Abstract
Dissociated cell cultures of the rodent hippocampus have become a standard model for studying many facets of neural development, including the development of polarity, axonal and dendritic growth, and synapse formation. The cultures are quite homogeneous—~90% of the cells are pyramidal neurons—and it is relatively easy to express green fluorescent protein (GFP)-tagged proteins by transfection. This article describes the cultures and the key features of the system used to image them. It also includes suggestions on labeling cells with GFP-tagged proteins. It concludes with a discussion of the advantages and disadvantages of this culture system.
INTRODUCTION
Dissociated cell cultures from rat or mouse hippocampus have become a standard model for studying the many facets of neural development (Goslin and Banker 1998; Kaech and Banker 2006). Culturing hippocampal neurons at low density results in a preparation in which axons and dendrites grow directly on a perfectly flat substrate, which is ideal for live cell imaging. Methods for imaging hippocampal cultures depend on the dynamics of the events under study. Developmental changes in cell morphology and protein localization occur over a time course of hours, whereas the cell biological events that underlie neuronal development often occur over seconds to minutes. Time-lapse studies of neural development require methods that permit cells to be imaged repeatedly over prolonged periods without compromising cell survival. Use of a computer-controlled microscope with an automated stage allows one to track changes in cells at multiple locations, greatly increasing the useful data that can be obtained from each imaging experiment. (See Long-Term Time-Lapse Imaging of Developing Hippocampal Neurons in Culture [Kaech et al. 2012a].) For capturing rapid subcellular events at high resolution, one must maximize signal and reduce noise. Because hippocampal cultures are essentially two dimensional, they are particularly well suited for imaging rapid events because there is no need to acquire images at multiple focal planes. (See Short-Term High-Resolution Imaging of Developing Hippocampal Neurons in Culture [Kaech et al. 2012b].)
The cellular composition of these cultures is comparatively simple. When prepared from late-stage embryos (before formation of the dentate gyrus), pyramidal neurons from the CA1 and CA3 regions account for >90% of the neuronal population. Because neurons harvested from late-stage embryos are largely postmitotic, antimitotic drugs can be used to prevent glial division. Although several different approaches are commonly used for making hippocampal cultures, the method established in our laboratory was designed with imaging in mind. We culture the neurons at low density, which makes it easier to see individual cells, and the neurites grow directly on the surface of polylysine-treated glass coverslips, rather than up, down, and over glial cells. However, these advantages come at a price. At the low densities we use, the neurons do not survive well on their own. Our work-around is to maintain the neuron-bearing coverslips in dishes that contain monolayer cultures of astroglia, which provide trophic support. We add tiny feet to the coverslips (actually small drops of paraffin) so they can be grown facing the glia but without contact between the two cell layers. When the neurons have reached the desired stage of development, the coverslips can be moved to an imaging chamber, and the glia are left behind. Although this method is a little complicated, it results in a preparation in which axons and dendrites grow directly on a perfectly flat substrate that is ideal for microscopy with high-numerical-aperture (high-NA) objectives. Details of our culture protocol were recently published (Kaech and Banker 2006) and are available on our website (www.ohsu.edu/xd/research/centers-institutes/neurology/jungers-center/research/banker-lab.cfm).
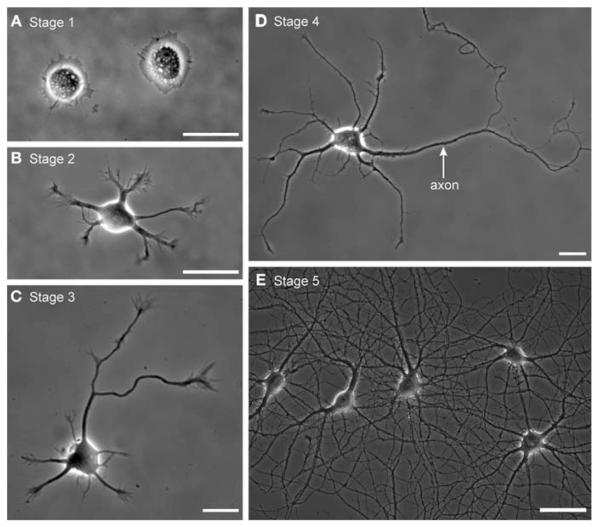
The stages of neuronal development in hippocampal cultures prepared according to this method have been well documented (Dotti et al. 1988; Fig. 1). Shortly after plating, hippocampal neurons extend a lamella all around the cell body (developmental stage 1 in the terminology of Dotti et al. 1988). Then this lamella coalesces at distinct spots around the cell periphery and minor neurites extend; this corresponds to developmental stage 2. The minor neurites, which undergo periods of elongation and retraction, have the capacity to become either axons or dendrites. After 12–36 h, one minor neurite undergoes a period of continuous elongation until it becomes distinctly longer than the rest. This change marks the beginning of developmental stage 3; the long neurite will continue to elongate rapidly and will acquire axonal characteristics. Over time, the remaining minor neurites grow and acquire the taper that is characteristic of dendrites, and synapses start to form. These events mark the beginning of developmental stage 4. With time, the dendrites become more highly branched, an extensive network of synaptic connections forms, and the dendrites become studded with spines. Such cells are said to be in stage 5 of development, although the transition between stages 4 and 5 is gradual, and there is no clear consensus on the characteristics that define a stage 5 cell. When everything goes well, these cultures survive for 3–5 wk.
