Summary
Generation of induced pluripotent stem cells (iPSCs) is a process whose mechanistic underpinnings are only beginning to emerge. Here, we applied in-depth quantitative proteomics to monitor proteome changes during the course of reprogramming of fibroblasts to iPSCs. We uncover a 2-step resetting of the proteome during the first and last three days of reprogramming, with multiple functionally related proteins changing in expression in a highly coordinated fashion. This comprised several biological processes with a previously unknown role in reprogramming, including changes in the stoichiometry of electron transport-chain complexes, repressed vesicle-mediated transport during the intermediate stage and an EMT-like process in the late phase. In addition, we demonstrate that the nucleoporin Nup210 is essential for reprogramming by permitting rapid cellular proliferation and subsequent progression through MET. Along with the identification of proteins expressed in a stage-specific manner, this study provides a rich resource towards an enhanced mechanistic understanding of cellular reprogramming.
Introduction
Somatic cells can be reprogrammed to induced pluripotent stem cells (iPSCs) by the forced expression of only four transcription factors (TFs): Oct4, Klf4, Sox2 and c-Myc (OKSM) (Park et al., 2008; Takahashi and Yamanaka, 2006; Yu et al., 2007). iPCS share many properties with embryonic stem cells (ESCs), offering great potential for clinical and medical applications such as patient-specific regenerative medicine (Wu and Hochedlinger, 2011). To fulfill these prospects, and to design strategies improving the efficiency of iPSC generation, a better understanding of the reprogramming process is required at the molecular level. Recent studies have shown that reprogramming is accompanied by remodeling of the somatic cell transcription and chromatin programs (Maherali et al., 2007; Mikkelsen et al., 2008) and that it proceeds via intermediate steps (Brambrink et al., 2008; Plath and Lowry, 2011; Stadtfeld et al., 2008), characterized by the rapid induction of proliferation and downregulation of somatic genes, followed by a mesenchymal-to-epithelial transition (MET) (Li et al., 2010; Samavarchi-Tehrani et al., 2010). Only in the late stage the regulators of the pluripotent state (Oct4, Nanog) are expressed (Brambrink et al., 2008; Stadtfeld et al., 2008). In addition, several individual parameters not directly related to the composition of the transcription factor-cocktail have been demonstrated to affect efficiency or kinetics of reprogramming, e.g. miRNAs acting on the cell cycle, inhibition of p53, chemical inhibition of histone deacetylase (HDAC) and hypoxic culture conditions (Feng et al., 2009b; Huangfu et al., 2008; Krizhanovsky and Lowe, 2009; Wang et al., 2008; Zhu et al., 2010). Collectively, these studies have substantiated the notion that reprogramming is a multi-factorial process, where multiple fundamental cellular processes act synergistically in a sequential manner to reach pluripotency (Hanna et al., 2009; Stadtfeld et al., 2008).
Intermediate cells are still poorly characterized. Their investigation has been hampered mainly by the low efficiency of reprogramming, and by the heterogeneity of the cells undergoing reprogramming. In addition, there is a limited availability of protein markers that can be used as hallmarks for reprogramming status, and for isolation of distinct cell populations. This has been addressed in a recently developed model, now facilitating the enrichment of intermediate cells destined to become iPSCs based on the expression of Thy1, SSEA-1 and Oct4-GFP (Stadtfeld et al., 2010; Stadtfeld et al., 2008). Extending recent proteomic studies that have compared fibroblasts, ESCs and iPSCs (Huang et al., 2012; Munoz et al., 2011; Phanstiel et al., 2011), we have now exploited this system to perform an in-depth quantitative proteomic analysis for the first time spanning the entire course of reprogramming, aiming to study the order, timing and magnitude of proteome changes of fibroblasts reverting to pluripotency.
Results
In-depth quantitative proteome analysis of cellular reprogramming
Reprogramming was initiated in secondary mouse embryonic fibroblasts (MEFs) by doxycycline-induced expression of Oct4, Klf4, Sox2 and c-Myc (Stadtfeld et al., 2010). Commitment to a stable pluripotent cell fate was observed by days 9–12 and iPSCs were identified at day 15 (Polo and Hochedlinger, unpublished data). Cells were isolated over 15 days at 3-day intervals by FACS sorting, based on Thy1, SSEA-1 and Oct4-GFP expression, to enrich for cells with the potential to become iPSCs (Stadtfeld et al., 2008) (Figure 1, Figure S1A). For in-depth quantitative proteomic profiling, protein extracts from two biological replicates of the six time-points were digested, and peptides were labeled with stable isotopes via reductive methylation. Differentially labeled peptides from two consecutive time-points were combined and fractionated using isoelectric focusing. Peptide fractions were then analyzed by high-resolution nano-LC-MS/MS, and quantification of the abundance changes was based on MS signal intensities of the isotopically labeled peptide pairs (Figure 1, Figure S1A).
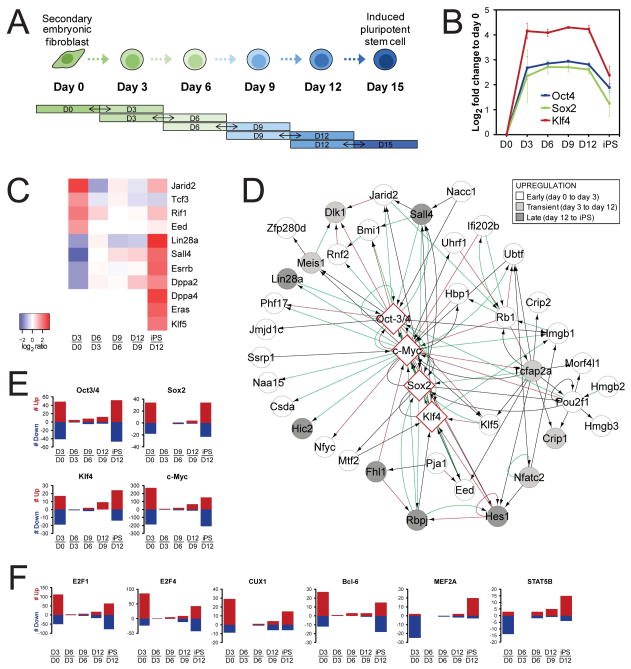
Figure 1. (A) Experimental design of the study.
Reprogramming was induced by expressing Oct4, Klf4, Sox2 and c-Myc. Cells were isolated at 3-day intervals by FACS sorting based on Thy1, SSEA-1 and GFP-Oct4 expression, followed by a quantitative proteomic analysis by pairwise comparison of two consecutive time-points. See also Fig. S1A. (B–F) Expression of transcription factors during reprogramming. (B) Expression profiles of the reprogramming factors. The average log2 expression change to day 0 +/− SEM for the transcription factors that were used to initiate reprogramming. C-Myc was not covered by the data, possibly because it is refractory to tryptic digestion. (C) Expression patterns of proteins in the core regulatory circuit associated with pluripotency. (D) Network of induced transcription factors that interact with the reprogramming factors. Increased expression early, transient or late is depicted in gray-scale. (E–F) Number of differentially expressed proteins that are targets of Oct4, Sox2, Klf4 or c-Myc (E) and other TFs (F). See also Figure S1.
