Abstract
Bacteria such as Escherichia coli swim along circular trajectories adjacent to surfaces. Thereby, the orientation (clockwise, counterclockwise) and the curvature depend on the surface properties. We employ mesoscale hydrodynamic simulations of a mechano-elastic model of E. coli, with a spherocylindrical body propelled by a bundle of rotating helical flagella, to study quantitatively the curvature of the appearing circular trajectories. We demonstrate that the cell is sensitive to nanoscale changes in the surface slip length. The results are employed to propose a novel approach to directing bacterial motion on striped surfaces with different slip lengths, which implies a transformation of the circular motion into a snaking motion along the stripe boundaries. The feasibility of this approach is demonstrated by a simulation of active Brownian rods, which also reveals a dependence of directional motion on the stripe width.
Motility is a particular property of flagellated prokaryotic microorganisms, which are ubiquitous in nature. Most bacteria exploit helical filaments for propulsion, driven by rotary motors located in their cell membrane. Prominent examples of peritrichous bacteria, which possess numerous flagella, are Escherichia coli, Salmonella typhimurium, Rhizobium lupini, or Proteus mirabilis to name just a few. In a bulk fluid, the bacteria move in a straight manner (run), with all flagella forming a bundle, interrupted by abrupt changes of the swimming direction (tumble) induced by disintegration of the bundle.1 The presence of a surface drastically alters the swimming behavior. For instance, the non-tumbling mutant of E. coli swims in a clockwise (CW) circular trajectory close to a solid boundary2 and a counterclockwise (CCW) trajectory close to a liquid-air interface.3 Hence, bacteria are able to “sense” the properties of a nearby surface, an aspect of paramount importance for surface selection and attachment in the early stages of biofilm formation or infection.2,4,5
The swimming behavior of bacteria near surfaces is governed by hydrodynamic forces6,7 and, hence, the CW and CCW circular trajectories of E. coli have been explained in terms of hydrodynamic interactions.2,3 Physically, the resulting trajectory is controlled by the fluid slip on the surface—CW trajectories follow for no-slip and CCW for perfect-slip boundary conditions. Usually, a surface exhibits partial slip due to adsorbents, microstructures, and hydrophobicity.8,9 A quantified measure of slip is the slip length b as the extrapolated distance below the surface, where the fluid velocity vanishes.10 By definition, b = 0 for no-slip and b = ∞ for perfect-slip surfaces. Intuitively, a trajectory can be expected to switch from CW to CCW (or vice versa) when b reaches some characteristic value b0. Such a transition has been observed experimentally for E. coli swimming near glass surfaces upon addition of alginate, and has been attributed to changes in the slip length.11 An attempt of a unified description has been presented, based on the far-field approximation of hydrodynamic interactions.12 Yet, there is no quantitative theoretical or simulation study on the effect of slip on the swimming behavior of bacteria at surfaces. A theoretical understanding of hydrodynamic interactions between swimming bacteria and surfaces not only sheds light on selective surface attachment, but opens an avenue for the design of microfluidic devices to control and guide bacterial motion13 for separation, trapping, stirring, etc.
Here, we exploit a mesoscale model of a non-tumbling E. coli-type bacterium to study its motion near surfaces, and present results for its circular dynamics at surfaces of various slip lengths, see Fig. 1. The simulation fully accounts for hydrodynamic interactions on length scales much smaller than the diameter of the cell, i.e., near-field hydrodynamics, which is very important for a quantitative theoretical description of the phenomenon. We find that E. coli senses the nanoscale slip length of the surface and responds with a circular trajectory of a particular radius. The obtained dependence of the curvature on the slip length is well described by a simple derived theoretical expression. Moreover, we employ these insights to suggest a novel route to direct bacterial motion by patterning a surface with stripes of different slip lengths and corresponding CW and CCW trajectories, respectively. This leads to an preferential snaking motion along the stripe boundaries for sufficiently wide stripes. We demonstrate the viability of this approach by a simulation of active Brownian rods, and elucidate the dependence of the diffusion anisotropy on the stripe width.
Figure 1. Swimming bacteria sense the slip of its nearby surface.
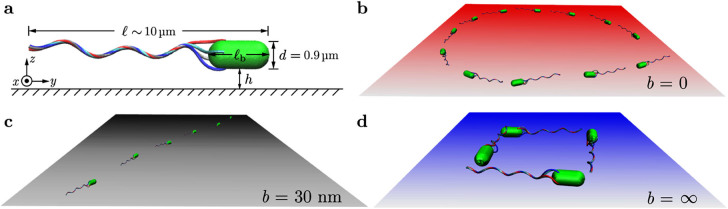
(a) The model bacterium of length  consists of a spherocylindrical body of length
consists of a spherocylindrical body of length 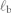 and diameter d and four helical flagella
each turned by a motor torque. The bacterial geometry and flagellar
properties are in agreement with experiments of E. coli (Methods and
SI). The body the flagellar bundle counter rotate. h is
the gap width between the body and the surface. (b) CW, (c) noisy straight,
and (d) CCW trajectories from hydrodynamic simulations of a bacterium
swimming near homogeneous surfaces with different slip lengths b as
indicated.
and diameter d and four helical flagella
each turned by a motor torque. The bacterial geometry and flagellar
properties are in agreement with experiments of E. coli (Methods and
SI). The body the flagellar bundle counter rotate. h is
the gap width between the body and the surface. (b) CW, (c) noisy straight,
and (d) CCW trajectories from hydrodynamic simulations of a bacterium
swimming near homogeneous surfaces with different slip lengths b as
indicated.
Results
Bacterial swimming near homogeneous surfaces
In our mesoscale hydrodynamic simulations, we model the bacterium E. coli by a spherocylindrical body with four attached helical flagella, see Fig. 1(a). The bacterium is immersed in a mesoscale fluid, described by the multiparticle collision dynamics (MPC) technique,14,15 which itself is confined between two parallel surfaces with tunable slip length. The MPC method includes thermal fluctuations and fully accounts for hydrodynamic interactions down to 100 nm, a length scale much less than the cell-body diameter d = 0.9 µm. Rotation of the flagella by applied torques leads to bundle formation16,17 and swimming motion18 (Movie S1 in SI). Further details are provided in the Methods section.