FIGURE 1.
Developmental stages of cultured hippocampal neurons imaged in phase contrast. (A) Cells in stage 1 are round and are encircled by a prominent lamella. (B) By stage 2, short neurites have formed. (C) In stage 3, the symmetry of the short neurites has been broken; one neurite has grown longer than the rest and has become specified as the axon. (D) In stage 4, the remaining minor neurites begin to grow and to acquire the tapered morphology characteristic of dendrites. This image represents an unusual case in which the somatodendritic domain of the neuron has not been contacted by axonal branches of any neighboring cell. (E) By stage 5, the network has become so dense that individual neurites are difficult to follow. The proximal portions of the dendrites that arise from each cell are obvious, but it is not possible to identify their axons without some additional method for selectively labeling them (see Kaech and Banker 2006, for further details). Scale bars, 25 μm in A–C, 50 μm in D. (Reprinted, with permission, from Kaech and Banker 2006.)
In our laboratory, we use hippocampal cultures to elucidate the molecular events that underlie the initial development of neuronal polarity during the transition from stage 2 to 3 and to analyze the cell biological machinery that selectively delivers membrane proteins to axons or dendrites (Burack et al. 2000; Jacobson et al. 2006). For both projects, we rely heavily on live cell imaging of proteins tagged with GFP or its color variants. In the remainder of this article, we will describe the imaging methods we use to follow developmental changes in cell morphology and protein localization (which typically involves long-term microscopy with periodic image acquisition) and to image vesicle trafficking (which involves short-term, high-resolution imaging). Hippocampal cultures are also widely used to study axonal and dendritic growth and synapse formation, including live cell imaging of synapse assembly (Friedman et al. 2000), receptor trafficking (Yudowski et al. 2007; Triller and Choquet 2008), and spine plasticity (Fischer et al. 1998). The methods we describe are equally applicable to such work.
GENERAL IMAGING CONSIDERATIONS
Imaging Modalities
Because, in our method, hippocampal neurons are grown directly on glass coverslips, a large field of view can be captured in a single focal plane. Axons and dendrites are quite thin so that optical sectioning techniques result in little improvement in resolution. Thus, conventional wide-field microscopy is well suited for imaging this preparation and is the approach we use for most applications.
We prefer confocal microscopy for high-resolution imaging of faint structures. When signal to noise is low, confocal imaging can reduce background by minimizing out-of-focus signal and out-of-focus autofluorescence. Culture media contain autofluorescent compounds such as riboflavin (Ludin et al. 1996) and folic acid, which contribute to noise in the image. Point-scanning confocal microscopy is not well suited for our applications because it can take several seconds to obtain images. Instead, we use a spinning-disk confocal microscope, which directs a laser beam through many pinholes to simultaneously scan multiple points in the specimen. The emitted light is captured with a charge-coupled device (CCD) camera. Each point in the specimen is illuminated intermittently, which prevents saturation of the fluorophore, reduces bleaching, and limits generation of reactive oxygen species, thereby lowering phototoxicity (Wang et al. 2005). The patented Yokogawa microlens design focuses an expanded laser beam into the pinholes on the spinning disk, which greatly increases light throughput of the system.
Microscope Configuration and Automation
For imaging cells in closed chambers (the configuration we use most frequently), either upright or inverted stands are suitable. To work with open chambers, an inverted stand is required. Another advantage of inverted stands is that they can be purchased with focus-stabilizing devices. These devices use an infrared laser beam to measure the distance between objective and coverslip and to correct for focal drift on a millisecond timescale. We have found that these devices greatly increase the yield of usable data in high-resolution imaging, such as imaging vesicle transport (see Short-Term High-Resolution Imaging of Developing Hippocampal Neurons in Culture [Kaech et al. 2012b]) in which focus drift is otherwise hard to eliminate.