From a total of 6,670,289 MS/MS spectra collected over 240 LC-MS/MS runs, 7918 unique protein groups were confidently identified with a false discovery rate of 1% (Table S1). Of these, 94% were identified on the basis of at least 2 peptides with an average of 8 unique peptides per protein (median 5). The approximate protein abundance spanned 7 orders of magnitude (Figure S1B), affirming that also very low-abundant proteins were detected. Importantly, the data cover large numbers of regulatory proteins, including 576 transcription factors (TFs), 357 kinases, 108 phosphatases, and 869 proteins involved in the cell cycle (Figure S1C, S1D).
In each of the sampled time-point comparisons, between 6262 and 6904 proteins were quantified in both replicates (Table S1). The accuracy of quantification was supported by the high number of quantitative events per protein (average ratio count 35, median 14). Overall, 5601 proteins were quantified at all time-points and in both replicates, while proteins not detected at one or more time-points suggest expression in a stage-specific manner.
The pluripotency network is induced rapidly
As expected, the three reprogramming factors that could be detected (Oct4, Sox2 and Klf4) showed enhanced expression along the course of reprogramming (Figure 1B). The data covered 65 Oct4-interacting proteins, including a large number of transcriptional regulators, chromatin modifiers, and general TFs that are thought to be important to maintain pluripotency in ESCs (Loh et al., 2011; Pardo et al., 2010; van den Berg et al., 2010) and that therefore may support a role in imposing pluripotency during reprogramming. The median 2-fold upregulation of these network components indicates a global and immediate activation upon induced Oct4 expression at day 3 (Figure S1E, Table S2). Their maintained expression during the reprogramming process suggests a sustained need for these proteins, such as proteins in the core regulatory circuit (Jarid2, Rif1, Tcf3, Eed) as well as protein complexes with a general role in transcription regulation (e.g. Mediator, TAF, RNA Pol II, Nurd) (Figure 1C and S1E, Table S2). In sharp contrast to this were several TFs whose expression was only induced at a late stage of reprogramming (Figure 1C), including proteins that have been defined as bona fide reprogramming factors themselves (Esrrb, Sall4, Klf5, Lin28) (Feng et al., 2009a; Jiang et al., 2008; Wong et al., 2008; Yu et al., 2007).
The network around the other core factors (Sox2, Klf4 and c-Myc) revealed many additional TFs with less established roles in the context of pluripotency (Figure 1D). Their early (Hmgb2, Hmgb3, Nfyc, Ssrp1, Oct1, Crip2), late (Jade1, Hic2) or transient expression (Meis1, Nfatc2, Crip1) (Table S1) may indicate stage-specific control of gene expression. Furthermore, exploring targets of TFs disclosed the differential expression of multiple OKSM targets in the first and last stage of reprogramming (Figure 1E). The stage-specific regulation of targets of various other TFs, such as the transcriptional repressor Bcl6, the homeobox protein Cux1 and the E2F cell cycle regulators, suggests transient activation of a diverse set of TFs (Figure 1F, S1F, S1G).
Proteome re-organization across multiple biological processes is highly coordinated and occurs in two oppositely regulated steps
The global proteomic changes of the 5601 proteins observed at all time points revealed that the large majority changed in expression at some point during reprogramming, most notably during the first and last 3 days (Figure 2A). At these time points, 1208 and 834 proteins were found to have a greater than 2-fold change in expression, respectively, while this was true for only 16, 43 and 77 proteins during intermediate stages. In addition, a strong anti-correlation (R = −0.90) was found for protein expression changes occurring early (transition from day 0 to day 3) and late (day 12 to iPS) (Figure 2B, Figure S2A). This shows that many proteins with a reduced expression in the early phase were upregulated in the late phase, and vice versa, suggesting a drastic resetting of the proteome at intermediate stages. Interestingly, a corresponding transcriptome dataset (Polo and Hochedlinger, unpublished data) showed a high degree of correlation with proteome changes within the first, but not the last 3 days of reprogramming. In fact, protein changes from day 12 to iPS correlated as well with mRNA changes from day 9 to day 12 (Figure S2B), suggesting that at least some of the proteome changes at day 12 may be the effect of transcriptional activation at an earlier time-point.
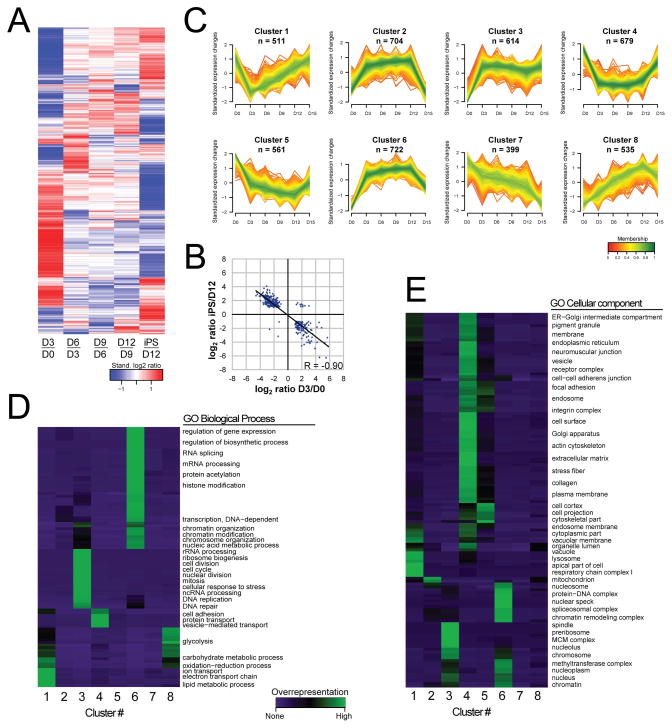
Figure 2. Proteome dynamics during reprogramming. (A) Heat map showing expression changes along reprogramming.
The row-clustered heatmap represents standardized average protein log2 ratios for all 5601 proteins quantified at all time-point comparisons, in both replicates. Note that most changes occur early and late. (B) Strong expression changes early and late show opposing direction. Log2 ratios of proteins with strong expression change both early and late during reprogramming (fold change >2 and ratio count ≥2 in both replicates from day 0 to day 3 and day 12 to day 15) are shown, together with the Pearson correlation coefficient. (C) Clusters of the protein dynamics along reprogramming. For the 5601 proteins quantified at all time-points (in both replicates), the ratio relative to day 0 was standardized and proteins were subjected to unsupervised clustering. An upper and lower ratio limit of log2(0.5) and log2(−0.5) was used for inclusion into a cluster. ‘n’ indicates the number of proteins within each cluster. Membership value represents how well the protein profile fit the average cluster profile. (D–E) Representative overrepresented biological processes and cellular components of the clusters. Each cluster from C was tested for overrepresented GO Biological Processes (D) and GO Cellular Components (E) compared to unregulated proteins. See also Figure S2.
