Abstract
Prolyl hydroxylation is a post-translational modification (PTM) that plays an important role in the formation of collagen fibrils and in the oxygen-dependent regulation of Hypoxia Inducible Factor-α (HIF-α). While this modification has been well characterized in the context of these proteins, it remains unclear to what extent it occurs in the remaining mammalian proteome. We explored this question using mass spectrometry to analyze cellular extracts subjected to various fractionation strategies. In one strategy, we employed the von Hippel Lindau tumor suppressor protein (VHL), which recognizes prolyl hydroxylated HIF-α, as a scaffold for generating hydroxyproline capture reagents. We report novel sites of prolyl hydroxylation within five proteins: FK506-binding protein 10 (FKBP10), Myosin heavy chain 10 (MYH10), Hexokinase 2 (HK2), Pyruvate Kinase (PKM), and C-1 Tetrahydrofolate synthase (MTHFD1). Furthermore, we show that identification of prolyl hydroxylation presents a significant technical challenge owing to widespread isobaric methionine oxidation, and that manual inspection of spectra of modified peptides in this context is critical for validation.
Keywords: mass spectrometry, posttranslational modification, Prolyl Hydroxylase Domain protein, prolyl hydroxylation
1 Introduction
Prolyl hydroxylation is a well-recognized PTM, but the extent of its occurrence in mammalian cells has not been well characterized. In collagen, upwards of 30% of prolines may be hydroxylated, and subsequent studies have shown that additional secreted or membrane bound proteins possess this modification including elastin, complement C1q, acetylcholinesterase, and others [1–8]. Importantly, prolyl hydroxylation has been found to be the critical PTM that regulates the protein turnover of Hypoxia Inducible Factor-α (HIF-α), the master transcriptional regulator of the hypoxic response [9]. Prolyl Hydroxylase Domain protein (PHD) site-specifically prolyl hydroxylates HIF-α in an oxygen-dependent manner. This modification provides a binding platform for the von Hippel Lindau protein (VHL)-containing E3 ubiquitin ligase complex that specifically targets hydroxylated HIF-α for degradation.
The discovery of regulatory prolyl hydroxylation on HIF-α raises the question of whether this distinctive modification may exist on other proteins. To date, little systematic research has been performed to determine the extent of this modification in the proteome. To address this, we employed high-resolution MS in combination with a number of fractionation strategies.
As a PTM, prolyl hydroxylation has a number of features that would be predicted to facilitate its identification, namely its relative stability and the absence of known dehydroxylases that could reverse this modification. However, at the same time, its identification presents considerable technical challenges, not the least of which is the presence of abundant methionine oxidation which yields a mass shift isobaric to proline hydroxylation. Isobaric modifications lessen the degree to which high mass accuracy (>1 ppm) spectrometers and associated bioinformatics tools can effectively differentiate between these types of modification in the absence of high quality fragmentation spectra. These factors can make the identification of hydroxylation particularly difficult.
Despite these considerable challenges, through a careful systematic search in conjunction with novel confirmation strategies, we describe the occurrence of site-specific prolyl hydroxylations on several proteins, none of which, to our knowledge, have been previously reported. We also identify limitations and discuss critical considerations in large-scale interrogation of protein oxidation events. We examine the differences in identifications made between two alternate analysis algorithms and contend that manual inspection of individual spectra is essential to proper interpretation of data.
2 Materials and Methods
2.1 Cell lines
Hela S3 cells were obtained from the ATCC and were maintained in spinner flasks in Joklik's modified Eagle's medium supplemented with 5% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin. The source of the HEK293FT cells has been described [10], and the cells were maintained in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin.
2.2 Cell extract preparation
Cells were treated with the proteasome inhibitor MG132 (10 μM, Sigma) for 4 hr, washed with PBS, and then lysed. Cells were lysed in one of two ways. In one, they were lysed with RIPA buffer (20 mM Tris, pH 8, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) supplemented with protease inhibitor cocktail (Sigma). Alternatively, cells were lysed directly in ST (100 mM Tris, pH 7.6, 4% SDS), and sonicated. In the latter case, following protein concentration measurements, DTT was added to a final concentration of 100 mM. Protein concentrations were determined using the DC Protein Assay kit (Bio-Rad).
2.3 Tryptic peptide preparation
Samples were dissolved in SDT (4% SDS, 100 mM DTT, 100 mM Tris-Cl, pH 7.6), and then subjected to alkylation and trypsin digestion using Filter Aided Sample Preparation (FASP) and Microcon YM-30 filtration units [11,12]. The sample was acidified by adding TFA to a final concentration of 0.1% to inhibit any further trypsin activity.
2.4 Isoelectric focusing (IEF)
Tryptic peptides obtained by FASP were fractionated on an Agilent 3100 OFFGEL IEF apparatus according the manufacturer's instructions.
