In this cross sectional study, one-fourth of LTCF residents were colonized intestinally with ST131 E. coli. The most dependent residents were at highest risk. Strain transmission occurred within and between LTCFs. Molecular typing showed similarities to clinical isolates.
Keywords: E. coli, E. coli resistance, long-term care facilities, ST131
Abstract
Background. Emerging data implicate long-term care facilities (LTCFs) as reservoirs of fluoroquinolone-resistant (FQ-R) Escherichia coli of sequence type 131 (ST131). We screened for ST131 among LTCF residents, characterized isolates molecularly, and identified risk factors for colonization.
Methods. We conducted a cross-sectional study using a single perianal swab or stool sample per resident in 2 LTCFs in Olmsted County, Minnesota, from April to July 2013. Confirmed FQ-R E. coli isolates underwent polymerase chain reaction-based phylotyping, detection of ST131 and its H30 and H30-Rx subclones, extended virulence genotyping, and pulsed-field gel electrophoresis (PFGE) analysis. Epidemiological data were collected from medical records.
Results. Of 133 fecal samples, 33 (25%) yielded FQ-R E. coli, 32 (97%) of which were ST131. The overall proportion with ST131 intestinal colonization was 32 of 133 (24%), which differed by facility: 17 of 41 (42%) in facility 1 vs 15 of 92 (16%) in facility 2 (P = .002). All ST131 isolates represented the H30 subclone, with virulence gene and PFGE profiles resembling those of previously described ST131 clinical isolates. By PFGE, certain isolates clustered both within and across LTCFs. Multivariable predictors of ST131 colonization included inability to sign consent (odds ratio [OR], 4.16 [P = .005]), decubitus ulcer (OR, 4.87 [ P = .04]), and fecal incontinence (OR, 2.59 [P = .06]).
Conclusions. Approximately one fourth of LTCF residents carried FQ-R ST131 E. coli resembling ST131 clinical isolates. Pulsed-field gel electrophoresis suggested intra- and interfacility transmission. The identified risk factors suggest that LTCF residents who require increased nursing care are at greatest risk for ST131 colonization, possibly due to healthcare-associated transmission.
The global emergence of antimicrobial-resistant sequence type 131 (ST131) Escherichia coli has been well documented across the entire spectrum of healthcare settings [1–4]. Sequence type 131 E. coli causes diverse types of extraintestinal infection, including bacteremia, pneumonia, intraabdominal infection, meningitis, epididymo-orchitis, prostatitis, musculoskeletal infection, wound infection, and most commonly, urinary tract infection (UTI).
The prevalence of ST131 among human clinical E. coli isolates varies by geographic region and host population, ranging from 12.5% to nearly 40% [2, 4–6]. Furthermore, ST131 and, more specifically, its H30 subclone (so-named for containing allele 30 of fimH, the type-1 fimbriae adhesin gene) account for the vast majority of fluoroquinolone-resistant (FQ-R) E. coli infections in the United States, whereas the H30-Rx subset within H30 is the main repository of the CTX-M-15 extended-spectrum-beta-lactamase (ESBL) globally [7, 8].
It is not clear what has enabled ST131 to disseminate so widely and rapidly. Sequence type 131 is clearly more extensively antimicrobial-resistant than other E. coli and contains an abundance of virulence factors (VFs) [9]. However, whether increased virulence plays a role in ST131's success is controversial, because in animal models ST131 is not consistently more virulent than other E. coli clonal groups [10–13]. Alternatively, ST131 may have superior ability to be transmitted from host to host or to colonize the host intestine, a process that commonly precedes UTI [14, 15].
The prevalence and risk factors for intestinal colonization with ST131 in different host populations, possible differences between colonizing versus clinical ST131 isolates, and the reservoirs and transmission routes of ST131 have not been rigorously assessed [6]. Emerging evidence from the United States and Europe suggests that long-term care facilities (LTCFs) may be reservoirs of ST131 [16–20]. A population-based study performed in Olmsted County, Minnesota, found that the prevalence of ST131 as a cause of E. coli infections increases progressively with age [2]. In that study, ST131 accounted for half of all E. coli infections in persons >90 years old and for 76% of E. coli isolates from LTCF residents. Studies from Ireland, the United Kingdom, and France have identified a high prevalence of ST131 among LTCF residents [19, 21, 22].
To determine whether LTCF residents in Olmsted County are a reservoir for ST131, we conducted a cross-sectional study of intestinal colonization with ST131 E. coli in 2 local LTCFs. We also assessed for risk factors for ST131 intestinal colonization among LTCF residents and compared the VF and pulsed-field gel electrophoresis (PFGE) profiles of colonizing ST131 isolates with those of previously reported clinical ST131 isolates.
METHODS
Subjects and Specimens
We collected perianal swabs or stool specimens from residents of 2 LTCFs in Olmsted County from April 2013 to July 2013. Facility 1 had 108 residents housed within a single building, including a subacute care wing, a long-term care wing, and a locked memory care unit. Facility 2 had 182 residents housed in 2 different buildings, including a subacute care building and a long-term care building. All subjects or a legal authorized representative provided consent. Consent was obtained from 133 residents, including 100 who provided consent themselves and 33 whose legal authorized representative provided consent. Specimens were collected from 40 of 108 (37%) residents in facility 1 and 93 of 182 (51%) residents in facility 2.
