Abstract
Dynamic remodeling of the actin cytoskeleton is required for both development and tissue homeostasis. While fixed image analysis has provided significant insight into such events, a complete understanding of cytoskeletal dynamics requires live imaging. Numerous tools for the live imaging of actin have been generated by fusing the actin-binding domain from an actin-interacting protein to a fluorescent protein. Here we comparatively assess the utility of three such tools – Utrophin, Lifeact, and F-tractin – for characterizing the actin remodeling events occurring within the germline-derived nurse cells during Drosophila mid-oogenesis or follicle development. Specifically, we used the UAS/GAL4 system to express these tools at different levels and in different cells, and analyzed these tools for effects on fertility, alterations in the actin cytoskeleton, and ability to label filamentous actin (F-actin) structures by both fixed and live imaging. While both Utrophin and Lifeact robustly label F-actin structures within the Drosophila germline, when strongly expressed they cause sterility and severe actin defects including cortical actin breakdown resulting in multi-nucleate nurse cells, early F-actin filament and aggregate formation during stage 9 (S9), and disorganized parallel actin filament bundles during stage 10B (S10B). However, by using a weaker germline GAL4 driver in combination with a higher temperature, Utrophin can label F-actin with minimal defects. Additionally, strong Utrophin expression within the germline causes F-actin formation in the nurse cell nuclei and germinal vesicle during mid-oogenesis. Similarly, Lifeact expression results in nuclear F-actin only within the germinal vesicle. F-tractin expresses at a lower level than the other two labeling tools, but labels cytoplasmic F-actin structures well without causing sterility or striking actin defects. Together these studies reveal how critical it is to evaluate the utility of each actin labeling tool within the tissue and cell type of interest in order to identify the tool that represents the best compromise between acceptable labeling and minimal disruption of the phenomenon being observed. In this case, we find that F-tractin, and perhaps Utrophin, when Utrophin expression levels are optimized to label efficiently without causing actin defects, can be used to study F-actin dynamics within the Drosophila nurse cells.
Keywords: actin, live imaging, oogenesis
Introduction
Drosophila oogenesis or follicle development consists of fourteen morphologically defined stages (reviewed in (Spradling, 1993)). Each egg chamber or follicle is comprised of 16 interconnected germline cells – 15 nurse cells and one oocyte – and approximately 1000 somatic cells termed follicle cells. Production of a viable egg requires dynamic remodeling of the actin cytoskeleton in both the somatic and germline cells. Thus, Drosophila oogenesis is an ideal system for studying the actin cytoskeleton and has been widely used to identify and elucidate the functions of actin binding proteins and regulatory factors (reviewed in (Hudson and Cooley, 2002)). Here we focus on events occurring within the germline-derived nurse cells. The main purpose of the nurse cells is to supply organelles, mRNA, and proteins to the oocyte, thereby providing the oocyte with everything needed to complete embryogenesis.
Spatial and temporal regulation of actin remodeling is required for this multi-stage transport process that is essential for the production of a viable egg. Transport occurs through the ring canals, remnants of incomplete cytokinesis, that connect the nurse cells to each other and the oocyte. In the earlier stages of follicle development (prior to stage 10B (S10B)) the transport of materials from the nurse cells into the oocyte is slow (reviewed in (Hudson and Cooley, 2002)). Little actin remodeling is seen within the nurse cells during these stages. Intriguingly, we have observed that some remodeling is occurring at the ring canals connected to the oocyte in the posterior nurse cells during stage 9 (S9) (Spracklen et al., 2014). This normally results in minimal filaments near the ring canals (see Fig. 1A-A’), but occasionally results in more extensive actin filament and/or aggregate formation (see Fig. 1B-B’). Such remodeling is regulated as we have found that genetic loss of the Drosophila COX-like enzyme, Pxt, results in a significant increase in the prevalence of these early actin structures (Spracklen et al., 2014).
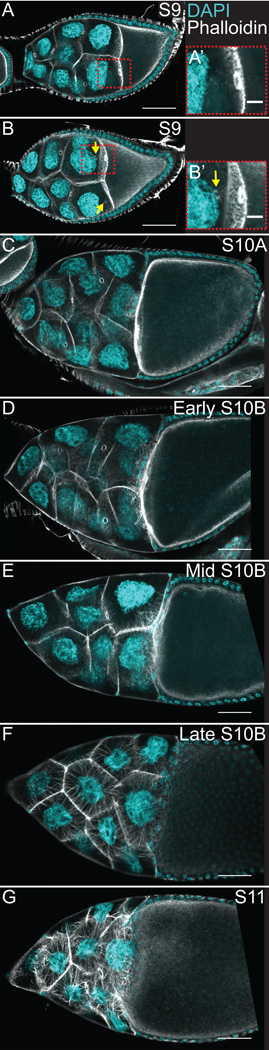
Figure 1. Actin remodeling is temporally and spatially regulated during mid-oogenesis.
(A–G) Maximum projections of 3–5 confocal slices of fixed and stained wild-type (yw) follicles, staged as indicated, taken at 20X magnification. Anterior is to the left. F-actin (phalloidin) = white, DNA (DAPI) = cyan. (A-B’) S9. (C) S10A. (D-F) S10B. (G) S11. During S9, the border cells and main-body follicle cells are undergoing migration and the nurse cell cytoplasm is largely devoid of actin filament structures (A-A’), but occasionally there are actin filament and aggregate structures emanating from the ring canals in the posterior nurse cells adjacent to the oocyte (B-B’). Both the border cell and main-body follicle cell migration are completed by S10A and the nurse cells lack cytoplasmic F-actin structures (C). During S10B, dynamic actin remodeling is occurring within the nurse cells. Actin filament bundles first form in the posterior nurse cells, at the nurse cell-oocyte boundary (D). These bundles continue to elongate and bundle formation initiates on all of the nurse cell membranes that are directly attached to their neighboring nurse cell by a ring canal (E). At the completion of S10B the bundles are uniformly distributed along the nurse cell membranes and extend all the way to the nucleus (F). During S11, the cortical actin contracts to squeeze the cytoplasmic contents of the nurse cells into the growing oocyte (G). Scale bars = 50 μm, except in A’ and B’ where scale bars = 10 μm.
During S10B, the nurse cells undergo dynamic actin remodeling to facilitate the second, rapid phase of transport that occurs during stage 11 (S11), termed nurse cell dumping. Nurse cell dumping requires two distinct actin remodeling events during S10B: 1) the strengthening of nurse cell cortical actin, which ultimately undergoes an actomyosin based contraction to squeeze the nurse cell cytoplasm into the oocyte (Wheatley et al., 1995) and 2) the generation of parallel actin filament bundles that traverse from the nurse cell membrane, inward towards the nucleus, to form a cage that prevents the nucleus from plugging the ring canals during contraction (Guild et al., 1997; Huelsmann et al., 2013). The system of nurse cell dumping has been widely used to both identify actin regulators and define their functions in cytoskeletal remodeling (Cooley et al., 1992; Gates et al., 2009; Mahajan-Miklos and Cooley, 1994a, b). We have shown that Pxt, and thus prostaglandins (PGs), are required to regulate these dynamic remodeling events (Groen et al., 2012; Tootle and Spradling, 2008). Such studies have relied on analyses of actin structures using fixed samples labeled with phalloidin to visualize filamentous actin (F-actin).
While fixed imaging has provided important insights into the actin cytoskeletal remodeling events occurring within the Drosophila germline, many questions remain regarding the spatial and temporal regulation of this remodeling. For example, it has previously been shown that the remodeling at S10B begins along the nurse cell oocyte boundary and then progresses anteriorly (see Fig. 1D–F) (Guild et al., 1997). However, this spatial and temporal progression is not widely appreciated and the mechanisms underlying this regulation remain poorly understood. Specifically, the roles of particular actin regulators in establishing spatial and temporal regulation over these cytoplasmic actin remodeling events remain unknown. Additionally, fixed analysis cannot be used to assess nurse cell actin filament bundle dynamics, or how pharmacologic or genetic perturbations alter those dynamics. One means of addressing these knowledge gaps is to utilize live imaging.
One actin cytoskeleton live imaging reagent routinely used in Drosophila is the GFP-tagged actin binding domain of Moesin, GFP-Moe (Edwards et al., 1997). GFP-Moe has successfully been used to examine actin dynamics in the Drosophila embryo during dorsal closure (Peralta et al., 2007; Toyama et al., 2008), tracheal morphogenesis (Kato et al., 2004), and hemocyte migration (Zanet et al., 2009). It has also been used to label actin dynamics within the somatic cells of the developing Drosophila follicles (Boyle et al., 2010; Edwards et al., 1997), and was recently used to label cortical actin structures within the germline (Weil et al., 2010). We have found that GFP-Moe (sqh promoter driven GFP-Moe) strongly labels both germline and follicle cell cortical actin during oogenesis, but fails to label the initial stages of bundle formation during S10B and only weakly labels the fully formed bundles (Spracklen and Tootle, unpublished observations), reducing the utility of this tool for assessing germline actin dynamics during mid-to-late Drosophila oogenesis.
Numerous additional tools for live imaging of actin dynamics have been developed and utilized in other systems and, in some cases, in Drosophila. These include directly tagging the N-terminus of actin with a fluorescent protein, i.e. GFP-Actin. While GFP-Actin does label actin filaments, such labeling has been associated with alterations in actin structure and dynamics (Aizawa et al., 1997; Hird, 1996; Kovar et al., 2006; Roper et al., 2005; Wu and Pollard, 2005). An alternative means of labeling actin is by fusing a fluorescent protein to the actin-binding domain from a known actin binding protein. It is thought that this “indirect” method of labeling actin filaments is less likely to disrupt the actin cytoskeleton. Examples of this type of tool include Utrophin, Lifeact, and F-Tractin (see Table 1). Utrophin is composed of the calponin homology domains of human ubiquitous dystrophin (Utrophin) (Burkel et al., 2007), Lifeact is the actin-binding domain from yeast ABP140 (Riedl et al., 2008), and F-tractin is the actin-binding domain from rat inositol triphosphate 3-kinase (Johnson and Schell, 2009).
Table 1. Actin labeling tools.
Summary of the actin binding domain fusions to fluorescent proteins that were used to generate transgenic Drosophila lines.
| Reagent | Actin binding domain |
Promoter | Tag | Reagent reference |
Fly reference |
|---|---|---|---|---|---|
| Utrophin | Human ubiquitous dystrophin | UASp and sqh | EGFP | Burkel et al. 2007. | Rauzi et al. 2010. |
| Lifeact | Yeast ABP140 | UASp | mEGFP | Riedl et al. 2008 | This work |
| F-tractin | Rat inositol triphosphate 3-kinase A (ITPKA) | UASp | mEGFP tdTomato |
Johnson and Schell. 2009 | This work |
While numerous studies have shown the utility of these tools for assessing actin dynamics in other systems (Brock et al., 2012; Hatan et al., 2011; Rauzi et al., 2010) and some of these tools have been used to label actin within the Drosophila germline (Huelsmann et al., 2013; Zanet et al., 2012; Zanet et al., 2009), the tools have not been comparatively assessed for their utility in monitoring the extensive cytoplasmic F-actin remodeling events occurring within the Drosophila germline. We have generated transgenic lines that express Lifeact-mEGFP, F-tractin-mEGFP and F-tractin-tdTomato (tdTom) under the control of the UASp promoter, which can be expressed in the germline. Additionally, we have obtained a similar GFP-Utrophin line (Rauzi et al., 2010) (Table 1). Here we compare the utility of these three labeling tools for examining the actin remodeling occurring within the nurse cells. Specifically, we assess whether actin remodeling occurs normally, using both fertility and F-actin morphology as readouts. Additionally, we assess the extent to which each marker colocalizes with F-actin by phalloidin staining, and whether these tools are effective for live imaging of actin dynamics within the Drosophila germline.
Results
Actin cytoskeletal remodeling events occurring within the Drosophila nurse cells during mid-oogenesis
Fixed image analysis has revealed that tight temporal and spatial actin remodeling within the germline-derived nurse cells is required for Drosophila follicle development and female fertility (reviewed in (Hudson and Cooley, 2002)). During S9, besides the cortical actin, there is little F-actin observed within the nurse cells (Fig. 1A-A’). Fixation conditions designed to preserve actin structures reveal that there are small actin structures at the posterior ring canals leading into the oocyte. Occasionally, more extensive actin structures, including elongated actin filament bundles and aggregates, are observed (Fig. 1B-B’, yellow arrows) (Spracklen et al., 2014). At S10A, these “early” actin structures are no longer apparent (Fig. 1C). During S10B, rapid cytoplasmic actin remodeling occurs (Fig 1D–F) (Cooley et al., 1992; Gates et al., 2009; Mahajan-Miklos and Cooley, 1994a, b). In early S10B, parallel actin filament bundles (subsequently referred to as actin bundles) begin to form along the nurse cell/oocyte boundary (Fig. 1D) (Guild et al., 1997; Huelsmann et al., 2013); the barbed ends of the filaments are positioned at the nurse cell membrane with the pointed ends proximal to the nucleus. By mid-S10B, these posterior bundles have elongated toward the nucleus and bundle formation has initiated along all of the nurse cell membranes sharing a ring canal with an adjacent nurse cell (Fig. 1E). These bundles continue to elongate throughout S10B, such that the bundles extend all the way to the nucleus by late S10B (Fig. 1F) (Guild et al., 1997; Huelsmann et al., 2013). During S10B, the cortical actin is also strengthened, and, during S11, the cortical actin contracts in an acto-myosin dependent manner to squeeze all of the nurse cell cytoplasmic contents into the oocyte in a process termed nurse cell dumping (Fig. 1G) (Wheatley et al., 1995). It is important to note that we assume that the actin cytoskeletal structures observed in fixed sample analyses accurately recapitulate what is occurring. In all the fixed experimental analyses presented here, the actin structures are being compared to controls (both GAL4 and UASp only) raised under the same conditions and processed in the same manner.