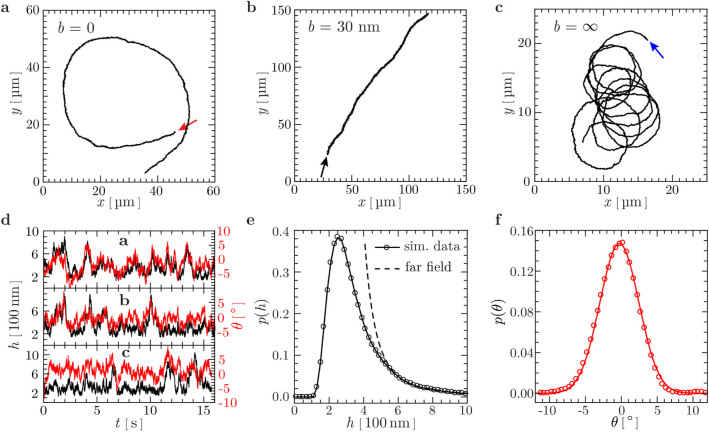
Near surfaces, bacteria generically swim on circular trajectories due to the
counter-rotation of the body and the flagellar bundle. Figures
2(a)–(c) illustrate the simulation trajectories of the
E. coli model for different slip lengths b. The trajectory
changes from a CW circle (Fig. 1(b) and Movie S2 in
SI) to a noisy straight line (Fig. 1(c)) and a
CCW circle (Fig. 1(d)) as b increases. Figure 2(d) shows that the distance h of the body to
the surface fluctuates around the average 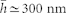 with standard deviation
with standard deviation 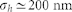 . The
probability distribution p(h) is presented in Fig.
2(e). It clearly deviates from the far-field prediction19
. The
probability distribution p(h) is presented in Fig.
2(e). It clearly deviates from the far-field prediction19
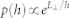 , which strongly overestimates the
probability for the bacterium to be very close to the surface. The simulation
data for h > 500 nm are well described by the far-field
expression, which provides the length scale
, which strongly overestimates the
probability for the bacterium to be very close to the surface. The simulation
data for h > 500 nm are well described by the far-field
expression, which provides the length scale 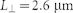 .
.
Figure 2.
(a)–(c) Simulated trajectories for the E. coli model with
body length 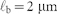 , viewed from above the
surface with slip length b and starting point indicated by the arrow.
(d) Time series of the gap width h and the inclination angle
θ between the bacterial swimming direction (pointing
from the flagellar to the body center of mass) and the surface.
θ < 0 if the bacterium swims toward the
surface. (e) Comparison of the gap-width distribution obtained from our
simulations to the far-field prediction19
, viewed from above the
surface with slip length b and starting point indicated by the arrow.
(d) Time series of the gap width h and the inclination angle
θ between the bacterial swimming direction (pointing
from the flagellar to the body center of mass) and the surface.
θ < 0 if the bacterium swims toward the
surface. (e) Comparison of the gap-width distribution obtained from our
simulations to the far-field prediction19
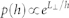 with fit parameter
with fit parameter 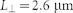 . (f) Distribution of the inclination
angle θ, fitted by a Gaussian distribution with mean
. (f) Distribution of the inclination
angle θ, fitted by a Gaussian distribution with mean 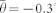 and standard deviation
σθ = 2.7°.
and standard deviation
σθ = 2.7°.
Our results demonstrate that the far-field approximation fails to quantitatively
describe the swimming behavior of bacteria near surfaces. The average distance
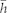 results from the balance of
hydrodynamic attraction19 and short-range repulsion (which mimics
the effect of additional surface-bacterium interactions, see Methods section),
independent of the initial position or orientation of E. coli. Thus, the
resulting height distribution is determined by near-field hydrodynamics, rather
than far-field effects. This conclusion is supported by a study of bacterial
motion near slip surfaces,3 which yields a similar
bacterium-surface distance
results from the balance of
hydrodynamic attraction19 and short-range repulsion (which mimics
the effect of additional surface-bacterium interactions, see Methods section),
independent of the initial position or orientation of E. coli. Thus, the
resulting height distribution is determined by near-field hydrodynamics, rather
than far-field effects. This conclusion is supported by a study of bacterial
motion near slip surfaces,3 which yields a similar
bacterium-surface distance 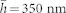 from
comparison of experimental data with theoretical results based on mobility
tensors of cylindrical and helical bodies. We note that the bacterium has the
chance to escape from the surface due to orientational fluctuations, because we
see some rare events where it moves from one wall to the other. These events are
observed more frequently for larger body lengths, which may be related to the
additional active noise due to an increased wobbling motion of the body (compare
Movie S1 in SI) with increasing
from
comparison of experimental data with theoretical results based on mobility
tensors of cylindrical and helical bodies. We note that the bacterium has the
chance to escape from the surface due to orientational fluctuations, because we
see some rare events where it moves from one wall to the other. These events are
observed more frequently for larger body lengths, which may be related to the
additional active noise due to an increased wobbling motion of the body (compare
Movie S1 in SI) with increasing 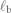 .
The angle θ between the bacterial swimming direction and the
surface in Fig. 2(d) deviates by at most 10°
from the average, and is found to follow a Gaussian distribution with mean
.
The angle θ between the bacterial swimming direction and the
surface in Fig. 2(d) deviates by at most 10°
from the average, and is found to follow a Gaussian distribution with mean 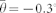 as shown in Fig.
2(f), indicating that the bacterium swims nearly parallel to the
surface. These results reflect the importance of noise on the swimming motion of
bacteria near surfaces.
as shown in Fig.
2(f), indicating that the bacterium swims nearly parallel to the
surface. These results reflect the importance of noise on the swimming motion of
bacteria near surfaces.
The quantitative dependence of swimming trajectories on the slip length is
displayed in Fig. 3(a) for different cell-body lengths 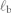 . The curvature κ for
each slip length is an average over up to 10 independent trajectories from
extensive hydrodynamic simulations. For each trajectory, the average curvature
is the inverse of the circle radius obtained from a least-square fit. The
curvature exhibits a smooth crossover from CW trajectories (κ
< 0) for no-slip surface to CCW trajectories (κ
> 0). The data are very well described by the analytical expression
. The curvature κ for
each slip length is an average over up to 10 independent trajectories from
extensive hydrodynamic simulations. For each trajectory, the average curvature
is the inverse of the circle radius obtained from a least-square fit. The
curvature exhibits a smooth crossover from CW trajectories (κ
< 0) for no-slip surface to CCW trajectories (κ
> 0). The data are very well described by the analytical expression
 |
which is based on the
consideration of the hydrodynamic lubrication forces on the cell body, which
consist of the competing shear force and the force due to the pressure
imbalance, as illustrated in Fig. 3(b); see Methods
section for a derivation. Here, κ0 < 0
is the curvature for a no-slip surface (b = 0),
κ∞ > 0 for a
perfect-slip surface (b = ∞), and heff the
effective gap width between the cell and the surface. The values of the fit
parameters κ0,
κ∞, and heff are
listed in Table 1. The fitted height
heff is comparable to the mean gap width 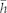 , whereas the latter values are somewhat
larger, which is attributed to the wobbling motion of the body. The radii of
curvature |κ0|−1 and
|κ∞|−1
obtained from the fits agree with the values directly measured from our
simulations as well as the experimental results.2,11 As shown in
the inset of Fig. 3(a), with the fitted values of
heff at different
, whereas the latter values are somewhat
larger, which is attributed to the wobbling motion of the body. The radii of
curvature |κ0|−1 and
|κ∞|−1
obtained from the fits agree with the values directly measured from our
simulations as well as the experimental results.2,11 As shown in
the inset of Fig. 3(a), with the fitted values of
heff at different 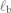 , all data points measured from different systems collapse onto a
single line (κ0 −
κ)/(κ −
κ∞) =
b/heff, which is equivalent to Eq.