Specimen Processing
Specimens were cultured directly onto MacConkey agar plates containing 1 µg/mL ciprofloxacin, HardyCHROM ESBL agar plates, and HardyCHROM carbapenem-containing agar plates (Hardy Diagnostics). All presumptive FQ-R E. coli were definitively identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Bruker Daltonics).
Molecular Typing
Established polymerase chain reaction-based assays were used to detect major E. coli phylogenetic groups [23], ST131 and its H30 and H30-Rx ST131 subclones [4, 24], and 49 extraintestinal virulence genes [24, 25]. XbaI PFGE was done according to the PulseNet protocol [23], and pulsotypes were assigned based on 94% similarity to reference strains within a large private PFGE library [9]. Dendrograms were created within BioNumerics (Applied Maths) according to the unweighted pair group method based on pairwise Dice similarity coefficients. Dendrograms were pulsotype-specific and contained, for each LTCF isolate, 4 randomly selected comparison isolates of the same pulsotype from the private PFGE library.
Epidemiological Data
Demographic and clinical data were collected through medical record review. Medication administration data, including antibiotics, for the 6 months before specimen collection were obtained from hospitalization records and the LTCF's medication administration record.
Statistical Analysis
Subject characteristics were summarized using descriptive statistics. Logistic regression was used to test the individual effects of candidate risk factors on the likelihood of FQ resistance to ST131, with those factors demonstrating a significant association from univariable modeling carried forward in a final multivariable model. For the multivariable model, any of the selected variables with data redundancy were identified and reduced to avoid collinearity. Continuous variables with skewed distributions were transformed (eg, logarithmic) to satisfy regression assumptions. We report adjusted odds ratios ([ORs] with 95% confidence interval) from the multivariable model to convey the magnitude of association between individual factors and ST131 colonization, and the model c-statistic was used as a measure of the overall discrimination. Statistical significance was set at an alpha level of 0.05. All analyses were performed using the SAS statistical software package (version 9.2; SAS Institute Inc., Cary, North Carolina).
RESULTS
Prevalence of Sequence Type 131 and Its H30 and H30-Rx Subclones
A total of 131 perianal swabs and 2 stool samples were collected from 133 LTCF residents (1 specimen per resident) from April 2013 to June 2013. The proportion of subjects with FQ-R E. coli was 33 of 133 (25%) overall, but this varied significantly by facility, ie, 18 of 41 (44%) in facility 1 vs 15 of 92 (16%) in facility 2 (P = .001). Sequence type 131 accounted for 32 of 33 (97%) of the FQ-R isolates; therefore, 32 of 133 (24%) of subjects overall were colonized intestinally with ST131 E. coli. All 32 ST131 isolates (100%) represented the H30 ST131 subclone, and 9 (28%) belonged to the H30-Rx subset within H30. Four of the FQ-R E. coli isolates, all of which were H30-Rx subclone members, were presumptive ESBL-producers (as evidenced by growth on the HardyCHROM ESBL plate), but none was resistant to carbapenems.
Comparison of Colonizing Versus Clinical Sequence Type 131 Isolates
The 32 colonizing ST131 isolates exhibited similar virulence gene profiles to those reported previously for ST131 clinical isolates (Table 1). By PFGE analysis, the 32 LTCF ST131 isolates represented 11 unique pulsotypes, most commonly types 968 (31%) and 800 (22%), which also dominated in previous studies involving primarily clinical ST131 isolates [2, 9]. Whereas isolates from both facilities represented pulsotypes 968 and 800, only isolates from facility 1 represented less common pulsotypes.
Table 1.
Molecular Characteristics of Colonizing ST131 Isolates From Long-Term Care Facility Residents in Olmsted County, Minnesota
| Category | Specific Characteristic (Definition)a | Prevalence Within ST131, No. of Isolates (% of 32) |
|---|---|---|
| O type | O25b (rfb variant; lipopolysaccharide synthesis) | 30 (94) |
| ST131 subclonal group | H30 (fluoroquinolone resistance-associated) | 32 (100) |
| H30Rx (ESBL-associated; subset of H30) | 9 (28) | |
| Adhesin | iha (adhesin-siderophore receptor) | 26 (81) |
| fimH (type 1 fimbriae adhesin) | 32 (100) | |
| Toxin | cnf1 (cytotoxic necrotizing factor) | 2 (6) |
| sat (secreted autotransporter toxin) | 26 (81) | |
| Siderophore receptor | fyuA (yersiniabactin receptor) | 32 (100) |
| iutA (aerobactin receptor) | 26 (81) | |
| Protectin | kpsM II (group 2 capsules) | 17 (53) |
| kfiC (K5 capsule) | 16 (50) | |
| K2/K100 (capsule variants) | 1 (3) | |
| traT (serum resistance-associated) | 25 (78) | |
| Other virulence genes | usp (uropathogenic-specific protein) | 32 (100) |
| ompT (outer membrane protease) | 32 (100) | |
| malX (pathogenicity island marker) | 30 (94) |
Abbreviations: ESBL, extended-spectrum-beta-lactamase; ST131, sequence type 131.