For long-term live cell imaging, full automation of the microscope setup is highly desirable. A motorized stage allows rapid scanning of the entire culture to find and to record the positions of several cells, which can then be imaged in parallel by programming the microscope to return to each position in sequence. z-axis automation can be used for autofocusing. Most image acquisition software includes an autofocus algorithm that steps through a series of z-positions, acquires test images, and determines which z-position gives optimal focus, based on maximization of image contrast. Software-based autofocusing works well for time-lapse imaging but is too slow and exposes the cells to too much light to be useful for high-resolution imaging. Automation is also needed to shutter the transmitted and fluorescence light sources when images are not being acquired and to switch between the transmitted and epifluorescence light paths.
Several approaches can be used to configure and to automate the microscope for two-color imaging, depending on the application. For long-term two-color imaging, a motorized turret can be used to switch the fluorescent filter cubes. When image acquisition speed is critical, switching filter cubes is too slow. For rapid sequential imaging in two wavelengths, it is preferable to use a double dichroic beam splitter (which remains in place) together with filter wheels to rapidly switch the excitation and emission filters. Alternatively, the emitted light can be split with a dichroic mirror onto two cameras or onto two halves of a single camera. Laser-based illumination, as used in confocal microscopes, or the new light-emitting diode light sources can switch excitation wavelengths in microseconds. When combined with emission separation, this approach results in essentially simultaneous image acquisition in two colors. When using simultaneous two-color excitation, it is critical to check for bleed-through between the two fluorophores.
This degree of automation requires sophisticated image acquisition software that allows imaging parameters to be separately defined for each region to be imaged and that integrates control of all of these devices. Different commercially available software packages usually support only a limited set of devices, so the choice of software is an important consideration in designing a system for automated time-lapse imaging. Alternatively, knowledgeable programmers can take advantage of the Open Source μManager software (www.micro-manager.org) to coordinate device control during imaging.
Objectives
Cells plated on coverslips are well within the working distance of all microscope objectives, including high-NA PlanApo objectives, which offer the greatest optical correction and superior light gathering ability. These high-NA objectives are ideal for live cell fluorescence imaging. Phase contrast is the method we find most useful for imaging cell morphology because it shows fine neurites more clearly than differential interference contrast. Thus, we use phase-contrast objectives, although the phase ring in the rear focal plane of the objective slightly reduces its light throughput.
Cameras
We generally use cooled 12-bit CCD cameras with high-resolution chips and dual-speed readout rates because they offer the flexibility of acquiring images either with high quality and very little noise (lower readout speed, 1–2 frames/sec) or with significantly noisier images (rapid readout) at much higher frequencies. Image acquisition rate can also be increased by using only a subarray at full resolution or keeping the same field of view but binning neighboring pixels, which also increases the signal-to-noise ratio.
Environmental Control
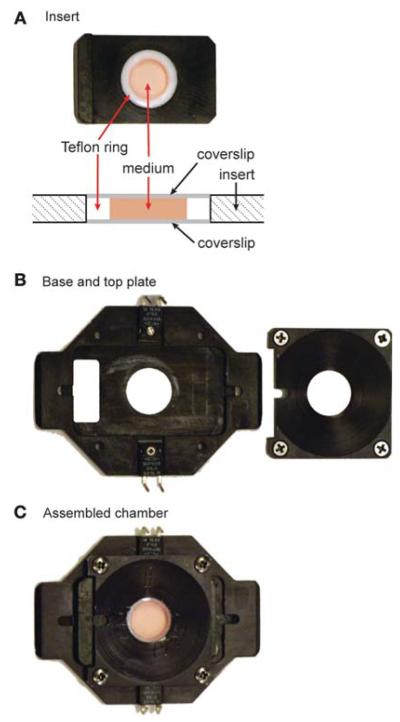
For long-term recordings, we prefer to use sealed chambers, which allow cells to be maintained in bicarbonate-based media without the need for maintaining a humidified CO2-enriched atmosphere. For good long-term growth, we have never found a substitute for the traditional bicarbonate-based culture media. We use the media from the dish the cells are grown in (so that it has been conditioned) and prepare the dishes with medium lacking phenol red, a substance suspected to give rise to toxic breakdown products when exposed to light. We use chambers manufactured by Warner Instruments, with custom-built inserts suitable for the 18-mm diameter coverslips we prefer (Fig. 2). In these chambers, the coverslip with cells attached rests face-up on a base plate. A Teflon ring above the coverslip forms a well to hold the culture medium, and a second, empty coverslip rests on this ring. Silicon grease applied to the Teflon ring forms a seal with the coverslips, and the entire coverslip sandwich is held in place between the top plate and the base, which are assembled together with screws. We have never encountered problems with toxicity from the Teflon or silicon grease but are suspicious of chambers that allow other materials such as rubber, to contact the culture medium. Thermistors regulate the temperature of the base, and heat from the base transfers to the outer edge of the coverslip. In our experience, this method maintains a reasonably constant temperature gradient across the coverslip (although it may not work well with large diameter coverslips). For accurate temperature control when using oil-immersion objectives, an objective heater is also essential.