To gain further insight into not only the temporal and spatial regulation of actin remodeling, but the dynamic nature of theses remodeling events within the nurse cells, we have obtained or generated transgenic lines to indirectly label F-actin live, including: GFP-Utrophin (Burkel et al., 2007; Rauzi et al., 2010); Lifeact-mEGFP (Riedl et al., 2008); and F-tractin-mEGFP and td-Tomato (Johnson and Schell, 2009) (Table 1). These transgenic lines express the actin labeling tool under the control of the UASp promoter, allowing us to use a variety of GAL4 lines to express the tools in either the soma or germline (Brand and Perrimon, 1993; Rorth, 1998). In our initial experiments we tested a number of GAL4 lines to determine their expression patterns during follicle development (STable 1). Based on those experiments, we chose to utilize: c355 GAL4 to drive expression in the somatic follicle cells (Skora and Spradling, 2010); nanos-VP16 GAL4 to drive weak germline expression during mid-oogenesis; and either maternal α-tubulin (3rd chromosome insert or 2nd chromosome insert [MK]; referred to as mat3 and mat2MK, respectively) or oskar GAL4 (3rd chromosome insert, referred to as osk3) (Telley et al., 2012), interchangeably, to drive strong germline expression during mid-oogenesis. Importantly, heterozygosity for these GAL4 drivers does not result in any significant actin remodeling abnormalities (SFig. 1), with the exception that two of the strong germline drivers, mat3 and osk3 GAL4, exhibit an increased frequency of early F-actin structures during S9 (SFig. 1D–E, arrows).
Strong germline expression of GFP-Utrophin results in female sterility and severe actin remodeling defects
Actin remodeling, both within the soma and the germline of developing Drosophila follicles, is required for female fertility. Transgenic lines expressing GFP-Utrophin (the first 261 aa residues, containing the calponin homology domains, of Utrophin (UtrCH); also known as UTR261 or UtrNT) (Burkel et al., 2007) under the control of the UASp promoter were previously generated by Rauzi and colleagues (Rauzi et al., 2010). We assessed fertility of GAL4 driven GFP-Utrophin expressing females by mating them to wild-type males (yw) at room temperature (~21°C) and quantifying the resulting number of progeny produced per female from a 24hr egg lay. We find that while somatic (c355) or weak germline (nanos-VP16) expression of GFP-Utrophin has little to no effect on female fertility, strong germline expression results in almost complete sterility (Table 2, SFig. 2A). This finding suggested that high levels of Utrophin within the germline might cause actin abnormalities that preclude normal follicle development.
Table 2. The relative effects of the different actin labeling tools on female fertility.
Fertility was assessed by scoring the number of adult progeny per female from a 24hr egg collection.
| GAL4 | Actin Labeling Tool | % Relative Fertility* |
|---|---|---|
| none | Utrophin-EGFP | 78.0% |
| Lifeact-mEGFP 13a | 151% | |
| Lifeact-mEGFP 14a | 85.8% | |
| Ftractin-tdTomato 10C | 158% | |
| Ftractin-tdTomato 15A | 171% | |
| c355 | Utrophin-EGFP | 70.9% |
| Lifeact-mEGFP 13a | 72.3% | |
| Lifeact-mEGFP 14a | 80.9% | |
| Ftractin-tdTomato 10C | 118% | |
| Ftractin-tdTomato 15A | 104% | |
| nanos VP16 | Utrophin-EGFP | 93.8% |
| Lifeact-mEGFP 13a | 109% | |
| Lifeact-mEGFP 14a | 96.6% | |
| Ftractin-tdTomato 10C | 70.1% | |
| Ftractin-tdTomato 15A | 71.0% | |
| osk3 | Utrophin-EGFP | 5.94% |
| Lifeact-mEGFP 13a | 0.78% | |
| Lifeact-mEGFP 14a | 0% | |
| Ftractin-tdTomato 10C | 137.5% | |
| Ftractin-tdTomato 15A | 72.2% | |
| mat3 | Utrophin-EGFP | 1.77% |
| Lifeact-mEGFP 13a | 0% | |
| Lifeact-mEGFP 14a | 0% | |
| Ftractin-tdTomato 10C | 81.2% | |
| Ftractin-tdTomato 15A | 102% |
% Relative Fertility is compared to GAL4 only controls
To determine the cause of the fertility defects, we used fixed image analysis to assess if Utrophin expression resulted in any actin abnormalities and the extent to which Utrophin labels F-actin structures during follicle development. Specifically we focused on the actin remodeling events occurring during S9 and S10B. As the UAS/GAL4 system is known to exhibit a temperature dependence, with higher temperatures resulting in higher levels of expression, the crosses and the resulting progeny were reared at three temperatures: 18, 21, and 25°C. Western blot analysis reveals that GFP-Utrophin expression is indeed highly temperature dependent with c355 and nanos-VP16 GAL4 drivers, but exhibits a smaller change in expression level at the different temperatures with the strong germline GAL4 drivers (mat3, mat2MK, and oskar GAL4) (SFig. 2B). Similarly, the actin defects observed by phalloidin staining are more severe and occur at a higher frequency in flies raised at 25°C compared to either 18 or 21°C (data not shown). Please note that both GAL4 and UASp only controls were utilized at each temperature to define what represented the “normal” actin phenotype.
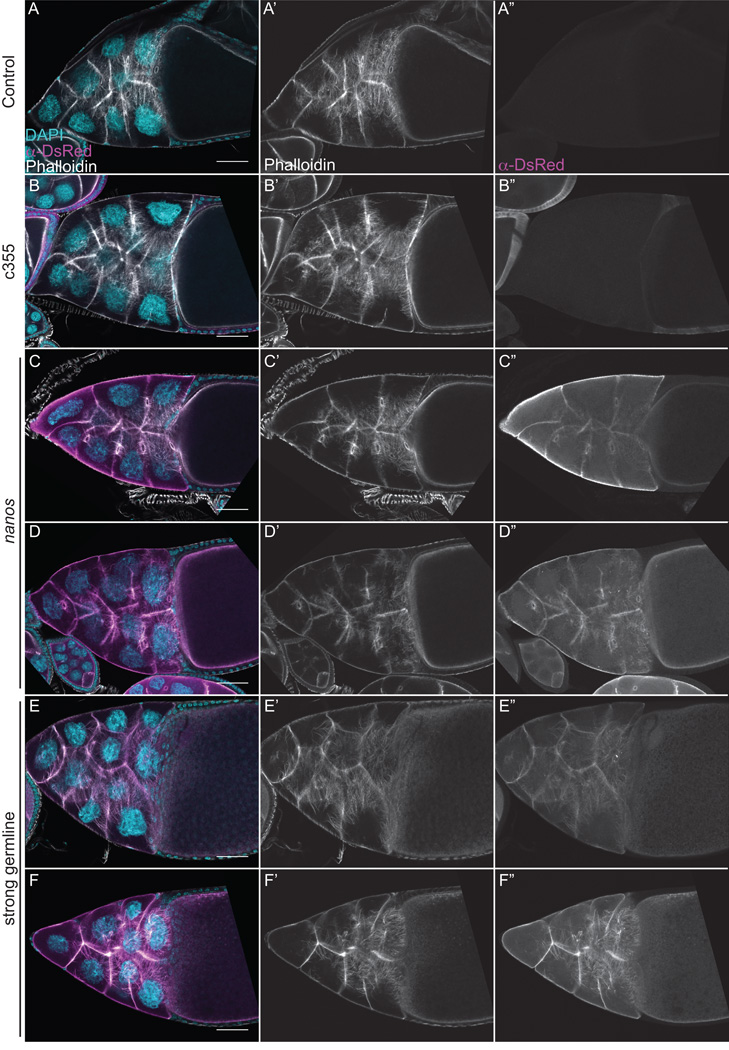
Fixed image analysis (phalloidin staining and anti-GFP immunofluorescence) reveals that expression of GFP-Utrophin results in F-actin defects during S9. While somatic expression of GFP-Utrophin does not result in any F-actin abnormalities within the nurse cells, when GFP-Utrophin is expressed in the border cells (c355 GAL4 driving GFP-Utrophin, 25°C) a delay in migration is often observed (Fig. 2B-B”, yellow arrow). Weak germline expression of GFP-Utrophin during S9 (nanos-VP16 GAL4) results in no obvious F-actin defects in the nurse cells and border cell migration appears normal (Fig. 2C-C”). However, the expression is patchy and often weak. When GFP-Utrophin is expressed strongly in the germline of S9 follicles, the F-actin structures are labeled well with GFP-Utrophin; however, severe actin defects are observed. In particular, clumps of ring canals and multi-nucleate nurse cells (Fig. 2 D–F”, red arrowheads and red circles, respectively) are observed. These defects are not due to incomplete cytokinesis but are the result of nurse cell cortical actin break down, as the cortical actin is intact in the earlier stages of oogenesis (data not shown). Early actin filaments and aggregates are also seen in the posterior nurse cells adjacent to the oocyte when GFP-Utrophin is strongly expressed in the germline at a much higher frequency and severity than observed in control follicles (Fig. 2D–F”, orange boxed regions); this suggests that GFP-Utrophin expression may stabilize and/or promote such cytoskeletal remodeling. Interestingly, similar, but less severe early actin structures are observed when PG signaling is lost (Spracklen et al., 2014).
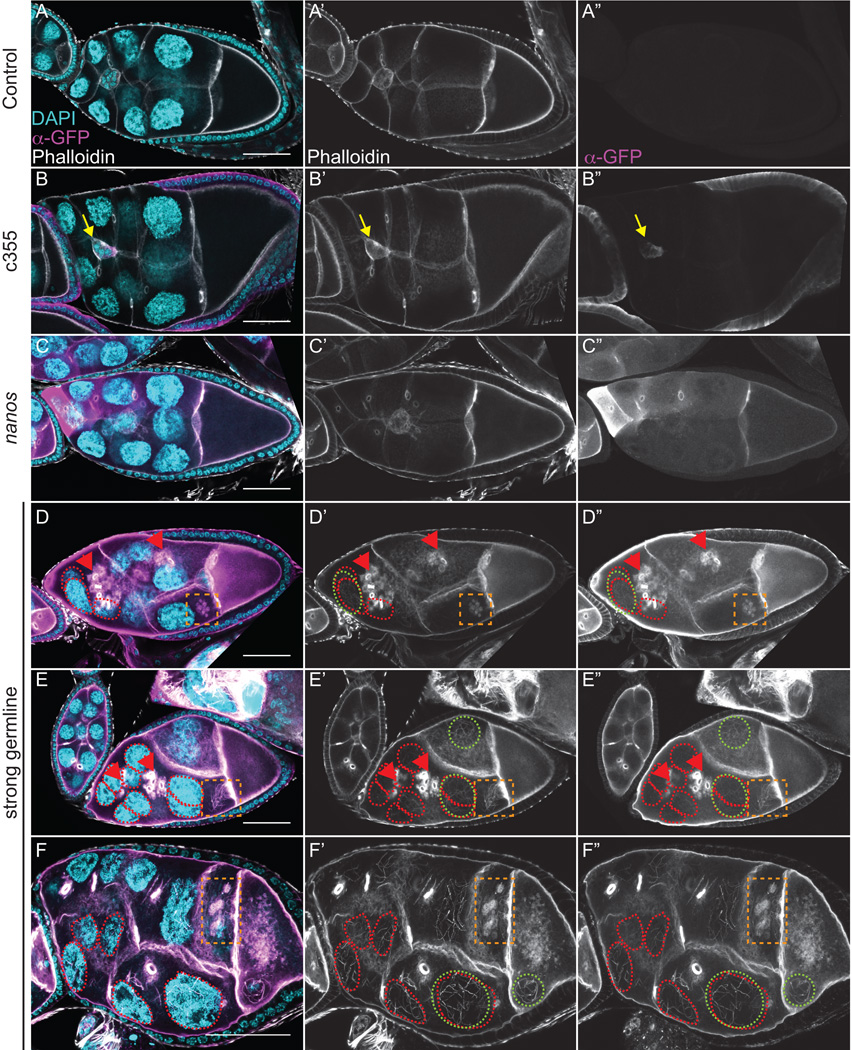
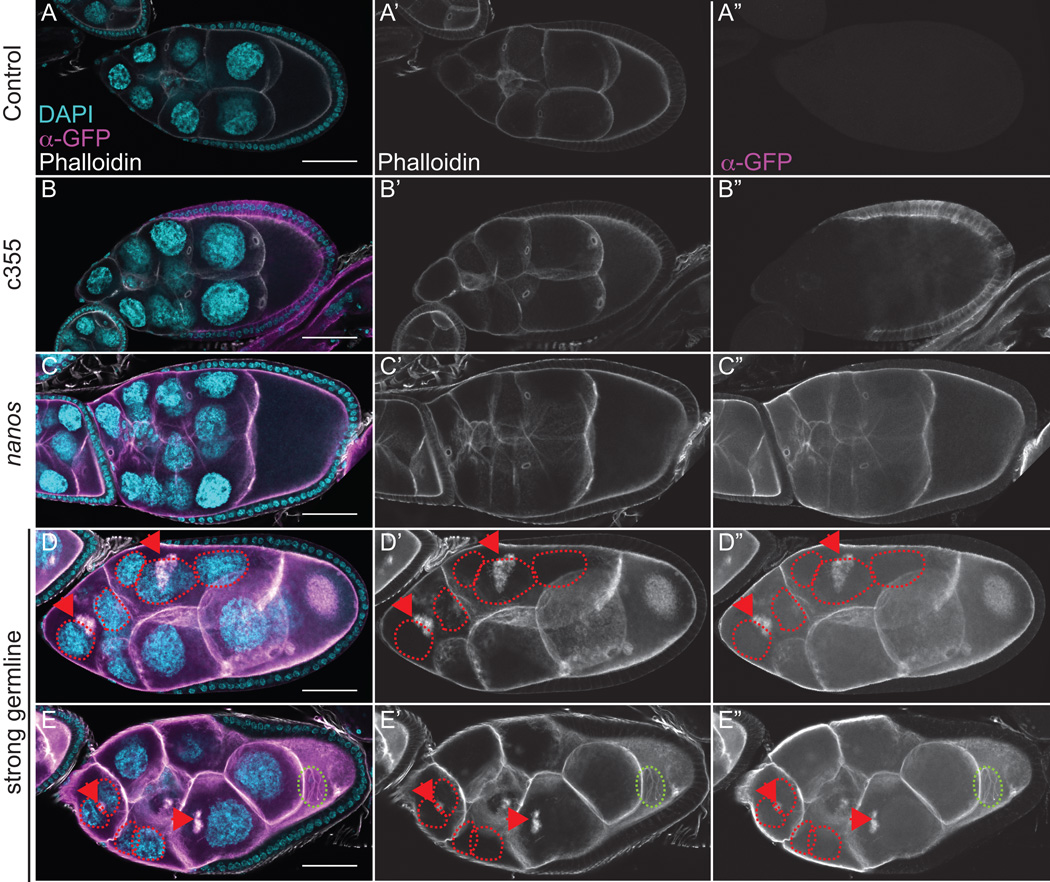
Figure 2. Strong germline expression of GFP-Utrophin results in cortical actin breakdown and the accumulation of F-actin in the nurse cell nuclei during S9.
