Abstract
Purpose
Lung cancer stem cells (CSCs) with elevated aldehyde dehydrogenase (ALDH) activity are self-renewing, clonogenic and tumorigenic. The purpose of our study is to elucidate the mechanisms by which lung CSCs are regulated.
Experimental Design
A genome-wide gene expression analysis was performed to identify genes differentially expressed in the ALDH+ vs. ALDH− cells. RT-PCR, western blot and Aldefluor assay were used to validate identified genes. To explore the function in CSCs we manipulated their expression followed by colony and tumor formation assays.
Results
We identified a subset of genes that were differentially expressed in common in ALDH+ cells, among which ALDH1A3 was the most upregulated gene in ALDH+ vs. ALDH− cells. ShRNA-mediated knockdown of ALDH1A3 in NSCLCs resulted in a dramatic reduction in ALDH activity, clonogenicity and tumorigenicity, indicating that ALDH1A3 is required for tumorigenic properties. By contrast, overexpression of ALDH1A3 by itself it was not sufficient to increase tumorigenicity. The ALDH+ cells also expressed more activated Signal Transducers and Activators of Transcription 3 (STAT3) than ALDH− cells. Inhibition of STAT3 or its activator EZH2 genetically or pharmacologically diminished the level of ALDH+ cells and clonogenicity. Unexpectedly, ALDH1A3 was highly expressed in female, never smokers, well differentiated tumors, or adenocarcinoma. ALDH1A3 low expression was associated with poor overall survival.
Conclusion
Our data show that ALDH1A3 is the predominant ALDH isozyme responsible for ALDH activity and tumorigenicity in most NSCLCs, and that inhibiting either ALDH1A3 or the STAT3 pathway are potential therapeutic strategies to eliminate the ALDH+ subpopulation in NSCLCs.
Keywords: Lung cancer, cancer stem cells, ALDH1A3, STAT3, Stattic
Introduction
The “cancer stem cell (CSC)” model proposes that tumor progression, drug resistance, metastasis, and relapse after therapy may be driven by a subset of cells within a tumor representing a functionally important example of intra-tumor heterogeneity (1, 2). Since the discovery of CSCs in breast cancer pleural effusions, accumulating evidence has supported the existence of CSCs in many solid tumors, which share important characteristics with embryonic and normal tissue stem cells, such as self-renewal capability and the competency to undergo differentiation (3). In non-small cell lung cancers (NSCLCs), several populations of CSCs have been identified and characterized by the expression of CSC markers including CD133, CD44, aldehyde dehydrogenase (ALDH), and Side Population (SP) (4–7). Ho et al. showed that as few as 1000 isolated SP cells from 6 lung cancer lines could generate xenograft tumors in NOD-SCID mice whereas the bulk of the SP− tumor cells could not (5). Eramo et al. found that CD133+ subpopulations in some NSCLC and SCLC tumors are self-renewing and highly tumorigenic, and are capable of efficiently propagating the disease both in vitro and in vivo (8). However several follow-up studies using SP and CD133 as identifiers of lung CSCs indicated these markers frequently identify non-CSC subpopulations, signifying a need for more reliable methods to identify and isolate lung CSCs (9, 10).
More recently, elevated ALDH activity has been employed as a CSC marker in multiple tumor types (11–13). We and others have identified a subpopulation of ALDH+ NSCLC cells with increased malignant behavior in many tumor cell lines and patient samples using the flow cytometry-based Aldefluor assay (6, 14, 15). Similar to findings in other types of cancers, ALDH+ tumor cells isolated from patient lung tumors and lung cancer cell lines are enriched in highly tumorigenic and clonogenic cells which are capable of self-renewal (6, 16–18). Of the 19 isozymes in this family, class one aldehyde dehydrogenases (ALDH1) are frequently associated with alcohol metabolism, retinoic acid synthesis, drug resistance, and stem cell homeostasis (19–21). Recently, expression of the ALDH1A1 isozyme was shown to be a biomarker of poor prognosis in tumors of the breast, colon, ovary and lung (22–24). However, additional evidence in metastatic breast and colon cancers implicated another ALDH isozyme, ALDH1A3, and other class one ALDH isozymes as putative CSC markers (18, 25, 26). Therefore, a thorough understanding of the expression and function of the role of specific ALDH isozymes in lung CSCs is necessary for clinical translation of CSCs identified by ALDH activity in lung cancer.
Transducers and Activators of Transcription 3 (STAT3) was originally identified as acute-phase response factor which bound to IL-6-response elements within the promoter region of various acute-phase response genes. Cytokines and growth factors are able to trigger STAT3 activation and constitutively active STAT3 is found in various tumors. A series of reports showed that the STAT3 pathway preferentially regulate CSC self-renewal, survival, and tumor initiation in many solid tumors (27–29). This led to studies showing that STAT3 pathway blockade causes a decrease in CSCs and to a significant reduction of tumorigenicity in mouse xenograft models (28–30). Thus, we investigated which ALDH isozyme was associated with the NSCLC stem cell subpopulation and if there was a connection between such ALDH+ cells and the STAT3 pathway.
In this study, we characterized the expression profile of ALDH+ and ALDH− tumor cells in a panel of NSCLC lines and found the expression of ALDH1A3 to be the most commonly elevated of all ALDH isozymes in the ALDH+ NSCLC subpopulations. We found that knockdown of ALDH1A3 significantly reduces the clonogenicity, tumorigenicity, and ALDH activity of lung cancer cells. Following this we were able to show the STAT3 pathway is more activated in ALDH+ cells than in ALDH− lung cancer cells and inhibition of the STAT3 pathway also impaired the maintenance of lung CSCs. Together, the data show that ALDH1A3 is functionally important for NSCLC malignant behavior and that ALDH1A3 and STAT3 are promising therapeutic targets for NSCLC through their significant role in the ALDH+ subpopulation of tumor cells.
Material and Methods
Cell culture
All NSCLC lines used in this study were obtained from the Hamon Cancer Center Collection (University of Texas Southwestern Medical Center) and maintained in RPMI-1640 (Life Technologies) supplemented with 5% fetal calf serum at 37°C in a humidified atmosphere containing 5% CO2 and 95% air. All cell lines have been DNA fingerprinted using the PowerPlex 1.2 kit (Promega) and are mycoplasma free using the e-Myco kit (Boca Scientific).
Aldefluor assay and FACS
The Aldefluor assay (Stem Cell Technologies) was used to profile and sort cells based on ALDH activity as previous described (14). ALDH+ and ALDH− cells were sorted by BD Aria (BD Biosciences) cell sorters and the purity was usually >90% confirmed by post-sort analyses. Flow cytometric profiling was performed on a FACScan flow cytometer (BD Biosciences) and analyzed using FlowJo software (Treestar).
Colony formation assay
Anchorage-dependent (liquid) and -independent (soft agar) colony formation assays were done as described (14). The inhibitors used in the study were Stattic (Calbiochem), Ruxolitinib (LC Laboratories), and GSK126 (Xcessbio).