FIGURE 2.
Closed imaging chamber. (A) A customized insert designed to hold 18-mm coverslips. The drawing shows a side view of the assembled insert. Note that the thickness of the Teflon ring is such that when the coverslips are in place, the assembly is flush with the top and bottom of the insert. To obtain an air-tight seal, a thin layer of silicon grease is applied to all areas of the ring in contact with the coverslips. When using the chamber in an open configuration, we use a thicker Teflon ring to compensate for the absence of the top coverslip. (B) The base and the top plate of the commercial platform (Series 20, Warner Instruments) are used to clamp the assembled insert in place. The base plate has a pair of resistive heater elements that are connected to a temperature controller. (C) The assembled chamber. For this illustration, the chamber volume was filled with phenol red-containing medium. For long-term imaging, we use medium lacking phenol red.
In short-term experiments, it is not necessary to optimize the medium for long-term cell survival. We prefer media with low background fluorescence (achieved by eliminating riboflavin and folic acid); and, for simplicity, we usually use media that maintain the correct pH in an air atmosphere. So long as there is no need to change the culture medium during the experiments, we use the same sealed chambers described above. Alternatively, open chambers can be used along with an imaging medium that maintains the correct pH in an air environment.
Labeling Cells with GFP-Tagged Proteins
For long-term imaging, our preferred approach is to transfect cells at the time of plating by Nucleofection, a method that combines electroporation with a cell-specific transfection medium that enhances nuclear delivery of plasmids. Using this approach, we routinely achieve transfection efficiencies in the double digits. However, there are caveats. As might be expected, transfection efficiency and level of expression are reduced for larger plasmids. We also find that the commonly used cytomegalovirus promoter does not give good long-term expression with Nucleofection. Instead, we prefer the chicken β-actin promoter (Niwa et al. 1991), which gives levels of expression that are lower but that persist for weeks.
For rapid image acquisition, the signal should be as high as possible. For this application, we use lipid-mediated transfection methods because they generally result in brighter cells.
ADVANTAGES AND DISADVANTAGES OF THE SYSTEM
The ability to follow individual cultured hippocampal neurons over time has greatly enhanced our understanding of the development of neuronal polarity and the way in which axons and dendrites grow and branch. For example, it is possible to follow dynamic changes in motor protein translocation in different neurites and to visualize transient changes in the accumulation of GFP-tagged signaling proteins in different growth cones during the period of competition that precedes axon specification (Nakata and Hirokawa 2003; Jacobson et al. 2006; Toriyama et al. 2006). Because hippocampal cultures are essentially two dimensional, it is much easier to image events with rapid dynamics, such as microtubule-based organelle trafficking (Burack et al. 2000; Kaether et al. 2000; Ruthel and Hollenbeck 2003; Wang and Schwarz 2009) and the dynamics of actin and microtubules (Fischer et al. 1998; Stepanova et al. 2003). Another major advantage of cultured cells is the unhindered accessibility to the cell’s surface. Single-molecule tracking using colloidal gold or nanoparticles has allowed analysis of receptor movements at synapses and has enabled localization of barriers to the diffusion of both proteins and lipids (Nakada et al. 2003; Triller and Choquet 2008). Because the cells grow directly on the surface of the coverslip, without intervening glial processes, hippocampal cultures also lend themselves to total internal reflection fluorescence (TIRF) microscopy, which detects events that occur within ~100 nm of the coverslip surface. This approach enhances the ability to detect weak signals, allowing detection of the release of single synaptic vesicles and the delivery of glutamate receptors to the cell surface (Balaji and Ryan 2007; Yudowski et al. 2007; Chen et al. 2008).