2.5 Gel filtration
Hela S3 cells were pelleted, washed in buffer A (10 mM Hepes, pH 7.9, 10 mM KCl), resuspended in three packed cell volumes of buffer A supplemented with mammalian protease inhibitor cocktail, allowed to swell on ice for 10 min, and then lysed by Dounce homogenization. The cytoplasmic extract was obtained following centrifugation at 15,800 × g for 15 min at 4 °C. The extract was loaded onto a Superdex 200 HR 10/30 gel filtration column equilibrated with PBS on an Akta FPLC system consisting of a P-920 pump, a UPC-900 monitor, and a Frac-950 fraction collector. The flow rate was 0.25 ml/min. Fractions of 2 ml volume were collected, and proteins precipitated by the addition of 1/10 volume of trichloroacetic acid. Gel filtration standards (1.35 kDa to 670 kDa) were from Bio-Rad.
2.6 Mass spectrometry
Samples were desalted and concentrated using C18 StageTips [13]. Peptides were separated on 50 cm × 75 μM Picofrit column that was packed with 4 μM C12 Jupiter Proteo beads (Phenomenex) that was maintained at 50 °C (Phoenix S&T). Nanoflow chromatography was performed at 300 nL/min using a 90 minute linear gradient from 2% to 42% ACN in 0.5% acetic acid. ESI was done at 2.2 kV with a Phoenix S&T μAutoNano spray source coupled to a Thermo Scientific LTQ-Orbitrap hybrid mass spectrometer. Full mass range of m/z 370-2000 was acquired in the Orbitrap at 60,000 resolution at 400 m/z, and the five most intense ions were selected for MS/MS fragmentation in the linear ion trap with a normalized collision energy of 35%. The Orbitrap had a target fill count of 3×106 ions, and the ion trap had a target fill count of 1×104 ions.
2.7 Protein identification and data analysis
Data analysis was performed using two software packages. One was the Tide software package [14], which provides a highly optimized algorithm for generating SEQUEST Xcorr scores [15]. Briefly, normalized spectra sorted by precursor mass were compared against a pre-compiled in silico digestion of the human proteome based on the Uniref100 database (downloaded August 14, 2012) using a 0.05 Th precursor mass window. Carbamidomethylation of cysteine was specified as a fixed modification. Additionally, we specified up to 2 variable modifications/peptide allowing potential single oxidations at any of the sites previously reported in Unimod [16], along with single or double oxidation of methionine or tryptophan. Filtering on a q-value of 0.01 was performed using the Q-ranker software package [17] and a scrambled protein database.
Data was also analyzed using MaxQuant software v. 1.3.0.5 [18], and spectra were searched using the MaxQuant search engine, Andromeda, against the human proteome in the Uniref 100 database. Fixed and variable modifications were set as described for Tide based analyses above. Filtering of both protein and peptide identification and PTM site localization were set at 1% false discovery rate (FDR). Search space was limited to a maximum of 3 modifications per peptide. All PTM identifications, from both Tide and MaxQuant, were post-hoc filtered based on deviation from the predicted precursor mass to those within 1 ppm. Visual analysis of identified fragmentation spectra, obtained from either Tide or MaxQuant analysis, was performed using the Molecular Weight Calculator software package (v. 6.49, PNNL, US DOE) or the Viewer component of the MaxQuant software package, respectively.
2.8 Peptide Synthesis and Identification
Synthetic peptides containing 4-hydroxyproline were obtained from GenScript (Piscataway, NJ). On account of a cysteine residue, the synthetic peptide corresponding to the PKM fragment was alkylated with iodoacetamide as described above. All peptides were dissolved in LC running buffer, and 30-300 ng was subjected to LC-MS/MS analysis as described. Fragmentation spectra were assessed manually as described above.
3 Results
3.1 Overall strategy
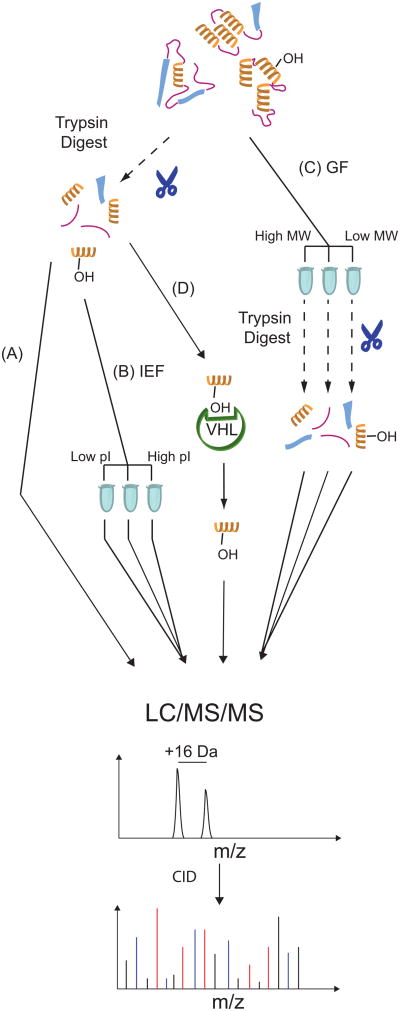
First, whole cell extracts were prepared, subjected to trypsin digestion, and analyzed using LC/MS/MS (Figure 1A). Next, we undertook a number of fractionation approaches in an effort to allow deeper coverage of the proteome. These included tryptic digestion of extracts followed by isoelectric focusing (Figure 1B), or alternatively, gel filtration of extracts followed by tryptic digestion (Figure 1C).
