Abstract
We identified 37 hematopoietic cell transplantation recipients with human herpesvirus 6 (HHV-6) central nervous system dysfunction and tested donors/recipients for chromosomally integrated (ci)HHV-6. One patient had ciHHV-6A with possible HHV-6A reactivation and encephalitis. There was no ciHHV-6 enrichment in this group, but larger studies are needed to determine if patients with ciHHV-6 are at increased risk for HHV-6-associated diseases or other complications.
Keywords: herpesvirus, hhv-6, integration, transplant, CNS
INTRODUCTION
Human herpesvirus 6 (HHV-6) has a unique ability to integrate into chromosomal telomeres of infected cells1. When this occurs in germ cells, Mendelian inheritance results in offspring with chromosomally integrated (ci)HHV-6 in every nucleated cell. This condition is present in ~1% of people1, can be the source of viral reactivation2,3, and has been implicated in HHV-6 central nervous system (CNS) disease4,5. To explore whether patients with ciHHV-6 have increased risk for HHV-6-associated disease, we tested the prevalence of ciHHV-6 in allogeneic hematopoietic cell transplantation (HCT) recipients with HHV-6 CNS dysfunction.
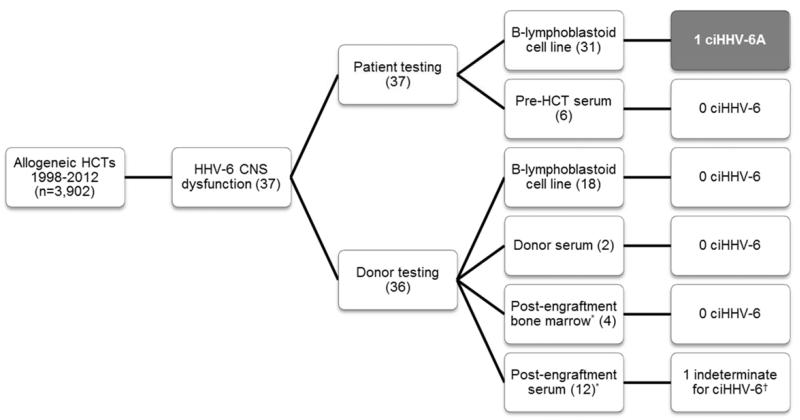
We identified 37 allogeneic HCT recipients at our center with HHV-6 CNS dysfunction as previously described6. We tested archived samples from donor/recipient pairs for ciHHV-6 using a novel method to detect HHV-6 and human ribonuclease P (RPP30, a reference gene for cell count) DNA by droplet digital PCR (Fig. 1)7. CiHHV-6 was ruled out if the ratio of HHV-6 DNA to cell genome equivalents (two RPP30/cell) fell outside the range of 1 ± 0.07.
Figure 1.
Flow diagram for chromosomally integrated HHV-6 testing. Archived pre-HCT patient and donor Beta-lymphoblastoid cell lines were the primary source for ciHHV-6 testing. When unavailable, other appropriate specimens (serum or bone marrow) were used as surrogates.
*Surrogate for donor sample in cases that were missing pre-HCT donor sample. These were obtained post-engraftment.
†One patient had a HHV-6/cellular DNA ratio significantly >1, making it impossible to rule out ciHHV-6 coupled with reactivation.
Abbreviations: HCT, hematopoietic cell transplantation; HHV-6, human herpesvirus 6; CNS, central nervous system; B-lymphoblastoid, Beta-lymphoblastoid; ciHHV-6, chromosomally integrated HHV-6.
CiHHV-6 species A was detected in one pre-HCT patient sample (prevalence 2.7%; 95% confidence interval, 0.07-14.5%) from a 53 year old man who developed findings consistent with HHV-6 encephalitis after a myeloablative matched, related donor HCT. He initially developed hallucinations and agitation on day 11 after HCT (D+11), one day after starting methylprednisolone 2 mg/kg/day for skin rash. Cerebrospinal fluid (CSF) on D+12 had two lymphocytes, four red blood cells, and protein 43 mg/dl. Bacterial, mycobacterial, and fungal stains and cultures were negative, as was PCR for other herpesviruses. Semi-quantitative PCR for HHV-6 DNA was positive with 2,500 copies/ml. The patient was started on foscarnet 90 mg/kg IV Q12h on D+14 for HHV-6 encephalitis.
The patient had initial improvement in mental status followed by progressive encephalopathy. He engrafted on D+17 and was weaned off steroids by D+23. Repeat CSF testing on D+26 had 2 lymphocytes, 54 red blood cells, and 2,500 copies/ml HHV-6 DNA by semi-quantitative PCR. Additional testing as above was negative. Foscarnet was discontinued on D+29 due to acute kidney injury. Chimerism and flow cytometry studies of bone marrow and blood on D+29 were of donor origin, and PCR for HHV-6 in bone marrow was negative at that time. Brain magnetic resonance imaging was negative on D+33. The patient developed hypoxia on D+38 attributed to aspiration in the setting of obtundation and died on D+40 due to progressive pulmonary disease. Autopsy revealed increased brain microglia and perivascular lymphocyte cuffing, as well as scattered focal hippocampal neuronal dropout; although non-specific, this could be consistent viral encephalitis.