a Molecular characteristics not observed include the following (trait definition): afa/draBC (Dr-binding adhesins), afaE8 (variant afimbrial adhesin), astA (toxin of enteroaggregative Escherichia coli), cdtB (cytolethal distending toxin), bmaE (M fimbriae), clbB and clbN (colibactin polyketide synthesis), clpG (variant adhesin), cvaC (microcin V), F17 (variant adhesin), focG (F1C fimbriae adhesin), gafD (G fimbriae), H7 fliC (flagellar variant), hlyD (alpha hemolysin), hlyF (variant hemolysin), hra (heat-resistant agglutinin), ibeA (invasion of brain endothelium), ireA (siderophore receptor), iroN (salmochelin receptor), iss (increased serum survival), K1 and K15 (group 2 capsule variants), kpsMT III (group 3 capsules), papAH, papC, papEF, papG, and papG alleles I, II, and III (P fimbriae structural subunits, assembly, and tip adhesin variants), pic (protein associated with intestinal colonization), rfc (O4 lipopolysaccharide synthesis), sfa/focDE (S and F1C fimbriae), sfaS (S fimbriae adhesin), tsh (temperature-sensitive hemagglutinin), vat (vacuolating toxin).
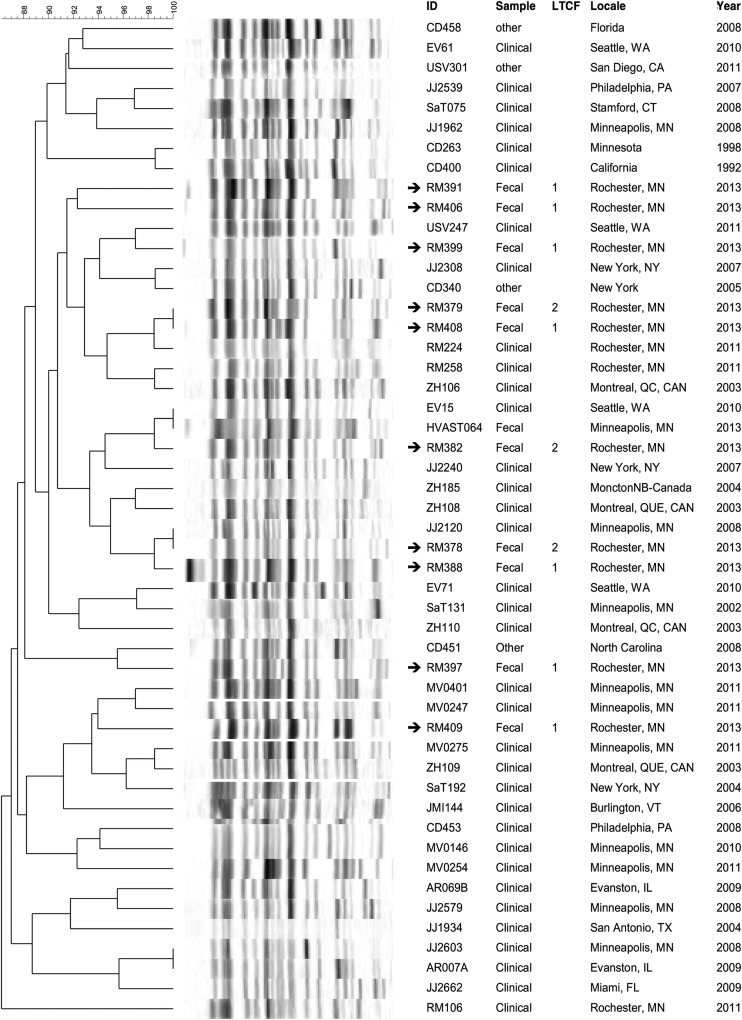
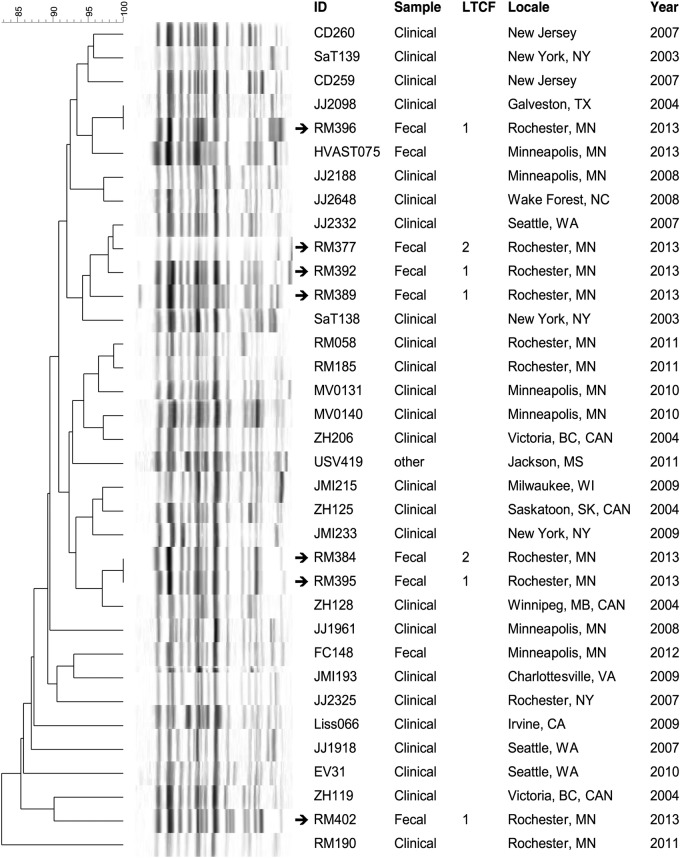
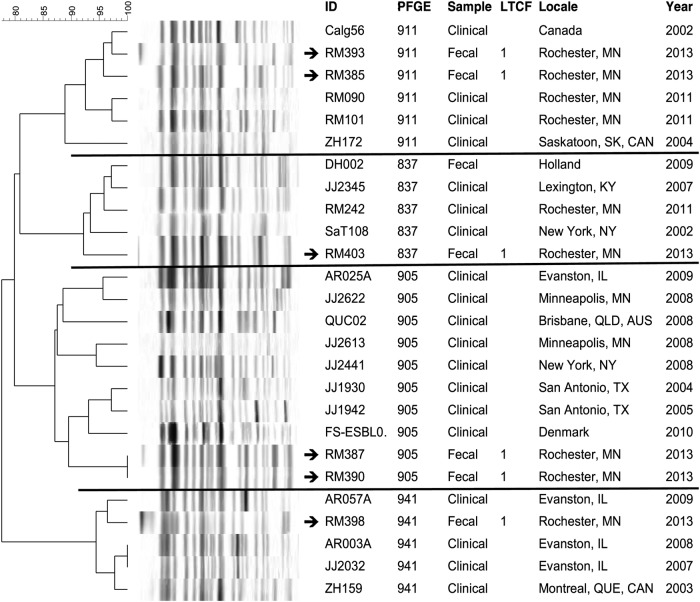
Pulsotype-specific dendrograms were created to compare the PFGE profiles of the present colonizing ST131 isolates with those of previously characterized clinical isolates of similar pulsotypes from humans and companion animals (Figures 1–3). In each dendrogram, the colonizing isolates intermingled with clinical isolates of the same pulsotype. In addition, among the LTCF isolates, some closely related profiles were confined to a single LTCF, whereas others crossed facilities. That is, 4 clusters each comprised 2 to 3 highly similar profiles of isolates from different residents in the same LTCF (in each instance facility 1), suggesting local clonal spread within that LTCF. In contrast, in 4 other clusters (in the dendrograms for pulsotypes 968 and 800), isolates from facilities 1 and 2 were nearest neighbors, suggesting transmission between LTCFs.