Figure 1.
Outline of sample preparation and enrichment strategies. (A) Direct analysis of unfractionated tryptic digests. (B) Following tryptic digests but prior to LC injection, peptides are fractionated by isoelectric focusing (IEF). (C) Fractionation of intact proteins by gel filtration chromatography, followed by tryptic digest prior to LC injection (GF). (D) VHL capture probe-based enrichment of hydroxylated peptides.
3.2 Initial Results
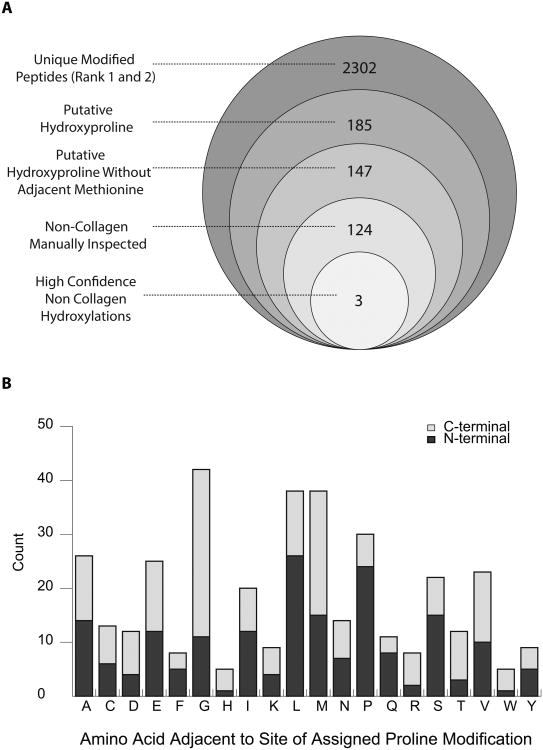
We identified peptide spectral matches obtained from these various approaches using two different strategies, Tide and Maxquant/Andromeda. The initial characterization of the dataset showed abundant oxidation events (2,302 events, Figure 2A), including that of methionine, which is known to readily oxidize during sample handling. Specifically, we observed that within the set of modified peptides identified by Tide, 64% had oxidation localized to methionine residues. Confounding cases of cysteine and tryptophan oxidation were also observed and were similarly excluded from further analysis, leaving 185 putative oxidation events. Further analysis revealed that many of the peptides identified with putative hydroxylated proline residues can be accounted for by proximal methionine oxidation and incidental, non-diagnostic intervening ions. In particular, within the called proline hydroxylation events identified by the Tide package, about 20% have a methionine in either the +1 or -1 position relative to the proline oxidation site (Figure 2B). In no instance were we able to identify a proline residue with an adjacent methionine in which the fragmentation spectra unambiguously indicated proline hydroxylation. Furthermore, since methionine oxidation is quite common, it is probable that the true oxidation was on the neighboring methionine residue rather than on the assigned proline. Exclusion of these oxidation events left 147 putative hydroxyproline containing peptides (Figure 2A).
Figure 2.
(A) Outline of results of computational analyses. Diagram shows results of Tide-based hydroxyproline search. Largest circle indicates the set of unique peptides that are called with at least one oxidation event (i.e. Proline, Methionine, etc.) as either the most likely, or when applicable, the second most likely candidate for a given spectra (i.e. first and second ranked peptide by XCorr score). Successive filtering steps are indicated. (B) Patterns of enrichment for amino acids immediately adjacent to primary site of assigned prolyl hydroxylation. Values presented are derived from the set of 185 unique peptides identified by Tide analyses as being likely prolyl hydroxylated.
3.3 Hydroxyproline Identification
Despite the prevalence of methionine oxidation, we identified a significant number of peptides with potential prolyl hydroxylation events. The MS2s of these peptides (124 unique non-collagen identifications, Figure 2A) were each then examined manually for spectral quality and accuracy of PTM localization. Many identified peptides suffered from MS2 spectra in which low intensity product ions, critical for PTM localization, were just barely above a reasonable signal to noise threshold.