Of course, dissociated cell cultures cannot reproduce the precise alignment of dendritic arbors and the laminar organization of afferent projections that typify the hippocampus in vivo. The spatially organized interactions between neurons and glia that underlie neurogenesis and neuronal migration are also lost in dissociated cultures, and myelination does not occur. Many of these aspects of tissue organization are better maintained in hippocampal slice cultures, which are also widely used for live cell imaging. In this method, tissue slices (typically a few hundred micrometers thick) are cultured on membrane filters that are suspended at the air-medium interface. Slice cultures prepared from postnatal animals have been widely used to monitor changes in dendritic spine morphology, often by combining live cell imaging with electrophysiology (Zito et al. 2004). GFP-tagged proteins can be expressed in slice cultures using biolistics transfection; alternatively, cultures can be prepared from transgenic animals that express tagged proteins. More recently, slice cultures from late-stage embryos have been used to follow early events in neuronal migration and axon formation, using time-lapse imaging (Tabata and Nakajima 2003; Noctor et al. 2004; LoTurco and Bai 2006). In utero or ex utero electroporation offers a reliable method to express GFP-tagged proteins in embryonic slices. Although slice cultures are well suited to time-lapse imaging, they are not ideal for high-resolution imaging or for following very rapid events. Slice cultures may be imaged through the membrane filter from below (using long-working-distance objectives) or from above (using dipping lenses); neither approach is suitable for using the highest-NA objectives. Because cells in slice cultures grow in three dimensions, it is usually necessary to capture z-stacks, which adds significantly to the time needed for image acquisition.
Given all of the things that are lacking in dissociated cultures, it is somewhat surprising that so many aspects of neuronal development are faithfully reproduced in hippocampal cultures. For those of us whose research relies heavily on dissociated cultures, it is reassuring to note that several novel phenomena first described in such cultures have subsequently been confirmed in slice cultures or in living animals. The competition among multiple neurites that occurs before axonal specification (Dotti et al. 1988; Tabata and Nakajima 2003; Noctor et al. 2004), the anterograde transport of actin via growth-cone-like waves (Ruthel and Banker 1998, 1999; Flynn et al. 2009; Tint et al. 2009), and the rapid shape changes that occur in dendritic spines (Fischer et al. 1998; Dunaevsky et al. 1999; Zito et al. 2004) were all discovered first in hippocampal cultures.
ACKNOWLEDGMENTS
Research in our laboratory is supported by National Institutes of Health Grants MH66179 and NS17112. The Advanced Light Microscopy Core is supported in part by P30 Center Grant NS061800.
REFERENCES
- Balaji J, Ryan TA. Single-vesicle imaging reveals that synaptic vesicle exocytosis and endocytosis are coupled by a single stochastic mode. Proc Natl Acad Sci. 2007;104:20576–20581. doi: 10.1073/pnas.0707574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burack MA, Silverman MA, Banker G. The role of selective transport in neuronal protein sorting. Neuron. 2000;26:465–472. doi: 10.1016/s0896-6273(00)81178-2. [DOI] [PubMed] [Google Scholar]
- Chen X, Barg S, Almers W. Release of the styryl dyes from single synaptic vesicles in hippocampal neurons. J Neurosci. 2008;28:1894–1903. doi: 10.1523/JNEUROSCI.4518-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaevsky A, Tashiro A, Majewska A, Mason C, Yuste R. Developmental regulation of spine motility in the mammalian central nervous system. Proc Natl Acad Sci. 1999;96:13438–13443. doi: 10.1073/pnas.96.23.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- Flynn KC, Pak CW, Shaw AE, Bradke F, Bamburg JR. Growth cone-like waves transport actin and promote axonogenesis and neurite branching. Dev Neurobiol. 2009;69:761–779. doi: 10.1002/dneu.20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman HV, Bresler T, Garner CC, Ziv NE. Assembly of new individual excitatory synapses: Time course and temporal order of synaptic molecule recruitment. Neuron. 2000;27:57–69. doi: 10.1016/s0896-6273(00)00009-x. [DOI] [PubMed] [Google Scholar]
- Goslin K, Banker G. Rat hippocampal neurons in low-density culture. In: Banker G, Goslin K, editors. Culturing nerve cells. MIT University Press; Cambridge, MA: 1998. pp. 339–370. [Google Scholar]