Unsupervised clustering partitioned the temporal profiles of the 5601 proteins quantified at all time points into 8 clusters with distinct expression patterns (Figure 2C). Intriguingly, the clusters revealed stage-specific expression of many functionally related proteins and protein classes, representing biological processes occurring at distinct time-points or intervals (Figure 2D, 2E). For instance, proteins related to regulation of gene expression, RNA processing and chromatin organization are strongly induced at an early stage with a slight decrease at the final step to pluripotency (Cluster 6, Figure 2D). This included several subunits of RNA polymerase II as well as histones of all five main histone families (Table S1). This is in line with the well-recognized role of the chromatin state during reprogramming (Onder et al., 2012) and shows that upregulation of proteins involved in regulating gene expression is an early event. This is supported by proteins of similar functionality (general TFs and histone modifiers) in cluster 2, showing elevated expression in the early phase, and switching back to original levels at the final step to pluripotency. In addition, overrepresentation of mitochondria in this cluster (Figure 2E) suggests temporal elevation of mitochondrial proteins during the course of reprogramming. Cluster 3 indicates that many processes related to cell cycle and DNA repair are strongly induced and maintained from day 3 onwards (Figure 2D), including cell cycle proteins such as Cdk1, Cdk2, Cyclin B1, Plk1 and NPAT. The latter protein, required for progression through G1 and S phase, along with the strong increase in expression of Mki67 and Rbl1 (P107) at day 3, indicate the onset toward fast proliferation at the earliest stage of reprogramming. This supports the recent notion that for successful reprogramming the barrier of cell-cycle arrest needs to be overcome early (Ruiz et al., 2011; Smith et al., 2010).
The metabolic switch to glycolysis is preceded by a stoichiometric change in the electron transport system
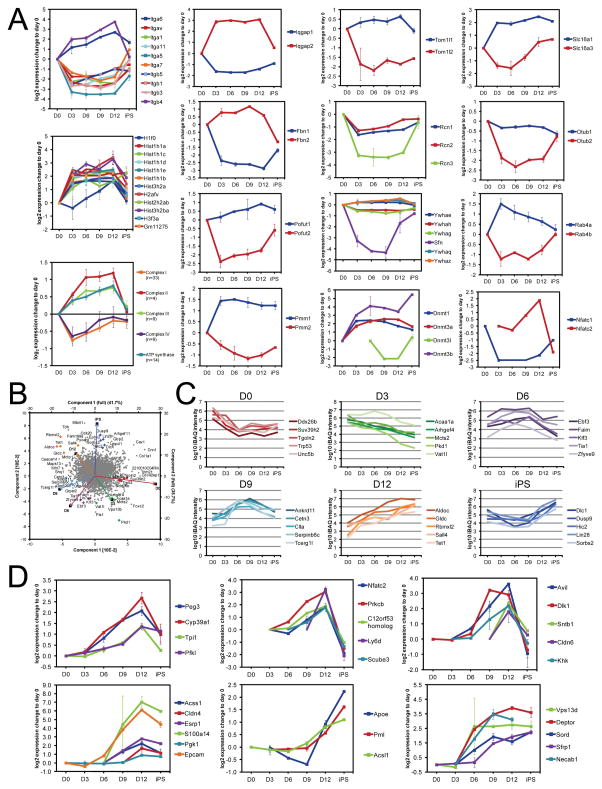
Of the 8 clusters, only two (7 and 8) showed gradual trends during the intermediate phase, with a decrease and increase over time, respectively (Figure 2C). Cluster 8 was found to be enriched for glycolytic proteins (Figure 2D), including e.g. Pfkl, Gpi and Pgk1. The overall gradual modest increase of expression of glycolytic proteins from day 0 to iPS (Fig S2C) suggests that the metabolic switch from mitochondrial oxidation to glycolysis, characteristic for fast-proliferating cells (Vander Heiden et al., 2009), is a gradual process. The early downregulation of proteins of the electron transport system, overrepresented in cluster 1 (Figure 2E), supports this idea. Interestingly, only complex I and IV were represented in cluster 1, while complexes II, III and V showed an opposite (i.e. increased) expression (Figure 5A). This indicates a change in the stoichiometry of the major components of the oxidative phosphorylation system, suggesting a change in the efficiency of oxidative phosphorylation (Boekema and Braun, 2007; van Raam et al., 2008). Together with a gradual increase in glycolytic activity noted above, this may provide a mechanism underlying the metabolic switch during reprogramming.
Figure 5. (A) Functionally related proteins with opposing expression profile along reprogramming.
Curves show mean+/− SEM of log2 expression change relative to day 0. (B–C) Principal component analysis reveals candidate proteins contributing particularly to a specific time-point. A PCA was applied to the approximated abundances (iBAQ) of each protein at each time-point. Proteins with probable high contribution to each time-point are highlighted in the biplot (B) and their temporal expression profiles shown in (C), with the same color-coding as in (B). (D) Proteins with a stage-specific expression pattern. Temporal expression profiles for examples of proteins with strong change (>fold 1.5) in at least one of the intermediate time-point comparisons, and only low change (<1.3-fold) or absence day 0 to day 3. Curves show mean +−/SEM of log2 expression change to day 0.