(1). Thus, the results from hydrodynamics simulations are clearly
consistent with our theoretical description and experimental results. We like to
stress that Eq. (1) is a general result and applies also to
other bacteria which exploit rotating flagella for swimming, such as Bacillus
subtilis, Salmonella typhimurium and Rhodobacter
sphaeroides.
, all data points measured from different systems collapse onto a
single line (κ0 −
κ)/(κ −
κ∞) =
b/heff, which is equivalent to Eq.
(1). Thus, the results from hydrodynamics simulations are clearly
consistent with our theoretical description and experimental results. We like to
stress that Eq. (1) is a general result and applies also to
other bacteria which exploit rotating flagella for swimming, such as Bacillus
subtilis, Salmonella typhimurium and Rhodobacter
sphaeroides.
Figure 3.
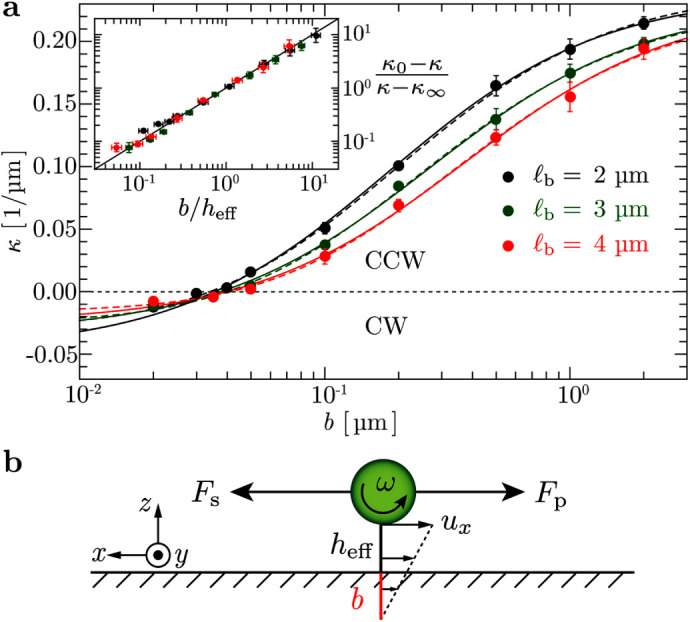
(a) Average curvature κ of E. coli swimming trajectories vs. surface slip length b.The solid and dashed lines are least-square fits of simulation data points to Eq. (1) and the far-field prediction in Ref. 12, respectively. Inset: (κ0 − κ)/(κ − κ∞) vs. b/heff, with the effective gap width heff obtained from the solid fitted lines in the main plot. The solid line in the inset corresponds to (κ0 − κ)/(κ − κ∞) = b/heff. (b) Schematic of the model used for analytical calculations for circular swimming trajectories. For a bacterium swimming along the y-axis as shown in Fig. 1(a), the cell body rotating around the y-axis experiences two forces along the x-axis: (i) the shear force Fs due to the fluid velocity gradient ∂zux ≈ (−ωd/2)/(heff + b), and (ii) the force Fp due to pressure difference between the converging (left) and diverging (right) flows in the gap. The bacterium runs CW with κ < 0 when F = Fs + Fp > 0, and CCW with κ > 0 when F < 0.
Table 1. Properties of model E. coli swimming near a surface, as obtained from
mesoscale hydrodynamic simulations. ℓb: body length;
ℓ/d: bacterial length to body diameter; U:
swimming velocity; 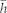 : mean gap width;
heff: effective gap width;
|κ0|−1 and
|κ∞|−1:
radius of curvature at b = 0 and b = ∞, respectively;
b0: slip length of the surface, at which E. coli
swims in a straight line. The ‘sim’ values for
|κ0|−1 and
|κ∞|−1
are directly measured from our simulations, ‘fit’ values
are obtained from the fits in Fig. 3(a), and
‘exp’ are from experiments of E. coli.
: mean gap width;
heff: effective gap width;
|κ0|−1 and
|κ∞|−1:
radius of curvature at b = 0 and b = ∞, respectively;
b0: slip length of the surface, at which E. coli
swims in a straight line. The ‘sim’ values for
|κ0|−1 and
|κ∞|−1
are directly measured from our simulations, ‘fit’ values
are obtained from the fits in Fig. 3(a), and
‘exp’ are from experiments of E. coli.
|
|
U[µm/s] |
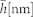
|
heff[nm] | |κ0|−1[µm] | |κ∞|−1[µm] | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sim | Fit | exp2 | Sim | Fit | exp11 | b0[nm] | |||||
| 2 | 8.7 | 9.5 ± 0.8 | 307 | 182 ± 22 | 19 ± 1 | 21 ± 3 | 4.1 ± 0.1 | 4.2 ± 0.1 | 36 ± 3 | ||
| 3 | 9.8 | 9.1 ± 0.4 | 366 | 267 ± 23 | 33 ± 2 | 31 ± 3 | ≃10–50 | 4.2 ± 0.1 | 4.3 ± 0.1 | ≃5 | 38 ± 2 |
| 4 | 10.9 | 8.4 ± 0.2 | 390 | 370 ± 50 | 39 ± 3 | 40 ± 4 | 4.3 ± 0.1 | 4.3 ± 0.3 | 40 ± 3 | ||
The dependence of the swimming-trajectory curvature on the slip length has been
analysed very recently within the far-field approximation for a rotlet dipole, a
simplified model for flagellated bacteria with counter-rotation of cell body and
flagella.12 The trajectory curvature κ is
found to be a complex function of the dipole strength q, the fluid gap
width h, and the bacterial aspect ratio γ; see Eq.
(71) in Ref. 12. As displayed in Fig. 3(a), our simulation data can be fitted very well by the
far-field prediction, shown by the dashed lines with the fit parameters
(q/[pN µm2],
h/[nm], γ) = (0.002, 259,
2.86), (0.005, 322, 3.00), and (0.016, 441, 3.79) for cell-body length 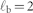 , 3, and 4 µm,
respectively. The values of h are consistent with the mean gap widths in
Table 1, whereas the values of γ
are about three times less than the bacterial aspect ratio
, 3, and 4 µm,
respectively. The values of h are consistent with the mean gap widths in
Table 1, whereas the values of γ
are about three times less than the bacterial aspect ratio 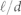 , but close to the body aspect ratio
, but close to the body aspect ratio 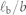 . From dimensional analysis, a reasonable
estimate of q is q ≈ 1.5 pN µm2, obtained from the product of the net torque
. From dimensional analysis, a reasonable
estimate of q is q ≈ 1.5 pN µm2, obtained from the product of the net torque 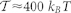 (thermal energy
(thermal energy 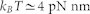 ) rotating the flagellar bundle and the body diameter d = 0.9
µm. However, this value of q is about two to three orders of
magnitude larger than the fitted values in the far-field expression! Note that
the above torque is close to the experimental value of 500 pN nm.27 The comparison reveals a discrepancy between our simulation results (and
experimental estimates of the dipole strength) and the rotlet-dipole approach,
which may arise from the overestimation of hydrodynamic interactions at short
distances in the far-field approximation.