Microarray analysis
Total RNA of sorted ALDH+ and ALDH− cells from 8 NSCLC lines was extracted using an RNeasy kit (Qiagen). Similarly, we prepared total RNA from H358 and H2087 cells expressing shGFP or shALDH1A3. Gene expression profiling on each sample was performed using Illumina HumanWG-6 V3 BeadArrays (for the 8 sorted NSCLC lines) or Illumina HumanHT-12 V4 BeadArrays (for the shRNA-expressing H358 and H2087). Bead-level data were obtained and pre-processed using the R package mbcb for background correction and probe summarization (31). Pre-processed data were then quartile-normalized and log-transformed.
Quantitative RT-PCR
cDNA was generated with an iScript cDNA synthesis kit (BioRad). Gene specific Taq-Man probes (Life Technologies) were utilized for quantitative analyses of mRNA transcript levels with the GAPDH gene as an internal reference. PCR reactions were run using the ABI 7300 Real-time PCR System and analyzed with the included software. The comparative CT method was used to calculate relative mRNA expression levels.
Western blot analysis
Whole-cell extracts were analyzed as described (18). Primary antibodies against ALDH1A3 (Abgent), ALDH1A1 (Abcam), STAT3, phospho-Y705 STAT3, GAPDH, and Hsp90 (Cell Signaling) were used in the study.
shRNA stable expression in lung cancer lines
Four pLKO.1 lentiviral shRNA constructs targeting against ALDH1A3 and pLKO.1-shGFP were purchased from Open Biosystems. Lentiviruses were packaged in 293T cells. Briefly, 293T cells were cultured in DMEM containing 10% FBS and transiently transfected with shRNA vector together with pMDG-VSVG and pCMV-ΔR8.91 plasmids using Fugene6 (Roche). After overnight incubation, the viral supernatant was collected, filtered, and used for the transduction of lung cancer cells in the presence of 8 μg/ml polybrene (Sigma-Aldrich). Stable shRNA expressing lung cancer cells were generated after a two-week selection in 1.5 μg/ml puromycin. To generate a stable ALDH1A3 overexpressing cell line, H2009 cells transfected with pCMV6 (Origene) or pCMV6-ALDH1A3 were grown under G418 selection (800 μg/ml) for two weeks.
In vivo xenograft growth
Limiting dilutions of H358 or H2087 cells infected with pLKO.1-shGFP or pLKO.1-shALDH1A3 were subcutaneously injected into the right flank of five female NOD/SCID mice per group. Tumor growth was monitored by caliper measurements and tumor volume was calculated by width × length2 × π/6. Two months later, mice were sacrificed and tumors were dissected. A single cell suspension of xenograft tumors was confirmed by microscopy after 4 hour incubation with 1 mg/ml Collagenase I in HBSS (Gibco) at 37°C with intermittent vortexing. Tumor cell lysate was generated using TissueLyser II (Qiagen).
Transient transfection of NSCLCs with siRNA
Endogenous STAT3 in NSCLC lines were silenced using four STAT3 siRNAs (Dharmacon) according to the manufacturer’s instructions. The silencing efficiency was detected using real-time PCR and western blotting.
Tissue microarray preparation and immunohistochemistry staining
Formalin-fixed and paraffin-embedded (FFPE) tissues from clinically annotated, surgically resected lung cancer specimens were obtained from the Lung Cancer Specialized Program of Research Excellence (SPORE) Tissue Bank at MD Anderson Cancer Center used to construct NSCLC tissue microarray (TMA). Tissue sections (5 μm) were stained with ALDH1A3 antibody (1:100, Abcam) and assigned expression scores. NSCLCs were dichotomized into ALDH1A3 high and low expression classes on the basis of its median expression scores.
Statistical analysis
Simple linear regression analysis was carried out to determine correlation between ALDH1A3 expression and ALDH activity in NSCLC lines. Analysis of variance, chi-square, and Student’s t-test were performed using GraphPad Prism 5 software to test for significant differences in RT-PCR, tumor growth, tumor incidence, % of ALDH+ cells, colony formation assays, and the correlation between ALDH1A3 expression and clinicopathological characteristics. The survival curves were plotted using the Kaplan-Meier method and long-rank test. The difference was statistically significant when P value was lower than 0.05.
Results
Gene expression profiling of ALDH+ and ALDH− subpopulations
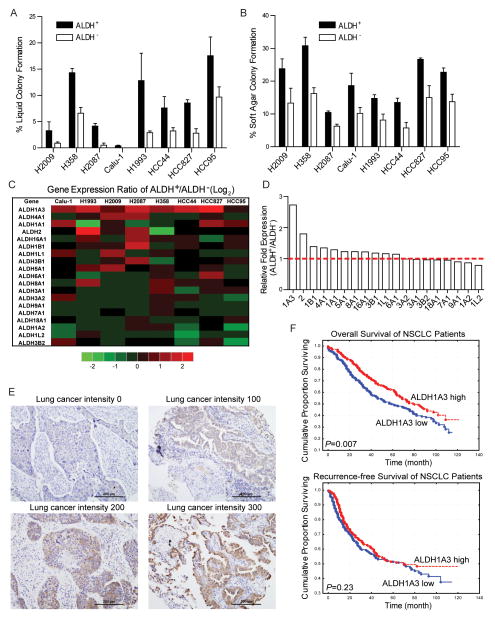
Previously we demonstrated that ALDH+ cells isolated from both patient tumor samples and NSCLC lines possess key CSC features including enhanced tumorigenicity and self-renewal, as well as elevated expression of Notch signaling components (14), To expand on these findings, we compared the global gene microarray expression studies of ALDH+ and ALDH− cells isolated from the same NSCLCs to determine common gene expression differences between ALDH+ and ALDH− tumor cell subpopulations. The Aldefluor assay was employed to separate ALDH+ and ALDH− cells from eight NSCLC lines representing a variety of oncogenotypes: Calu-1, H358, H1993, H2009, H2087, HCC44, HCC95, and HCC827. The ALDH+ fraction of these cell lines varied from 1% up to ~15% (Supplementary Fig S1). Anchorage dependent and independent colony formation assays were carried out to confirm the enhanced colony forming ability of ALDH+ cells compared to ALDH− cells (Fig 1A, 1B, and Supplementary Fig S2). The mRNA transcripts of sorted cells were analyzed using the HumanWG-6 genome wide microarray platform. Some gene expression differences between ALDH+ and ALDH− sorted cell populations were common across all cell lines. Chief among these differences was the upregulated expression of ALDH1A3 transcript in ALDH+ cells. Since there are 19 isozymes in the human ALDH superfamily, we compared their expression and found that ALDH1A3 was the most upregulated gene in the family in ALDH+ populations (Fig 1C, 1D). In light of this commonality between sorted cell transcript expression, we hypothesized that ALDH1A3 is the principal CSC-associated ALDH isozyme in NSCLC.
Figure 1.