Opposing expression changes early and late point to altered protein transport and an epithelial-to-mesenchymal transition
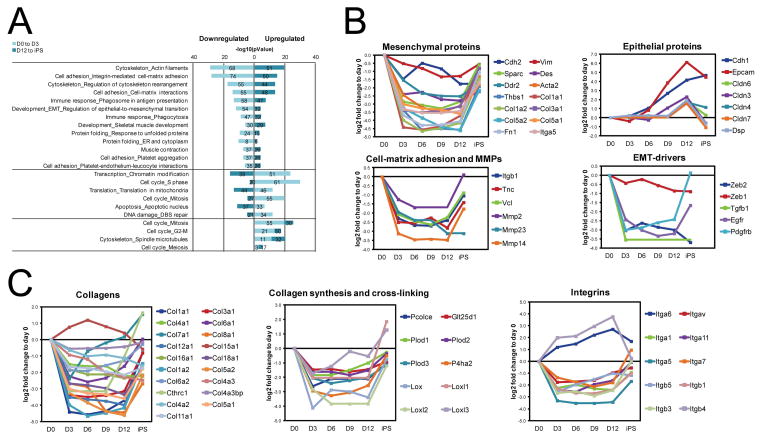
In the cluster reflecting strongly reduced expression from day 0 to day 3 and elevated expression from day 12 to the iPS stage (cluster 4), extracellular matrix (ECM) and cell adhesion proteins, including 158 plasma membrane proteins, were clearly overrepresented (Figure 2D). Intriguingly, this cluster was also enriched for proteins involved in vesicle-mediated transport (e.g. Rab34, Cd63, Mrc2, Sec24d), and included many proteins of the Golgi apparatus (e.g. Gosr2, Stx4, Scamp1, Scamp2) (Figure 2D and 2E). The co-regulation of these protein classes raises the possibility that reduced endosome recycling provides a mechanism to regulate surface-exposed signaling and adhesion proteins in intermediate stages. Interestingly, among the several processes that were found oppositely regulated early and late was ‘Regulation of Epithelial-to-mesenchymal transition’ (Figure 3A), a process strongly associated with altered cell adhesion and motility. This reflects the demonstrated role of MET in the early stage of reprogramming (Li et al., 2010; Samavarchi-Tehrani et al., 2010), and suggests an opposite process (EMT) occurring before the iPS state is reached. To further explore this idea, the expression characteristics of the key proteins in MET and EMT were examined in more detail. Consistent with an early MET, mesenchymal markers like N-cadherin, Fibronectin, Vimentin and Sparc, as well as cell-matrix adhesion proteins (e.g. Vinculin) and matrix metalloproteases (MMPs, e.g. Mmp14) were strongly reduced in expression at day 3, along with well-known inducers of EMT such as TGF-β, Zeb1 and Zeb2 (Figure 3B, Table S3). Furthermore, epithelial markers such as E-cadherin, claudins and Epcam, all showed an increased expression after day 3 or 6 (Figure 3B, Table S3), meaning that the repression of mesenchymal proteins precedes the upregulation of epithelial proteins. Strikingly, at day 12, most of the mesenchymal markers, cell-matrix adhesion proteins and MMPs were strongly upregulated, while the epithelial markers started to decrease in expression (Figure 3B), supporting an EMT-like process before reaching the iPS state. Although the key EMT transcription factor Snail remained undetected in our study, several other proteins known to induce EMT followed a pattern consistent with the induction of EMT around day 12 (e.g. Pdgfr, Egfr) (Ahmed et al., 2006; Yang et al., 2006). This also applies to several collagens and integrins (Figure 3C), protein classes that are thought to promote EMT and contribute to a gain in stem-cell properties (Hayashi et al., 2007; Imamichi and Menke, 2007). Interestingly, while MET at day 3 was reflected both at the transcript and protein level, a late EMT was only apparent from changes in protein levels (Figure 3B, S3B and S3C, Table S3), suggesting that the EMT-like process may be regulated at the post-transcriptional level.
Figure 3. (A) Overrepresented network processes for protein expression changes early and late.
Proteins with expression change (fold change >1.4 (|0.5| on log2-scale)) both early and late during reprogramming, were tested for overrepresented network processes. Displayed network processes were found overrepresented (p<0.01) both early and late. Note that no process was overrepresented for proteins whose expression decreased both from day 0 to day 3 and day 12 to iPS. (B) Temporal expression profiles of proteins related to EMT and MET. (C) Temporal expression profiles of proteins related to cytoskeletal organization and ECM proteins. See also Figure S3.
Proteins within complexes are tightly co-regulated during reprogramming
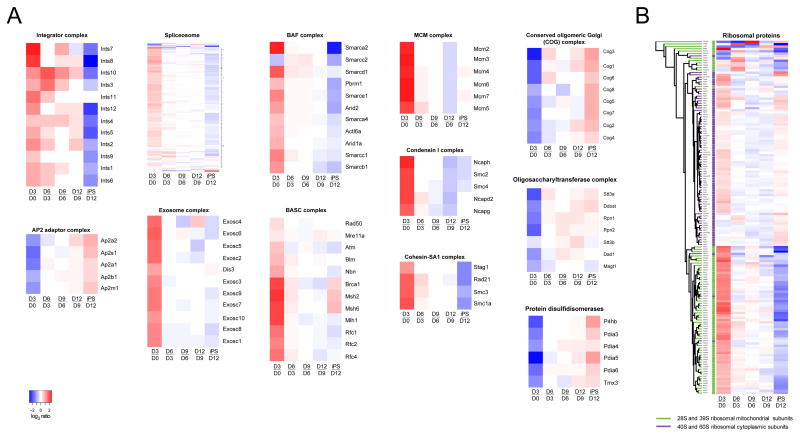
The coordinated proteome changes observed for biological processes (Figure 2) were also evidenced among components of protein complexes and protein families, showing concerted temporal dynamics of proteins within a large number of complexes with statistical significance (Table S4). These encompassed a range of functionalities, e.g. RNA processing (spliceosome and integrator complex), mRNA surveillance (exosome), DNA damage and replication (BASC and MCM complexes), vesicular transport (AP2 adaptor complex), protein degradation (COP9 signalosome), and glycosylation (COG and OST complexes), each of which showed a distinct expression profile (Figure 4A). For many complexes, our data fully covered all known subunits. While some of the complexes have previously been shown to play a role during reprogramming (e.g. BAF (Singhal et al., 2010)) or ESC-specific gene programs (e.g. Cohesin complex (Nitzsche et al., 2011)), many others have not. Interestingly, the data substantiate that protein folding in the ER (115 proteins with KEGG term ‘Protein processing in endoplasmic reticulum’, including disulfide isomerases), glycosylation in the Golgi and ER (COG and OST complexes) and protein transport (AP2 complex, Golgin subfamily A and KEGG term ‘Protein export’) are inhibited at intermediate stages (Table S4, Figure 4A). In addition, we observed subtle but consistent differences between the chromosome segregation complexes Cohesin and Condensin, suggesting tightly controlled protein expression (Figure 4A). This is also reflected by cytoplasmic and mitochondrial ribosomes each having a distinct expression profile, supported by the demonstration that unsupervised clustering could reconstitute 28S and 39S ribosomes on one hand, and 40S and 60S on the other (Figure 4B). Similarly, the 19S (PA700 complex) and the 20S subcomponents of the proteasome show distinct and even slightly opposing profiles (Table S4). All together, the data point to the tight and specific regulation of functionally distinct subgroups of the proteome at specific stages of reprogramming.
Figure 4. Proteins within complexes or with related function showing exceptionally similar dynamics along reprogramming.
The significance of expression profile similarities within groups of interest was assessed using the R package proteinProfiles. (A) Examples of protein complexes or protein families for which the proteins show similar dynamics with statistical significance (all examples have p<0.015). (B) Clustering of mitochondrial and cytoplasmic ribosomal proteins. Similarities in expression profiles showed statistical significance within all four ribosomal subunits (28S and 39S (mitochondrial) and 40S and 60S (cytoplasmic)) (maximal p-value was 0.0005). The proteins of all four subunits were grouped by unsupervised clustering based on expression changes. Mitochondrial and cytoplasmic are indicated in green and purple, respectively.