Figure 1.
Dendrogram of pulsotype 968 sequence type 131 isolates. XbaI pulsed-gel electrophoresis (PFGE) profiles of long-term care facility (LTCF) colonization isolates are compared with those of previously characterized clinical isolates from a large private PFGE library (J. R. J.). Arrows denote isolates from LTCF residents. ID, strain identifier. Sample, specimen type or context.
Figure 2.
Dendrogram of pulsotype 800 sequence type 131 isolates. XbaI pulsed-gel electrophoresis (PFGE) profiles of long-term care facility (LTCF) colonization isolates are compared with those of previously characterized clinical isolates from a large private PFGE library (J. R. J.). Arrows denote isolates from LTCF residents. ID, strain identifier. Sample, specimen type or context.
Figure 3.
Dendrogram of pulsotype 911, 837, 905, and 941 sequence type 131 isolates. XbaI pulsed-gel electrophoresis (PFGE) profiles of long-term care facility (LTCF) colonization isolates are compared with those of previously characterized clinical isolates from a large private PFGE library (J. R. J.). Arrows denote isolates from LTCF residents. ID, strain identifier. Sample, specimen type or context.
Risk Factors for Sequence Type 131 Intestinal Colonization
Univariable analysis on 132 subjects (excluding the one subject with non-ST131 FQ-R E. coli) identified several factors significantly associated with ST131 colonization, including specific LTCF facility (P = .002), time in LTCF (P = .03), level of care (ie, subacute, long-term, or memory unit) (P = .01), inability to sign consent (P < .001), urinary incontinence (P = .02), fecal incontinence (P < .001), and decubitus ulcer (P = .01) (Table 2). In contrast, antibiotic use and hospitalization within the prior 6 months as well as age were not significantly associated with ST131 colonization.
Table 2.