(A-E”) Maximum projections of 35 confocal slices of fixed and stained S9 follicles taken at 20X magnification. (F-F”) Maximum projections of 4–8 confocal slices of a fixed and stained S9 follicle taken at 63X magnification. (A-F) Merged images: DNA (DAPI) = cyan, F-actin (phalloidin) = white, Utrophin (anti-GFP) = magenta. (A’-F’) F-actin (phalloidin) = white. (A”-F”) Utrophin (anti-GFP) = white. (A-A”) GFP-Utrophin/+. (B-B”) c355 GAL4; GFP-Utrophin. (C-C”) GFP-Utrophin; nanos-VP16 GAL4. (D-F”) Strong germline GAL4 (either mat2MK, mat3, or oskar GAL4) driving GFP-Utrophin. Somatic expression of GFP-Utrophin has no effect on the nurse cell actin cytoskeleton or follicle cell morphology (B-B’’ compared to A-A’’), but expression within the border cells results in delayed migration (B-B”, yellow arrow). Weak germline expression of GFP-Utrophin is patchy and variable in the level of expression and ability to label cortical and ring canal F-actin, and does not alter F-actin morphology (C-C’’ compared to A-A’’). While cortical and ring canal F-actin label well when GFP-Utrophin is strongly expressed in the germline, high levels of GFP-Utrophin result in numerous defects including cortical actin breakdown with aggregates of ring canals (D-E”, red arrowheads and multi-nucleate nurse cells (D-F”, red circles), and early actin filaments and aggregates emanating from the ring canals in the posterior nurse cells (D-F”, orange boxes). Additionally, phalloidin and Utrophin positive F-actin structures that we term threads accumulate in the nurse cell nuclei and germinal vesicle when GFP-Utrophin is strongly expressed in the germline (E-F”, green circles). Scale bars = 50 μm.
The most surprising and striking F-actin defect observed by fixed image analysis when GFP-Utrophin is strongly expressed in the germline line is the presence of GFP-Utrophin and phalloidin positive F-actin in the nurse cell nuclei and germinal vesicle (Fig. 2D–F”, green circles). To verify that the F-actin is within and not on the outside of the nucleus we used wheat germ agglutinin (WGA) to label the nuclear envelope. We find that the F-actin is indeed in the nurse cell nuclei (SFig. 3A-A”’). Given the coiled appearance of the nuclear F-actin and its unknown structure, we have termed these nuclear F-actin structures “threads.” Utrophin has been previously observed on nuclear F-actin structures and nuclear-targeted Utrophin may cause F-actin polymerization or stabilization (reviewed in (Grosse and Vartiainen, 2013)). Interestingly, the nuclear actin threads we observe are only seen in particular stages of follicle development. Specifically, S6-S9 follicles exhibit the highest frequency of these structures, with a few nurse cell nuclei retaining them into S10A-B (fixed image data not shown and Fig. 4E, live imaging). The temporal presence of these nuclear actin threads suggests that there may be a different concentration or composition of nuclear actin during these stages. Further supporting this idea, we find that strong germline expression of particular Drosophila GFP-Actins (Roper et al., 2005), including 42A and 57B, results in shorter nuclear F-actin, termed “rods”, during S6-9 (Fagan and Tootle, unpublished observation).
We next wanted to assess by fixed image analysis (phalloidin staining and anti-GFP immunofluorescence) if GFP-Utrophin labels the actin bundles that form during S10B and whether expression of GFP-Utrophin alters these F-actin structures. As expected, somatic expression of GFP-Utrophin has no effect on bundle formation during S10B (Fig 3B-B”). Weak germline expression of GFP-Utrophin results in poor labeling of the actin bundles by Utrophin, and the actin bundles are relatively normal (Fig. 3C-C”), although some disorganization is observed. However, while strong germline expression of GFP-Utrophin results in good labeling of the actin bundles, severe actin defects are observed, including cortical actin breakdown with aggregates of ring canals (red arrowheads) and multi-nucleate nurse cells (red circles), and overly elongated and disorganized actin bundles (Fig. 3D-D”). Together, these data suggest that strong germline expression of GFP-Utrophin is detrimental to F-actin dynamics and therefore, to the morphogenic events necessary for follicle development.
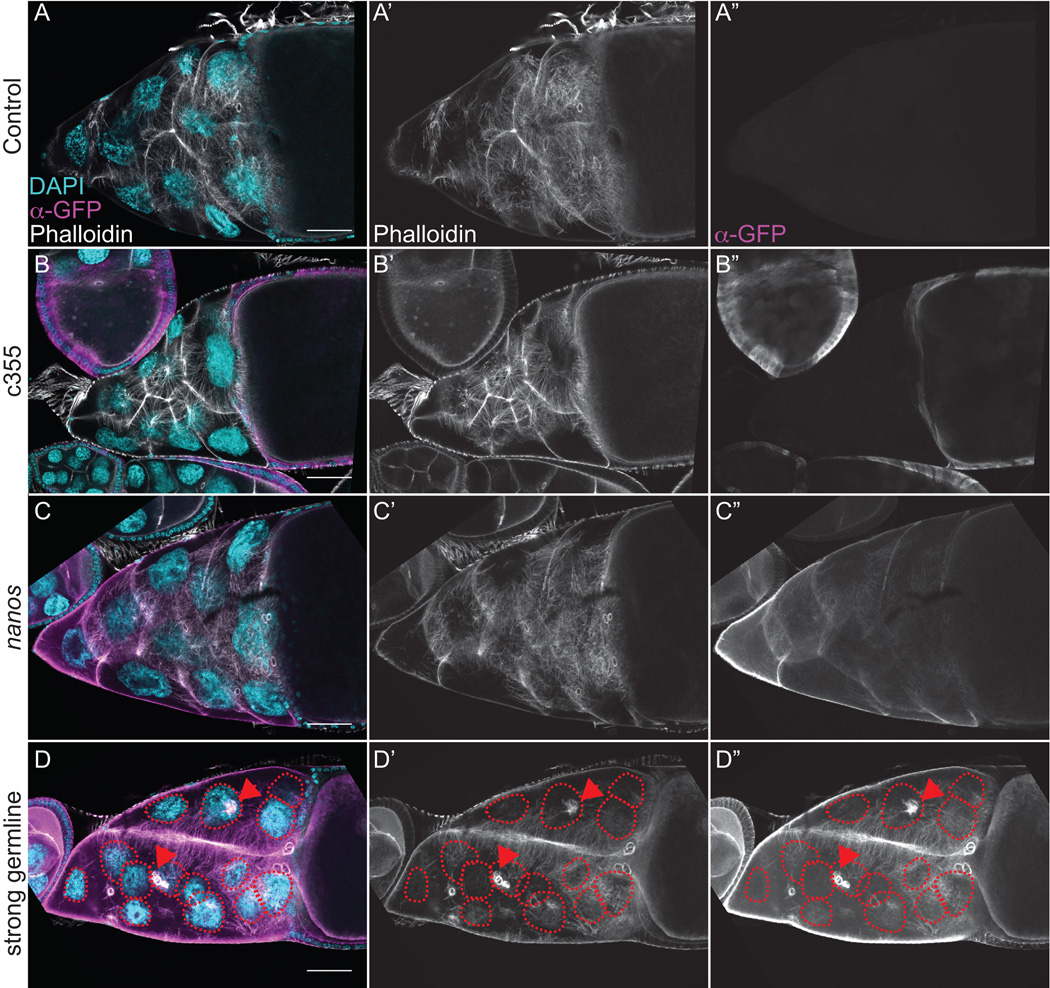
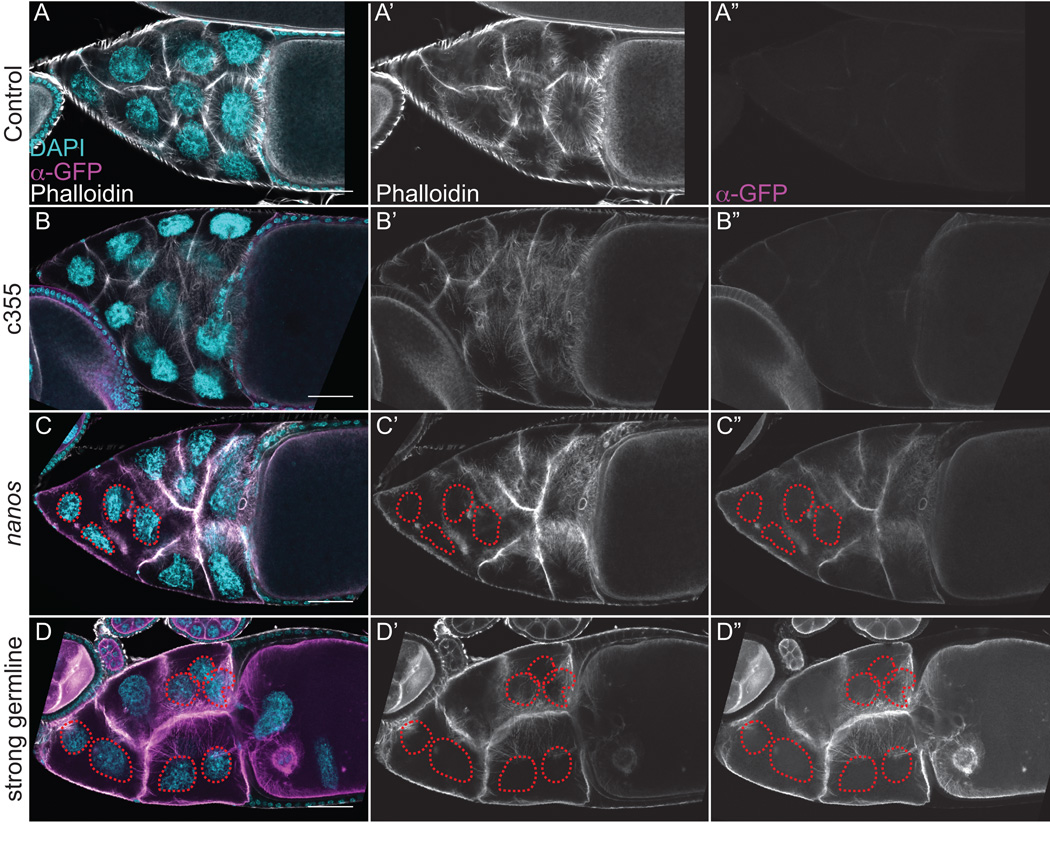
Figure 3. GFP-Utrophin labels actin filament bundles during S10B, but strong germline expression results in cortical actin breakdown.
(A-D”) Maximum projections of 3–5 confocal slices of fixed and stained S10B follicles taken at 20X magnification. (A-D) Merged images: DNA (DAPI) = cyan, F-actin (phalloidin) = white, Utrophin (anti-GFP) = magenta. (A’-D’) F-actin (phalloidin) = white. (A”-D”) Utrophin (anti-GFP) = white. (A-A”) GFP-Utrophin/+. (B-B”) c355 GAL4; GFP-Utrophin. (C-C”) GFP-Utrophin; nanos-VP16 GAL4. (D-D”) Strong germline GAL4 (either mat2MK, mat3, or oskar GAL4) driving GFP-Utrophin. Follicle cell morphology and nurse cell actin remodeling is not altered by either somatic expression or weak germline expression of GFP-Utrophin during S10B (B-B’’ and C-C”, respectively, compared to A-A’’). Both cytoplasmic actin filament bundles and cortical actin deposits are labeled by either weak or strong germline expression of GFP-Utrophin (C-D”). However, strong germline expression of GFP-Utrophin results in cortical actin breakdown with ring canal aggregates (red arrowheads) and multi-nucleate nurse cells (red circles), as well as elongated and disorganized actin bundles (D-D”). Scale bars = 50 μm.