) rotating the flagellar bundle and the body diameter d = 0.9
µm. However, this value of q is about two to three orders of
magnitude larger than the fitted values in the far-field expression! Note that
the above torque is close to the experimental value of 500 pN nm.27 The comparison reveals a discrepancy between our simulation results (and
experimental estimates of the dipole strength) and the rotlet-dipole approach,
which may arise from the overestimation of hydrodynamic interactions at short
distances in the far-field approximation.
Equation (1) provides the characteristic slip length b0, which separates CW and CCW trajectories, i.e., yields a trajectory with vanishing average curvature. By setting κ = 0, we find
 |
For the E. coli
model with body length 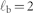 , 3, and 4
µm at a distance of
, 3, and 4
µm at a distance of 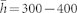 from
the surface, b0 is about 40 nm and nearly independent of
from
the surface, b0 is about 40 nm and nearly independent of 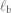 , see Table 1.
It is very interesting to see that a slip length of a few tens of nanometers can
substantially alter the swimming behavior of E. coli. Therefore, we
propose that E. coli can be employed as a natural sensor for the slip
length of surfaces.
, see Table 1.
It is very interesting to see that a slip length of a few tens of nanometers can
substantially alter the swimming behavior of E. coli. Therefore, we
propose that E. coli can be employed as a natural sensor for the slip
length of surfaces.
Directed bacterial motion near patterned surfaces
Motivated by the results for homogeneous surfaces, we propose that striped
surfaces with different slip lengths can be used to direct bacterial motion. The
alternating slip lengths of the stripes are chosen according to Eq. (1) such that E. coli swims in circles with opposite sign
of curvature on neighboring stripes. This should result in an alternating
trajectory along a stripe border, i.e., a 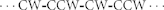 motion, see Fig. 4(b) and Movie S3 in
SI. The dynamical behavior depends on the trajectory curvatures on the
two types of stripes and the stripe width, as discussed in the following.
motion, see Fig. 4(b) and Movie S3 in
SI. The dynamical behavior depends on the trajectory curvatures on the
two types of stripes and the stripe width, as discussed in the following.
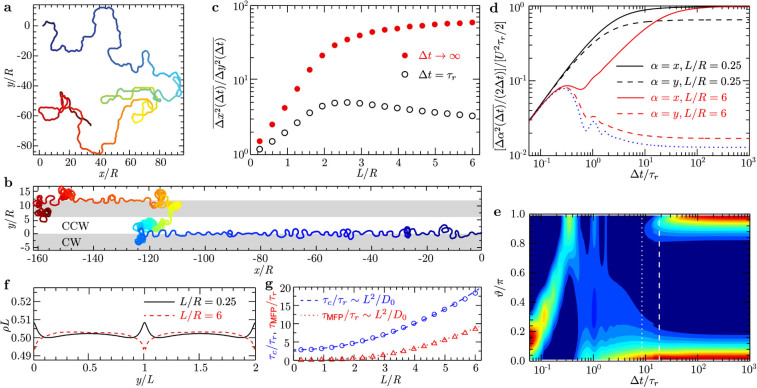
Figure 4. Active Brownian rod swimming on a ‘symmetric’ striped surface with curvature radii equal in magnitude but opposite in sign.
Trajectories for (a) narrow (L/R = 0.25) and (b) broad (L/R = 6) stripes from simulations of duration t = 80τr with starting points indicated in dark blue. The rotational diffusion time is τr = 1/Dr = 18 s (see Methods section). (c) The ratio of mean square displacement (MSD) parallel (x) and perpendicular (y) to the stripes as a function of stripe width L for Δt = τr and Δt → ∞. (d) Rescaled MSD. The blue dotted line corresponds to a free circle swimmer.34 (e) Probability distribution of turning angle ϑ defined in Eq. (6) between successive trajectory segments of duration Δt for L/R = 6. High and low probabilities are shown in red and blue, respectively. (f) Density profile ρ across two stripes. (g) Crossover time τc between ballistic and diffusive regime along the stripes and the mean first-passage time τMFP for the rod to cross the stripe scale both quadratically with L. The white dashed and dotted lines in (e) indicate the corresponding τc and τMFP, respectively.
For the demonstration of the emergence of directed motion, we first consider
stripes of equal width L and infinite length parallel to the
x-axis, with curvatures |κ| = 1/R of equal
magnitude. We model the bacterium as a self-propelled rod33,34
of length  and swimming velocity
U along its axis e = (cos φ, sin
φ) with the orientation angle
and swimming velocity
U along its axis e = (cos φ, sin
φ) with the orientation angle 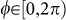 . This allows us to reach the required large length-
and time-scales in the simulations. For the symmetric setup of trajectory
curvatures, the bacterium rotates at a rate Ω = Ω0
= U/R when it swims within CCW stripes and Ω =
–Ω0 within CW stripes. When the bacterium
crosses a CW-CCW border (e.g., at y = 0), we assume a linear profile
Ω = Ω0y/λ with the width
. This allows us to reach the required large length-
and time-scales in the simulations. For the symmetric setup of trajectory
curvatures, the bacterium rotates at a rate Ω = Ω0
= U/R when it swims within CCW stripes and Ω =
–Ω0 within CW stripes. When the bacterium
crosses a CW-CCW border (e.g., at y = 0), we assume a linear profile
Ω = Ω0y/λ with the width
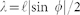 taking into account the
orientation and finite size of the bacterium. The bacterium also exhibits
translational and rotational diffusion with coefficients D||,
taking into account the
orientation and finite size of the bacterium. The bacterium also exhibits
translational and rotational diffusion with coefficients D||,
 , and Dr. Further
simulation details are described in the Methods section.
, and Dr. Further
simulation details are described in the Methods section.