Univariable Analysis of Potential Risk Factors for Escherichia coli ST131 Colonization Among 132 Long-Term Care Facility Residents in Olmsted County, Minnesota
| Variable | FQ-R ST131 E. coli (n = 32) | No FQ-R E. coli (n = 100) | Odds Ratio (95% CI) | P Value |
|---|---|---|---|---|
| Age (years), mean (SD) | 83.2 ± 8.5 | 81.6 ± 14.1 | 1.01 (0.98, 1.05) | .56 |
| Male gender | 11 (34%) | 35 (35%) | 0.97 (0.42, 2.25) | .95 |
| Specific facility | .002 | |||
| Facility 1 | 17 (53%) | 23 (23%) | 3.79 (1.64, 8.75) | |
| Facility 2 | 15 (47%) | 77 (77%) | 1.0 (Referent) | |
| Time in long-term care facility (years)a | 1.8 (1.1, 4.3) | 1.2 (0.3, 2.4) | 1.49 (1.05, 2.11) | .03 |
| Level of care: | .01 | |||
| Long-term care | 24 (75%) | 74 (74%) | 1.0 (Referent) | |
| Subacute care | 3 (9%) | 25 (25%) | 0.37 (0.10, 1.33) | |
| Memory unit | 5 (16%) | 1 (1%) | 15.42 (1.72, 138.56) | |
| Unable to sign consent | 16 (50%) | 16 (16%) | 5.25 (2.19, 12.60) | <.001 |
| Urinary catheter | 3 (9%) | 8 (8%) | 1.19 (0.30, 4.78) | .81 |
| Urinary incontinence | 23 (72%) | 48 (48%) | 2.77 (1.17, 6.57) | .02 |
| Fecal incontinence | 18 (56%) | 19 (19%) | 5.48 (2.32, 12.94) | <.001 |
| Dialysis | 1 (3%) | 3 (3%) | 1.33 (0.13, 8.43) | .78b |
| Corticosteroids in prior 6 mo | 7 (22%) | 15 (15%) | 1.59 (0.58, 4.32) | .37 |
| Clostridium difficile in prior 6 mo | 0 (0%) | 1 (1%) | 1.03 (<0.01, 19.62) | .99b |
| Chronic kidney disease | 5 (16%) | 25 (25%) | 0.56 (0.19, 1.60) | .28 |
| Diabetes mellitus | 5 (16%) | 26 (26%) | 0.53 (0.18, 1.51) | .23 |
| Ostomy tube | 3 (9%) | 4 (4%) | 2.54 (0.54, 11.06) | .22b |
| Decubitus ulcer | 6 (19%) | 4 (4%) | 5.54 (1.45, 21.09) | .01 |
| Chronic wound | 2 (6%) | 10 (10%) | 0.60 (0.12, 2.89) | .53 |
| Hospitalized in prior 6 mo | 13 (41%) | 48 (48%) | 0.74 (0.33, 1.66) | .47 |
| Antibiotics in prior 6 mo | 18 (56%) | 56 (56%) | 1.01 (0.45, 2.25) | .98 |
| Days of antibiotics in prior 6 mo: | .32 | |||
| 0 | 14 (44%) | 44 (44%) | 1.17 (0.39, 3.45) | |
| 1–7 | 6 (19%) | 22 (22%) | 1.0 (Referent) | |
| 8–14 | 4 (13%) | 17 (17%) | 0.86 (0.21, 3.55) | |
| 15–28 | 6 (19%) | 6 (6%) | 3.67 (0.86, 15.59) | |
| >28 | 2 (6%) | 10 (10%) | 0.73 (0.13, 4.29) |
Abbreviations: CI, confidence interval; FQ-R, fluoroquinolone-resistant; IQR, interquartile range; SD, standard deviation; ST131, sequence type 131.
a Median (IQR) reported due to skewed distribution; effect measured in logistic regression based on log-transformed data.
b Due to sparse data, we used Firth's correction to estimate the odds ratio (displayed along with profile likelihood-based 95% CIs) and the likelihood ratio test to assess the effect's significance.
A multivariable model was constructed using noncollinear factors that were significant in the univariable model (model c-statistic, 0.797), which included time in LTCF, inability to sign consent, decubitus ulcer, and fecal incontinence as candidate predictors (Table 3). Inability to sign consent (P = .005) and presence of a decubitus ulcer (P = .04) were both independently associated with ST131 colonization, with ORs greater than 4.0, and fecal incontinence approached significance (P = .06), with an OR of approximately 2.6. In contrast, length of time in the LTCF was not significantly associated with ST131 colonization (P = .36), with an OR of only 1.2.
Table 3.
Multivariable Analysis of Risk Factors for Intestinal Colonization With ST131 Among LTCF Residents in Olmsted County, Minnesota
| Variable | OR (95% CI) | P Value |
|---|---|---|
| Time in LTCFa (log-years) | 1.20 (0.81, 1.77) | .36 |
| Unable to sign consent | 4.16 (1.53, 11.30) | .005 |
| Decubitus ulcer | 4.87 (1.06, 22.26) | .04 |
| Fecal incontinence | 2.59 (0.95, 7.06) | .06 |
Abbreviations: CI, confidence interval; LTCF, long-term care facility; OR, odds ratio.
a Data were log-transformed to approximate a normal distribution.
DISCUSSION
In this cross-sectional survey of colonization with FQ-R E. coli among residents of 2 LTCFs in Olmsted County, Minnesota, we found that nearly one fourth of residents carried ST131 E. coli, but that this varied by LTCF, and that colonizing ST131 isolates closely resembled clinical ST131 isolates. These findings suggest that certain LTCFs may be important reservoirs for ST131 in our community.
Although LTCFs are well known as reservoirs of other antimicrobial resistant pathogens [19, 20, 26–28] and have been implicated as reservoirs of ST131 in Europe [20, 29], this study is among the first to describe LTCFs as reservoirs of ST131 in the United States. Our PFGE analysis demonstrated both clustering of ST131 genotypes within 1 LTCF and commonality of genotypes across facilities, suggesting both intrafacility and interfacility transmission. We also identified inability to sign consent and presence of a decubitus ulcer as independent risk factors for ST131 intestinal colonization, suggesting that the most dependent LTCF residents are at greatest risk for colonization.