Genome wide gene expression profiling of ALDH+ and ALDH− subpopulations isolated from 8 NSCLC lines. A, ALDH+ and ALDH− lung cancer cells were sorted by FACS based on ALDH activity. Colony formation efficiency was determined by growing sorted cells for 2 weeks in liquid culture. B, sorted cells were plated in 0.33% agar in 12-well plates. Soft agar colonies were stained and counted after 3 weeks. C, fold change of 19 ALDH isozymes in sorted ALDH+ vs. ALDH− cells isolated from 8 NSCLC lines by microarray analysis and their average values in D. E, different level of ALDH1A3 in NSCLC TMA samples were detected by IHC staining. F, Kaplan-Meier plots of NSCLC patient survival based on tumors expressing high (red: n=239, IHC score ≥ 150) and low (blue: n=216, IHC score < 150) ALDH1A3 showed ALDH1A3 high expression was associated with better overall survival (upper penal, P = 0.007) but not recurrence-free survival (lower penal, P = 0.23).
ALDH1A3 protein expression in NSCLC patient tumor samples
Previously, we reported that ALDH1A1 high expression is associated with poor overall survival in NSCLCs, which is consistent with the hypothesis that lung cancers enriched in CSCs would have a worse prognosis (14). To investigate whether NSCLCs with different ALDH1A3 expression were associated with clinicopathological characteristics or clinical outcome, we analyzed ALDH1A3 protein expression by IHC in 455 NSCLC specimens. Representative different level of ALDH1A3 staining was shown in Fig 1E. While ALDH1A3 cancer cell subpopulations were found in a substantial number of NSCLC patients, statistical analysis revealed that ALDH1A3 high expression was significantly associated with female (P < 0.019), never smokers (P < 0.0005), adenocarcinoma histology (P < 0.0001), and well differentiated lung tumors (P < 0.0001, Supplementary Table S3). Kaplan-Meier survival analysis was carried out to examine the prognostic value of tumor ALDH1A3 expression. We found that ALDH1A3 high expression was associated with better overall survival but not recurrence-free survival in the whole cohort (Fig 1F).
ALDH1A3 expression is associated with ALDH+ Lung CSCs
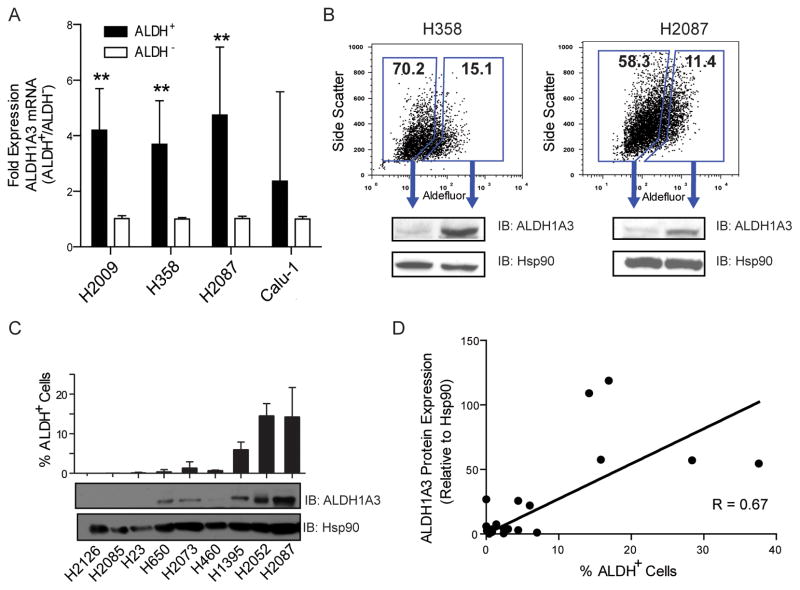
To test our hypothesis, we examined the expression of ALDH1A3 in sorted cells from H2087, H358, H2009, and Calu-1. ALDH1A3 messenger RNA was significantly higher in ALDH+ compared to ALDH− cells (Fig 2A). Western blot also confirmed that the ALDH+ subpopulation contained significantly more ALDH1A3 protein compared to ALDH− cells (Fig 2B). Furthermore, a strong positive correlation was observed between ALDH1A3 protein expression and the percent of ALDH+ cells in a large panel of NSCLC lines (r = 0.67, P < 0.05), suggesting a connection between the percentage of lung CSCs and ALDH1A3 expression in a given cell line (Fig 2C, 2D, Supplementary Fig S3, and Table S1). We also knocked down ALDH1A3 using siRNAs followed by liquid colony formation assays in NSCLC lines with a variety of important different driver mutations, such as KRAS, EGFR, EML4-ALK fusion, PTEN, PIK3CA, BRAF, and LKB1 mutation (Supplementary Fig S3C). We found that ALDH1A3 depletion significantly impaired liquid colony forming ability in all of the tested NSCLC lines except H3122 cells (which contain EML4-ALK fusion mutation), indicating that the role of ALDH1A3 is predominant in most NSCLC lines with variable driver mutations.
Figure 2.
ALDH+ NSCLC cells contain more ALDH1A3 transcript and protein. A, qRT-PCR analysis revealed an approximate 4 fold increase of ALDH1A3 expression in ALDH+ subpopulation compared to their ALDH− counterparts in H2009, H358, and H2087 lung cancer lines (**P < 0.01). B, western blots showed significantly higher ALDH1A3 protein expression in ALDH+ cells than in ALDH− H358 and H2087 cells. Hsp90 served as loading control. C, ALDH1A3 protein levels in a set of NSCLC lines were detected by western blots. The corresponding percentage of ALDH+ cells was analyzed by the Aldefluor assay. D, a correlation between ALDH1A3 protein expression and ALDH activity was analyzed by the linear regression in a large panel of NSCLC lines.
Previous analyses have indicated ALDH1A1 as the principal CSC-associated ALDH isozyme in lung cancer (14, 32, 33). To determine the relationship between ALDH1A1 and ALDH1A3 expression in lung cancer we assayed their expression in a panel of lung cancer lines. Interestingly, significant ALDH1A1 expression was detected in SCLC lines and a small number of NSCLC lines, whereas ALDH1A3 was detected in most NSCLC lines with the exception of those that highly express ALDH1A1 (Supplementary Fig S3A, S3B). Together, these data indicate that either ALDH1A3 or ALDH1A1 are responsible for the ALDH+ phenotype with ALDH1A3 being significantly more frequent than ALDH1A1 in NSCLC.