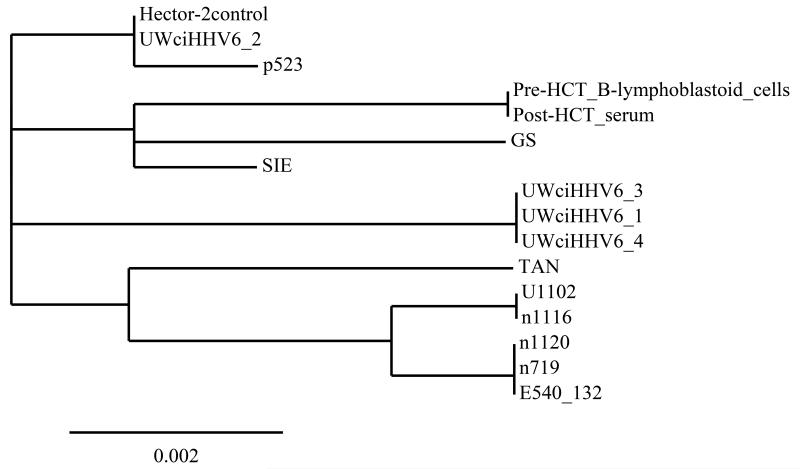
Retrospective testing of multiple archived serum samples with droplet digital PCR detected HHV-6 DNA with decreasing HHV-6/cellular DNA ratios of 0.5 to 0.02 (day 5 through 39). This could be consistent with allograft replacement of recipient ciHHV-6 hematopoietic cells or treatment of HHV-6 reactivation with foscarnet. Sanger sequencing of a 900 base pair region of the HHV-6 envelope glycoprotein B (gB) gene in a pre-HCT cell sample and post-HCT serum sample (D+39) revealed complete homology but divergence from 14 other strains8 (Fig. 2). A fresh-frozen brain section of the inferior temporal lobe had HHV-6/cellular DNA ratio of 1.0, consistent with ciHHV-6 but without evidence of additional HHV-6 replication. Testing of formalin-fixed, paraffin-embedded samples from the medial temporal lobes (the predominant site of HHV-6 CNS disease) yielded inconclusive results due to poor preservation. RNA testing and viral culture could not be performed on archived specimens.
Figure 2.
Phylogenetic analysis of HHV-6A gB gene sequences from pre and post-HCT samples of the described patient with ciHHV-6A compared to ciHHV-6A from 4 patients at our center, the Hector-2 cell line (Bioworld Consulting Laboratories), and 9 published sequences8.
This figure demonstrates that the sequence of an ~900 base pair region of the HHV-6A gB gene in the discussed patient’s pre-HCT Beta-lymphoblastoid cell line and post-HCT (D+39) serum sample were identical and diverged from 14 other available sequences.
Bar = number of nucleotide changes per 100 sites.
The gB gene sequences of 9 HHV-6A primary or laboratory-adapted isolates were obtained from Dr Henri Agut (Paris, France), and the geographical origins were as follows: GS, United States; U1102, Uganda; SIE, Côte d’Ivoire; TAN, Congo; and 1120, E540_132, 1116, 719 and, p523, France. An additional 4 isolates were obtained from patients at the University of Washington with chromosomally integrated HHV-6: UWciHHV6_1, UWciHHV6_2, UWciHHV6_3, UWciHHV6_4.
The tree was constructed from the comparison of gB gene nucleotide sequences using the free Phylogeny.fr webservice, available at http://www.phylogeny.fr/version2_cgi/index.cgi.
This is the first epidemiologic study of the prevalence of ciHHV-6 in patients with HHV-6-associated CNS dysfunction. Convincing evidence of HHV-6 reactivation and pathogenicity from ciHHV-6 cell lines is well-described2-5. We did not identify a clear overrepresentation of ciHHV-6 in this cohort of 37 patients compared to population-based studies1, but the confidence interval was wide and suggests an incidence as high as 14%. We also describe the first case of a patient with ciHHV-6A and HHV-6A reactivation as a possible cause of post-HCT encephalitis.
Although HHV-6A DNA detected in post-HCT samples may have represented cells with ciHHV-6A rather than active replication, and alternative causes of encephalitis were possible, we think the findings are best explained by HHV-6A reactivation. De novo HHV-6 infection was unlikely given sequence homology between pre and post-HCT HHV-6 strains. Recipient hematopoietic cells with ciHHV-6A would be an unlikely source of persistent HHV-6A DNA in serum samples after a myeloablative HCT, especially given evidence of full donor chimerism and absence of HHV-6 DNA detection in the D+29 bone marrow biopsy. HHV-6A DNA in CSF samples was also unlikely to be from CNS or residual hematopoietic cells with ciHHV-6A given the low ratio of nucleated cells (2) to HHV-6 copies (2,500); a study of HHV-6 levels in CSF from patients with ciHHV-6 and unrelated CNS disease showed close correlation between leukocyte count and viral load9. Unfortunately, RNA testing and viral culture of saved specimens was not possible due to prior processing and extended storage.