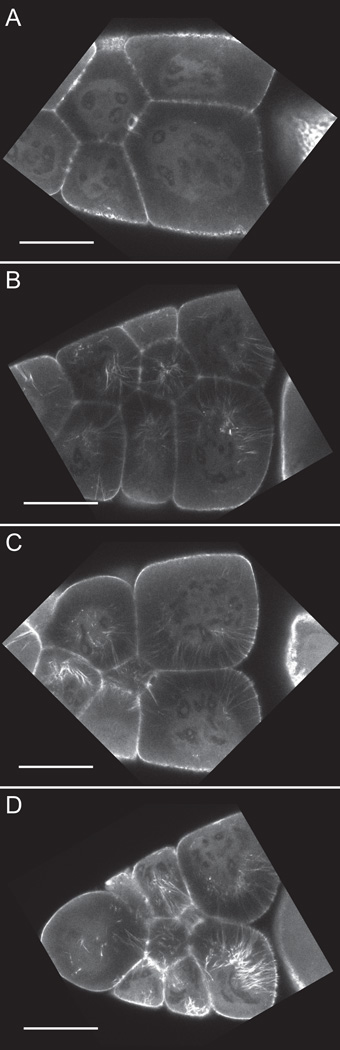
In addition to examining GFP-Utrophin by immunofluorescence, we also assessed whether this tool is useful for live imaging of actin dynamics during mid-oogenesis. As expected based on our fixed image analyses (Fig. 3), nanos-VP16 GAL4 driven GFP-Utrophin (25°C) exhibits patchy expression and poor filament labeling (data not shown). In attempt to overcome these issues, progeny from room temperature crosses were shifted to 29°C to increase expression levels. We find that these conditions are suitable for live imaging of actin dynamics as actin bundle formation (data not shown), elongation (Fig. 4A–B), and contraction (Fig. 4C–D) can be visualized. It is important to note that these conditions also result in cortical actin breakdown and filament disorganization in some of the follicles (data not shown). We also examined strong germline GAL4 driven GFP-Utrophin expression live, and find that, similar to what we observed by fixed imaging, that severe actin defects including cortical actin breakdown (Fig 4E–G, red asterisks) and disorganized bundles are readily observed (Fig. 4E–G). Additionally, the nuclear actin threads are also apparent in live follicles, indicating that their presence is not an artifact of fixation (Fig 4E, green circles). Together, these live-imaging studies reveal that tightly titrating the level of GFP-Utrophin expression, achieved by the combination of a weaker driver and a higher temperature, may allow one to visualize the actin remodeling events occurring within the Drosophila germline.
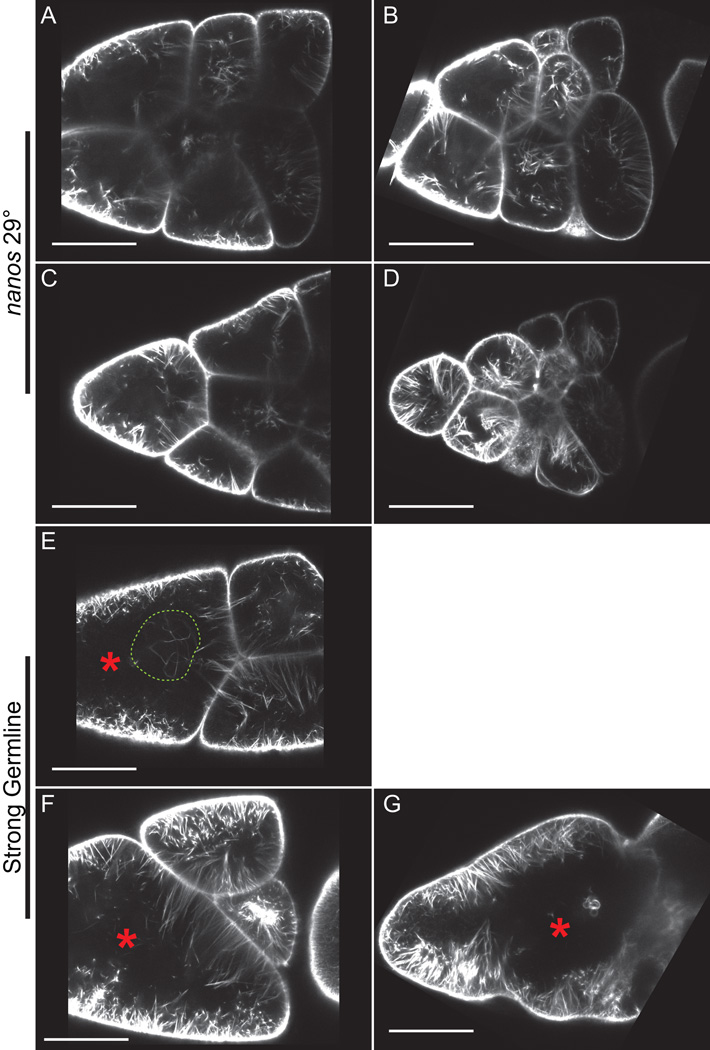
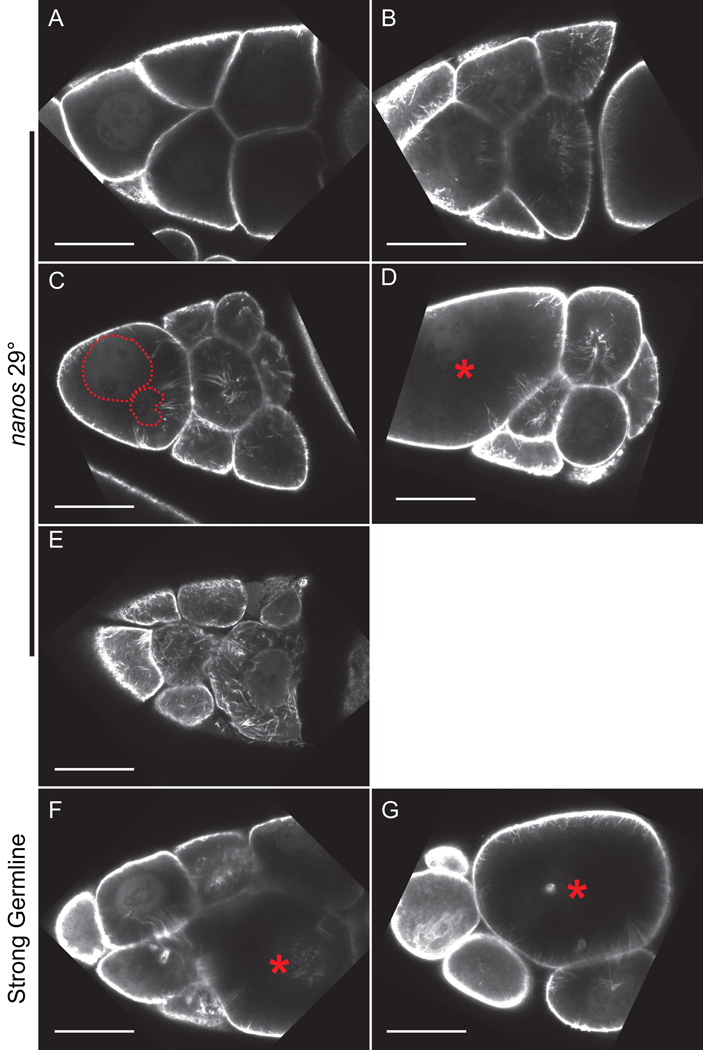
Figure 4. Live imaging of S10B GFP-Utrophin expressing follicles.
(A-G) Single focal plane from confocal z-stack of live S10B-S11 follicles taken at 40X magnification. (A-G) GFP-Utrophin = white. (A-D) GFP-Utrophin; nanos-VP16 GAL4 at 29°C. (E-G) Strong germline GAL4 (mat3 or oskar GAL4) driving GFP-Utrophin (room temperature). Cortical actin deposits and cytoplasmic actin filament bundles can be readily visualized when GFP-Utrophin is driven by nanos-VP16 GAL4 at 29° (A-D) or a strong germline GAL4 at room temperature (E-G). However, strong expression of GFP-Utrophin results in severe actin defects including cortical actin breakdown indicated by floating ring canals and oversized nurse cells (red asterisks, E-G), the formation of nuclear threads (E, dashed green circle), and disorganized bundles (F-G). Scale bars = 50 μm.
Germline expression of Lifeact results in female sterility and severe actin remodeling defects
We generated transgenic flies expressing Lifeact-mEGFP, the first 17 aa residues of yeast actin binding protein 140 fused to mEGFP (Riedl et al., 2008), under the control of the UASp promoter. We tested numerous transgenic insertion sites and found that all insertions tested exhibited similar expression patterns and phenotypes. Here we present data from two independent insertions referred to as 13A (2nd chromosome insertion) and 14A (3rd chromosome insertion), which exhibit robust expression with the different GAL4 drivers tested (SFig. 4B). As expected, both somatic (c355 GAL4) and weak germline (nanos-VP16 GAL4) expression of Lifeact-mEGFP are temperature dependent. However, more uniform expression of Lifeact-mEGFP is observed with the three strong germline GAL4 drivers (mat3, mat2MK and oskar GAL4).
To begin to assess if Lifeact-mEGFP is a useful tool for examining actin dynamics during Drosophila oogenesis, we first wanted to determine if expression of Lifeact-mEGFP affects female fertility. We find that somatic or weak germline expression of either Lifeact insertion has little to no effect of female fertility. Conversely, strong germline expression of Lifeact results in female sterility (Table 2, SFig. 4A).
To determine if the sterility observed is due to F-actin defects caused by expression of Lifeact-mEGFP, we examined S9 and S10B follicles by fixed image analysis (phalloidin staining and anti-GFP immunofluorescence) for actin abnormalities. During S9, somatic and weak germline expression of Lifeact-mEGFP caused no apparent defects in F-actin (Fig. 5B-C” compared to A-A”) and labeled F-actin structures well. Interestingly, Lifeact-mEGFP exhibits a more uniform expression pattern compared to GFP-Utrophin (Fig. 2B-C”), suggesting that perhaps Lifeact-mEGFP is more stable at the protein level than GFP-Utrophin. Indeed, Lifeact-mEGFP expression seems less temperature dependent than GFP-Utrophin (SFig. 4B compared to SFig. 2B). Strong germline expression of Lifeact-mEGFP during S9 results in severe defects in F-actin and follicle morphology, including the loss of cortical actin integrity, which results in aggregates of ring canals and multi-nucleate nurse cells (Fig. 5D-E”, red arrowheads and red circles, respectively), altered nurse cell and oocyte shape, and the formation of phalloidin and Lifeact positive threads in the germinal vesicle (Fig. 5E-E”, green circle, and SFig. 3B-B”’).
Figure 5. Strong germline expression of Lifeact-mEGFP results in cortical actin breakdown during S9.
(A-E”) Maximum projections of 3–5 confocal slices of fixed and stained S9 follicles taken at 20X magnification. (A-E) Merged images: DNA (DAPI) = cyan, F-actin (phalloidin) = white, Lifeact (anti-GFP) = magenta. (A’-E’) F-actin (phalloidin) = white. (A”-E”) Lifeact (anti-GFP) = white. (A-A”) Lifeact-mEGFP /+. (B-B”) c355 GAL4; Lifeact-mEGFP. (C-C”) Lifeact-mEGFP; nanos-VP16 GAL4. (D-E”) Strong germline GAL4 (either mat2MK, mat3, or oskar GAL4) driving Lifeact-mEGFP. Neither somatic nor weak germline expression of Lifeact-mEGFP alters the nurse cell cytoskeleton or follicle cell morphology (B-B” and C-C”, respectively, compared to A-A”). Strong germline expression of Lifeact-mEGFP results in morphologic defects (D-E”), including misshapen nurse cells and oocyte, and cortical actin breakdown with aggregates of ring canals (red arrowheads) and multi-nucleate nurse cells (red circles). Additionally, phalloidin and Lifeact-mEGFP positive threads accumulate within the germinal vesicle following strong germline expression of Lifeact-mEGFP (E-E”, green circle). Scale bars = 50 μm.
During S10B fixed image analysis reveals that expression of Lifeact-mEGFP in the soma causes no apparent defects in follicle cell or nurse cell F-actin (Fig. 6B-B” compared to A-A”), while defects are widely observed when it is expressed in the germline (Fig. 6C-D”). Both the cortical actin and actin bundles in the nurse cells are well labeled by germline expressed Lifeact-mEGFP. Weak germline expression of Lifeact does not significantly alter female fertility (Table 2, SFig. 4A), yet mild defects in cortical actin integrity and actin bundle disorganization are observed (Fig. 6C-C”). Strong germline expression of Lifeact severely compromises the cortical actin, as multinucleate nurse cells are always observed (Fig. 6D-D”, red circles); indeed, when such flies are reared at 25°C, often all 15 nurse cell nuclei are contained within one or two “cells” as determined by cortical actin appearance (data not shown). Interestingly, aberrant F-actin structures are also observed around the germinal vesicle (Fig. 6D-D”). Together these fixed image analyses reveal that strong germline expression of Lifeact-mEGFP has severe consequences on the actin cytoskeleton, and therefore, follicle development. Furthermore, the defects observed due to Lifeact-mEGFP expression seem more severe than those observed due to the expression of GFP-Utrophin.
Figure 6. Lifeact-mEGFP labels actin filament bundles during S10B, but germline expression results in cortical actin breakdown.
(A-D”) Maximum projections of 3–5 confocal slices of fixed and stained S10B follicles taken at 20X magnification. (A-D) Merged images: DNA (DAPI) = cyan, F-actin (phalloidin) = white, Lifeact (anti-GFP) = magenta. (A’-D’) F-actin (phalloidin) = white. (A”-D”) Lifeact (anti-GFP) = white. (A-A”) Lifeact-mEGFP /+. (B-B”) c355 GAL4; Lifeact-mEGFP. (C-C”) Lifeact-mEGFP; nanos-VP16 GAL4. (D-D”) Strong germline GAL4 (either mat2MK, mat3, or oskar GAL4) driving Lifeact-mEGFP. Somatic expression of Lifeact-mEGFP does not alter nurse cell actin remodeling or follicle cell morphology during S10B (B-B” compared to A-A”). While Lifeact-mEGFP clearly labels cytoplasmic actin filament bundles within the nurse cells, both weak and strong germline expression of Lifeact-mEGFP results in cortical actin defects causing multi-nucleate nurse cells (C-D”, red circles). Scale bars = 50 μm.