ALDH1A3 knockdown reduces NSCLC ALDH activity and tumor cell clonogenicity
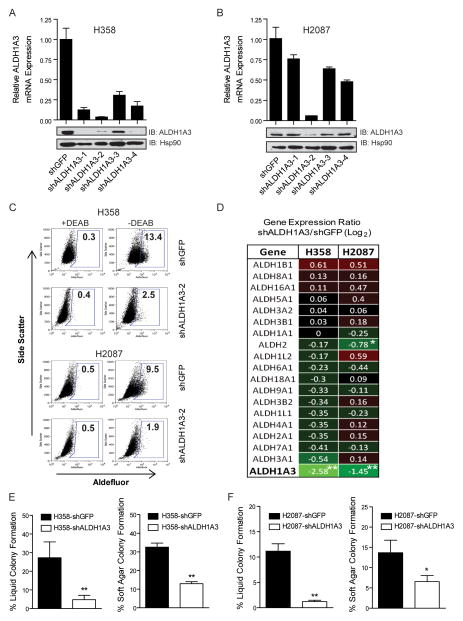
ALDH mediated reduction of cellular aldehydes has been shown to be important in a variety of cellular functions including cell detoxification, growth, differentiation, and self-renewal (34, 35). To examine the function of ALDH1A3 in the context of lung CSCs, we evaluated the effect of suppressing ALDH1A3 in NSCLC line H358 and H2087. We tested 4 short hairpin RNA (shRNA) targeting ALDH1A3 to achieve stable knockdown of ALDH1A3 via lentiviral delivery and found a shRNA clone that could effectively reduce ALDH1A3 expression in two lines compared with control cells expressing shGFP (Fig 3A, 3B). The control H358 and H2087 cells contained 13% and 9% ALDH+ cells, respectively, which were comparable to their uninfected parental cells. In contrast, expression of shALDH1A3 resulted in a 5-fold reduction in the percent of H358 and H2087 ALDH+ cells (Fig 3C). To test whether ALDH1A3 was required for the clonogenicity of lung CSCs in vitro, colony formation assays revealed that knockdown of ALDH1A3 in H358 cells decreased the clonogenic capacity by 3-fold in anchorage dependent and 5-fold in anchorage independent assays, respectively, compared to control cells (P < 0.01) (Fig 3E). Similar effects were observed in H2087 cells (Fig 3F). Consistently, depletion of ALDH1A3 using siRNAs significantly reduced liquid colony forming ability of sorted ALDH+ H358 cells (Supplementary Fig S3D). The data suggest that the impaired colony forming ability was due to reduction of the ALDH+ subpopulation in lung cancer cells and that ALDH1A3 is necessary for NSCLC cells to form robust colonies in vitro.
Figure 3.
shRNA mediated knockdown of ALDH1A3 reduces ALDH activity and clonogenicity of lung cancer cells. A, qRT-PCR analysis showed a significant reduction of ALDH1A3 expression in H358 cells expressing 4 different shALDH1A3 constructs compared to control H358-shGFP cells (upper panel). Decreased ALDH1A3 protein level was confirmed by western blot (lower panel). B, similarly, knockdown of ALDH1A3 in H2087 cells was achieved using the shALDH1A3-2 construct. C, Aldefluor assays of H358- and H2087- shGFP or shALDH1A3-2 cells showed a dramatic reduction of ALDH+ population in shALDH1A3 expressing cells compared to control cells. D, the specificity of ALDH1A3 knockdown was confirmed by microarray analysis and shown in a heat map of 19 ALDH isozyme gene expression ratios in shALDH1A3 vs. shGFP expressing H358 and H2087 cells (*P < 0.05, **P < 0.01). E and F, anchorage dependent (left) and independent (right) colony formation assays revealed a significant reduction of clonogenic ability due to depletion of ALDH1A3 in H358 and H2087 cells (*P < 0.05, **P < 0.01).
To confirm that these results were not due to off-target effects on other ALDH isozymes, microarray analysis was performed on control and shALDH1A3 expressing H358 and H2087 cells to examine the mRNA expression of the 19 members in ALDH family. We observed that shRNA mediated knockdown of ALDH1A3 reduced its transcript expression by approximately 3- to 5-fold in H2087- and H358-shALDH1A3 cells, respectively, whereas the expression levels of most other ALDH isozymes remained relatively unchanged compared to shGFP cells (Fig 3D). These data suggest that ALDH1A3 is the key ALDH isozyme that is functionally important for maintaining NSCLC ALDH+ cells and clonogenic growth in vitro.
ALDH1A3 knockdown impairs lung cancer cell tumorigenicity
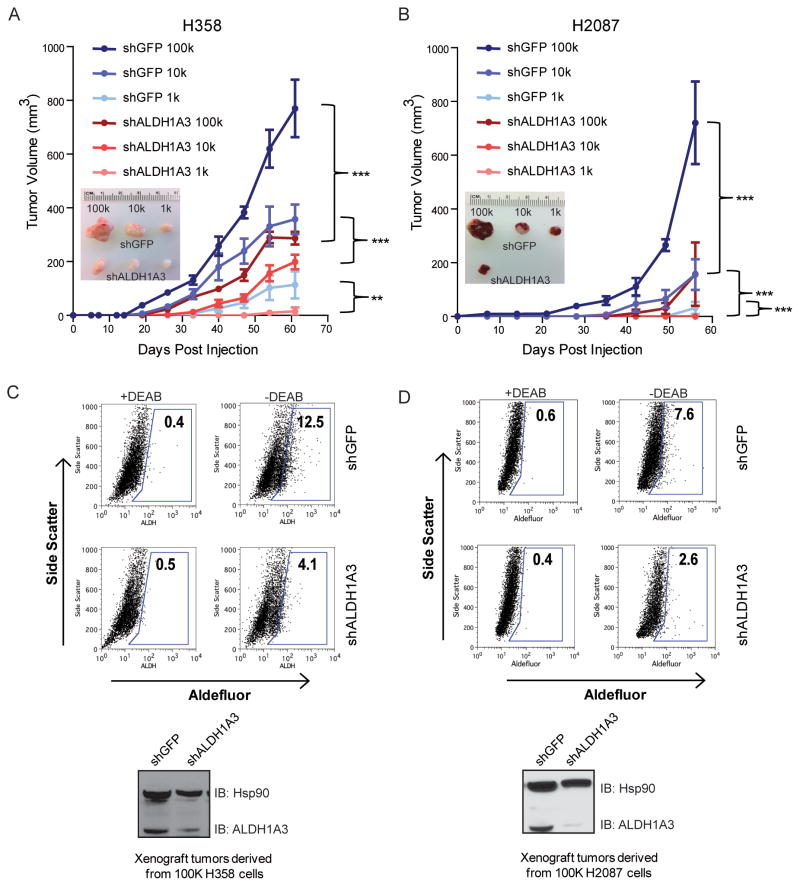
To investigate whether ALDH1A3 is essential for the tumorigenicity of lung CSC in vivo, we assayed the tumor-forming ability of limiting dilutions of stable shALDH1A3 expressing H358 and H2087 cells. Six groups of five female NOD/SCID mice were subcutaneously injected with 105, 104, or 103 shALDH1A3 or control shGFP-expressing H358 and H2087 cells. We observed that suppression of ALDH1A3 expression in H358 and H2087 resulted in a significant decrease in tumor-forming ability relative to control cells. The greatest reduction in H358- and H2087-shALDH1A3 cell tumorigenicity was observed at the lowest (103) cell dilution (Fig 4A, 4B and Supplementary Table S2). Of the mice that did form tumors from shALDH1A3 cells, tumor volumes were significantly reduced and their growth rates significantly diminished compared to shGFP-derived tumors from the same inoculation group. shALDH1A3-expressing H358 cells were dramatically smaller than those from control cells (105 and 104 injected cell groups, P < 0.001, 103 group, P < 0.01); likewise, shALDH1A3-expressing H2087 cells generated significantly smaller tumors than control H2087 cells (P < 0.001).