The swimming trajectories in Figs. 4(a) and 4(b) illustrate
that the bacterial motion is rather isotropic for narrow stripes
(L/R = 0.25), but becomes highly anisotropic along the
x-axis for broad stripes (L/R = 6). In the latter case,
we observe an oscillatory motion along stripe borders. To understand this
behavior, we approximate the width λ by its average 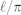 . By neglecting noise terms in the
equations of motion (5), we then obtain a pendulum equation
. By neglecting noise terms in the
equations of motion (5), we then obtain a pendulum equation
 |
which implies a stable oscillation of the
orientation angle φ around φ =
π (i.e., the negative x-direction) at CW-CCW
borders. Similarly, at CCW-CW borders (e.g., at y = L)
oscillations around φ = 0 (i.e., the positive
x-direction) are stable. The oscillatory motion is also seen with other
choices of Ω(y), e.g., where 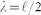 , indicating that our results are robust with respect to modeling
details. Figure 4(d) shows the mean square displacements
(MSD) parallel and perpendicular to the stripes,
, indicating that our results are robust with respect to modeling
details. Figure 4(d) shows the mean square displacements
(MSD) parallel and perpendicular to the stripes, 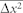 and
and 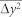 ,
respectively. For narrow stripes,
,
respectively. For narrow stripes, 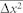 and
and
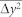 are almost equal and resemble the
MSD of a persistent random walk, i.e., the MSD is ballistic for
Δt < τr =
1/Dr and diffusive for
are almost equal and resemble the
MSD of a persistent random walk, i.e., the MSD is ballistic for
Δt < τr =
1/Dr and diffusive for 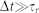 . Independent of the stripe width,
. Independent of the stripe width, 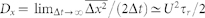 . For broad stripes, however,
. For broad stripes, however, 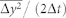 is orders of magnitude smaller than ±
is orders of magnitude smaller than ±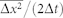 , and Dy converges
with increasing L/R to the diffusion coefficient
, and Dy converges
with increasing L/R to the diffusion coefficient 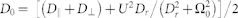 of a free circle swimmer.34 The transport on a substrate with broad stripes may be understood as follows:
a bacterium diffuses with D0 within a stripe; once it hits the
stripe border, it performs a directed snaking motion at the velocity U
along x; the average duration of snaking is
τr, because there is no limit-cycle and
snaking is neutrally stable; therefore, after τr
(on average) the bacterium starts to diffuse again within a stripe; as soon as
the particle reach the neighboring border, motions along the positive and
negative x-directions are equally probable and the transport along
x gets diffusive. This becomes apparent from the distribution of the
turning angles ϑ between successive displacements of duration
Δt (see Methods section), shown in Fig.
4(e) for L/R = 6, which characterizes the geometrical
properties of a trajectory on various time scales. At short times
Δt < τr, the
characteristic angle with maximal probability drifts from
ϑmax≈0 to
ϑmax≈π due to
free circular motion. On larger time scales, the motion gets first
unidirectional (ϑmax≈0) and finally
bidirectional (ϑmax≈0 and
ϑmax≈π are
equally probable). We calculate the crossover time
τc between the ballistic and diffusive regime
along x as the time where the exponent α ≈
1.4 in
of a free circle swimmer.34 The transport on a substrate with broad stripes may be understood as follows:
a bacterium diffuses with D0 within a stripe; once it hits the
stripe border, it performs a directed snaking motion at the velocity U
along x; the average duration of snaking is
τr, because there is no limit-cycle and
snaking is neutrally stable; therefore, after τr
(on average) the bacterium starts to diffuse again within a stripe; as soon as
the particle reach the neighboring border, motions along the positive and
negative x-directions are equally probable and the transport along
x gets diffusive. This becomes apparent from the distribution of the
turning angles ϑ between successive displacements of duration
Δt (see Methods section), shown in Fig.
4(e) for L/R = 6, which characterizes the geometrical
properties of a trajectory on various time scales. At short times
Δt < τr, the
characteristic angle with maximal probability drifts from
ϑmax≈0 to
ϑmax≈π due to
free circular motion. On larger time scales, the motion gets first
unidirectional (ϑmax≈0) and finally
bidirectional (ϑmax≈0 and
ϑmax≈π are
equally probable). We calculate the crossover time
τc between the ballistic and diffusive regime
along x as the time where the exponent α ≈
1.4 in 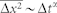 . Moreover, we determine the
mean first-passage time τMFP, i.e., the
mean time to cross the stripe. As can be seen from Fig.
4(g), both τc and
τMFP are proportional to
L2/D0 and
τc >
τMFP, consistent with our
conjecture; furthermore, τc corresponds to the
appearance of bidirectional motion, see the vertical white lines in Fig. 4(e). The absence of a limit-cycle at the stripe border
becomes obvious from the nearly flat density distribution
ρ(y) in Fig. 4(f). Finally,
we show in Fig. 4(c) the ratio
. Moreover, we determine the
mean first-passage time τMFP, i.e., the
mean time to cross the stripe. As can be seen from Fig.
4(g), both τc and
τMFP are proportional to
L2/D0 and
τc >
τMFP, consistent with our
conjecture; furthermore, τc corresponds to the
appearance of bidirectional motion, see the vertical white lines in Fig. 4(e). The absence of a limit-cycle at the stripe border
becomes obvious from the nearly flat density distribution
ρ(y) in Fig. 4(f). Finally,
we show in Fig. 4(c) the ratio 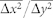 as a function of L at Δt =
τr and Δt →
∞. At the short time scale Δt =
τr, the anisotropy assumes a maximum at
as a function of L at Δt =
τr and Δt →
∞. At the short time scale Δt =
τr, the anisotropy assumes a maximum at 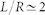 . However, at long time scales, the
diffusion anisotropy increases monotonically with L, exponentially for
L/R ≤ 2 and then saturates for
. However, at long time scales, the
diffusion anisotropy increases monotonically with L, exponentially for
L/R ≤ 2 and then saturates for 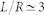 .
.
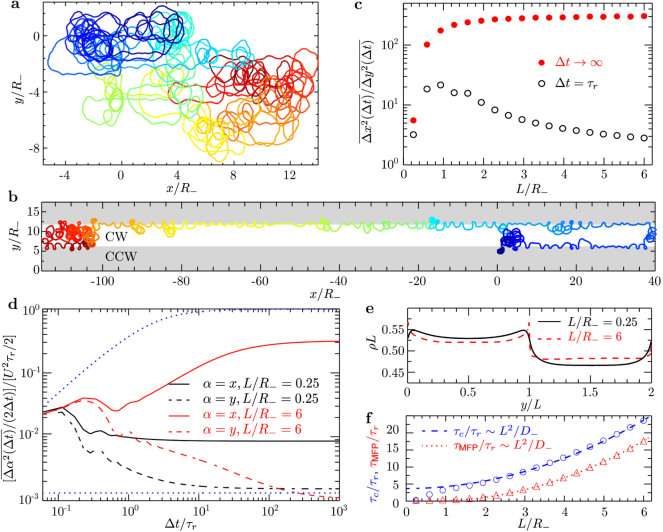
The setup of a symmetrically striped surface, with equal magnitude of the
trajectory radius R on both stripes, is very special, because it would
require a fine-tuning of slip lengths on the two kinds of stripes, and therefore
a very particular selection of wall materials. Therefore, we study how
asymmetrically striped surfaces modify the behavior. More precisely, we set
R− = 40 µm on CW stripes and
R+ = R–/3 on CCW stripes. The
trajectories in Figs. 5(a) and 5(b) show a similar
behavior as in the case of a symmetric pattern, i.e., isotropic motion for
narrow stripes and highly anisotropic along the x-axis for broad stripes.