Live imaging of germline expression of Lifeact-mEGFP recapitulates what we observed in fixed samples. We find that nanos-VP16 GAL4 driven Lifeact-mEGFP (25°C) is not suitable for live imaging due to weak and patchy expression (data not shown), therefore we assessed if increasing the temperature at which the adults were maintained to 29°C would improve expression. While the higher temperature did result in more uniform expression and better labeling of the filaments, early bundle formation could not be visualized (Fig. 7A). Additionally, while elongated bundles were labeled well (Fig. 7B), cortical actin breakdown (Fig 7C, multi-nucleate nurse cells nuclei circled in red, and Fig 7D, red asterisk) and variable nurse cell contraction (Fig. 7D, i.e. some nurse cells contract while others do not) were observed. Roughly one third of the follicles examined exhibited robust, but extremely abnormal bundle formation (Fig. 7E). We find that strong germline expression of Lifeact-mEGFP (room temperature) fails to robustly label bundles and cortical actin defects are readily apparent (Fig. 7F–G, red asterisks). Together these studies suggest utilizing Lifeact-mEGFP to visualize actin dynamics within the Drosophila germline will require significant optimization.
Figure 7. Live imaging of S10B Lifeact-mEGFP expressing follicles.
(A-G) Single focal plane from confocal z-stack of live S10B-S11 follicles taken at 40X magnification. (A-G) Lifeact-mEGFP = white. (A-D) Lifeact-mEGFP; nanos-VP16 GAL4 at 29°. (E-G) Strong germline GAL4 (mat3) driving Lifeact-mEGFP (room temperature). Cortical actin deposits and cytoplasmic actin filament bundles can be readily visualized when Lifeact-mEGFP is driven by nanos-VP16 GAL4 at 29° or a strong germline GAL4 at room temperature (A-G). Weakly driving expression of Lifeact-mEGFP fails to label actin filament bundle initiation (A), but does label actin bundle elongation (B), and contraction (C-D). Occasionally, cortical actin breakdown (C, nuclei within multi-nucleate nurse cell outlined with dashed red lines, and D, red asterisk) and disrupted actin filament bundle morphology (E) are observed under these conditions. In contrast, strong germline expression of Lifeact-mEGFP results in better labeling of F-actin structures, but also results in severe actin defects including cortical actin breakdown (red asterisks, F-G) and disorganized actin bundles (F-G). Scale bars = 50 μm.
F-tractin robustly labels F-actin structures during mid-oogenesis without causing fertility defects
We generated transgenic lines bearing two different F-tractin variants: F-tractin-mEGFP (comprised of ITPK residues 9–52; linker sequence: DPPVAT) (Johnson and Schell, 2009) and F-tractin-tdTom (comprised of ITPK residues 9–40; linker sequence: GGSGSDPPVAT) (Michael Schell, unpublished). Unfortunately, multiple insertions of F-tractin-mEGFP generally fail to label F-actin structures within Drosophila follicles (data not shown); this weak labeling is likely due to the different linker region in this construct. Only the F-tractin-tdTom transgenics were utilized in the subsequent analyses. We present data from two independent insertions referred to as 10C (3rd chromosome insertion) and 15A (2nd chromosome insertion), which exhibit extremely weak expression with both c355 and nanos-VP16 GAL4, low expression with oskar GAL4, and relatively stronger expression with either mat3 or mat2MK GAL4 (SFig. 5B). It is important to note that the DsRed antibody utilized cross reacts with a number of proteins by immunoblot, including one with a similar molecular weight to F-tractin-tdTom.
Once again, we utilized fertility assays as the first means of assessing the utility of F-tractin-tdTom as a tool for labeling the actin cytoskeleton during oogenesis. We find that expression of either F-tractin-tdTom insertion in the soma or the germline (either weakly or strongly) has no effect on female fertility (Table 2, SFig. 5A). This suggests that unlike GFP-Utrophin and Lifeact-GFP, F-tractin-tdTom does not cause F-actin defects that prevent follicle development.
We next assessed the utility of F-tractin-tdTom in labeling F-actin structures using fixed image analysis (phalloidin staining and anti-DsRed immunofluorescence). In general, the strength of the labeling of F-actin by F-tractin-tdTom is considerably weaker than the other two labeling tools, however, less actin defects are observed. Given the weak expression level of F-tractin-tdTom, the fixed images presented are from flies reared at 25°C. During S9, both somatic and weak germline expression of F-tractin-tdTom cause no apparent defects in F-actin within either the soma or the germline (Fig. 8B-C” compared to A-A”). However, weak germline expression of F-tractin-tdTom also does not label the cortical actin well compared to the cytoplasmic background staining (Fig. 8C-C”). Conversely, strong germline expression of F-tractin-tdTom during S9 labels the nurse cell cortical actin well (Fig. 8D-E”), but aberrant, early F-actin filaments (Fig. 8E-E”, orange box), and, at a low frequency, aggregates (data not shown) are sometimes observed. Additionally, F-tractin-tdTom puncta that are phalloidin negative are also seen throughout the nurse cell cytoplasm and at the nuclear periphery (Fig. 8D-E” and data not shown).
Figure 8. F-tractin-tdTom labels F-actin structures during S9 without causing severe actin defects.
(A-E”) Maximum projections of 3–5 confocal slices of fixed and stained S9 follicles taken at 20X magnification. (A-E) Merged images: DNA (DAPI) = cyan, F-actin (phalloidin) = white, F-tractin (anti-DsRed) = magenta. (A’-E’) F-actin (phalloidin) = white. (A”-E”) F-tractin (anti-DsRed)= white. (A-A”) F-tractin-tdTom/+. (B-B”) c355 GAL4; F-tractin-tdTom. (C-C”) F-tractin-tdTom; nanos-VP16 GAL4. (D-E”) Strong germline GAL4 (either mat2MK, mat3, or oskar GAL4) driving F-tractin-tdTom. Somatic expression of F-tractin-tdTom does not alter the nurse cell cytoskeleton or follicle cell morphology (B-B’’ compared to A-A’’). Similarly, follicles weakly expressing F-tractin-tdTom in the germline exhibit normal F-actin structures during S9 (C-C” compared to A-A”). While strong germline expression of F-tractin-tdTom does not cause morphologic defects (D-E”), it results in an increased frequency of early F-actin filaments in the posterior nurse cells (E-E”, orange box). Additionally, strong germline expression of F-tractin-tdTom results in the appearance of anti-DsRed positive, but phalloidin negative, puncta in the nurse cell cytoplasm (D”, E”). Scale bars = 50 μm.
Fixed image (phalloidin staining and anti-DsRed immunofluorescence) analysis reveals S10B follicles expressing F-tractin-tdTom are generally normal in morphology and F-actin structures. While somatic expression of F-tractin-tdTom in mid-to-late oogenesis is very weak and patchy, the actin remodeling occurring within the S10B nurse cells appears normal (Fig. 9B-B” compared to A-A”). Weak germline expression also does not alter actin remodeling, however, the actin bundles are not labeled (Fig. 9C-C”) or only weakly labeled (Fig. 9D-D”). Strong germline expression of F-tractin-tdTom reveals that it labels the actin bundles at varying intensities while not causing actin defects (Fig. 9E-F”). Occasionally, actin bundles exhibit a slight disorganization, but nurse cell dumping occurs and females are fertile, suggesting that the level of actin defects caused by strong germline expression of F-tractin-tdTom are not enough to adversely affect follicle development.
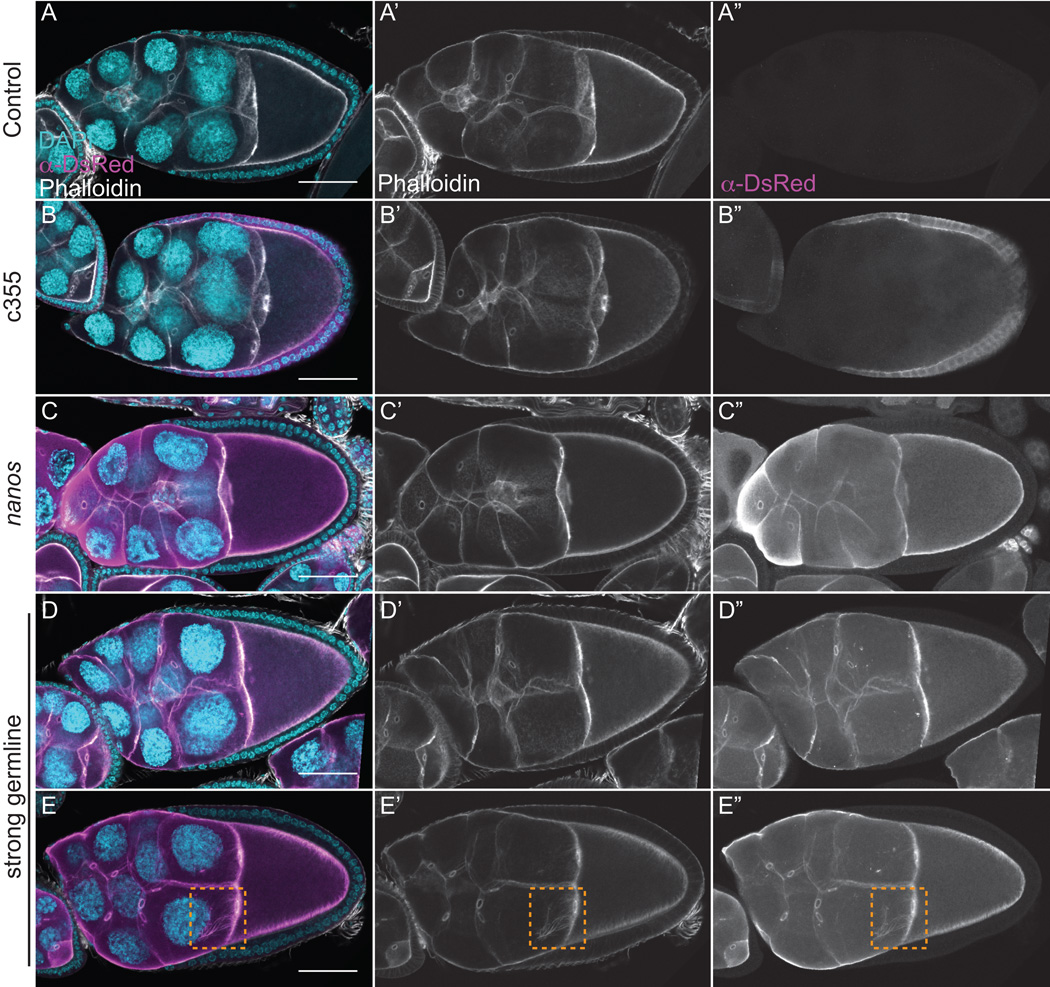
Figure 9. F-tractin-tdTom labels F-actin structures during S10B without causing actin defects.
(A-F”) Maximum projections of 3–5 confocal slices of fixed and stained S10B follicles taken at 20X magnification. (A-F) Merged images: DNA (DAPI) = cyan, F-actin (phalloidin) = white, F-tractin (anti-DsRed) = magenta. (A’-F’) F-actin (phalloidin) = white. (A”-F”) F-tractin (anti-DsRed)= white. (A-A”) F-tractin-tdTom/+. (B-B”) c355 GAL4; F-tractin-tdTom/+. (C-D”) F-tractin-tdTom/+; nanos-VP16 GAL4. (E-F”) Strong germline GAL4 (either mat2MK, mat3, or oskar GAL4) driving F-tractin-tdTom. Somatic expression of F-tractin-tdTom does not alter nurse cell actin remodeling or follicle cell morphology during S10B (B-B” compared to A-A”). Follicles weakly expressing F-tractin-tdTom in the germline exhibit normal nurse cell actin filament bundles and cortical actin deposits (C-D” compared to A-A”). However, the actin filament bundles within the nurse cells are unlabeled (C-C”) or only weakly labeled (D-D”) when F-tractin-tdTom is weakly expressed using nanos-VP16 GAL4. Strong germline expression of F-tractin-tdTom does not cause striking defects in nurse cell actin filament bundles and cortical actin deposits, and actin bundles are labeled, although to varying degrees (E-F”). Scale bars = 50 μm.
We next assessed whether F-tractin-tdTom can be utilized for live imaging of the actin dynamics occurring within the Drosophila germline. Given that nanos-VP16 GAL4 driven expression of F-tractin-tdTom poorly labels filaments by fixed analyses, we used strong germline GAL4 driven F-tractin-tdTom for live imaging. We find that F-tractin-tdTom is difficult to observe by live imaging when the flies are reared at 25°C (data not shown), so we performed live imaging of follicles from flies incubated at 29°C as adults. We find that while F-tractin-tdTom is expressed at a weaker level than the other two tools, it can be used to observe bundle formation (Fig. 10A), elongation (Fig. 10B), and compaction (Fig. 10C–D) via live-imaging without observing any striking actin defects. Indeed, time-lapse imaging (SMovies 1–2) reveals that the actin cytoskeletal dynamics during S10B-S11 can be visualized with strong germline GAL4 driven F-tractin-tdTom (29°C). It is important to point out that in order to clearly observe the actin bundles it is necessary to overexpose the F-tractin-tdTom cytoplasmic aggregates. In conclusion, F-tractin-tdTom is a useful actin labeling tool for examining actin dynamics occurring within the Drosophila germline during mid-oogenesis.
Figure 10. Live imaging of S10B F-tractin-tdTom expressing follicles.