Functionally related proteins with opposing expression profiles point to a potential role in reprogramming
While co-regulation of functionally related proteins reflects tight coordination of protein expression, deviation from this pattern by one or more proteins may indicate a specific functionality during reprogramming. Several of such profiles were observed in the data (Figure 5A). This included integrins alpha 6 and beta 4, which associate to form a receptor for laminin and have a suggested role in proliferation and migration properties (Guo and Giancotti, 2004); the epithelial marker 14-3-3 protein sigma (Sfn) regulating ESC proliferation (Chang et al., 2012); and Histone H1.0, which is differentially incorporated into chromatin during reprogramming (Terme et al., 2011).
Interestingly, opposed expression profiles were found for several protein pairs involved in ECM remodeling and EMT, e.g. Nfatc1 and Nfatc2, as well as Iqgap1 and Iqgap2 (Figure 5A). This suggests functionality in reprogramming by opposing roles of these proteins in the regulation of cell cycle and apoptosis, as well as in inhibiting EMT and promoting MET (Kuroda et al., 1998; Robbs et al., 2008; White et al., 2009). Analogously, the opposing expression profiles of Fibrillin-1 and -2 may control differential availability of TGF-β and bone morphogenetic protein signaling, which inhibit and activate EMT, respectively (Maherali and Hochedlinger, 2009; Nistala et al., 2010; Samavarchi-Tehrani et al., 2010). In addition, the opposing expression profile of the two O-fucosyltransferases Pofut1 and Pofut2 possibly reflects the fact that Pofut2, but not Pofut1, targets components of the ECM thereby modulating ECM synthesis and remodeling (Du et al., 2010).
The data also covered the quantification of all three mammalian DNA methyltransfereases Dnmt1, Dnmt3a and Dnmt3b, critical for altering the epigenetic landscape during reprogramming (Lister et al., 2011). While Dnmt1 and Dnmt3a showed highest expression in the intermediate phase, Dnmt3b was strongly upregulated from day 12 to day 15, to a final iPS expression more than 40-fold higher than in fibroblasts (Figure 5A). This was concomitant with a sharp increase of Dnmt3l, a catalytic activator of both Dnmt3a and Dnmt3b (Gowher et al., 2005). The results fit a model where the differential regulation of the two de novo DNA methyltransferases controls non-CpG methylation along reprogramming (Ziller et al., 2011).
Nup210 is essential for reprogramming
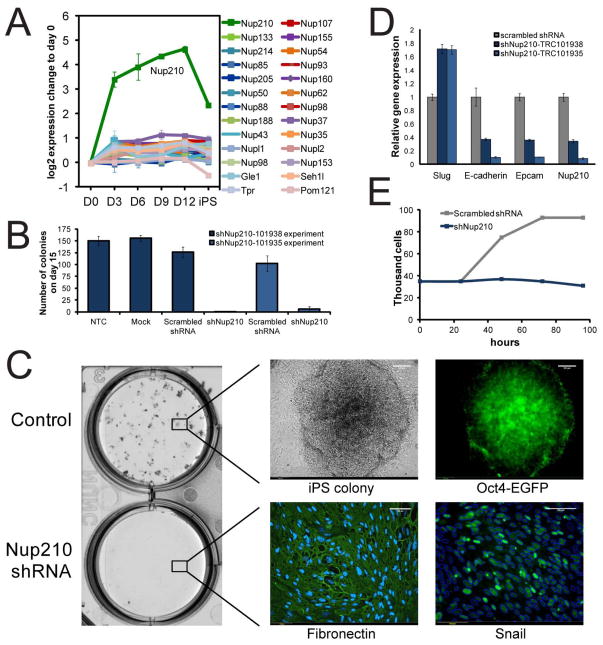
Of the 26 nucleoporins that were quantified, 25 showed a coherent modestly increased expression, while in sharp contrast the transmembrane nucleoporin Nup210 showed a more than 10-fold increase from day 3 onwards (Figure 6A). The recent observation that Nup210 shows a very similar profile during differentiation, regulating the expression of pluripotency and somatic genes (D’Angelo et al., 2012), raises the possibility that Nup210 fulfills a critical role during reprogramming as well. To address this question, secondary fibroblasts were transduced with two independent Nup210 shRNAs followed by the induction of reprogramming. Strikingly, reprogramming and iPSC colony formation was completely blocked in the absence of Nup210 (Figure 6B and 6C). In fact most of the cells retained their mesenchymal-like morphology even 15 days after OKSM induction, and expressed markers highly expressed in mesenchymal cells (Fibronectin and Snail) (Figure 6C). In addition, appearance of epithelial markers (Epcam and E-cadherin) was interrupted (Figure 6D). Although Nup210 is expressed at a very low level in fibroblast cells (Figure S4E), its repression completely halted cellular proliferation of the cells (Figure 6E). These results indicate that increased expression of Nup210 at the onset of reprogramming is required to permit rapid cell proliferation, and hence progression through MET.
Figure 6. Nup210 is required for reprogramming. (A) Protein xxpression profile of nucleoporins.
Curves show mean+/− SEM of log2 expression change relative to day 0. (B–E) Nup210 was depleted (using shRNA) in secondary MEF cells during reprogramming to iPS cells. (B) Number of colonies (mean+/− SD) in controls and Nup210 knocked-down cells are shown, for two independent Nup210 shRNAs. (C) While many Oct4-EGFP colonies developed in the controls, Nup210-knock down cells showed expression of Fibronectin and the mesenchymal marker Snail after 15 days of reprogramming. Upper panel shows iPS colony formation for control cells, and lower panel the Nup210 knock-down cells and their expression of Fibronectin and Snail at day 15 after induction of reprogramming. Scale bar represents 100 μm. For Snail expression, the border of the nucleus (defined by Hoechst staining, see Figure S4) is shown in blue, demonstrating that virtually all cells retain a mesenchymal phenotype. (D) Relative gene expression (mean+/− SD) in control and Nup210 knocked-down cells 15 days after reprogramming. (E) Growth curve for control and Nup210-knock down MEFs.
Stage-specific protein expression characterizes intermediate cells
The identification of proteins that are expressed in a stage-specific manner may help to explain the mechanism of reprogramming, or to elucidate markers for intermediate cells. To nominate such proteins, a Principle Component Analysis (PCA) was applied to the approximated abundance of each protein observed at each time-point. The overall pattern, displayed as a biplot (Figure 5B), positions the sampled time points in chronological order, pointing to a successive change in proteome composition. Most importantly, the proteins that contributed most prominently in the PCA analysis provide candidate proteins that best represent the individual time points (e.g. p53, Mcts2, Tia, Serpinb6c, Aldoc and Lin28 for day 0, 3, 6, 9, 12 and 15, respectively, Figure 5C). This collective set of proteins may provide a signature for subsequent states of reprogramming.