Our overall colonization prevalence estimates among LTCF residents of 25% for FQ-R E. coli and 24% for ST131 are similar to data from a study in the Netherlands, which recovered ciprofloxacin-resistant E. coli from 23% of LTCF residents [16]. Another recent US study found that 47.5% of LTCF residents acquired colonization with FQ-R E. coli over 12 months [30]. However, neither of these prior studies specifically evaluated ST131 E. coli. The few studies that have evaluated intestinal colonization with ST131 in different populations found a wide range in ST131 carriage rates, ranging from 0% to 4% among healthy children and adults in Australia [31, 32] to approximately 15% among US adult men before prostate biopsy [33]. Sequence type 131 colonization rates clearly vary greatly by host group, suggesting that preventative interventions can be focused on the highest-risk populations.
We found that the molecular characteristics of colonizing ST131 isolates were similar to those reported previously for clinical ST131 isolates with respect to prevalence of the H30 subclone, virulence profiles, and predominant pulsotypes [4, 24, 34]. This suggests that the colonizing intestinal isolates are an integral part of the total ST131 clonal ecology rather than being distinct from invasive, clinical isolates. This congruence supports the fecal reservoir hypothesis [14, 35] and suggests that ST131, like other uropathogenic E. coli, causes disease by first colonizing the intestine and then migrating to extraintestinal sites.
Several of the observed risk factors for ST131 intestinal colonization correspond with a requirement for increased nursing care, which we speculate may reflect healthcare worker-related transmission. These include urinary and fecal incontinence, decubitus ulcers, inability to sign consent, and memory unit residence. For example, LTCF residents with urine or fecal incontinence require assistance with cleansing and topical care to prevent skin breakdown. Decubitus ulcers occur mostly among residents who are less mobile and require assistance with transfers. Residents who cannot sign consent and who require memory unit care are more likely to need assistance with activities of daily living. Notably, memory care unit residents were at highest risk for ST131 colonization. Alternatively, host-specific factors associated with dependency may directly promote intestinal colonization or transmission, independent of healthcare workers. Further study is needed to clarify the basis for the observed associations.
We did not find that antibiotic use in the prior 6 months was associated with ST131 colonization. The antibiotic use data were obtained from the medication administration record in the resident's LTCF chart. This information should be complete for antibiotics administered at the facility, but it may not have included all the antibiotics administered during hospitalization stays. Hospitalization summaries and patients' electronic medical records also were reviewed to complete the data collection from hospitalization stays, although these were not always available for all patients. Alternatively, the relevant time frame for assessing prior antibiotic use or contacts with the acute care system may be multiple years, or even the subject's total lifetime, rather than shorter intervals such as the past 6 months. Chronically ill individuals with increased comorbidities typically have had more lifetime exposure to antibiotics and to acute care, which would also correlate with our finding that residents requiring increased nursing care were at greatest risk for ST131 colonization.
Intrafacility transmission of ST131 was suggested by the finding that isolates from different individuals within the same LTCF were often nearest neighbors in the PFGE dendrogram. The underlying mechanism(s) for such transmission is not well understood. Intrafacility transmission conceivably could occur due to environmental contamination of common-use areas, direct personal contact among residents, and/or healthcare workers (as discussed above). In contrast, interfacility transmission of ST131 was suggested by the finding that isolates from different LTCFs were sometimes nearest neighbors in the PFGE dendrogram. Possible mechanisms for this phenomenon include direct transfer of residents between LTCFs and transfer of residents to either LTCF from a common third facility, such as an acute care hospital. We lack evidence to support or refute these hypotheses; collecting relevant evidence should be a goal for future research.
Our findings have potentially important clinical implications. First, medical providers who care for LTCF residents require increased awareness about ST131, including its high prevalence among LTCF residents and its multidrug-resistant phenotype, in order to select appropriate empiric antibiotics for infections possibly due to E. coli. Second, infection control measures within LTCFs should be strengthened to prevent transmission of ST131 and other multidrug-resistant organisms both within the facility and across the healthcare continuum, because LTCF residents transfer frequently between community and hospital settings. Infection control practices within LTCFs vary by facility and are generally more relaxed than in acute care hospitals due to limited resources and conflicting priorities [36]. For example, at the 2 study facilities, hand sanitizers, soap and water, and medical examination gloves were not available near each room. Furthermore, isolation precautions in LTCFs for residents colonized with multidrug-resistant organisms are controversial due to the risk of reducing quality of life and quality of care by decreasing interpersonal interactions [37, 38]. Third, despite individual-level antibiotic exposure not being confirmed here as a significant risk factor for colonization with ST131, effective antimicrobial stewardship interventions should be pursued to reduce overuse of antimicrobials within LTCFs, which should reduce overall selection pressure for ST131 and other multidrug-resistant pathogens. Design and implementation of effective infection control and stewardship interventions in LTCFs deserve further study.