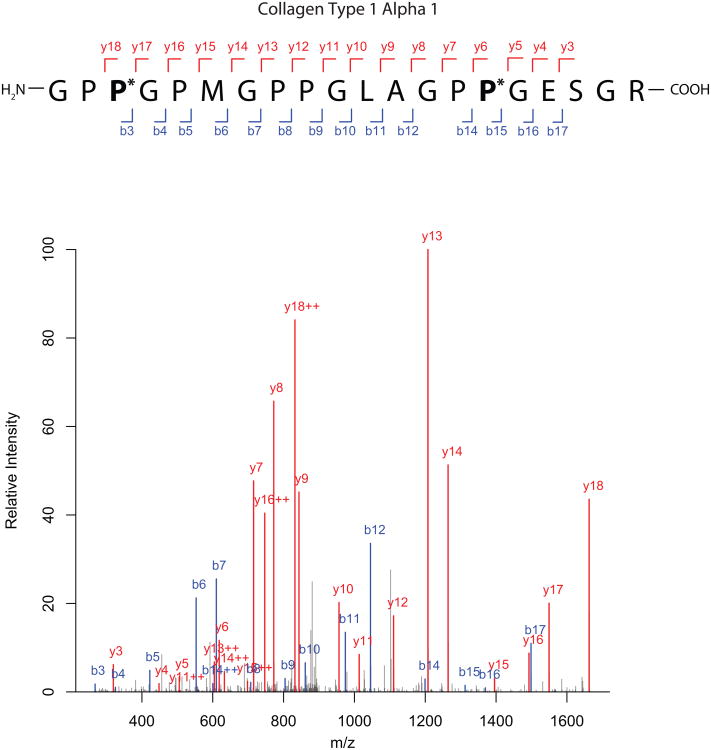
It is likely that we have excluded true positives on account of low-abundance ions that did not meet our stringent criteria for unambiguous identification for prolyl hydroxylation. In this regard, it has been observed that charge-induced dissociation of precursor ions to generate product ions is somewhat less effective towards the C-terminal as opposed to N-terminal side of proline residues [19]. We observed this phenomenon when examining collagen peptides, which are known to be prolyl hydroxylated, wherein the intensity of y-ions generated on the N-terminal side of proline were regularly 2-3 fold higher than the y-ion generated on the C-terminal side (for example, compare ions y13 to y12, and y7 to y6 in Figure 3). This phenomenon would tend to produce spectra with weak diagnostic product ions on the C-terminal side of prolyl residues.
Figure 3.
Representative fragmentation spectra (MS2) for Collagen type I, α1 chain. The b and y ions are indicated only when ions appeared above a 2-fold signal to noise ratio. Ions appearing in +2 charge states are indicated with “++”; all others are singly charged species.
3.4 Results Filtering and Refinement
These stringent criteria eliminate the majority of PTM IDs despite initial search parameters and filtering on q-values <1%. Peptides thus identified were next queried, where appropriate, against the NCBI protein database of known polymorphisms to exclude possible misidentifications resulting from naturally occurring Ala > Ser and Phe > Tyr substitutions, both of which are inherently indistinguishable from post-translational hydroxylation events. Indeed, by including Ala and Phe oxidation as a variable modification on our initial screens, a number of known Ala > Ser and Phe > Tyr polymorphisms were identified (data not shown), confirming the ability of our approach to accurately identify the hydroxylation mass shift and to correctly identify MS2 spectra of modified peptides. With this in mind, we examined the peptides containing putative hydroxyproline residues, and identified a number of assigned hydroxylation events that appear more likely to be isobaric Ala > Ser or Phe > Tyr substitutions rather than true prolyl hydroxylation events, due to lack of intervening diagnostic ions (∼10-15% of the 124 putative non-collagen hydroxylation events from the Tide analysis). In pursuing the approach outlined above, we identified a number of collagen peptides, including ones from collagen I (Figure 3) as well as numerous other collagen II and collagen IV peptides (data not shown), validating the overall methodology.
3.5 FDR Rate Estimations
Our data also highlight an important general consideration regarding the reporting of PTMs. While we computationally filtered all of our identifications to a 1% FDR rate based on accepted methodologies [20], we observed that in the end, only a small fraction of these peptides (<10% of the 185 original Tide derived identifications) actually appear to surpass reasonable levels of scrutiny as to either peptide identity and/or localization of hydroxylation. This suggests that calculation of FDR or q-values for post-translational modifications based on precursor mass shifts that turn out to be identical to confounding mass shifts (i.e. methionine, cysteine, or tryptophan oxidation), in conjunction with incomplete fragmentation spectra, may lead to significant underestimations of FDR.