As an alternative approach, individual proteins that changed in expression specifically at intermediate stages were selected (Figure 5D). Several of these had emerged in the context of MET (e.g. Epcam, Claudin 4 and 6) or glycolysis (e.g. Pgk1, Pfkl, Tpi1), but they also include several other transiently expressed proteins (e.g. Dlk1, Ly6d, Peg3, Avil). Interestingly, Dlk1 was recently shown to be a stem cell protein, whose loss in expression results in enhanced differentiation (Begum et al., 2012), and Epcam has been shown to be useful for isolating iPSCs (Chen et al., 2011; Gundry et al., 2012). This confirms the usefulness of the approach and indicates that the presented proteins may help to distinguish intermediate cellular populations that are prone to reach the pluripotent state.
Finally, we compiled inventories of additional protein classes based on distinct expression patterns. Specifically, Table S5 shows proteins that were not identified until the fully reprogrammed iPS state, thus representing proteins that may be regarded as iPS-cell specific (e.g. Sod3 and Eras), as well as proteins that disappear directly after the start of reprogramming (e.g. Tgfb1 and Slc6a9). Table S6 includes membrane proteins that are changing strongly between any two consecutive time-points, and attain their highest expression at the iPS stage. These proteins, e.g. Cav1 and Gbp2, may be strong candidates for FACS-based sorting of partially and fully reprogrammed cells.
Discussion
Since the pioneering finding in 2006 that pluripotency can be induced in somatic cells (Takahashi and Yamanaka, 2006) a comprehensive view of the underlying mechanisms that drives reprogramming to the induced pluripotent state remains incomplete. Since there is a notable lack of understanding of reprogramming at the protein level, we applied quantitative proteomics to profile dynamic changes in protein abundance during the course of reprogramming of fibroblasts to pluripotency, resulting in a proteome sampled at great depth (close to 8000 proteins) and spanning a wide dynamic range (7 orders of magnitude).
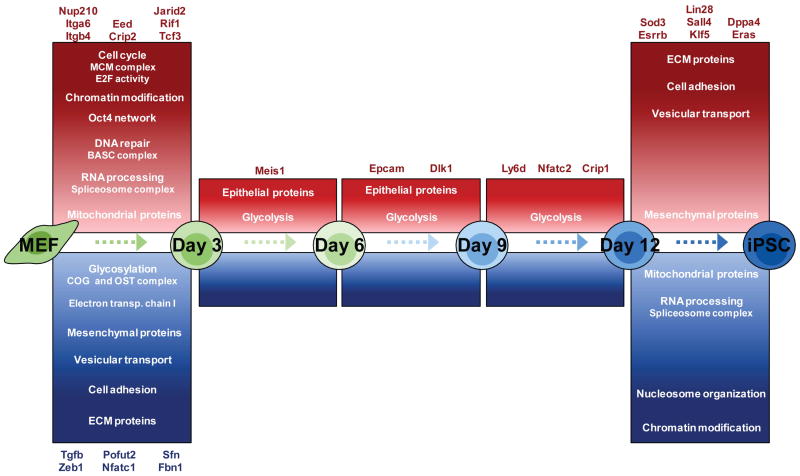
While it has become evident that successful reprogramming is a multistep process, most studies have focused on the initiation phase, leaving the changes required for the final transition poorly characterized (Plath and Lowry, 2011). The capability to obtain intermediate cells destined to become iPSCs (Stadtfeld et al., 2010; Stadtfeld et al., 2008) enabled us to generate a detailed proteomic view of the events taking place across the entire process of reprogramming. We found that major reorganization of the proteome takes place during the first three days of reprogramming, as well as in the final step after day 12, while more subtle changes occur in the intermediate phase (Figure 7). This applied to multiple gene ontologies, suggesting a global and coordinated 2-step rearrangement of the proteome. The pronounced changes of the proteome within the first three days reflect the kinetics observed in gene expression analyses (Samavarchi-Tehrani et al., 2010) (Polo and Hochedlinger, unpublished data). A striking finding was the opposing direction of protein expression changes during the first and last step of reprogramming. Indeed, our data clearly show that a large portion of the proteins have their extreme expression (either maximal or minimal) in the intermediate cells (Figure 2), indicating a highly particular proteomic identity very different from their origin (fibroblasts) or destination (iPSC).
Figure 7. Model of highly coordinated proteome dynamics during reprogramming.
Protein upregulation and downregulation is shown in red and blue, respectively, where color intensity reflects the degree of regulation. Examples are given of individual proteins that are expressed or repressed in a stage-specific manner.
Highly coordinated proteome changes were apparent from synchronized biological processes (Figure 2), but even more from the tight co-regulation of subunits in protein complexes indicating the concerted involvement of multiple processes across multiple cellular compartments (Figure 4, Table S4). The strong upregulation at day 3 of numerous proteins related to cell division, DNA replication, chromatin modification and DNA damage response can all be related to accelerated cell cycle progression, by shortening of the G1 phase for iPSCs compared to somatic cells (Ghule et al., 2011). It is striking that the machinery to effectuate and proofread this program is implemented already at the earliest stage of reprogramming. This was concomitant with elevated levels of proteins involved in transcription, post-transcriptional processes (mRNA processing, splicing, and degradation, Figure 2D and 4A) and translation (translation factors and ribosomes, Table S1, Figure 4B). In contrast to these processes, glycolytic enzymes increased progressively in the intermediate phase, suggesting a gradual transformation of energy metabolism (Figure 2D and S2C). Strikingly, this was preceded by the decreased expression of complex I and IV in the electron transport chain at day 3 (Figure 5A), which was in sharp contrast to the rest of the complexes within the electron transport chain, as well as to most mitochondrial proteins, showing a temporal elevated expression in the intermediate phase (Figure 2E). This change in stoichiometry of the electron transport chain complexes is likely to affect the composition of respiratory supercomplexes, leading to a change in efficiency of oxidative phosphorylation and suggests reduced or even uncoupling of ATP generation via oxidative phosphorylation (Boekema and Braun, 2007; van Raam et al., 2008). This is in accordance with the notion that high proliferation rates are fuelled by a shift from oxidative phosphorylation to glycolysis (DeBerardinis et al., 2008; Vander Heiden et al., 2009). Indeed, stimulation of the glycolytic flux has been shown to increase the efficiency of reprogramming (Folmes et al., 2011; Zhu et al., 2010), where glycolysis and oxidative phosphorylation are compensatory mechanisms (Folmes et al., 2011). This suggests that a decrease in oxidative phosphorylation via a reduction of complex I early during reprogramming may be one of the driving forces to enhance glycolysis.