This study has limitations. First, sampling bias could have occurred because subjects who consented to participate in the study may be inherently different from those who did not consent. Second, the adequacy of data retrieval via medical record review was dependent on the documentation entered by LTCF nursing and medical staff, which is not standardized. Third, the cross-sectional study design precluded longitudinal assessment for possible transmission events; transmission was inferred based solely on PFGE profile similarity, which can be misleading. Finally, we surveyed only 2 LTCFs, both in a single county; therefore, our results may not be generalizable to facilities in different geographic regions or with different resident populations.
CONCLUSIONS
In conclusion, colonization with FQ-R ST131 E. coli that resemble clinical ST131 isolates is common among LTCF residents in Olmsted County; thus, LTCF residents may constitute a potentially important reservoir of ST131 that could cause infections in LTCF residents and the general public alike. Intensified infection control efforts within LTCFs and screening of LTCF residents upon hospital admission may be needed to prevent the spread of antimicrobial-resistant gram-negative bacilli such as ST131 E. coli into communities and hospitals, analogous to the screening for methicillin-resistant Staphylococcus aureus performed in many hospitals. Further studies are needed to determine whether active surveillance for ST131 among LTCF residents is useful or cost effective.
Acknowledgments
We thank administrators, staff, and residents at both LTCFs for participation. We also thank the numerous nurses from Mayo Clinic's Clinical Research Unit for assistance with specimen collection. Providers of reference library isolates included Ritu Banerjee (Mayo Clinic, Rochester, Minnesota), Mariana Castanheira (JMI Laboratories, North Liberty, Iowa), Chitrita DebRoy (Escherichia coli Reference Center, The Pennsylvania State University, University Park, Pennsylvania), Michael Liss (University of Texas Health Science Center, San Antonio, Texas), Bente Olesen (Herlev Hospital, Herlev, Denmark), Johann Pitout (Calgary Laboratory Services, Calgary, Alberta, Canada), Joanne Platell (Biosecurity Queensland, Coopers Plains, QLD, Australia), Ari Robicsek (Northshore University HealthSystem, Evanston, Illinois), Daniel Sahm (Eurofins-Medinet, Chantilly, Virginia), Flemming Scheutz (Statens Serum Institut, Copenhagen, Denmark), Evgeni Sokurenko (University of Washington, Seattle, Washington), and George Zhanel (University of Manitoba, Winnipeg, Manitoba, Canada).
Financial support. This publication was supported by Clinical and Transitional Science Award Grant Number KL2 TR000136 from the National Center for Advancing Translational Science (to R. B). It also is based in part upon work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs, Grant Number 1 I01 CX000192 01 (to J. R. J.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Johnson JR, Tchesnokova V, Johnston B, et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis. 2013;207:919–28. doi: 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee R, Johnston B, Lohse C, et al. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect Control Hosp Epidemiol. 2013;34:361–9. doi: 10.1086/669865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen AE, Lautenbach E, Morales KH, et al. Fluoroquinolone-resistant Escherichia coli in the long-term care setting. Am J Med. 2006;119:958–63. doi: 10.1016/j.amjmed.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 4.Colpan A, Johnston B, Porter S, et al. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis. 2013;57:1256–65. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Cerero L, Bellido Mdel M, Serrano L, et al. Escherichia coli O25b:H4/ST131 are prevalent in Spain and are often not associated with ESBL or quinolone resistance. Enferm Infecc Microbiol Clin. 2013;31:385–8. doi: 10.1016/j.eimc.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27:543–74. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011;66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 8.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother. 2008;61:273–81. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JR, Nicolas-Chanoine MH, DebRoy C, et al. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967–2009. Emerg Infect Dis. 2012;18:598–607. doi: 10.3201/eid1804.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson JR, Porter SB, Zhanel G, et al. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect Immun. 2012;80:1554–62. doi: 10.1128/IAI.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clermont O, Lavollay M, Vimont S, et al. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J Antimicrob Chemother. 2008;61:1024–8. doi: 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- 12.Mora A, Dahbi G, Lopez C, et al. Virulence patterns in a murine sepsis model of ST131 Escherichia coli clinical isolates belonging to serotypes O25b:H4 and O16:H5 are associated to specific virotypes. PLoS One. 2014;9:e87025. doi: 10.1371/journal.pone.0087025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavigne JP, Vergunst AC, Goret L, et al. Virulence potential and genomic mapping of the worldwide clone Escherichia coli ST131. PLoS One. 2012;7:e34294. doi: 10.1371/journal.pone.0034294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno E, Andreu A, Perez T, et al. Relationship between Escherichia coli strains causing urinary tract infection in women and the dominant faecal flora of the same hosts. Epidemiol Infect. 2006;134:1015–23. doi: 10.1017/S0950268806005917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee R, Johnson JR. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother. 2014;58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Donk CF, Schols JM, Driessen CJ, et al. Prevalence and spread of multidrug resistant Escherichia coli isolates among nursing home residents in the southern part of The Netherlands. J Am Med Dir Assoc. 2013;14:199–203. doi: 10.1016/j.jamda.2012.09.026. [DOI] [PubMed] [Google Scholar]