Figure 4.
shRNA mediated knockdown of ALDH1A3 reduces tumorigenicity of lung cancer cells. A, limiting dilutions of H358-shALDH1A3 and H358-shGFP cells were injected subcutaneously into NOD/SCID mice and tumor growth was monitored for 9 weeks (upper panel). Representative dissected tumors from each group are shown in the bottom panel. B, similarly, tumors derived from H2087-shALDH1A3 cells were smaller than tumors generated from H2087-shGFP cells (**P < 0.01, ***P < 0.001). C and D, ALDH activity was assayed in tumors derived from shGFP or shALDH1A3 expressing H358 and H2087 cells, indicating that H358/H2087-shALDH1A3 cell derived tumors possessed a smaller percentage of ALDH+ cells than shGFP-expressing cell derived tumors. Reduced ALDH1A3 protein level in shALDH1A3 expressing cell derived tumors was confirmed by western blot (bottom panel).
To determine if the reduction in H358- and H2087-shALDH1A3 cells was associated with a decrease in ALDH activity, Aldefluor assay of disassociated tumor cells revealed that the percentage of ALDH+ cells was about 3-fold lower in both H358-and H2087-shALDH1A3 tumors compared to their corresponding control xenografts (Fig 4C and 4D). To ensure that ALDH1A3 expression was suppressed in H358- and H2087-shALDH1A3 cell derived xenografts, western blots confirmed the reduction of ALDH1A3 protein in shALDH1A3 cell derived xenografts compared to control xenografts, with the greatest reduction of ALDH1A3 protein observed in H2087-shALDH1A3 tumors (Fig 4C and 4D). Together, these results support the hypothesis that ALDH1A3 expression is required for lung cancer ALDH activity and tumorigenicity in vivo.
Overexpression of ALDH1A3 is not sufficient to promote lung cancer cell tumorigenicity
To determine if ALDH1A3 was not only necessary but sufficient to promote CSC activity, we generated stable ALDH1A3 overexpressing H2009 cells as well as an empty vector-transfected control H2009 cell line (Supplementary Fig S5A). Ectopic overexpression of ALDH1A3 significantly increased the proportion of ALDH+ cells in H2009 from 4% to 59% (Supplementary Fig S5B). However, in vitro colony formation assays revealed no significant difference in clonogenicity between ALDH1A3 overexpressing H2009 cells and control cells (Supplementary Fig S5C). Four groups of five female NOD/SCID mice were subcutaneously injected with 105 or 104 H2009-pCMV6-ALDH1A3 or H2009-pCMV6 cells, and after eight weeks, no significant difference in tumor engraftment or growth rate was observed between limiting dilutions of ALDH1A3-overexpressing and control groups (Supplementary Fig S5D). These data indicate that ALDH1A3 alone is not sufficient to enhance tumor initiating ability of NSCLC cells.
STAT3 signaling pathway regulates ALDH activity in NSCLC stem cells
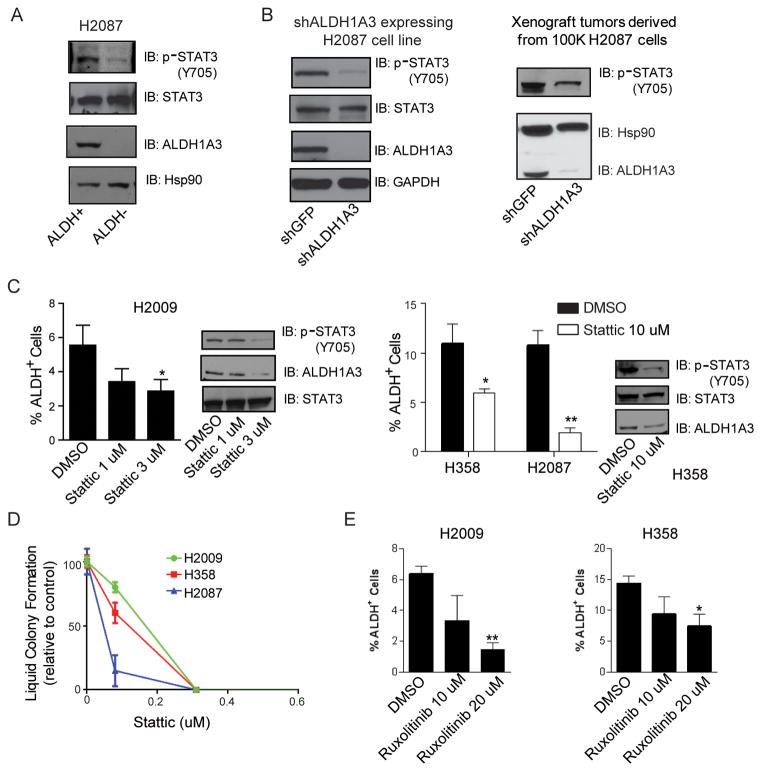
To further investigate how ALDH activity in NSCLC stem cells is regulated, we performed a siRNA screen in which 40 genes related to stem cell self-renewal pathways were knocked down followed by cell viability and liquid colony formation assays. We found that the Notch pathway components Hey1 were involved in the regulation of lung cancer colony formation, which is consistent with our prior report showing the requirement of the Notch signaling in colony formation for lung cancer ALDH+ cells (14). The data also identified STAT3 as a potential target for lung cancer ALDH+ clonogenic cells. Immunoblot analysis showed that sorted ALDH+ H2087 cells contained more phospho-Tyr705 STAT3 than ALDH− cells (Fig 5A), while H2087-shALDH1A3 cells expressed much less activated STAT3 compared to control cells. Similarly, phospho-STAT3 was less abundant in xenograft tumors derived from H2087-shALDH1A3 cells compared to control tumors (Fig 5B). To examine the role of STAT3 pathway in ALDH+ lung cancer cells, we targeted STAT3 pharmacologically with Stattic, and assessed ALDH activity. Stattic has been reported as a potent STAT3 specific inhibitor. We found that treatment with 1 μM or 3 μM Stattic diminished ALDH+ populations compared to control H2009 NSCLC cells (P < 0.05, Fig 5C). Similarly, Stattic treatment also reduced ALDH1A3 expression and ALDH+ cells in H358 and H2087 cells (Fig 5C). Liquid colony formation assay revealed that Stattic significantly reduced anchorage-dependent colony formation in several NSCLC lines at very low concentrations (Fig 5D). To test the potential role of JAKs, common upstream activators of STAT3, in ALDH+ lung cancer cells, H358 and H2009 cells were exposed to two JAK inhibitors, Ruxolitinib (JAK1/2 inhibitor) or Tofacitinib (JAK1/3 inhibitor), followed by Aldefluor assays. We found that Ruxolitinib but not Tofacinitib reduced ALDH+ lung cancer cells indicating the role of JAK2 in this process (Figure 5E and data not shown).