In conclusion, we did not find evidence of ciHHV-6 enrichment in a cohort of HCT patients with HHV-6 CNS dysfunction compared to the general population, but larger studies are needed to comprehensively determine the true incidence and prevalence of reactivation in high-risk patients. Our findings begin to address the call to reevaluate transplant practices in the setting of ciHHV-610 and underscore the need for large, multicenter collaborations to determine the impact of ciHHV-6 on patient outcomes.
Acknowledgements
We would like to thank the Research Cell Bank at Fred Hutchinson Cancer Research Center for facilitating acquisition of Beta-lymphoblastoid cell lines. These cells were previously collected and studied under the International Histocompatibility Workshops and Conferences; they were obtained directly from the International Histocompatibility Working Group in Seattle, WA. We also thank Henri Agut for generously sharing the nucleotide sequences for the gB gene region of 9 HHV-6A strains.
Financial support: This work was supported by a Pilot Grant from the HHV-6 Foundation (J.A.H.) and by the National Institutes of Health (grant numbers CA18029 and HL093294, M.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential conflicts of interest: M.B. has served as a consultant and has received research support from Chimerix Inc., Genentech/Roche, and Gilead in addition to consulting for Clinigen.
References
- 1.Pellett P, Ablashi D. [Accessed July 16, 2014];Chromosomally integrated human herpesvirus 6: questions and answers. Rev. Med. Virol. 2012 (22):144–155. doi: 10.1002/rmv.715. Available at: http://onlinelibrary.wiley.com/doi/10.1002/rmv.715/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbuckle JH, Medveczky MM, Luka J, et al. [Accessed November 9, 2012];The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 2010 107(12):5563–8. doi: 10.1073/pnas.0913586107. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2851814&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endo A, Watanabe K, Ohye T, et al. [Accessed May 8, 2014];Molecular and virological evidence of viral activation from chromosomally integrated HHV-6A in a patient with X-SCID. Clin. Infect. Dis. 2014 doi: 10.1093/cid/ciu323. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24803376. [DOI] [PubMed] [Google Scholar]
- 4.Wittekindt B, Berger A, Porto L, et al. [Accessed September 24, 2013];Human herpes virus-6 DNA in cerebrospinal fluid of children undergoing therapy for acute leukaemia. Br. J. Haematol. 2009 145(4):542–5. doi: 10.1111/j.1365-2141.2009.07641.x. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19344404. [DOI] [PubMed] [Google Scholar]
- 5.Troy SB, Blackburn BG, Yeom K, Caulfield AKF, Bhangoo MS, Montoya JG. [Accessed November 9, 2012];Severe encephalomyelitis in an immunocompetent adult with chromosomally integrated human herpesvirus 6 and clinical response to treatment with foscarnet plus ganciclovir. Clin. Infect. Dis. 2008 47(12):e93–6. doi: 10.1086/593315. Available at: http://www.ncbi.nlm.nih.gov/pubmed/18991511. [DOI] [PubMed] [Google Scholar]
- 6.Hill JA, Boeckh MJ, Sedlak RH, Jerome KR, Zerr DM. [Accessed July 30, 2014];Human herpesvirus 6 can be detected in cerebrospinal fluid without associated symptoms after allogeneic hematopoietic cell transplantation. J. Clin. Virol. 2014 doi: 10.1016/j.jcv.2014.07.001. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25066883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sedlak RH, Cook L, Huang M-L, et al. [Accessed July 11, 2014];Identification of chromosomally integrated human herpesvirus 6 by droplet digital PCR. Clin. Chem. 2014 60(5):765–72. doi: 10.1373/clinchem.2013.217240. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24594780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Achour A, Malet I, Le Gal F, et al. [Accessed May 21, 2014];Variability of gB and gH genes of human herpesvirus-6 among clinical specimens. J. Med. Virol. 2008 80(7):1211–21. doi: 10.1002/jmv.21205. Available at: http://www.ncbi.nlm.nih.gov/pubmed/18461623. [DOI] [PubMed] [Google Scholar]
- 9.Ward KN, Leong HN, Thiruchelvam AD, Atkinson CE, Clark D. [Accessed November 9, 2012];a. Human herpesvirus 6 DNA levels in cerebrospinal fluid due to primary infection differ from those due to chromosomal viral integration and have implications for diagnosis of encephalitis. J. Clin. Microbiol. 2007 45(4):1298–304. doi: 10.1128/JCM.02115-06. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1865851&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flamand L. [Accessed May 8, 2014];Pathogenesis from the reactivation of chromosomally-integrated HHV-6: Facts rather than fiction. Clin. Infect. Dis. 2014 doi: 10.1093/cid/ciu326. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24803375. [DOI] [PubMed] [Google Scholar]