However, the anisotropy of the diffusion increases nearly five-fold, see Fig. 5(c). Furthermore, 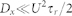 and Dx decreases with decreasing L, see
Fig. 5(d); Dy converges to
D−D+/(D−
+ D+) with increasing L
(D− and D+ are the diffusion
coefficients of a free circle swimmer in the CW and CCW stripe, respectively).
The crossover time τc ~
L2/D− and mean
first-passage time τMFP ~
L2/D− are dominated by
the faster diffusion process on the CW stripes with larger trajectory radius. In
addition, the density distribution now displays different values in the two
kinds of stripes because
and Dx decreases with decreasing L, see
Fig. 5(d); Dy converges to
D−D+/(D−
+ D+) with increasing L
(D− and D+ are the diffusion
coefficients of a free circle swimmer in the CW and CCW stripe, respectively).
The crossover time τc ~
L2/D− and mean
first-passage time τMFP ~
L2/D− are dominated by
the faster diffusion process on the CW stripes with larger trajectory radius. In
addition, the density distribution now displays different values in the two
kinds of stripes because 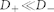 , see Fig. 5(e). We conclude that the anisotropic transport over a
striped surface is a very robust phenomenon.
, see Fig. 5(e). We conclude that the anisotropic transport over a
striped surface is a very robust phenomenon.
Figure 5. Active Brownian rod swimming on an ‘asymmetric’ striped surface with stripes of different curvatures (R−/R+ = 3).
Trajectories for (a) narrow (L/R− = 0.25) and (b) broad (L/R− = 6) stripes from simulations of duration t = 80τr with starting points indicated in dark blue. (c) Parallel-to-perpendicular MSD ratio as a function of stripe width L for Δt = τr and Δt → ∞. (d) Rescaled MSD. The upper and lower blue dotted lines correspond to a persistent random walk and the diffusion coefficient D−D+/(D− + D+), respectively. D− and D+ are the diffusion coefficients of a free circle swimmer on the CW and CCW stripes. (e) Crossover time τc between ballistic and diffusive regime along the stripes and the mean first-passage time τMFP as a function of L. (f) Density profile ρ across two stripes.
Discussion
We have presented a theoretical and numerical investigation of the effect of partial slip at surfaces on the curvature of swimming trajectories of flagellated bacteria. From a combination of mesoscale hydrodynamic simulations and scaling arguments, we predict that E. coli bacteria are able to sense surface slip on the nanoscale. An implication of our results is to use E. coli as a biosensor of surface slip. Moreover, an increasing slip length, which can be achieved experimentally by addition of grafted polymers, implies CCW circles of decreasing radius. Such tighter circles may enhance bacteria surface adsorption due to an increased local residence time; compare Figs. 1(b) and (d). Hence, physical sensing of high slip lengths may be paramount for biofilm formation and infection.
We further demonstrate that striped surfaces can be designed to direct bacterial motion along the stripe boundaries. The proposed striped surfaces with slip length smaller and larger than about 30 nm and width of order of 100 µm are accessible in laboratory experiments. Very importantly, the directed motion and anisotropic diffusion does not require a fine-tuning of slip lengths on the alternating stripe patterns, but is very robust and seems to only require different trajectory curvatures. Anisotropic transport even exists for stripes yielding the same sign of trajectory curvature (both CW or CCW) but different magnitudes. The resulting trajectory along the stripe border resembles a prolate cycloid. Therefore, transport along stripe boundaries is a very generic phenomenon.
First, it is important to emphasize that the anisotropic diffusion will persist at finite bacterial densities. In this case, a persistent multi-lane motion of bacteria can be predicted, in which individual bacteria occasionally change lanes, very similar as predicted above. Second, since the curvature radius of the swimming trajectory depends on the geometric parameters of the bacterium shape, this approach could also be employed for sorting bacteria according to size by an appropriate choice of stripe width. Bacteria with a smaller trajectory curvature would diffuse more isotropically, whereas bacteria with a larger trajectory curvature would diffuse more anisotropically along the stripe boundaries. Thus, after placing a mixture of both bacteria types on a small spot on the striped surface, bacteria with small and large trajectory curvatures will be found preferentially in a location in the y- and x-directions, respectively, at some distance from the initial location of the drop. In addition, a bacterial motion rectifier could be constructed from a transversally confined two-stripe system. In this case, there is only a single stripe boundary, so that all bacteria would move in the same direction in the center of the channel. Since there would also be directed motion along the wall,13 a more detailed study of such possible devices is required.
Methods
Mesoscale hydrodynamic simulations
The mesoscale hydrodynamic simulations combine coarse-grained molecular dynamics
simulations for the bacterium and multiparticle collision dynamics method14,15 for the solvent. The model bacterium is constructed by
coarse-grained particles, and consists of a spherocylindrical body and four
helical flagellar filaments, see Fig. 1(a). The flagellar
filament is based on the helical worm-like chain model.24,25 We
choose the bacterial geometry and flagellar elastic properties as extracted from
experiments of E. coli (SI). Each flagellum is driven by a motor
torque 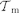 and the opposite torque is
applied to the body to ensure that the bacterium is torque-free. We choose
and the opposite torque is
applied to the body to ensure that the bacterium is torque-free. We choose 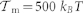 , i.e., 2100 pN nm with Boltzmann
constant kB and temperature T = 300 K, smaller than the
stall torque of approximately 4500 pN nm26 of the flagellar
motor. The flagellar bundle rotates at a frequency around 100 Hz and generates a
propulsion force of about 0.6 pN, consistent with the experimental value of 0.57
pN.27
, i.e., 2100 pN nm with Boltzmann
constant kB and temperature T = 300 K, smaller than the
stall torque of approximately 4500 pN nm26 of the flagellar
motor. The flagellar bundle rotates at a frequency around 100 Hz and generates a
propulsion force of about 0.6 pN, consistent with the experimental value of 0.57
pN.27
The surface is modeled by a hard wall with tunable boundary conditions. No-slip
condition is implemented by applying bounce-back collision and virtual wall
particles,28 whereas perfect-slip condition is obtained with
specular reflection of fluid particles at the wall. The velocity of the fluid
particle at the wall is inverted in bounce-back collision; in specular
reflection, only the normal component of the velocity is inverted. A random mix
of these two boundary conditions leads to partial slip.29 We
systematically vary the mixing ratio to obtain slip lengths from 20 nm to 2
µm, a range readily accessible in experiments.8,9 We
mimic the effect of additional surface-bacterium interactions such as van der
Waals, electrostatic and steric interactions30,31 by a
short-range repulsive potential 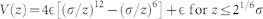 and
V(z) = 0 otherwise, where
and
V(z) = 0 otherwise, where 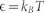 and the interaction range σ = 100 nm is set to
avoid close contact of the bacterium with the surface. This short-range
repulsion automatically takes care of possible bacterium-wall collision, which
has been shown to be a possible cause of the accumulation of bacteria near
walls.20,32,33 There is no attractive interaction potential
between the bacterium and the surface. The simulations are done in cubic boxes
of length
and the interaction range σ = 100 nm is set to
avoid close contact of the bacterium with the surface. This short-range
repulsion automatically takes care of possible bacterium-wall collision, which
has been shown to be a possible cause of the accumulation of bacteria near
walls.20,32,33 There is no attractive interaction potential
between the bacterium and the surface. The simulations are done in cubic boxes
of length 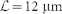 with periodic boundaries in
x- and y-directions and two walls at z = 0 and
with periodic boundaries in
x- and y-directions and two walls at z = 0 and 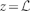 .