(A-D) Single focal plane from confocal z-stack of live S10B-S11 follicles taken at 40X magnification. (A-D) F-tractin-tdTom = white. (A-D) F-tractin-tdTom; mat3 GAL4 at 29°. Cortical actin deposits and cytoplasmic actin filament bundles can be readily visualized when F-tractin-tdTomato is driven using a strong germline GAL4 at 29°. Cytoplasmic actin filament bundle initiation (A), elongation (B-C), and contraction (D) are visualized. Scale bars = 50 μm.
Discussion
Here, we provide the first side-by-side comparison of three common F-actin labeling tools—Utrophin, Lifeact, and F-tractin—during mid-to-late Drosophila oogenesis. While both Utrophin and Lifeact are strongly expressed using the UAS/GAL4 system and robustly label F-actin structures during follicle development, their germline expression results in severe actin defects including cortical actin breakdown, early actin filament and aggregate formation during S9, disorganized actin bundle formation during S10B, and female sterility. Intriguingly, we also find that Utrophin results in phalloidin positive F-actin thread formation in the nurse cell nuclei and germinal vesicle during S6-9, while Lifeact causes F-actin threads to form only within the germinal vesicle. In contrast, F-tractin is expressed more weakly and labels F-actin structures without causing female sterility or striking actin defects.
In order to achieve sufficient live labeling of F-actin structures with minimal actin defects, expression of either Utrophin or Ftractin had to be modulated by utilizing particular GAL4 drivers and incubating the flies at particular temperatures. Specifically, nanos-VP16 GAL4 driven GFP-Utrophin (29°C) has a low enough expression level to sufficiently reduce the actin defects observed, while strong germline GAL4 (mat3, mat2MK, or osk3) driven F-tractin-tdTom (29°C) has a strong enough expression level to allow actin dynamics in the germline during mid-oogenesis to be observed by live-imaging. It is important to note that both the rate of fly and follicle development increase with temperature, and thus, cellular dynamics including actin cytoskeletal remodeling may be affected by temperature. Thus, live imaging of actin dynamics using either Utrophin or Ftractin in flies maintained at 29°C may not completely reflect the process occurring at lower temperatures. However, flies are fertile at 29°C, and as accurate actin remodeling is required for nurse cell dumping and fertility, it is likely that the majority of the actin cytoskeletal events are occurring normally and can be dissected at this temperature.
The extent of the actin defects that we observe when these tools are expressed during Drosophila oogenesis seems surprising given the broad use of these labeling tools in other systems and/or cell types. Indeed, Lifeact (Deibler et al., 2011; Riedl et al., 2008; Saengsawang et al., 2013; Zanet et al., 2012), Utrophin (Belin et al., 2013), and F-tractin (Case and Waterman, 2011; Saengsawang et al., 2013; Yi et al., 2012) have been widely used to visualize actin dynamics in cell-culture models. Additionally, Lifeact has been successfully employed in vivo in developmental models including zebrafish (Phng et al., 2013; Xu et al., 2014), Arabidopsis (van der Honing et al., 2011; Vidali et al., 2009), and even mouse models (Riedl et al., 2010). Utrophin has also been successfully used to visualize actin dynamics during embryonic germband extension in Drosophila (Rauzi et al., 2010), wound healing in Xenopus oocytes (Burkel et al., 2007), and cytokinesis in zebrafish embryos (Wuhr et al., 2011).
However, not all cell types tolerate these tools well. Transient transfection of C2C12 myoblasts with either Lifeact or Utrophin causes defects in cell shape and cell size, while lentiviral infection is cytotoxic to C2C12 myoblasts, but not 293T cells (Mary Baylies, personal communication). Additionally, it has also been reported that high levels of Lifeact expression in Arabidopsis result in subtle alterations to actin filament dynamics within root hair cells, but does not appear to cause gross defects in plant development (van der Honing et al., 2011). Similarly, expression of either Lifeact, Utrophin, or F-tractin, using the same lines utilized in our study, have subtle effects on actin dynamics within Drosophila hemocytes, resulting in increased filopodia length compared to GAL4 controls (Soichi Tanda, personal communication). Thus, some cell types and tissues may be more sensitive to alterations in F-actin dynamics caused by these labeling tools.
Our data suggests that the Drosophila germline is particularly sensitive to the F-actin labeling tools tested, with both Lifeact and Utrophin resulting in nurse cell cortical actin breakdown and altered actin bundle morphology, but F-tractin having little to no effect. There are a number of reasons why this may be the case, including intrinsic properties of the Drosophila germline, how these labeling tools interact with actin, and the relative level of expression of the different tools examined.
One explanation for the observed defects due to Lifeact and Utrophin could be the very nature of the nurse cells themselves. These cells are very large, polyploid, and must produce an enormous amount of mRNA transcript and protein for the oocyte. It is possible that further stressing the nurse cell biosynthesis machinery, by strongly driving ectopic expression of these tools within the germline, using the bipartite UAS/GAL4 system, places an excess burden on this machinery and results in subsequent breakdown of the germline. However, we don’t favor this model as numerous prior studies have ectopically expressed various proteins, including F-actin binding proteins (Groen et al., 2012; Zanet et al., 2012; Zanet et al., 2009), in the Drosophila germline without resulting F-actin disruption or female sterility.
An alternative explanation for the deleterious effects caused by strong expression of Lifeact and Utrophin, but not F-tractin, in the Drosophila germline may be found in how these actin-labeling tools interact with F-actin. In vitro, Lifeact has been shown to bind to both G- and F-actin, with a much higher affinity for G-actin (~40 nM) compared to F-actin (~1.3 µM) (Riedl et al., 2008). Thus, the extremely severe cortical actin breakdown observed when Lifeact is strongly expressed in the Drosophila germline may be due to it sequestering G-actin. Unlike Lifeact, Utrophin does not bind G-actin and has a reduced affinity for F-actin in vitro (~19µM) (Winder et al., 1995) compared to Lifeact. Utrophin (Prochniewicz et al., 2009; Winder et al., 1995), like Lifeact, does not bundle actin filaments in vitro. Interestingly, both full length Utrophin as well as its N-terminal ABD1 increase actin filament resiliency, by decreasing the amplitude of actin’s rotational flexibility while increasing the rotational rate in vitro (Lin et al., 2012). Thus, it is possible that expression of Utrophin alters biomechanical properties of actin filaments and bundles during Drosophila oogenesis; these alterations could modulate F-actin dynamics, contributing to the severe cortical actin defects and altered actin bundle morphology observed when Utrophin is strongly expressed in the germline.
It is also possible that the binding of Lifeact and Utrophin to F-actin displaces or otherwise alters the ability of endogenous actin binding proteins to interact with and regulate actin bundle formation and dynamics during Drosophila follicle development. It is important to note that while Lifeact has previously been shown not to alter the ability of myosin II or α-actinin, two actin binding proteins that laterally bind to actin filaments, in vitro (Riedl et al., 2008), the extent to which Lifeact and similar actin labeling tools alter actin/actin-binding protein interactions in vivo remains unclear.
F-tractin, consisting of residues 9-52 of rat ITPKA, likely has a much lower affinity for F-actin than either Lifeact or Utrophin, does not alter actin polymerization or depolymerization rates and exhibits actin bundling activity in vitro (Johnson and Schell, 2009). F-tractin’s lower in vitro affinity for F-actin and inability to bind actin monomers likely explains why F-tractin does not perturb actin dynamics during follicle development or adversely effect female fertility. Thus, the deleterious effects observed when Lifeact and Utrophin, but not F-tractin, are strongly expressed in the germline may be indicative of these differences.
Finally, it is possible that differences in the relative expression levels or stability of the tools examined may explain their differential utility. Interestingly, Lifeact and Utrophin, which both result in striking F-actin defects and female sterility, express and/or accumulate at a much higher level during Drosophila oogenesis than F-tractin, which labels F-actin without altering F-actin dynamics or female fertility. Perhaps, Lifeact and Utrophin can be effectively used to label F-actin bundles during Drosophila oogenesis by carefully titrating their expression levels. Indeed, we find that using the weaker germline driver (nanos-VP16 GAL4) in combination with a higher temperature (29°C) to drive GFP-Utrophin can be used to visualize actin dynamics by live imaging. Other means of achieving a lower level of expression include adding destabilization signals to reduce protein stability, or driving expression directly with a weaker germline promoter rather than the UAS/GAL4 system. Additionally, better results may be achieved by adding a layer of temporal regulation on top of the UAS/GAL4 system such as by using a temperature sensitive GAL80 (GAL80ts), a transcriptional repressor of the UAS/GAL4 system (McGuire et al., 2003), or a drug-inducible GAL4 driver (reviewed in (Duffy, 2002; McGuire et al., 2004)). An alternative means of labeling actin dynamics within the Drosophila germline may be to generate transgenic lines that allow GFP-Moe (Edwards et al., 1997), the actin binding domain of Moesin, to be strongly expressed in the germline. In our hands sqh promoter driven GFP-Moe only weakly labels the actin bundles within the germline (Spracklen and Tootle, unpublished observation).
It is important to note that two other groups have previously used Lifeact to monitor actin bundles within the Drosophila female germline without reporting any deleterious effects (Huelsmann et al., 2013; Zanet et al., 2012). As the Lifeact constructs in all three studies were generated from the same original construct (Riedl et al., 2008), the proteins likely have the same activity and the different phenotypes observed are most likely a consequence of expression level. Huelsmann et al. used a low level of germline expression achieved by a drug-inducible, germline-specific GAL4 (mat-tub-gal4_GeneSwitch, matGS) in the absence of drug to express Lifeact (Huelsmann et al., 2013). Thus, the level of expression was likely considerably lower than we utilized. Indeed, they have also found that strong expression of their Lifeact construct using the same mat-GAL4 driver utilized in our study results in phenotypes (Sven Huelsmann, personal communication). Similarly, the level of Lifeact expression was likely lower in the studies by Zanet et al., as they utilized nanos-VP16 GAL4 to express Lifeact (Zanet et al., 2012). Notably, we see only minimal defects when we express our Lifeact-GFP construct using the same GAL4 driver. Thus, we suspect that the majority of the defects we observe are due to a higher level of Lifeact expression and not construct design.
Actin labeling tools, in addition to allowing the visualization of cytoplasmic F-actin structures, also provide insight into nuclear actin. In recent years it has become increasingly clear that there is nuclear actin (McDonald et al., 2006; Scheer et al., 1984) and that it has numerous roles including transcriptional regulation, chromatin remodeling, pre-mRNA processing, and nuclear organization and structure (reviewed in (Grosse and Vartiainen, 2013; Visa and Percipalle, 2010)). Nuclear-targeted Lifeact and Utrophin (full length and truncated forms) have been used to examine nuclear actin. Nuclear-targeted Lifeact in NIH3T3 cells is diffuse in the nucleus under normal conditions, but labels nuclear F-actin upon serum starvation or activation of formin (Baarlink et al., 2013). Conversely, another group found that nuclear-targeted Lifeact in U2OS cells induces aberrant, phalloidin positive, nuclear F-actin (Belin et al., 2013). Full length nuclear-targeted Utrophin also induces ectopic nuclear actin polymerization and/or filament stabilization in U2OS cells, while a truncated form (Utrophin230) labels submicron length F-actin that is excluded from chromatin (Belin et al., 2013).
Here we find that strong germline expression of either Lifeact or Utrophin results in phalloidin positive nuclear F-actin structures. Importantly, these are not nuclear-targeted constructs, the two tools exhibit different abilities to induce nuclear F-actin in the different cell types, and the nuclear F-actin structures are only present at specific stages of follicle development. These data are consistent with the idea that Lifeact and Utrophin may label distinct nuclear actin pools (reviewed in (Grosse and Vartiainen, 2013)). Additionally, our findings suggest that nuclear actin may play critical but currently unknown roles in the germline during S6-9 of Drosophila oogenesis. Further supporting this, we have observed that germline expression of specific Drosophila GFP-actin (Roper et al., 2005) constructs induces F-actin rod formation during these same stages of follicle development (Fagan and Tootle, unpublished observation).
In summary, we find that strong germline expression of both Lifeact and Utrophin disrupt F-actin remodeling during Drosophila oogenesis, resulting in severe cortical actin breakdown, altered actin bundle morphology, and female sterility. Conversely, while F-tractin is not expressed as highly and does not label actin bundles as strongly as Lifeact or Utrophin, it does not significantly alter F-actin remodeling during follicle development or adversely effect female fertility. Together these studies reveal the importance of evaluating the utility of a particular live imaging tool within the tissue and cell type of interest to identify the best compromise between acceptable labeling and minimal disruption of the process being observed.
Methods
Fly strains
Fly stocks were maintained at 18, 21 (room temperature), 25, or 29°C as indicated on standard cornmeal-agar-yeast food. Flies were fed with wet yeast paste daily for fertility assays and ovary analyses, including immunofluorescence, live imaging, and immunoblotting. The following stocks were obtained from the Bloomington Drosophila Stock Center: maternal α-tubulin (matα) GAL4 (3rd chromosome; mat3) and c355 GAL4. We obtained matα GAL4 mk (2nd chromosome; matα2MK) from Kim McKim; oskar GAL4 (3rd chromosome; osk3) from Anne Ephrussi (Telley et al., 2012); and UASp GFP-Utrophin from Thomas Lecuit (Rauzi et al., 2010).