3.6 MaxQuant and Tide Analysis Comparison
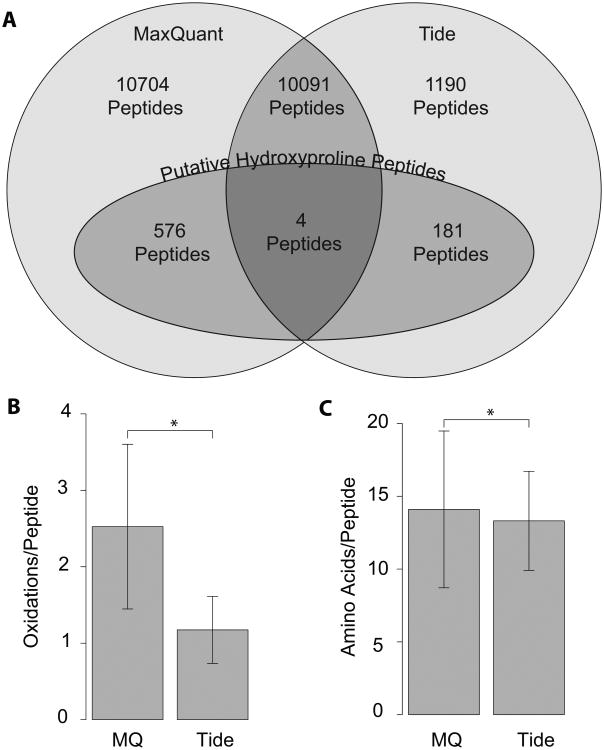
A number of different strategies exist to parse and identify peptide spectra from LC/MS/MS experiments. Within these, we have explored different software packages for the analysis of our data. The expansion of our search space to include such a large variety of possible sites of modifications poses a significant computational challenge, but the pre-compiled in silico digests associated with the Tide software package reduced the computional time needed to complete the analysis. We also subjected our data set to a parallel analysis by Maxquant/Andromeda and found that there was only a small intersection between the two data sets with regard to identification of hydroxylation events (Figure 4A). This was unexpected, since the overall overlap in identifications between the two methods was actually quite high, with greater than 90% of unmodified peptides identified by Tide also being identified by MaxQuant, and other studies of parallel analysis have showed similar levels of concordance [21]. As noted in Table 1 and Figure 4A, a subset of putatively prolyl hydroxylated peptides were identified in both analyses, but most were unique to one or the other. Maxquant yielded over twice the total number of identifications relative to Tide, although in the end it had a lower proportion of high confidence PTM identification. In general, Maxquant-specific candidate modified peptides were longer and more heavily modified (often containing 3-4 low confidence oxidation events on a single peptide) than those unique to the Tide ouput (Figure 4B and C). Peptides that were identified by both analysis packages were derived from high confidence fragmentation patterns and contained low numbers of modifications. While the majority of unmodified peptides are likely to be identified correctly by both search engines, modified peptides had a small degree of overlap between search engines decreasing confidence in the accuracy of these identifications. The discrepancy in this regard is likely due to the fundamental differences in the assumptions being made in regard to the two software packages, most notably the relative focus on precursor mass accuracy by MaxQuant relative to Tide. Conversely, the somewhat wider tolerance to precursor mass by Tide and reliance on initial identification by spectral cross-correlation with in silico predictions would likely yield more robust PTM localization in the presence of isobaric mass shifts.
Figure 4.
(A) Comparison of overall peptide and putative hydroxyproline peptide identification rates between Tide- and MaxQuant-based analyses platforms at the 1% FDR level. All values refer to numbers of unique peptides. “Putative Hydroxyproline Peptides” refers to the total set of peptides identified as being prolyl hydroxylated by the given analysis package prior to subtraction of likely confounding methionine oxidation and/or manual inspection of fragment spectra. Comparison of (B) average number of oxidations per unique modified peptide and (C) average length of both modified and unmodified peptides, as identified by MaxQuant and Tide analyses respectively. Bars indicate +/- 1 SD, and * indicates p < 0.05 by Student's t-Test.
Table 1.