Figure 5.
STAT3 activation is required for the maintenance of ALDH+ NSCLC cells. A, western blot showed that sorted ALDH+ H2087 cells contained more phospho-Tyr705 STAT3 than ALDH− cells. B, shALDH1A3 mediated knocking down of ALDH1A3 significantly reduced activated STAT3 level in H2087 cells. Xenograft tumors derived from H2087-shALDH1A3 cells contained less phospho-STAT3 than tumors generated from H2087-shGFP cells. C, Stattic treatment for 48 h attenuated ALDH+ populations in a dose-dependent manner in H2009 cells. Similar effect of 10 μM Stattic on ALDH activity was observed in H358 and H2087 cells (*P < 0.05, **P < 0.01). D, liquid colony formation assay showed that Stattic significantly reduced lung cancer cell colony formation at a low concentration. E, Ruxolitinib treatment for 48 h significantly reduced ALDH+ H2009 and H358 cells (*P < 0.05, **P < 0.01).
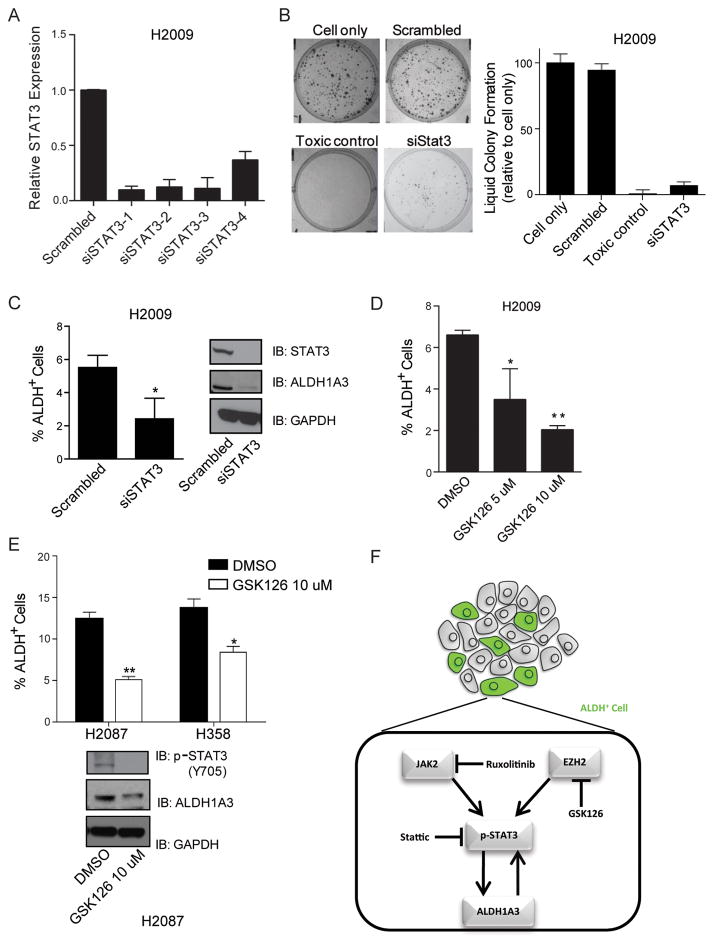
To further interrogate the effect of STAT3 on ALDH activity, we transiently transfected H2009 cells with four different siRNAs targeting STAT3. We found significant reductions of both STAT3 mRNA expression (Fig 6A) and clonogenicity (Fig 6B). Aldefluor assay and western blot revealed that knocking down of STAT3 by siRNA in H2009 cells caused a reduction of ALDH activity (Fig 6C). These data suggest that the STAT3 pathway is activated in ALDH+ compared to ALDH− lung cancer cells and abolishing STAT3 reduces tumor cell clonogenicity. In addition, Enhancer of Zeste Homolog 2 (EZH2) has recently been shown to bind to and methylate STAT3, leading to enhanced STAT3 activation in glioblastoma stem-like cells (28). We treated H2009, H358, and H2087 cells with 5 μM or 10 μM of the highly selective EZH2 inhibitor GSK126 and found that ALDH activity was decreased (Fig 6D, 6E). Thus, our data obtained from in vitro and in vivo experiments in NSCLCs support the hypothesis that ALDH1A3 is the major isozyme responsible for elevated ALDH activity in a subpopulation of NSCLC, and that the STAT3 pathway is involved in the regulation of ALDH activity, which is illustrated in our current working model (Fig 6F).
Figure 6.
STAT3 pathway regulates ALDH activity in NSCLC stem cells. A, real-time PCR analysis revealed that 4 siRNAs significantly reduced STAT3 mRNA expression in H2009 cells. B, liquid colony formation assay showed that H2009 cells transfected with siSTAT3 exhibited a reduction in clonogenicity. C, Aldefluor assay and western blot indicated that knocking down of STAT3 by siRNA in H2009 cells caused a reduction of ALDH+ cells. D and E, Aldefluor assay revealed that EZH2 inhibitor GSK126 (5 or 10 μM) significantly reduced ALDH+ cells in H2009, H2087, and H358 cell lines (*P < 0.05, **P < 0.01). F, a schematic model that illustrates the regulation of ALDH1A3 by STAT3 in ALDH+ lung cancer stem cells.
Discussion
In the current study, we isolated ALDH+ cells from 8 NSCLC lines and determined common gene expression signatures to identify the subpopulation of ALDH+ highly clonogenic and tumorigenic cells residing within the bulk tumor. We found that each cell line pair of isolated ALDH+ and ALDH− populations clustered together, indicating that the gene expression patterns of various ALDH+ subpopulations are different and that the gene expression difference among NSCLCs is greater than the difference between ALDH+ and ALDH− counterparts. Wicha and colleagues asked a similar question about breast CSCs and observed that only limited genes were differentially expressed between ALDH+ breast CSCs and their parental cells (17). One of the genes that demonstrated differential expression between the ALDH+ and ALDH−populations in lung cancers is the ALDH isozyme ALDH1A3 which we then studied in detail. We found that ALDH1A3 depletion in NSCLCs resulted in a significant reduction in ALDH activity, clonogenicity and tumorigenicity, suggesting that ALDH1A3 is indispensable for NSCLC cell survival and growth.