- Jacobson C, Schnapp B, Banker GA. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 2006;49:797–804. doi: 10.1016/j.neuron.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kaech S, Huang C-F, Banker G. Long-term time-lapse imaging of developing hippocampal neurons in culture. Cold Spring Harb Protoc. 2012a doi: 10.1101/pdb.prot068239. doi: 10.1101/pdb.prot068239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Huang C-F, Banker G. Short-term high-resolution imaging of developing hippocampal neurons in culture. Cold Spring Harb Protoc. 2012b doi: 10.1101/pdb.prot068247. doi: 10.1101/pdb.prot068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaether C, Skehel P, Dotti CG. Axonal membrane proteins are transported in distinct carriers: A two-color video microscopy study in cultured hippocampal neurons. Mol Biol Cell. 2000;11:1213–1224. doi: 10.1091/mbc.11.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoTurco JJ, Bai J. The multipolar stage and disruptions in neuronal migration. Trends Neurosci. 2006;29:407–413. doi: 10.1016/j.tins.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Ludin B, Doll T, Meili R, Kaech S, Matus A. Application of novel vectors for GFP-tagging of proteins to study microtubule-associated proteins. Gene. 1996;173:107–111. doi: 10.1016/0378-1119(95)00899-3. [DOI] [PubMed] [Google Scholar]
- Nakada C, Ritchie K, Oba Y, Nakamura M, Hotta Y, Iino R, Kasai RS, Yamaguchi K, Fujiwara T, Kusumi A. Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat Cell Biol. 2003;5:626–632. doi: 10.1038/ncb1009. [DOI] [PubMed] [Google Scholar]
- Nakata T, Hirokawa N. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J Cell Biol. 2003;162:1045–1055. doi: 10.1083/jcb.200302175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Ruthel G, Banker G. Actin-dependent anterograde movement of growth-cone-like structures along growing hippocampal axons: A novel form of axonal transport? Cell Motil Cytoskeleton. 1998;40:160–173. doi: 10.1002/(SICI)1097-0169(1998)40:2<160::AID-CM5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ruthel G, Banker G. Role of moving growth cone-like “wave” structures in the outgrowth of cultured hippocampal axons and dendrites. J Neurobiol. 1999;39:97–106. doi: 10.1002/(sici)1097-4695(199904)39:1<97::aid-neu8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Ruthel G, Hollenbeck PJ. Response of mitochondrial traffic to axon determination and differential branch growth. J Neurosci. 2003;23:8618–8624. doi: 10.1523/JNEUROSCI.23-24-08618.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein) J Neurosci. 2003;23:2655–2664. doi: 10.1523/JNEUROSCI.23-07-02655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Multipolar migration: The third mode of radial neuronal migration in the developing cerebral cortex. J Neurosci. 2003;23:9996–10001. doi: 10.1523/JNEUROSCI.23-31-09996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tint I, Jean D, Baas PW, Black MM. Doublecortin associates with microtubules preferentially in regions of the axon displaying actin-rich protrusive structures. J Neurosci. 2009;29:10995–11010. doi: 10.1523/JNEUROSCI.3399-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama M, Shimada T, Kim KB, Mitsuba M, Nomura E, Katsuta K, Sakumura Y, Roepstorff P, Inagaki N. Shootin1: A protein involved in the organization of an asymmetric signal for neuronal polarization. J Cell Biol. 2006;175:147–157. doi: 10.1083/jcb.200604160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triller A, Choquet D. New concepts in synaptic biology derived from single-molecule imaging. Neuron. 2008;59:359–374. doi: 10.1016/j.neuron.2008.06.022. [DOI] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136:163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Babbey CM, Dunn KW. Performance comparison between the high-speed Yokogawa spinning disc confocal system and single-point scanning confocal systems. J Microsc. 2005;218:148–159. doi: 10.1111/j.1365-2818.2005.01473.x. [DOI] [PubMed] [Google Scholar]
- Yudowski GA, Puthenveedu MA, Leonoudakis D, Panicker S, Thorn KS, Beattie EC, von Zastrow M. Real-time imaging of discrete exocytic events mediating surface delivery of AMPA receptors. J Neurosci. 2007;27:11112–11121. doi: 10.1523/JNEUROSCI.2465-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito K, Knott G, Shepherd GM, Shenolikar S, Svoboda K. Induction of spine growth and synapse formation by regulation of the spine actin cytoskeleton. Neuron. 2004;44:321–334. doi: 10.1016/j.neuron.2004.09.022. [DOI] [PubMed] [Google Scholar]