Hydroxyproline Containing Peptides and Cellular Sources.
| Modified Peptide Sequence (Modified Residue Number) | Protein/Accession/Gene | Mol. Weight Observed (m/z) | Mol. Weight Expected p.p.m. error | Xcorr Score | MQ Score | Acquisition Notes1 |
|---|---|---|---|---|---|---|
| GPP*GPMGPPGLAGPP*GESGR (997,1009) | Collagen, Type 1 NP_000079.2 COL1A1 | 1815.8570 (908.9358) | 1815.8573 -0.167 | 5.75 | 263.15 | HeLa, Ly |
| ASP*AGGPLEDVVIER (36) | FKBP10/Peptidyl-Prolyl cis-trans isomerase NP_068758.3 FKBP10 | 1524.7784 (763.3947) | 1524.7784 0.022 | 4.323 | 185.2 | HeLa, HEK GF, VHL |
| YEILTP*NAIPK (733) | Myosin Heavy Chain -10 NP_005955.3 MYH10 | 1273.6914 (637.853) | 1273.6918 -0.257 | 2.581 | 168.3 | HeLa, HEK Ly, VHL |
| LSP*ELLNTGR (326) | Hexokinase 2 NP_000180.2 HK2 | 1114.5980 (558.3063) | 1114.5982 -0.179 | 3.200 | ND2 | HeLa, HEK VHL |
| GIFPVLCKDP*VQEAWAEDVDLR (477) | Pyruvate Kinase NP_002645.3 PKM | 2572.26321 (858.4296) | 2572.26313 -0.017 | ND | 161.51 | HeLa, HEK GF |
| AAQAP*SSFQLLYDLK (809) | C-1 tetrahydro- folate synthase NP_005947.3 MTHFD1 | 1666.8561 (834.4353) | 1666.8566 -0.299 | ND | 218.21 | HeLa Ly, VHL |
Samples types under which high quality spectra were observed: HeLa: HeLa; HEK: Human Embryonic Kidney 293 FT; Ly: Unenriched Lysate; GF: Gel Filtration Fast Protein Liquid Chromatography Fractionation of HEK293 FT; VHL: VHL/VHL Mutant Immunoprecipitation.
ND, Not Determined.
Includes Carbamidomethylation of Cysteine 474 (+57.03404)
3.7 VHL Capture
In an attempt to confirm the presence of prolyl hydroxylated peptides, we used a novel VHL capture reagent-based approach (Figure 1D, detailed in Supplementary Information). Briefly, VHL recognizes only prolyl hydroxylated HIF, so we generated point mutants of VHL with the intent of potentially broadening hydroxyproline recognition (Supplemental Figure 1), and then used cocktails of these mutants in pulldowns of tryptic peptide digests. Hydroxylated peptides were identified using VHL-based capture probes, and they were significantly enriched when controlling for the proportion of total peptide identifications that were derived from these experiments. Specifically, even though VHL-based capture probes yielded only 10% (1,121 vs 11,281) of total peptide spectral matches and 30% of putative hydroxylated peptides (60 vs. 185), 80% of the hydroxylated peptides that we identified and confirmed as hydroxylated from traditional fractionation experiments (excluding collagen) were also obtained independently with VHL-capture based approaches. This represents a highly significant increase in prevalence of true hydroxylated peptides relative to the total peptides for VHL-capture experiments (Z-test, p<0.001). Importantly, this particular enrichment for hydroxylated peptides serves as independent evidence that these peptides identified by manual inspection from conventionally fractionated samples are likely to contain genuine prolyl hydroxylation events. Conversely, we did not observe any hydroxylated peptides using a negative control S111A VHL-based probe, which does not recognize hydroxyproline owing to a lack of a critical hydrogen bond.
3.8 Synthetic Peptide Confirmation
Guided by the considerations detailed above, manual inspection of MS2 spectra lead to the elimination of the vast majority of putative hydroxyproline identifications, resulting in a small number (7) of peptides with convincing MS2 spectra. To examine whether the peptide spectra that we observed were in fact reflective of actual hydroxylation, we obtained synthetic peptides corresponding to these peptides. The peptides, containing hydroxyproline at the relevant sites, were subjected to MS/MS analysis and experimental spectra were compared to those obtained from in vivo sources. Fragmentation spectra for five of the seven of these synthetic peptides were similar to experimental spectra (Compare panel A to B in Supplemental Figures 2-6).
While the total number of convincingly hydroxylated peptides is relatively small, we found that the proteins containing them are involved in diverse functions (Table 1). We identified the hydroxylation modification in a protein, FKBP10 (Supplemental Figure 2), that possesses peptidyl prolyl cis-trans isomerase activity and is critical to collagen processing in the endoplasmic reticulum; mutations in this protein can lead to osteogeneis imperfecta [22]. Our analysis identified prolyl hydroxylation of three proteins—hexokinase 2, pyruvate kinase (muscle isozyme), and C-1 tetrahydrofolate synthase—which are enzymes that participate in key metabolic pathways (Supplemental Figures 3, 4, and 5). Hexokinase 2 and pyruvate kinase catalyze the first and last steps of glycolysis, namely, the phosphorylation of glucose to glucose-6-phosphate and the terminal dephosphorylation of phosphoenolpyruvate to pyruvate, respectively. Additionally, hexokinase and pyruvate kinase are both overexpressed in many cancers [23,24]. C-1 tetrahydrofolate synthase catalyzes the interconversion of tetrahydrofolate derivatives, and as such, plays an important role in nucleotide and methionine synthesis [25]. Finally, we identified prolyl hydroxylation of myosin heavy chain 10 (Supplemental Figure 6), a non-muscle myosin that is essential for normal development of the central nervous system and heart [26].