- 17.Lautenbach E, Tolomeo P, Black N, et al. Risk factors for fecal colonization with multiple distinct strains of Escherichia coli among long-term care facility residents. Infect Control Hosp Epidemiol. 2009;30:491–3. doi: 10.1086/597234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maslow JN, Lautenbach E, Glaze T, et al. Colonization with extraintestinal pathogenic Escherichia coli among nursing home residents and its relationship to fluoroquinolone resistance. Antimicrob Agents Chemother. 2004;48:3618–20. doi: 10.1128/AAC.48.9.3618-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rooney PJ, O'Leary MC, Loughrey AC, et al. Nursing homes as a reservoir of extended-spectrum beta-lactamase (ESBL)-producing ciprofloxacin-resistant Escherichia coli. J Antimicrob Chemother. 2009;64:635–41. doi: 10.1093/jac/dkp220. [DOI] [PubMed] [Google Scholar]
- 20.Burke L, Humphreys H, Fitzgerald-Hughes D. The revolving door between hospital and community: extended-spectrum beta-lactamase-producing Escherichia coli in Dublin. J Hosp Infect. 2012;81:192–8. doi: 10.1016/j.jhin.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Nicolas-Chanoine MH, Robert J, Vigan M, et al. Different factors associated with CTX-M-producing ST131 and non-ST131 Escherichia coli clinical isolates. PLoS One. 2013;8:e72191. doi: 10.1371/journal.pone.0072191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhanji H, Doumith M, Rooney PJ, et al. Molecular epidemiology of fluoroquinolone-resistant ST131 Escherichia coli producing CTX-M extended-spectrum beta-lactamases in nursing homes in Belfast, UK. J Antimicrob Chemother. 2011;66:297–303. doi: 10.1093/jac/dkq463. [DOI] [PubMed] [Google Scholar]
- 23.Ribot EM, Fair MA, Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 24.Banerjee R, Robicsek A, Kuskowski MA, et al. Molecular epidemiology of Escherichia coli sequence type 131 and its H30 and H30-Rx subclones among extended-spectrum-beta-lactamase-positive and -negative E. coli clinical isolates from the Chicago region, 2007 to 2010. Antimicrob Agents Chemother. 2013;57:6385–8. doi: 10.1128/AAC.01604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–8. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.March A, Aschbacher R, Dhanji H, et al. Colonization of residents and staff of a long term care facility and adjacent acute-care hospital geriatric unit by multiresistant bacteria. Clin Microbiol Infect. 2009;16:934–44. doi: 10.1111/j.1469-0691.2009.03024.x. [DOI] [PubMed] [Google Scholar]
- 27.Lautenbach E, Han J, Santana E, et al. Colonization with extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella species in long-term care facility residents. Infect Cont Hosp Epidemiol. 2012;33:302–4. doi: 10.1086/664055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoo J, Byeon J, Yang J, et al. High prevalence of extended-spectrum beta-lactamases and plasmid-mediate AmpC beta-lactamases in Enterobacteriaceae isolated from long term care facilities in Korea. Diag Microbiol Infect Dis. 2010;67:261–5. doi: 10.1016/j.diagmicrobio.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Brisse S, Diancourt L, Laouenan C, et al. Phylogenetic distribution of CTX-M- and non-extended-spectrum-beta-lactamase-producing Escherichia coli isolates: group B2 isolates, except clone ST131, rarely produce CTX-M enzymes. J Clin Microbiol. 2012;50:2974–81. doi: 10.1128/JCM.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han JH, Maslow J, Han X, et al. Risk factors for the development of gastrointestinal colonization with fluoroquinolone-resistant Escherichia coli in residents of long-term care facilities. J Infect Dis. 2014;209:420–5. doi: 10.1093/infdis/jit471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudinha T, Johnson JR, Andrew SD, et al. Escherichia coli sequence type 131 as a prominent cause of antibiotic resistance among urinary Escherichia coli isolates from reproductive-age women. J Clin Microbiol. 2013;51:3270–6. doi: 10.1128/JCM.01315-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudinha T, Johnson JR, Andrew SD, et al. Genotypic and phenotypic characterization of Escherichia coli isolates from children with urinary tract infection and from healthy carriers. Pediatr Infect Dis J. 2013;32:543–8. doi: 10.1097/INF.0b013e31828ba3f1. [DOI] [PubMed] [Google Scholar]
- 33.Liss MA, Peterson EM, Johnston B, et al. Prevalence of ST131 among fluoroquinolone-resistant Escherichia coli obtained from rectal swabs before transrectal prostate biopsy. Urology. 2013;81:548–55. doi: 10.1016/j.urology.2012.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olesen B, Hansen DS, Nilsson F, et al. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J Clin Microbiol. 2013;51:1779–85. doi: 10.1128/JCM.00346-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto S, Tsukamoto T, Terai A, et al. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J Urol. 1997;157:1127–9. [PubMed] [Google Scholar]
- 36.Nicolle LE. Infection control in long-term care facilities. Clin Infect Dis. 2000;31:752–6. doi: 10.1086/314010. [DOI] [PubMed] [Google Scholar]
- 37.Morgan DJ, Diekema DJ, Sepkowitz K, et al. Adverse outcomes associated with contact precautions: a review of the literature. Am J Infect Control. 2009;37:85–93. doi: 10.1016/j.ajic.2008.04.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuno JP, Krein S, Lansing B, et al. Health care worker opinions on use of isolation precautions in long-term care facilities. Am J Infect Control. 2012;40:263–6. doi: 10.1016/j.ajic.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]