Transgenic generation
The Lifeact-mEGFP fusion was PCR amplified out of the parent plasmid (pmEGFP-N1-Lifeact-EcoR1-BamHI, a generous gift from Roland Wedlich-Soldner) (Riedl et al., 2008) using the following primers: Forward: 5’- CACCATGGGTGTCGCAGATTTGAT -3' Reverse: 5’ TTACTTGTACAGCTCGTCCAT -3’. The F-tractin-mEGFP fusion was PCR amplified out of the parent plasmid (ITPKN9-52mEGFPN1GNBk, a generous gift from Michael Schell, Uniformed Services University) (Johnson and Schell, 2009) using the following primers: Forward: 5’- CACCATGGGCATGGCGCGACC -3’, Reverse: 5’- TTTACTTGTACAGCTCGTCCAT -3’. The F-tractin-tdTom fusion was PCR amplified out of the parent plasmid (ITPKN9-40TomatoN1GNBk, a generous gift from Michael Schell, Uniformed Services University) using the following primers: Forward: 5’- CACCATGGGCATGGCGCGACC -3’, Reverse: 5’-TTTACTTGTACAGCTCGTCCAT -3’. The PCR products were gel extracted and used to generate Gateway entry clones using the pENTR/D-TOPO cloning kit (Invitrogen, Carlsbad, CA, USA). The Gateway LR Clonase II Enzyme mix (Invitrogen, Carlsbad, CA, USA) was used to swap the inserts into the UASp vector (pPW; The Drosophila Gateway Vector Collection, Carnegie), placing the fusion proteins under the control of a germline optimized, yeast upstream activating sequence (UASp). P-element transformations were performed by Genetic Services (Cambridge MA). Multiple insertion lines were generated for each element and their chromosomal locations were determined using standard crosses.
Fertility assays
Three females (~4 days old, fed wet yeast every day prior to mating) of the indicated genotypes were allowed to mate with two or three wild-type (yw) males for 2 days; matings were performed in triplicate for each genotype. Fresh wet yeast was provided daily. The flies were then transferred to a fresh vial, provided wet yeast, and allowed to lay eggs for 24 hrs. The adults were removed after 24 hrs and the resulting adult progeny were counted ~18 days later. The number of progeny per female was determined for each vial, and the average and standard deviation of the three independent vials per genotype was calculated. Fertility assays performed at room temperature (~21°C).
Immunoblotting
Standard immunoblotting techniques were used. The following primary antibodies were used: rabbit α-GFP (Torre Pines TP401; 1: 5000); rabbit α-DsRed (Clontech 632496; 1:1000); mouse α-αtubulin (T9026 Sigma 1:250); rat α−Vasa (DSHB, Spradling, A.C. and Williams, D.; 1:100 or 1:200). The following secondary antibodies were used: Peroxidase-AffiniPure Goat Anti-Rat IgG (H+L) (1:5000); Peroxidase-AffiniPure Goat Anti-Mouse IgG (H+L) (1:5000); Peroxidase-AffiniPure Goat Anti-Rabbit IgG (H+L) (1:5000) (Jackson ImmunoResearch Laboratories, West Grove, PA, USA). Blots were developed with SuperSignal West Pico or Femto Chemiluminescent Substrate (Thermo Scientific, Waltham, MA, USA) and imaged using ChemiDoc-It Imaging System and VisionWorksLS software (UVP, Upland, CA, USA). Bands were quantified by densitometry in ImageJ (Abramoff et al., 2004).
Follicle staging
Follicles were staged based on their morphological appearance as previously described (reviewed in (Spradling, 1993). Follicles were identified as S9 if they had undergone elongation, becoming ellipsoid in shape, and met the following criteria: 1) the border cells had visibly delaminated from the surrounding epithelia, forming a rosette structure at the anterior of the follicle, and/or 2) the main body follicle cells were in the process of their posterior migration, forming a clear anterior to posterior gradient of follicle cell thickness. Follicles were identified as S10B if half of the total follicle length was composed of the oocyte, the main body follicle cells had reached the nurse cell/oocyte boundary, the follicle length was equal to a S14 follicle, and the centripetal follicle cells had begun to migrate over the anterior surface of the oocyte.
Immunofluorescence
Whole-mount samples were fixed for 10 min at room temperature in 4% paraformaldehyde in Grace’s insect media (Lonza, Walkersville, MD, U.S.). Samples were processed using standard procedures (Cox and Spradling, 2003; Groen et al., 2012). The following primary antibodies were used: rabbit α-GFP (Torre Pines TP401; 1:2000–1:2500) and rabbit α-DsRed (Clontech 6324)96; 1:300). Additional stains used and their concentrations are as follows: rhodamine::phalloidin, 1:250- 1:500; Alexa Fluor (AF) 488::phalloidin, 1:250–1:500; Alexa Fluor (AF) 647::phalloidin, 1:100- 1:250; DAPI (5mg/ml), 1:5000–1:10,000; Alexa Fluor (AF) 555::wheat germ agglutinin (WGA), 1:500 in secondary only (all from Life Technologies, Carlsbad, CA, USA). The following secondary antibodies were used at 1:1000 as appropriate: AF488::goat α-rabbit and AF568::goat α-rabbit (Life Technologies, Carlsbad, CA, USA).
Live imaging
Individual S10B follicles were isolated in room temperature in vitro egg maturation (IVEM) media (10% FBS and 1X Pen/Strep in Grace’s insect media). Follicles were spotted onto a coverslip-bottom dish in a droplet of IVEM media for short-term live imaging. For long-term live imaging, follicles were immobilized by imbedding in low-melt agarose on coverslip-bottom dishes using a protocol adapted from (Westerfield, 2000). In short, molten 1% low melt agarose in Grace’s insect media was mixed 1:1 with 2X IVEM media (20% FBS and 2X Pen/Strep in Grace’s insect media) media to generate the embedding agarose and held at 42°C. Molten embedding agarose was used to cover the coverslip of a coverslip-bottom dish and isolated follicles were transferred into embedding agarose using a Pasteur pipet and gravity. Once the embedding media had solidified, the follicles were subjected to live imaging. Imaging was performed using Zen software on a Zeiss 700 LSM mounted on an Axio Observer.Z1 using a LD C-APO 40X/1.1 W/0 objective (Carl Zeiss Microscopy, Thornwood, NY, U.S.). Maximum projections, image rotation, cropping, and generation of movie files was performed in ImageJ (Abramoff et al., 2004). Figure panels were assembled in Illustrator (Adobe, San Jose, CA, U.S.).
Confocal microscope image acquisition and processing
All fixed microscopy images were acquired via LAS AF SPE Core software on a Leica TCS SPE mounted on a Leica DM2500 using an ACS APO 20X/0.60 IMM CORR -/D or an ACS APO 63X/1.30 Oil CS 0.17/E objective (Leica Microsystems, Buffalo Grove, IL, USA). Maximum projections, image rotation, and cropping were performed in ImageJ (Abramoff et al., 2004). Figure panels were assembled in Illustrator (Adobe, San Jose, CA, U.S.). To aid in visualization, all fixed images were brightened by 30% using Photoshop (Adobe, San Jose, CA, U.S.).
Supplementary Material
(A-J) Maximum projections of 3–5 confocal slices of fixed and stained follicles of the indicated genotypes taken at 20X magnification. Anterior is to the left. F-actin (phalloidin) = white, DNA (DAPI) = cyan. (A-E) S9 follicles. (F-J) S10B follicles. (A, F) c355 GAL4/+. (B, G) nanos-VP16 GAL4/+. (C, H) mat2MK GAL4/+. (D, I) mat3 GAL4/+. (E, J) oskar GAL4/+. Heterozygosity for the GAL4 lines utilized in this study have little to no apparent effect on F-actin dynamics during mid-oogenesis (A-J), with the exception that strong germline GAL4 drivers increase the incidence of early F-actin remodeling in the posterior nurse cells of S9 follicles (D-E, yellow arrows). Scale bars = 50 μm.
(A) Chart depicting the number of adult progeny produced per female from a 24 hour egg collection; error bars represent standard deviation. (B) Representative immunoblots of whole ovary lysates of the given genotypes and raised at the indicated temperatures (°C, RT is approximately 21°C). The anti-GFP blots reveal the expression level of GFP-Utrophin (UAS-UT; ~56.9 kDa) compared to the loading control anti-Vasa. Somatic (c355 GAL4) or weak germline (nanos-VP16 GAL4) expression of GFP-Utrophin has no striking effect on female fertility. However, strong germline expression results in severely reduced fertility (mat2MK and oskar GAL4) or sterility (mat3 GAL4) (A). Expression of UAS GFP-Utrophin exhibits the expected temperature dependence with all of the GAL4 lines, with weaker expression at 18 degrees and strongest expression at 25°C (B).
(A-B”’) Maximum projections of 3–7 confocal slices of fixed and stained follicles of the indicated genotypes taken at 63X magnification. Anterior is to the left. (A-B) Merged images: Nuclear envelope (WGA) = magenta, anti-GFP = cyan, and F-actin (phalloidin) = white. (A’, B’) Nuclear envelope (WGA) = white. (A”, B”) anti-GFP = white. (A”’, B”’) F-actin (phalloidin) = white. (A-A”) GFP-Utrophin; mat3 GAL4. (B-B”) Lifeact-meGFP; mat3 GAL4. Nuclear GFP and phalloidin positive F-actin threads are induced in the nurse cells when GFP-Utrophin is strongly expressed in the germline (A-A”’), while such threads are only seen in the germinal vesicle when Lifeact-mEGFP is expressed (B-B”’, yellow arrow head). Scale bars = 50 μm.
(A) Chart depicting the number of adult progeny produced per female from a 24 hour egg collection; error bars represent standard deviation. (B) Representative immunoblots of whole ovary lysates of the given genotypes and raised at the indicated temperatures (°C, RT is approximately 21°C). The anti-GFP blots reveal the expression level of Lifeact-mEGFP (~ 29.5 kDa; two different inserts: LA 14A and LA 13A) compared to the loading control anti-Vasa. Somatic (c355 GAL4) or weak germline (nanos-VP16 GAL4) expression of either insert of Lifeact-mEGFP has no striking effects on female fertility. However, strong germline expression results in severely reduced fertility (mat2mk GAL4) or sterility (mat3 and osk3 GAL4) (A). Expression of either UAS Lifeact-mEGFP insertion exhibits the expected temperature dependence with the c355 and nanos-VP16 GAL4 lines, with weaker expression at 18 degrees and strongest expression at 25°C (B). However, this temperature dependence is severely diminished with the strong germline GAL4 drivers (B; mat3, mat2mk, and osk3 Gal4).
(A) Chart depicting the number of adult progeny produced per female from a 24 hour egg collection; error bars represent standard deviation. (B) Representative immunoblots of whole ovary lysates of the given genotypes and raised at the indicated temperatures (°C, RT is approximately 21°C). The anti-DsRed blots reveal the expression level of F-tractin-tdTom (~58.2 kDa; two different inserts: Ft 10C and Ft 15A) compared to the loading control anti-Vasa (note that 2x the amount of sample was loaded onto the gels probed for DsRed vs the gels probed for Vasa). Expression of F-tractin-tdTom with any of the GAL4 drivers has no striking effects on female fertility (A). Expression of either UAS F-tractin-tdTom insertion exhibits fairly weak expression with all GAL4 drivers, and the DsRed antibody detects a number of cross reacting bands, one of which is at the same molecular weight as F-tractin-tdTom. In particular, F-tractin-tdTom levels are very low in ovary lysates where expression was driven with c355 or nanos-VP16 GAL4, is weak with osk3 GAL4, and is comparatively stronger with mat2MK and mat3 GAL4 (B).
Summary of the GAL4 lines utilizes in this study, if they exhibit temperature dependence, and when during oogenesis they are expressed. * GAL4 lines not used in the majority of the studies.
Movie of a single frame from confocal stack imaged every 10 min. Bundle elongation and contraction is observed. Scale bar = 50 μm.
Movie of a single frame from confocal stack imaged every 10 min. Filament and nurse cell contraction is observed. Scale bar = 50 μm.
Acknowledgements
We thank the Lin Lab for both useful discussions and assistance in mounting samples for long term live-imaging, the Frank Lab for helpful discussions, members of the Tootle lab for helpful discussions and careful review of the manuscript, Dr. Huelsmann for insight into Lifeact, and the Drosophila community for sharing reagents. A.J.S. was previously supported by the National Institutes of Health Predoctoral Training Grant in Pharmacological Sciences T32GM067795. National Science Foundation MCB-1158527 supports the project. Data storage support was provided by the ICTS, which is funded through the CTSA supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1RR024979.