4 Discussion
Hydroxylation is a PTM that is known to occur on proline, aspartic acid, asparagine, histidine, and lysine residues, among others. Prolyl hydroxylation plays important roles in protein structural stability as in collagen, and in protein turnover as in HIF-α. The present data extends the known occurrence of prolyl hydroxylation in the proteome. While the total number of prolyl hydroxylation events identified in this study was relatively small, there are a number of factors that could account for this. First, a finite number of cell lines and conditions were examined, so it is conceivable that there are additional cell-type specific prolyl hydroxylation events that occur. Second, we were conservative in our identification criteria, thus potentially leading us to exclude authentic prolyl hydroxylation events.
Initial studies of HIF prolyl hydroxylation drew attention to an LXXLAP motif, raising the possibility that there might be a readily identifiable consensus motif among prolyl hydroxylated proteins. However, such a consensus was not found. In this regard, it is conceivable that the PHDs do not abide by a strict consensus, and indeed, in vitro studies indicate that amino acid substitutions can be tolerated at the -1, -2, and -5 positions of this motif without adversely affecting hydroxylation [27,28]. In addition, it is possible that these hydroxylation events might, in fact, reflect modification by non-PHD prolyl hydroxylases, or depend on higher order structures beyond the primary sequence.
The data emphasizes the importance of significant proteomic depth for analysis, necessitating additional dimensions of separation in order to detect Hyp-containing peptides. Indeed, only a few of the Hyp-containing peptides were identified in single dimension separations, despite the fact that many of the hydroxylated peptides are components of relatively high-abundance proteins. Pre-HPLC fractionation yielded a significant increase in the number of identified hydroxylation events.
Also important, we observed a large number of assigned calls relative to what appear to be high confidence hydroxylation events, despite the FDR based pre-screening. This suggests that when using common search algorithms, there may be significant false positives that become apparent only on post-hoc screening, and that existing scoring paradigms are underestimating FDR for modifications of this nature. These factors, in conjunction with the stochastic nature of whole proteome MS, may account for us not observing a number of prolyl hydroxylation events that have previously been reported [2–7]. Also differing from previous reports, we used no specific enrichment strategy to target particular proteins or protein families.
The data also demonstrate other challenges in identifying prolyl hydroxylated peptides. Several points are worth noting. First, different detection algorithms can yield dramatically different results. The use of Tide, for example, yielded a Hyp dataset that showed little overlap with that obtained with MaxQuant. The differences that we observe between alternate search algorithms highlight both the richness of the underlying data and the inability of any single given search strategy to comprehensively identify all hydroxylation events. Second, even with a high quality MS2 spectrum, it is still possible and seemingly common that hydroxylation may be erroneously assigned to proline, especially when the b- and y-ions flanking the prolyl residue of interest are not particularly abundant. We contend that it is essential to manually inspect MS2 spectra to confirm their authenticity. In addition, it is potentially risky to rely on a single MS2 to infer the presence of a hydroxylation event (“one hit wonder”). This risk is exacerbated if inspection of a potential modification is conducted without considering other potential sources of an isobaric mass shift, such as a confounding oxidation (i.e., of Met, Cys, and Trp). Taken together, this emphasizes the need for caution in interpreting MS2 spectra that putatively identify prolyl hydroxylation. We believe that the critical bar for establishing genuine proline hydroxylation, in light of these factors, should be set high. Our VHL-IP capture strategy yielded identification of prolyl hydroxylation events far in excess of the proportion of total peptide identifications obtained with non-capture samples, supporting the utility of the VHL-based capture probe approach.
These results highlight the need for caution when conducting large-scale identification of any given post-translational modification in the absence of alternate verification either by multiple search and identification algorithms, manual MS2 inspection, or alternative experimental approaches. This study suggests that the number of prolyl hydroxylated proteins is larger than was previously identified, although it may also be restricted in terms of scope and abundance. Lastly, it remains to be determined what the functional roles of these hydroxylation events are.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01-GM090301 (FSL and SRM) and the Brody Family Medical Trust Fund (PRA). We thank Mr. Adrian Flores for valuable technical assistance in the early phase of this work.