Other studies have reported that ALDH activity measured by the Aldefluor assay is regulated by different isozymes in different types of cancer. For example, Levi et al. showed that ALDH2, ALDH3A1, and/or ALDH9A1 could be responsible for ALDH activity in ALDH1A1-deficient hematopoietic cells (36). Van den Hoogen et al. discovered that ALDH7A1 was highly expressed in prostate cancer cell lines and prostate cancer tissue, indicating that ALDH7A1 was responsible for the ALDH activity in prostate cancer cells (37). Chen et al. showed that ALDH1B1 was expressed in 98% of colon cancer samples (26). Luo et al. reported that ALDH+ melanoma cells, in which ALDH1A1 and ALDH1A3 were the predominant isozymes, were more tumorigenic compared to ALDH− cells isolated from human melanoma tumors (18). Therefore, we expected that one or a few ALDH isozymes could be upregulated in ALDH+ lung cancer cells. Our data revealed that the ALDH1A3 isozyme was highly expressed in ALDH+ compared to ALDH− lung tumor cells. Similarly, Marcato et al. demonstrated that ALDH1A3 was an important breast CSC marker through IHC, western blot, and shRNA-mediated knockdown assays (25). These findings suggest that different ALDH isozymes contribute to the elevated ALDH activity in CSCs of varied origin.
Our previous and current IHC staining showed that ALDH1A1 and ALDH1A3 are differentially expressed in lung squamous cell carcinomas (SCC) and adenocarcinomas, respectively (14). Likewise, Alamgeer et al. reported that ALDH1A1 is strongly expressed in lung SCC (P = 0.002) (38). Alamgeer et al. like our study found that the CSC CD133 was not associated with prognosis in adenocarcinoma histology, but that the tumors with both a high ALDH1A1 and CD133 score had the worse prognosis. Our current study showed that ALDH1A3 expression, while found in the majority of NSCLCs, was associated with adenocarcinoma well differentiated histology and female gender. Given the expected better prognosis of tumors with these clinical characteristics, when the group was tested as a whole, it should not be surprising that tumors with increased ALDH1A3 score actually had better survival. Overall, it appears that NSCLCs can have different classes of ALDH+ cell subpopulations – at least one driven by ALDH1A1 that has a worse prognosis than tumors driven by ALDH1A3. Why one particularly ALDH isozyme is selected for the cancer cell subpopulation remains to be determined.
We wanted to know the functional role of ALDH1A3 in the NSCLC malignant phenotype. shRNA mediated knockdown experiments revealed that ALDH1A3 was necessary in maintaining both the ALDH+ population and lung tumor growth in vitro and in vivo. However, exogenously overexpressed ALDH1A3 was not sufficient by itself to enhance NSCLC tumor growth. It is possible that other factors might be required along with ALDH1A3 expression to promote tumor progression. It is also possible that there is a negative feedback loop between retinoic acid and expression of ALDH1 family isozymes that might affect the ability of ALDH1A3 to promote tumor growth (39, 40). Recently, the ALDH1/retinoic acid signaling pathway has been shown to play a role in regulating ALDH1 expression in both normal and cancer stem cells (41–43). Moreb et al. reported that treating various lung cancer cell lines with all-trans retinoic acid (ATRA) led to a reduction of ALDH1 and increased their sensitivity to cyclophosphamide (44). In contrast, our retinoic acid studies did not lead to a significant growth inhibition in many NSCLC and SCLC lines (45). The potential clinical application of ATRA in solid tumors is still unclear and a small, phase I/II clinical trial designed to study the combination of ATRA and Tamoxifen in breast cancer patients did not provide a significant benefit (46). Going forward it will be critical to elucidate the biochemical targets of ALDH1A3 in lung cancer cell growth and survival.
In order to decipher underlying mechanisms by which ALDH activity is regulated, we knocked down the genes related to stem cell self-renewal and other potential regulators in a siRNA screen. We found that abolishing STAT3 impaired NSCLC cell growth and liquid colony formation. This also led us to find that inhibiting ALDH1A3 led to a decrease of p-STAT3. Finally we found that pharmacologically inhibiting EZH2, a recently described STAT3 activator, also diminished STAT3 and the number of ALDH+ tumor cells. Further studies are now required to precisely define how STAT3/EZH2 is preferentially activated in ALDH+ lung cancer cells in vivo.
In summary, we have shown that the ALDH1A3 isozyme is a robust marker for a subpopulation of highly clonogenic and tumorigenic NSCLC cells. It is also essential for these growth functions in vitro and in vivo. We found that lung cancers contain ALDH+ subpopulations with different mRNA expression profiles. However, a common feature was our discovery that in many cases of NSCLC, ALDH1A3 is the isozyme driving the ALDH+ phenotype, that STAT3 activation is essential for the maintenance of the subpopulation of ALDH+ lung cancer cells, and that in turn, this was driven by EZH2. The results presented here provide a working model toward developing targeted therapy for this biologically important ALDH1A3+ subset of lung cancer cells.
Supplementary Material
Translational Relevance.
The malignant phenotype appears driven by a subpopulation of cancer stem cells (CSCs), which self-renew, differentiate, and contribute to drug resistance and metastasis. Our group and others have identified CSCs in non-small cell lung cancers (NSCLCs) by their enhanced aldehyde dehydrogenase (ALDH) activity and found that the ALDH+ subpopulation contains highly tumorigenic and clonogenic cells. Here, we show that one of the 19 ALDH isozymes, ALDH1A3, is responsible for ALDH activity in a majority of NSCLCs despite oncogenotype, functionally required for lung CSC survival, and that pSTAT3 is tightly linked to ALDH1A3 activity. Inhibition of ALDH1A3 or STAT3 attenuated ALDH1A3 expression, ALDH+ lung cancer cells, and tumor cell clonogenicity. Collectively, our findings indicate that ALDH1A3 expression and STAT3 pathway activation are essential for the maintenance of NSCLC CSCs, and that both ALDH1A3 and the STAT3 pathway are rationale therapeutic targets for eliminating NSCLC stem cells.
Acknowledgments
Financial Support
This project was supported by CPRIT, NCI SPORE P50CA70907, UTSW Cancer Center Support Grant 5P30-CA142543, and the Gillson-Longenbaugh Foundation.
We would like to thank the UTSW Flow Cytometry and Microarray core staff for their assistance.
Footnotes
Disclosure of Potential Conflict of Interest
The authors indicate no potential conflicts of interest.