.
Analytical considerations
To derive an analytical expression for the curvature of the bacterial swimming
trajectory, we consider a bacterium of length  swimming with the velocity U parallel to the
surface at a distance h, see Fig. 1(a). As
illustrated in Fig. 3(b), the cell body of length
swimming with the velocity U parallel to the
surface at a distance h, see Fig. 1(a). As
illustrated in Fig. 3(b), the cell body of length 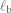 and diameter d rotates CCW
around the y-axis at a rate ω and experiences two
forces of hydrodynamic origin along the x-axis: (i) a pressure
difference21 generated by the converging and diverging flows
on either side of the gap between the body and the surface leads to a force
and diameter d rotates CCW
around the y-axis at a rate ω and experiences two
forces of hydrodynamic origin along the x-axis: (i) a pressure
difference21 generated by the converging and diverging flows
on either side of the gap between the body and the surface leads to a force 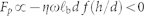 , where η is the
fluid viscosity, and the dimensionless function f has the properties
f(h/d) → 0 for
, where η is the
fluid viscosity, and the dimensionless function f has the properties
f(h/d) → 0 for 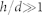 and f(h/d) →
constant for
and f(h/d) →
constant for 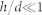 ;23 (ii)
the fluid is sheared in the gap region, implying a velocity gradient and thus a
shear force Fs. By assuming a linear velocity profile
∂zux ≈
(−ωd/2)/(heff +
b), where heff is the effective gap width and
b the slip length of the surface, we obtain
;23 (ii)
the fluid is sheared in the gap region, implying a velocity gradient and thus a
shear force Fs. By assuming a linear velocity profile
∂zux ≈
(−ωd/2)/(heff +
b), where heff is the effective gap width and
b the slip length of the surface, we obtain 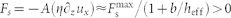 , where
, where 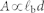 is
the shear area and
is
the shear area and 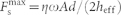 is the maximum
shear force. These two forces add up to
is the maximum
shear force. These two forces add up to
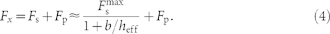 |
This expression describes the hydrodynamic interaction of rotating
spherical, ellipsoidal, and spherocylindrical bodies with surfaces over a wide
range of slip lengths very well, as shown by a comparison with existing
numerical data in the range of 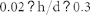 (Fig.
S1 and Table S1 in SI).
(Fig.
S1 and Table S1 in SI).
Since the swimming bacterium is force-free, a force equal and opposite to Eq. (4) acts on the flagellar bundle, producing a torque 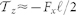 that drives the rotation of the cell
around the z-axis at a rate
that drives the rotation of the cell
around the z-axis at a rate 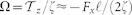 ,
with the rotational friction coefficient
,
with the rotational friction coefficient 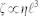 . For a no-slip surface, Faxén's
calculation22 implies that the force (4) translates the body
in the direction of rolling along the surface, i.e.
. For a no-slip surface, Faxén's
calculation22 implies that the force (4) translates the body
in the direction of rolling along the surface, i.e.
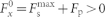 , which is responsible for CW motion of
the bacterium. For a perfect-slip surface, the force (4) is
, which is responsible for CW motion of
the bacterium. For a perfect-slip surface, the force (4) is 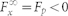 , causing CCW motion. The curvature of a trajectory,
κ = Ω/U, is then give by Eq. (1), which interpolates between
, causing CCW motion. The curvature of a trajectory,
κ = Ω/U, is then give by Eq. (1), which interpolates between 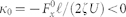 for no-slip surfaces (b = 0) and
for no-slip surfaces (b = 0) and 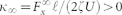 for perfect-slip surfaces (b =
∞). Here, κ < 0 denotes CW and
κ > 0 CCW trajectories. We note that
κ is independent of fluid viscosity η
and swimming velocity U, since ω ∝
U.
for perfect-slip surfaces (b =
∞). Here, κ < 0 denotes CW and
κ > 0 CCW trajectories. We note that
κ is independent of fluid viscosity η
and swimming velocity U, since ω ∝
U.
Brownian dynamics simulations
We consider a self-propelled rod swimming in two dimensions over a patterned
surface and neglect the fluctuations of its distance from the surface, since the
variation in the distance h of the bacterium to the surface from our
hydrodynamic simulations is smaller than its length  , as shown in Fig. 2(e). The
translational and rotational motions of the active rod are described according
to
, as shown in Fig. 2(e). The
translational and rotational motions of the active rod are described according
to
 |
where r = (x,
y) is the center of mass position, U is the swimming velocity
along the orientation of the rod e = (cos φ, sin
φ) with the orientation angle 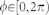 . Here, ξ and
τ are zero-mean Gaussian white noises of variance
. Here, ξ and
τ are zero-mean Gaussian white noises of variance 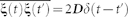 and
and 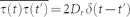 . The diffusion tensor
. The diffusion tensor 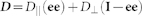 is given in terms of the parallel and perpendicular
translational diffusion coefficients D|| and
is given in terms of the parallel and perpendicular
translational diffusion coefficients D|| and 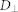 . Dr is the rotational
diffusion coefficient. Using experimental results for E. coli,2,20,27 we set
. Dr is the rotational
diffusion coefficient. Using experimental results for E. coli,2,20,27 we set 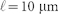 ,
U = 20 µm/s, R = U/|Ω| = 40
µm, D|| = 0.14 µm2/s,
,
U = 20 µm/s, R = U/|Ω| = 40
µm, D|| = 0.14 µm2/s,
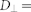 D||/2 and Dr = 0.057
s−1.
D||/2 and Dr = 0.057
s−1.
Geometrical characterization of trajectories
In order to characterize the geometrical properties of a swimming trajectory r(t) on various time scales, we examine the probability distribution function P(ϑ,Δt) of turning angles ϑ between successive trajectory segments defined as
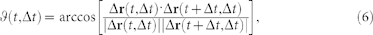 |
where Δr(t, Δt) = r(t + Δt) − r(t) is the vector between two successive positions separated by the lag time Δt;35 Δt controls the degree of temporal coarse graining. The distribution function P(ϑ,Δt) is flat for a homogenous diffusive motion due to the scale invariance of this process.
Author Contributions
J.H., A.W., R.G.W. and G.G. designed the research, analyzed the data, and wrote the paper; J.H. and A.W. performed the research.
Additional Information
How to cite this article: Hu, J., Wysocki, A., Winkler, R.G. & Gompper, G. Physical Sensing of Surface Properties by Microswimmers – Directing Bacterial Motion via Wall Slip. Sci. Rep. 5, 9586; doi: 10.1038/srep09586 (2015).