Abbreviations
- ABC
Ammonium bicarbonate
- FDR
false discovery rate
- HIF
Hypoxia Inducible Factor
- Hyp
Hydroxyproline
- MQ
MaxQuant
- PHD
Prolyl Hydroxylase Domain protein
- VBC
von Hippel Lindau tumor suppressor protein: Elongin B:Elongin C complex
- VHL
Von Hippel Lindau tumor suppressor protein
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
References
- 1.Gorres KL, Raines RT. Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol. 2010;45:106–124. doi: 10.3109/10409231003627991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummins EP, Berra E, Comerford KM, Ginouves A, et al. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc Natl Acad Sci USA. 2006;103:18154–18159. doi: 10.1073/pnas.0602235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo W, Hu H, Chang R, Zhong J, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moser SC, Bensaddek D, Ortmann B, Maure JF, et al. PHD1 Links Cell-Cycle Progression to Oxygen Sensing through Hydroxylation of the Centrosomal Protein Cep192. Developmental Cell. 2013;26:381–392. doi: 10.1016/j.devcel.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholz CC, Cavadas MAS, Tambuwala MM, Hams E, et al. Regulation of IL-1β–induced NF-κB by hydroxylases links key hypoxic and inflammatory signaling pathways. PNAS. 2013;110:18490–18495. doi: 10.1073/pnas.1309718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie L, Pi X, Mishra A, Fong G, et al. PHD3-dependent hydroxylation of HCLK2 promotes the DNA damage response. J Clin Invest. 2012;122:2827–2836. doi: 10.1172/JCI62374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie L, Xiao K, Whalen EJ, Forrester MT, et al. Oxygen-regulated beta(2)-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci Signal. 2009;2:ra33. doi: 10.1126/scisignal.2000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends in Genetics. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Song D, Li LS, Heaton-Johnson KJ, Arsenault PR, et al. Prolyl Hydroxylase Domain Protein 2 (PHD2) Binds a Pro-Xaa-Leu-Glu Motif, Linking it to the Heat Shock Protein 90 Pathway. Journal of Biological Chemistry. 2013;288:9662–9674. doi: 10.1074/jbc.M112.440552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Meth. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 12.Manza LL, Stamer SL, Ham AJL, Codreanu SG, Liebler DC. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5:1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 13.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protocols. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 14.Diament BJ, Noble WS. Faster SEQUEST Searching for Peptide Identification from Tandem Mass Spectra. J Proteome Res. 2011;10:3871–3879. doi: 10.1021/pr101196n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 16.Creasy DM, Cottrell JS. Unimod: Protein modifications for mass spectrometry. Proteomics. 2004;4:1534–1536. doi: 10.1002/pmic.200300744. [DOI] [PubMed] [Google Scholar]
- 17.Spivak M, Weston J, Bottou L, Käll L, Noble WS. Improvements to the Percolator Algorithm for Peptide Identification from Shotgun Proteomics Data Sets. J Proteome Res. 2009;8:3737–3745. doi: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 19.Qin J, Chait BT. Collision-induced dissociation of singly charged peptide ions in a matrix-assisted laser desorption ionization ion trap mass spectrometer. International Journal of Mass Spectrometry. 1999;190–191:313–320. [Google Scholar]
- 20.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Meth. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 21.Yu W, Taylor JA, Davis MT, Bonilla LE, et al. Maximizing the sensitivity and reliability of peptide identification in large-scale proteomic experiments by harnessing multiple search engines. Proteomics. 2010;10:1172–1189. doi: 10.1002/pmic.200900074. [DOI] [PubMed] [Google Scholar]
- 22.Alanay Y, Avaygan H, Camacho N, Utine GE, et al. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86:551–559. doi: 10.1016/j.ajhg.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ros S, Schulze A. Glycolysis Back in the Limelight: Systemic Targeting of HK2 Blocks Tumor Growth. Cancer Discovery. 2013;3:1105–1107. doi: 10.1158/2159-8290.CD-13-0565. [DOI] [PubMed] [Google Scholar]
- 24.Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Letters. 2015;356:184–191. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Watkins D, Rosenblatt DS. Update and new concepts in vitamin responsive disorders of folate transport and metabolism. J Inherit Metab Dis. 2012;35:665–670. doi: 10.1007/s10545-011-9418-1. [DOI] [PubMed] [Google Scholar]
- 26.Ma X, Adelstein RS. A point mutation in Myh10 causes major defects in heart development and body wall closure. Circ Cardiovasc Genet. 2014;7:257–265. doi: 10.1161/CIRCGENETICS.113.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Zhao Q, Mooney SM, Lee FS. Sequence Determinants in Hypoxia-inducible Factor-1? for Hydroxylation by the Prolyl Hydroxylases PHD1, PHD2, and PHD3. J Biol Chem. 2002;277:39792–39800. doi: 10.1074/jbc.M206955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landázuri MO, Vara-Vega A, Vitón M, Cuevas Y, del Peso L. Analysis of HIF-prolyl hydroxylases binding to substrates. Biochem Biophys Res Commun. 2006;351:313–320. doi: 10.1016/j.bbrc.2006.09.170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.