Another key event early during reprogramming is MET, recapitulated in our data by the sudden loss of multiple ECM proteins, including mesenchymal markers, and the gradual gain of epithelial proteins (Figure 3). Intriguingly, this pattern is almost completely reversed as reprogramming approaches completion, reminiscent of an EMT-like process. Although this notion as such is novel, it may bridge an incongruity between the observation that MET is required for reprogramming (Li et al., 2010; Samavarchi-Tehrani et al., 2010), and at the same time that the opposed process (i.e. EMT) confers stem cell properties to epithelial cells (Mani et al., 2008; Morel et al., 2008). Epithelial markers (e.g. Epcam) only started to revert to their initial levels by day 15, suggesting that only partial EMT occurs and that reprogrammed cells retain some epithelial characteristics. Therefore, the possibility remains that the observed effect may not be strictly defined as EMT but is simply a change in adhesion molecules. In conjunction, we observed the loss and regain of proteins in ER, Golgi and endosomal vesicles in a profile very similar to ECM proteins (Figure 2D and 4A). This may reflect a mechanistic link between protein folding, glycosylation and transport to regulate deposition of surface adhesion molecules, thereby modulating cellular interaction or morphology in intermediate stages to facilitate cellular reprogramming.
Despite few pronounced events between day 3 and 12, we identified multiple individual proteins that are transiently expressed at early, late or intermediate stages with a potential role in driving reprogramming to completion (Figure 5, Table S5, Table S6). A pivotal example is the nuclear pore protein Nup210 whose expression sharply increased at day 3 (Figure 6). Countering this by using shRNA-mediated inhibition of Nup210, we showed that MEFs fail to reprogram in the absence of Nup210 (Figure 6). Interestingly, earlier observations have indicated that Nup210 is dispensable for nuclear pore complex assembly (Eriksson et al., 2004; Stavru et al., 2006), showing preferred expression in epithelial cells while absent in mesenchymal cells, including fibroblasts (Eriksson et al., 2004; Olsson et al., 2004; Stavru et al., 2006). In spite of low expression of Nup210 in MEFs (Figure S4), we observed that its repression resulted in inhibition of cellular proliferation. Consequently, progression through MET was prevented and reprogramming blocked (Figure 6). Recent data have shown that Nup210 is also required for cellular differentiation by regulating the expression of pluripotency and somatic genes (D’Angelo et al., 2012). Intriguingly, among the genes that are upregulated upon Nup210 knock-down during cellular differentiation (i.e. that are repressed in the presence of Nup210 (D’Angelo et al., 2012)), most of the corresponding proteins in our data showed an upregulation during reprogramming (Figure S4D). This suggests that Nup210 may have opposing effects on the regulation of target genes depending on cellular context. All together, our observations indicate that Nup210 has a critical regulatory role in cell cycle progression and reprogramming. In addition, this indicates that factors affecting reprogramming may be sought beyond classical transcription factors and chromatin modifiers.
In summary, as the first analysis of temporal protein dynamics during reprogramming, the data presented here offer important insights into proteome dynamics underlying OKSM-induced reprogramming. We have identified many individual proteins, protein complexes and biological processes that accompany reprogramming, some of which in a causal manner as exemplified for Nup210. We therefore believe that these data constitute a rich resource that may assist to further our mechanistic understanding of cellular plasticity, and to advance practical applications of iPS technology.
Experimental procedures
Reprogramming experiments and cell collection
MEF cultures were established from E13.5 embryos from a reprogrammable mice strain carrying one copy of the OKSM cassette and Rosa26-M2rtTA allele (het/het) or carrying two copies of the OKSM cassette and Rosa26-M2rtTA allele (ho/ho), as well as the GFP reporter for Oct4. Reprogramming was performed in ESC medium in the presence of doxycycline. Cells were isolated at 3-day intervals from day 0 to day 15 by FACS sorting, based on Thy1, SSEA-1 and Oct4-GFP expression. For Nup210 knock-down experiment, secondary MEF cells were transduced by lentiviral vectors (two different Nup210 shRNAs, scrambled shRNA or mock), and reprogramming was induced as described above.
Peptide stable isotope labeling and fractionation
After lysis of cells, proteins were reduced/alkylated and digested with trypsin. Resulting peptides were differentially labeled with stable isotope dimethyl labeling on column as previously described (Boersema et al., 2009). Briefly, peptides from consecutive reprogramming time-points were labeled with a mixture of either formaldehyde-H2 and sodium cyanoborohydride (‘light’ reagent) or formaldehyde-D2 with cyanoborohydride (‘heavy’ reagent). In a second biological replicate, time-point reagents were swapped, and the ‘light’ and ‘heavy’ labeled samples were mixed in 1:1 ratio based on total peptide amount. Sample complexity was reduced by fractionating the peptides with OFFGEL isoelectric focusing (Agilent), into 12 fractions.
LC-ESI-MS/MS analysis
In technical duplicates, peptides were separated by nanoflow UPLC on a 120 min gradient and analyzed by ESI-MS/MS on an LTQ Orbitrap Velos or Orbitrap Velos Pro (Thermo Fisher Scientific). Full scan spectra from m/z 300 to 1700 at resolution 30,000 were acquired in the Orbitrap MS. The most intense ions (up to 15) from the full scan MS were selected for fragmentation in the ion trap.
Protein identification and quantification
MS raw data files were processed with MaxQuant (version 1.2.2.5) (Cox and Mann, 2008). The derived peak list was searched using the in-built Andromeda search engine (version 1.2.2.5) in MaxQuant against the Uniprot mouse database (2011.06.21). A 1% false discovery rate (FDR) was required at both the protein level and the peptide level. The protein identification was reported as an indistinguishable “protein group” if no unique peptide sequence to a single database entry was identified. The iBAQ algorithm was used for estimation of the abundance of different proteins within a single sample (proteome) (Schwanhausser et al., 2011).
Bioinformatic analysis
Protein classification was performed using PANTHER classification system (Mi et al., 2007). Network and transcription factor activity analysis was done using MetaCore (GeneGo Inc., (Nikolsky et al., 2005)). GproX was used for clustering and Gene Ontology (GO) enrichment analysis (Rigbolt et al., 2011). Principal component analysis was performed with Perseus (version 1.2.0.11, within the MaxQuant package). The significance of expression profile similarities within groups of interest was assessed using the R package proteinProfiles.
Acknowledgments
We gratefully acknowledge the EMBL Proteomics Core Facility for excellent technical support and Bernd Fischer for advice in statistics. We thank Ryan Walsh for secondary MEF cells and Martin W. Hetzer for Nup210 shRNA. This work was supported by the Netherlands Organisation for Scientific Research (NWO).