References
- Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with Image. J. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Aizawa H, Sameshima M, Yahara I. A green fluorescent protein-actin fusion protein dominantly inhibits cytokinesis, cell spreading, and locomotion in Dictyostelium. Cell structure and function. 1997;22:335–345. doi: 10.1247/csf.22.335. [DOI] [PubMed] [Google Scholar]
- Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340:864–867. doi: 10.1126/science.1235038. [DOI] [PubMed] [Google Scholar]
- Belin BJ, Cimini BA, Blackburn EH, Mullins RD. Visualization of actin filaments and monomers in somatic cell nuclei. Molecular biology of the cell. 2013;24:982–994. doi: 10.1091/mbc.E12-09-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle MJ, French RL, Cosand KA, Dorman JB, Kiehart DP, Berg CA. Division of labor: subsets of dorsal-appendage-forming cells control the shape of the entire tube. Developmental biology. 2010;346:68–79. doi: 10.1016/j.ydbio.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brock AR, Wang Y, Berger S, Renkawitz-Pohl R, Han VC, Wu Y, Galko MJ. Transcriptional regulation of Profilin during wound closure in Drosophila larvae. Journal of cell science. 2012;125:5667–5676. doi: 10.1242/jcs.107490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkel BM, von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell motility and the cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Waterman CM. Adhesive F-actin waves: a novel integrin-mediated adhesion complex coupled to ventral actin polymerization. PloS one. 2011;6:e26631. doi: 10.1371/journal.pone.0026631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Verheyen E, Ayers K. chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell. 1992;69:173–184. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development. 2003;130:1579–1590. doi: 10.1242/dev.00365. [DOI] [PubMed] [Google Scholar]
- Deibler M, Spatz JP, Kemkemer R. Actin fusion proteins alter the dynamics of mechanically induced cytoskeleton rearrangement. PloS one. 2011;6:e22941. doi: 10.1371/journal.pone.0022941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Developmental biology. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- Gates J, Nowotarski SH, Yin H, Mahaffey JP, Bridges T, Herrera C, Homem CC, Janody F, Montell DJ, Peifer M. Enabled and Capping protein play important roles in shaping cell behavior during Drosophila oogenesis. Developmental biology. 2009;333:90–107. doi: 10.1016/j.ydbio.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen CM, Spracklen AJ, Fagan TN, Tootle TL. Drosophila Fascin is a novel downstream target of prostaglandin signaling during actin remodeling. Molecular biology of the cell. 2012;23:4567–4578. doi: 10.1091/mbc.E12-05-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse R, Vartiainen MK. To be or not to be assembled: progressing into nuclear actin filaments. Nature reviews. Molecular cell biology. 2013;14:693–697. doi: 10.1038/nrm3681. [DOI] [PubMed] [Google Scholar]
- Guild GM, Connelly PS, Shaw MK, Tilney LG. Actin filament cables in Drosophila nurse cells are composed of modules that slide passively past one another during dumping. J Cell Biol. 1997;138:783–797. doi: 10.1083/jcb.138.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatan M, Shinder V, Israeli D, Schnorrer F, Volk T. The Drosophila blood brain barrier is maintained by GPCR-dependent dynamic actin structures. J Cell Biol. 2011;192:307–319. doi: 10.1083/jcb.201007095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird S. Cortical actin movements during the first cell cycle of the Caenorhabditis elegans embryo. Journal of cell science. 1996;109(Pt 2):525–533. doi: 10.1242/jcs.109.2.525. [DOI] [PubMed] [Google Scholar]
- Hudson AM, Cooley L. Understanding the function of actin-binding proteins through genetic analysis of Drosophila oogenesis. Annual review of genetics. 2002;36:455–488. doi: 10.1146/annurev.genet.36.052802.114101. [DOI] [PubMed] [Google Scholar]
- Huelsmann S, Ylanne J, Brown NH. Filopodia-like Actin Cables Position Nuclei in Association with Perinuclear Actin in Drosophila Nurse Cells. Developmental cell. 2013;26:604–615. doi: 10.1016/j.devcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HW, Schell MJ. Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Molecular biology of the cell. 2009;20:5166–5180. doi: 10.1091/mbc.E09-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Chihara T, Hayashi S. Hedgehog and Decapentaplegic instruct polarized growth of cell extensions in the Drosophila trachea. Development. 2004;131:5253–5261. doi: 10.1242/dev.01404. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Lin AY, Prochniewicz E, Henderson DM, Li B, Ervasti JM, Thomas DD. Impacts of dystrophin and utrophin domains on actin structural dynamics: implications for therapeutic design. Journal of molecular biology. 2012;420:87–98. doi: 10.1016/j.jmb.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Cooley L. Intercellular cytoplasm transport during Drosophila oogenesis. Developmental biology. 1994a;165:336–351. doi: 10.1006/dbio.1994.1257. [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Cooley L. The villin-like protein encoded by the Drosophila quail gene is required for actin bundle assembly during oogenesis. Cell. 1994b;78:291–301. doi: 10.1016/0092-8674(94)90298-4. [DOI] [PubMed] [Google Scholar]
- McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Science's STKE : signal transduction knowledge environment, 2004. 2004:l6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Peralta XG, Toyama Y, Hutson MS, Montague R, Venakides S, Kiehart DP, Edwards GS. Upregulation of forces and morphogenic asymmetries in dorsal closure during Drosophila development. Biophysical journal. 2007;92:2583–2596. doi: 10.1529/biophysj.106.094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phng LK, Stanchi F, Gerhardt H. Filopodia are dispensable for endothelial tip cell guidance. Development. 2013;140:4031–4040. doi: 10.1242/dev.097352. [DOI] [PubMed] [Google Scholar]
- Prochniewicz E, Henderson D, Ervasti JM, Thomas DD. Dystrophin and utrophin have distinct effects on the structural dynamics of actin. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7822–7827. doi: 10.1073/pnas.0812007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauzi M, Lenne PF, Lecuit T. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature. 2010;468:1110–1114. doi: 10.1038/nature09566. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nature methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Flynn KC, Raducanu A, Gartner F, Beck G, Bosl M, Bradke F, Massberg S, Aszodi A, Sixt M, Wedlich-Soldner R. Lifeact mice for studying F-actin dynamics. Nature methods. 2010;7:168–169. doi: 10.1038/nmeth0310-168. [DOI] [PubMed] [Google Scholar]
- Roper K, Mao Y, Brown NH. Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo. Journal of cell science. 2005;118:3937–3948. doi: 10.1242/jcs.02517. [DOI] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mechanisms of development. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Saengsawang W, Taylor KL, Lumbard DC, Mitok K, Price A, Pietila L, Gomez TM, Dent EW. CIP4 coordinates with phospholipids and actin-associated proteins to localize to the protruding edge and produce actin ribs and veils. Journal of cell science. 2013;126:2411–2423. doi: 10.1242/jcs.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Hinssen H, Franke WW, Jockusch BM. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 1984;39:111–122. doi: 10.1016/0092-8674(84)90196-x. [DOI] [PubMed] [Google Scholar]
- Skora AD, Spradling AC. Epigenetic stability increases extensively during Drosophila follicle stem cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7389–7394. doi: 10.1073/pnas.1003180107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklen AJ, Kelpsch DJ, Chen X, Spracklen CN, Tootle TL. Prostaglandins temporally regulate cytoplasmic actin bundle formation during Drosophila oogenesis. Molecular biology of the cell. 2014;25:397–411. doi: 10.1091/mbc.E13-07-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC. Developmental genetics of oogenesis. In: Martinez-Arias B, editor. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- Telley IA, Gaspar I, Ephrussi A, Surrey T. Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J Cell Biol. 2012;197:887–895. doi: 10.1083/jcb.201204019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootle TL, Spradling AC. Drosophila Pxt: a cyclooxygenase-like facilitator of follicle maturation. Development. 2008;135:839–847. doi: 10.1242/dev.017590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama Y, Peralta XG, Wells AR, Kiehart DP, Edwards GS. Apoptotic force and tissue dynamics during Drosophila embryogenesis. Science. 2008;321:1683–1686. doi: 10.1126/science.1157052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Honing HS, van Bezouwen LS, Emons AM, Ketelaar T. High expression of Lifeact in Arabidopsis thaliana reduces dynamic reorganization of actin filaments but does not affect plant development. Cytoskeleton. 2011;68:578–587. doi: 10.1002/cm.20534. [DOI] [PubMed] [Google Scholar]
- Vidali L, Rounds CM, Hepler PK, Bezanilla M. Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PloS one. 2009;4:e5744. doi: 10.1371/journal.pone.0005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harbor perspectives in biology. 2010;2:a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil TT, Xanthakis D, Parton R, Dobbie I, Rabouille C, Gavis ER, Davis I. Distinguishing direct from indirect roles for bicoid mRNA localization factors. Development. 2010;137:169–176. doi: 10.1242/dev.044867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio) 4 ed. ed. Eugene, OR: University of Oregon Press; 2000. [Google Scholar]
- Wheatley S, Kulkarni S, Karess R. Drosophila nonmuscle myosin II is required for rapid cytoplasmic transport during oogenesis and for axial nuclear migration in early embryos. Development. 1995;121:1937–1946. doi: 10.1242/dev.121.6.1937. [DOI] [PubMed] [Google Scholar]
- Winder SJ, Hemmings L, Maciver SK, Bolton SJ, Tinsley JM, Davies KE, Critchley DR, Kendrick-Jones J. Utrophin actin binding domain: analysis of actin binding and cellular targeting. Journal of cell science. 1995;108(Pt 1):63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wuhr M, Obholzer ND, Megason SG, Detrich HW, 3rd, Mitchison TJ. Live imaging of the cytoskeleton in early cleavage-stage zebrafish embryos. Methods in cell biology. 2011;101:1–18. doi: 10.1016/B978-0-12-387036-0.00001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Ye D, Behra M, Burgess S, Chen S, Lin F. Gbeta1 controls collective cell migration by regulating the protrusive activity of leader cells in the posterior lateral line primordium. Developmental biology. 2014;385:316–327. doi: 10.1016/j.ydbio.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Wu XS, Crites T, Hammer JA., 3rd Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Molecular biology of the cell. 2012;23:834–852. doi: 10.1091/mbc.E11-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J, Jayo A, Plaza S, Millard T, Parsons M, Stramer B. Fascin promotes filopodia formation independent of its role in actin bundling. J Cell Biol. 2012;197:477–486. doi: 10.1083/jcb.201110135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J, Stramer B, Millard T, Martin P, Payre F, Plaza S. Fascin is required for blood cell migration during Drosophila embryogenesis. Development. 2009;136:2557–2565. doi: 10.1242/dev.036517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A-J) Maximum projections of 3–5 confocal slices of fixed and stained follicles of the indicated genotypes taken at 20X magnification. Anterior is to the left. F-actin (phalloidin) = white, DNA (DAPI) = cyan. (A-E) S9 follicles. (F-J) S10B follicles. (A, F) c355 GAL4/+. (B, G) nanos-VP16 GAL4/+. (C, H) mat2MK GAL4/+. (D, I) mat3 GAL4/+. (E, J) oskar GAL4/+. Heterozygosity for the GAL4 lines utilized in this study have little to no apparent effect on F-actin dynamics during mid-oogenesis (A-J), with the exception that strong germline GAL4 drivers increase the incidence of early F-actin remodeling in the posterior nurse cells of S9 follicles (D-E, yellow arrows). Scale bars = 50 μm.
(A) Chart depicting the number of adult progeny produced per female from a 24 hour egg collection; error bars represent standard deviation. (B) Representative immunoblots of whole ovary lysates of the given genotypes and raised at the indicated temperatures (°C, RT is approximately 21°C). The anti-GFP blots reveal the expression level of GFP-Utrophin (UAS-UT; ~56.9 kDa) compared to the loading control anti-Vasa. Somatic (c355 GAL4) or weak germline (nanos-VP16 GAL4) expression of GFP-Utrophin has no striking effect on female fertility. However, strong germline expression results in severely reduced fertility (mat2MK and oskar GAL4) or sterility (mat3 GAL4) (A). Expression of UAS GFP-Utrophin exhibits the expected temperature dependence with all of the GAL4 lines, with weaker expression at 18 degrees and strongest expression at 25°C (B).
(A-B”’) Maximum projections of 3–7 confocal slices of fixed and stained follicles of the indicated genotypes taken at 63X magnification. Anterior is to the left. (A-B) Merged images: Nuclear envelope (WGA) = magenta, anti-GFP = cyan, and F-actin (phalloidin) = white. (A’, B’) Nuclear envelope (WGA) = white. (A”, B”) anti-GFP = white. (A”’, B”’) F-actin (phalloidin) = white. (A-A”) GFP-Utrophin; mat3 GAL4. (B-B”) Lifeact-meGFP; mat3 GAL4. Nuclear GFP and phalloidin positive F-actin threads are induced in the nurse cells when GFP-Utrophin is strongly expressed in the germline (A-A”’), while such threads are only seen in the germinal vesicle when Lifeact-mEGFP is expressed (B-B”’, yellow arrow head). Scale bars = 50 μm.
(A) Chart depicting the number of adult progeny produced per female from a 24 hour egg collection; error bars represent standard deviation. (B) Representative immunoblots of whole ovary lysates of the given genotypes and raised at the indicated temperatures (°C, RT is approximately 21°C). The anti-GFP blots reveal the expression level of Lifeact-mEGFP (~ 29.5 kDa; two different inserts: LA 14A and LA 13A) compared to the loading control anti-Vasa. Somatic (c355 GAL4) or weak germline (nanos-VP16 GAL4) expression of either insert of Lifeact-mEGFP has no striking effects on female fertility. However, strong germline expression results in severely reduced fertility (mat2mk GAL4) or sterility (mat3 and osk3 GAL4) (A). Expression of either UAS Lifeact-mEGFP insertion exhibits the expected temperature dependence with the c355 and nanos-VP16 GAL4 lines, with weaker expression at 18 degrees and strongest expression at 25°C (B). However, this temperature dependence is severely diminished with the strong germline GAL4 drivers (B; mat3, mat2mk, and osk3 Gal4).
(A) Chart depicting the number of adult progeny produced per female from a 24 hour egg collection; error bars represent standard deviation. (B) Representative immunoblots of whole ovary lysates of the given genotypes and raised at the indicated temperatures (°C, RT is approximately 21°C). The anti-DsRed blots reveal the expression level of F-tractin-tdTom (~58.2 kDa; two different inserts: Ft 10C and Ft 15A) compared to the loading control anti-Vasa (note that 2x the amount of sample was loaded onto the gels probed for DsRed vs the gels probed for Vasa). Expression of F-tractin-tdTom with any of the GAL4 drivers has no striking effects on female fertility (A). Expression of either UAS F-tractin-tdTom insertion exhibits fairly weak expression with all GAL4 drivers, and the DsRed antibody detects a number of cross reacting bands, one of which is at the same molecular weight as F-tractin-tdTom. In particular, F-tractin-tdTom levels are very low in ovary lysates where expression was driven with c355 or nanos-VP16 GAL4, is weak with osk3 GAL4, and is comparatively stronger with mat2MK and mat3 GAL4 (B).
Summary of the GAL4 lines utilizes in this study, if they exhibit temperature dependence, and when during oogenesis they are expressed. * GAL4 lines not used in the majority of the studies.
Movie of a single frame from confocal stack imaged every 10 min. Bundle elongation and contraction is observed. Scale bar = 50 μm.
Movie of a single frame from confocal stack imaged every 10 min. Filament and nurse cell contraction is observed. Scale bar = 50 μm.