyclopedia Of DNA Elements. http://www.genome.gov/encode/ (Accessed 01/12/2014).
- Kircher M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G. M. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome. Res. 15, 901–913 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas http://cancergenome.nih.gov/ (Accessed 01/12/2014).
- Gao J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCI pathway interaction database. Accessed 01/12/2014. http://pid.nci.nih.gov/ (2014).
- Boland C. R. et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 58, 5248–5257 (1998). [PubMed] [Google Scholar]
- Smith C. G. et al. Somatic profiling of the epidermal growth factor receptor pathway in tumors from patients with advanced colorectal cancer treated with chemotherapy +/- cetuximab. Clin. Cancer Res. 19, 4104–4113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasan H. et al. Intermittent chemotherapy plus either intermittent or continuous cetuximab for first-line treatment of patients with KRAS wild-type advanced colorectal cancer (COIN-B): a randomised phase 2 trial. Lancet Oncol. 15, 631–639 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. G. et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 37, 1243–1246 (2005). [DOI] [PubMed] [Google Scholar]
- Broderick P. et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer. Res. 69, 6633–6641 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorini G. et al. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood 122, 3298–3307 (2013). [DOI] [PubMed] [Google Scholar]
- Chubb D. et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nat. Genet. 45, 1221–1225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson M. et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum. Mol. Genet. 20, 2897–2904 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbins S. E. et al. Common variation at 10p12.31 near MLLT10 influences meningioma risk. Nat. Genet. 43, 825–827 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. et al. Wnt/beta-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 18, 2404–2417 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. J., Erickson J. W., Lin R. & Cerione R. A. The gamma-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature 405, 800–804 (2000). [DOI] [PubMed] [Google Scholar]
- Leve F. & Morgado-Diaz J. A. Rho GTPase signaling in the development of colorectal cancer. J. Cell Biochem. 113, 2549–2559 (2012). [DOI] [PubMed] [Google Scholar]
- Gao L., Bai L. & Nan Q. Activation of Rho GTPase Cdc42 promotes adhesion and invasion in colorectal cancer cells. Med. Sci. Monit Basic Res. 19, 201–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zins K., Gunawardhana S., Lucas T., Abraham D. & Aharinejad S. Targeting Cdc42 with the small molecule drug AZA197 suppresses primary colon cancer growth and prolongs survival in a preclinical mouse xenograft model by downregulation of PAK1 activity. J. Transl. Med. 11, 295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozyraki R. et al. The human intrinsic factor-vitamin B12 receptor, cubilin: molecular characterization and chromosomal mapping of the gene to 10p within the autosomal recessive megaloblastic anemia (MGA1) region. Blood 91, 3593–3600 (1998). [PubMed] [Google Scholar]
- Nykjaer A. et al. Cubilin dysfunction causes abnormal metabolism of the steroid hormone 25(OH) vitamin D(3). Proc. Natl. Acad. Sci. U S A 98, 13895–13900 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett J. K. et al. Dietary intake of B vitamins and methionine and colorectal cancer risk. Nutr. Cancer 65, 659–667 (2013). [DOI] [PubMed] [Google Scholar]
- Razzak A. A. et al. Associations between intake of folate and related micronutrients with molecularly defined colorectal cancer risks in the Iowa Women’s Health Study. Nutr. Cancer 64, 899–910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai H. et al. Clinical significance of long intergenic noncoding RNA-p21 in colorectal cancer. Clin. Colorectal. Cancer 12, 261–266 (2013). [DOI] [PubMed] [Google Scholar]
- Ling H. et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 23, 1446–1461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.