Authors’ Contributions
Conception and design: J.D. Minna, C. Shao
Development of methodology: C. Shao, J.P. Sullivan, J. Rodriguez, H. Liu
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): C. Shao, J.P. Sullivan, P. Yenerall, J. Rodriguez, H. Liu
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): L. Girard, C. Shao
Writing, review, and/or revision of the manuscript: J.D. Minna, C. Shao, A. Augustyn, J. Shay
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): C. Shao, L. Girard, C. Behrens, I.I. Wistuba,
Study supervision: J.D. Minna
References
- 1.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–5. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–28. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–6. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–33. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 6.Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330–8. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One. 2008;3:e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–14. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 9.Meng X, Li M, Wang X, Wang Y, Ma D. Both CD133+ and CD133- subpopulations of A549 and H446 cells contain cancer-initiating cells. Cancer Sci. 2009;100:1040–6. doi: 10.1111/j.1349-7006.2009.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salnikov AV, Gladkich J, Moldenhauer G, Volm M, Mattern J, Herr I. CD133 is indicative for a resistance phenotype but does not represent a prognostic marker for survival of non-small cell lung cancer patients. Int J Cancer. 2010;126:950–8. doi: 10.1002/ijc.24822. [DOI] [PubMed] [Google Scholar]
- 11.Alison MR, Guppy NJ, Lim SM, Nicholson LJ. Finding cancer stem cells: are aldehyde dehydrogenases fit for purpose? J Pathol. 2010;222:335–44. doi: 10.1002/path.2772. [DOI] [PubMed] [Google Scholar]
- 12.Cheung AM, Wan TS, Leung JC, Chan LY, Huang H, Kwong YL, et al. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia. 2007;21:1423–30. doi: 10.1038/sj.leu.2404721. [DOI] [PubMed] [Google Scholar]
- 13.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70:9937–48. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang CP, Tsai MF, Chang TH, Tang WC, Chen SY, Lai HH, et al. ALDH-positive lung cancer stem cells confer resistance to epidermal growth factor receptor tyrosine kinase inhibitors. Cancer Lett. 2013;328:144–51. doi: 10.1016/j.canlet.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 16.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–13. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Dallaglio K, Chen Y, Robinson WA, Robinson SE, McCarter MD, et al. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells. 2012;30:2100–13. doi: 10.1002/stem.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707–12. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreb J, Schweder M, Suresh A, Zucali JR. Overexpression of the human aldehyde dehydrogenase class I results in increased resistance to 4-hydroperoxycyclophosphamide. Cancer Gene Ther. 1996;3:24–30. [PubMed] [Google Scholar]
- 22.Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer. 2008;59:340–9. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: Its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10:1378–84. doi: 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- 24.Visus C, Wang Y, Lozano-Leon A, Ferris RL, Silver S, Szczepanski MJ, et al. Targeting ALDHbright Human Carcinoma-Initiating Cells with ALDH1A1-Specific CD8+ T Cells. Clin Cancer Res. 2011;17:6174–84. doi: 10.1158/1078-0432.CCR-11-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29:32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem Biophys Res Commun. 2011;405:173–9. doi: 10.1016/j.bbrc.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ginestier C, Liu S, Diebel ME, Korkaya H, Luo M, Brown M, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. The Journal of clinical investigation. 2010;120:485–97. doi: 10.1172/JCI39397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 Activates STAT3 Signaling via STAT3 Methylation and Promotes Tumorigenicity of Glioblastoma Stem-like Cells. Cancer cell. 2013;23:839–52. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin L, Liu A, Peng Z, Lin HJ, Li PK, Li C, et al. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res. 2011;71:7226–37. doi: 10.1158/0008-5472.CAN-10-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroon P, Berry PA, Stower MJ, Rodrigues G, Mann VM, Simms M, et al. JAK-STAT Blockade Inhibits Tumor Initiation and Clonogenic Recovery of Prostate Cancer Stem-like Cells. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-0874. [DOI] [PubMed] [Google Scholar]
- 31.Ding LH, Xie Y, Park S, Xiao G, Story MD. Enhanced identification and biological validation of differential gene expression via Illumina whole-genome expression arrays through the use of the model-based background correction methodology. Nucleic Acids Res. 2008;36:e58. doi: 10.1093/nar/gkn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreb JS, Baker HV, Chang LJ, Amaya M, Lopez MC, Ostmark B, et al. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol Cancer. 2008;7:87. doi: 10.1186/1476-4598-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreb JS, Zucali JR, Ostmark B, Benson NA. Heterogeneity of aldehyde dehydrogenase expression in lung cancer cell lines is revealed by Aldefluor flow cytometry-based assay. Cytometry B Clin Cytom. 2007;72:281–9. doi: 10.1002/cyto.b.20161. [DOI] [PubMed] [Google Scholar]
- 34.Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 35.Sladek NE, Kollander R, Sreerama L, Kiang DT. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: a retrospective study. Rational individualization of oxazaphosphorine-based cancer chemotherapeutic regimens. Cancer Chemother Pharmacol. 2002;49:309–21. doi: 10.1007/s00280-001-0412-4. [DOI] [PubMed] [Google Scholar]
- 36.Levi BP, Yilmaz OH, Duester G, Morrison SJ. Aldehyde dehydrogenase 1a1 is dispensable for stem cell function in the mouse hematopoietic and nervous systems. Blood. 2009;113:1670–80. doi: 10.1182/blood-2008-05-156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70:5163–73. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- 38.Alamgeer M, Ganju V, Szczepny A, Russell PA, Prodanovic Z, Kumar B, et al. The prognostic significance of aldehyde dehydrogenase 1A1 (ALDH1A1) and CD133 expression in early stage non-small cell lung cancer. Thorax. 2013;68:1095–104. doi: 10.1136/thoraxjnl-2012-203021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duester G, Mic FA, Molotkov A. Cytosolic retinoid dehydrogenases govern ubiquitous metabolism of retinol to retinaldehyde followed by tissue-specific metabolism to retinoic acid. Chem Biol Interact. 2003;143–144:201–10. doi: 10.1016/s0009-2797(02)00204-1. [DOI] [PubMed] [Google Scholar]
- 40.Elizondo G, Medina-Diaz IM, Cruz R, Gonzalez FJ, Vega L. Retinoic acid modulates retinaldehyde dehydrogenase 1 gene expression through the induction of GADD153-C/EBPbeta interaction. Biochem Pharmacol. 2009;77:248–57. doi: 10.1016/j.bcp.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ginestier C, Wicinski J, Cervera N, Monville F, Finetti P, Bertucci F, et al. Retinoid signaling regulates breast cancer stem cell differentiation. Cell Cycle. 2009;8:3297–302. doi: 10.4161/cc.8.20.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tallman MS, Andersen JW, Schiffer CA, Appelbaum FR, Feusner JH, Ogden A, et al. All-trans-retinoic acid in acute promyelocytic leukemia. N Engl J Med. 1997;337:1021–8. doi: 10.1056/NEJM199710093371501. [DOI] [PubMed] [Google Scholar]
- 43.Tonge PD, Andrews PW. Retinoic acid directs neuronal differentiation of human pluripotent stem cell lines in a non-cell-autonomous manner. Differentiation. 2010;80:20–30. doi: 10.1016/j.diff.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Moreb JS, Gabr A, Vartikar GR, Gowda S, Zucali JR, Mohuczy D. Retinoic acid down-regulates aldehyde dehydrogenase and increases cytotoxicity of 4-hydroperoxycyclophosphamide and acetaldehyde. J Pharmacol Exp Ther. 2005;312:339–45. doi: 10.1124/jpet.104.072496. [DOI] [PubMed] [Google Scholar]
- 45.Geradts J, Chen JY, Russell EK, Yankaskas JR, Nieves L, Minna JD. Human lung cancer cell lines exhibit resistance to retinoic acid treatment. Cell Growth Differ. 1993;4:799–809. [PubMed] [Google Scholar]
- 46.Budd GT, Adamson PC, Gupta M, Homayoun P, Sandstrom SK, Murphy RF, et al. Phase I/II trial of all-trans retinoic acid and tamoxifen in patients with advanced breast cancer. Clin Cancer Res. 1998;4:635–42. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.