References
- Ahmed N, Maines-Bandiera S, Quinn MA, Unger WG, Dedhar S, Auersperg N. Molecular pathways regulating EGF-induced epithelio-mesenchymal transition in human ovarian surface epithelium. Am J Physiol Cell Physiol. 2006;290:C1532–1542. doi: 10.1152/ajpcell.00478.2005. [DOI] [PubMed] [Google Scholar]
- Begum A, Kim Y, Lin Q, Yun Z. DLK1, delta-like 1 homolog (Drosophila), regulates tumor cell differentiation in vivo. Cancer Lett. 2012;318:26–33. doi: 10.1016/j.canlet.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema EJ, Braun HP. Supramolecular structure of the mitochondrial oxidative phosphorylation system. J Biol Chem. 2007;282:1–4. doi: 10.1074/jbc.R600031200. [DOI] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Liu CC, Hsing EW, Liang SM, Chi YH, Sung LY, Lin SP, Shen TL, Ko BS, Yen BL, et al. 14-3-3sigma Regulates beta-Catenin-Mediated Mouse Embryonic Stem Cell Proliferation by Sequestering GSK-3beta. PLoS One. 2012;7:e40193. doi: 10.1371/journal.pone.0040193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HF, Chuang CY, Lee WC, Huang HP, Wu HC, Ho HN, Chen YJ, Kuo HC. Surface marker epithelial cell adhesion molecule and E-cadherin facilitate the identification and selection of induced pluripotent stem cells. Stem Cell Rev. 2011;7:722–735. doi: 10.1007/s12015-011-9233-y. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A change in nuclear pore complex composition regulates cell differentiation. Dev Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Du J, Takeuchi H, Leonhard-Melief C, Shroyer KR, Dlugosz M, Haltiwanger RS, Holdener BC. O-fucosylation of thrombospondin type 1 repeats restricts epithelial to mesenchymal transition (EMT) and maintains epiblast pluripotency during mouse gastrulation. Dev Biol. 2010;346:25–38. doi: 10.1016/j.ydbio.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson C, Rustum C, Hallberg E. Dynamic properties of nuclear pore complex proteins in gp210 deficient cells. FEBS Lett. 2004;572:261–265. doi: 10.1016/j.febslet.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009a;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009b;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Medina R, Lengner CJ, Mandeville M, Qiao M, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Reprogramming the pluripotent cell cycle: restoration of an abbreviated G1 phase in human induced pluripotent stem (iPS) cells. J Cell Physiol. 2011;226:1149–1156. doi: 10.1002/jcp.22440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- Gundry RL, Riordon DR, Tarasova Y, Chuppa S, Bhattacharya S, Juhasz O, Wiedemeier O, Milanovich S, Noto FK, Tchernyshyov I, et al. A Cell Surfaceome Map for Immunophenotyping and Sorting Pluripotent Stem Cells. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M112.018135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816–826. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Furue MK, Okamoto T, Ohnuma K, Myoishi Y, Fukuhara Y, Abe T, Sato JD, Hata R, Asashima M. Integrins regulate mouse embryonic stem cell self-renewal. Stem Cells. 2007;25:3005–3015. doi: 10.1634/stemcells.2007-0103. [DOI] [PubMed] [Google Scholar]
- Huang X, Tian C, Liu M, Wang Y, Tolmachev AV, Sharma S, Yu F, Fu K, Zheng J, Ding SJ. Quantitative proteomic analysis of mouse embryonic fibroblasts and induced pluripotent stem cells using 16O/18O labeling. J Proteome Res. 2012;11:2091–2102. doi: 10.1021/pr300155r. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamichi Y, Menke A. Signaling pathways involved in collagen-induced disruption of the E-cadherin complex during epithelial-mesenchymal transition. Cells Tissues Organs. 2007;185:180–190. doi: 10.1159/000101319. [DOI] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–1086. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, et al. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin- mediated cell-cell adhesion. Science. 1998;281:832–835. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Yang L, Yang JC, Li H, Collins JJ, Daley GQ. Genomic approaches to deconstruct pluripotency. Annu Rev Genomics Hum Genet. 2011;12:165–185. doi: 10.1146/annurev-genom-082410-101506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz J, Low TY, Kok YJ, Chin A, Frese CK, Ding V, Choo A, Heck AJ. The quantitative proteomes of human-induced pluripotent stem cells and embryonic stem cells. Mol Syst Biol. 2011;7:550. doi: 10.1038/msb.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolsky Y, Ekins S, Nikolskaya T, Bugrim A. A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol Lett. 2005;158:20–29. doi: 10.1016/j.toxlet.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Nistala H, Lee-Arteaga S, Smaldone S, Siciliano G, Carta L, Ono RN, Sengle G, Arteaga-Solis E, Levasseur R, Ducy P, et al. Fibrillin-1 and -2 differentially modulate endogenous TGF-beta and BMP bioavailability during bone formation. J Cell Biol. 2010;190:1107–1121. doi: 10.1083/jcb.201003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L, Huebner N, Mann M, et al. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS One. 2011;6:e19470. doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M, Scheele S, Ekblom P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp Cell Res. 2004;292:359–370. doi: 10.1016/j.yexcr.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Phanstiel DH, Brumbaugh J, Wenger CD, Tian S, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, et al. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat Methods. 2011;8:821–827. doi: 10.1038/nmeth.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Vanselow JT, Blagoev B. GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol Cell Proteomics. 2011;10:O110 007450. doi: 10.1074/mcp.O110.007450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbs BK, Cruz AL, Werneck MB, Mognol GP, Viola JP. Dual roles for NFAT transcription factor genes as oncogenes and tumor suppressors. Mol Cell Biol. 2008;28:7168–7181. doi: 10.1128/MCB.00256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S, Panopoulos AD, Herrerias A, Bissig KD, Lutz M, Berggren WT, Verma IM, Izpisua Belmonte JC. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F, Nautrup-Pedersen G, Cordes VC, Gorlich D. Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells. J Cell Biol. 2006;173:477–483. doi: 10.1083/jcb.200601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Terme JM, Sese B, Millan-Arino L, Mayor R, Belmonte JC, Barrero MJ, Jordan A. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J Biol Chem. 2011;286:35347–35357. doi: 10.1074/jbc.M111.281923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raam BJ, Sluiter W, de Wit E, Roos D, Verhoeven AJ, Kuijpers TW. Mitochondrial membrane potential in human neutrophils is maintained by complex III activity in the absence of supercomplex organisation. PLoS One. 2008;3:e2013. doi: 10.1371/journal.pone.0002013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Gaspar-Maia A, Ramalho-Santos M, Reijo Pera RA. High-efficiency stem cell fusion-mediated assay reveals Sall4 as an enhancer of reprogramming. PLoS One. 2008;3:e1955. doi: 10.1371/journal.pone.0001955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu ZR. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell. 2006;127:139–155. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Muller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]