Supplementary Material
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Acknowledgments
Financial support by the VW Foundation (VolkswagenStiftung) within the program Computer Simulation of Molecular and Cellular Biosystems as well as Complex Soft Matter of the initiative New Conceptual Approaches to Modeling and Simulation of Complex Systems is gratefully acknowledged. We thank Gerhard Nðgele and Andrea Costanzo for helpful discussions.
References
- Berg, H. C. E. Coli in Motion (Springer-Verlag, Berlin, 2004)
- Lauga E., DiLuzio W. R., Whitesides G. M. & Stone H. A. Swimming in circles: motion of bacteria near solid boundaries. Biophys. J. 90, 400–412 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leonardo R., Dell'Arciprete D., Angelani L. & Iebba V. Swimming with an image. Phys. Rev. Lett. 106, 038101 (2011). [DOI] [PubMed] [Google Scholar]
- Ottemann K. M. & Miller J. F. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24, 1109 (1997). [DOI] [PubMed] [Google Scholar]
- Pratt L. A. & R. Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and Type I pili. Mol. Microbiol. 30, 285 (1998). [DOI] [PubMed] [Google Scholar]
- Lauga E. & Powers T. R. The hydrodynamics of swimming microorganisms. Rep. Progs. Phys. 72, 096601 (2009). [Google Scholar]
- Elgeti J., Winkler R. G. & Gompper G. Physics of Microswimmers – Single Particle Motion and Collective Behavior. Rep. Progs. Phys., to appear, arXiv:1412.2692 (2015). [DOI] [PubMed] [Google Scholar]
- Lauga E., Brenner M. & Stone H. in Springer Handbook of Experimental Fluid Mechanics (eds Tropea, C., Yarin, A. & Foss, J.) (Springer Berlin Heidelberg., 2007).
- Rothstein J. P. Slip on superhydrophobic surfaces. Annu. Rev. Fluid Mech. 42, 89–109 (2010). [Google Scholar]
- Navier C. L. M. H. Memoire sur les lois du mouvement des fluides. Mem. Acad. R. Sci. Inst. France 6, 389–440 (1823). [Google Scholar]
- Lemelle L., Palierne J.-F., Chartre E., Vaillant C. & Place C. Curvature reversal of the circular motion of swimming bacteria probes for slip at solid/liquid interfaces. Soft Matter 9, 9759–9762 (2013). [Google Scholar]
- Lopez D. & Lauga E. Dynamics of swimming bacteria at complex interfaces. Phys. Fluid 26, 071902 (2014). [Google Scholar]
- DiLuzio W. R. et al. Escherichia coli swim on the right-hand side. Nature. 435, 1271–1274 (2005). [DOI] [PubMed] [Google Scholar]
- Malevanets A. & Kapral R. Mesoscopic model for solvent dynamics. J. Chem. Phys. 110, 8605–8613 (1999). [Google Scholar]
- Gompper G., Ihle T., Kroll D. M. & Winkler R. G. Multi-particle collision dynamics: a particle-based mesoscale simulation approach to the hydrodynamics of complex fluids. Adv. Polym. Sci. 221, 1–87 (2009). [Google Scholar]
- Janssen P. J. A. & Graham M. D. Coexistence of tight and loose bundled states in a model of bacterial flagellar dynamics. Phys. Rev. E 84, 011910 (2011). [DOI] [PubMed] [Google Scholar]
- Reigh S. Y., Winkler R. G. & Gompper G. Synchronization and bundling of anchored bacterial flagella. Soft Matter 8, 4363–4372 (2012). [Google Scholar]
- Watari N. & Larson R. G. The hydrodynamics of a run-and-tumble bacterium propelled by polymorphic helical flagella. Biophys. J. 98, 12–17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke A. P., Turner L., Berg H. C. & Lauga E. Hydrodynamic attraction of swimming microorganisms by surfaces. Phys. Rev. Lett. 101, 038102 (2008). [DOI] [PubMed] [Google Scholar]
- Drescher K., Dunkel J., Cisneros L. H., Ganguly S. & Goldstein R. E. Fluid dynamics and noise in bacterial cell-cell and cell-surface scattering. Proc. Natl. Acad. Sci. USA 108, 10940–10945 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey D. J. & Onishi Y. The slow motion of a cylinder next to a plane wall. Q. J. Mech. Appl. Math. 34, 129–137 (1981). [Google Scholar]
- Happel J. & Brenner H. Low Reynolds number hydrodynamics (Martinus Nijhoff, The Hague., 1983). [Google Scholar]
- Davis A. M., Kezirian M. T. & Brenner H. . On the Stokes-Einstein model of surface diffusion along solid surfaces: slip boundary conditions. J. Colloid Interface Sci. 165, 129–140 (1994). [Google Scholar]
- Yamakawa H. Helical wormlike chains in polymer solutions (Springer-Verlag, Berlin Heidelberg., 1997). [Google Scholar]
- Vogel R. & Stark H. Force-extension curves of bacterial flagella. Eur. Phys. J. E 33, 259–271 (2010). [DOI] [PubMed] [Google Scholar]
- Berry R. M. & Berg H. C. Absence of a barrier to backwards rotation of the bacterial flagellar motor demonstrated with optical tweezers. Proc. Natl. Acad. Sci. USA 94, 14433–14437 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Moldovan R., Yeung C. & Wu X. L. Swimming efficiency of bacterium Escherichia coli. Proc. Natl. Acad. Sci. USA 103, 13712–13717 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamura A., Gompper G., Ihle T. & Kroll D. M. Multi-particle collision dynamics: Flow around a circular and a square cylinder. Europhys. Lett. 56, 319–325 (2001). [Google Scholar]
- Whitmer J. K. & Luijten E. Fluid-solid boundary conditions for multiparticle collision dynamics. J. Phys.: Condens. Matter 22, 104106 (2010). [DOI] [PubMed] [Google Scholar]
- Li G., Tam L.-K. & Tang J. X. Amplified effect of Brownian motion in bacterial near-surface swimming. Proc. Natl. Acad. Sci. USA 105, 18355–18359 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson H. H. & Weibel D. B. Bacteria-surface interactions. Soft Matter 9, 4368–4380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. & Tang J. X. Accumulation of microswimmers near a surface mediated by collision and rotational Brownian motion. Phys. Rev. Lett. 103, 078101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgeti J. & Gompper G. Self-propelled rods near surfaces. EPL 85, 38002 (2009). [Google Scholar]
- van Teeffelen S. & Löwen H. Dynamics of a Brownian circle swimmer. Phys. Rev. E 78, 020101 (2008). [DOI] [PubMed] [Google Scholar]
- Burov S., Tabei S. M. A., Huynh T., Murrell M. P., Philipson L. H., Rice S. A., Gardel M. L., Scherer N. F. & Dinner A. R. Distribution of directional change as a signature of complex dynamics. Proc. Natl. Acad. Sci. USA 110, 19689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information