Genome-similarity based codes were assigned to individual ebolavirus isolates. Codes were found to be informative of phylogenetic and epidemiological relationships. It is proposed that such codes should be assigned to every genome-sequenced virus to complement current viral taxonomy.
Keywords: average nucleotide identity, classification, ebolavirus, epidemiology, phylogeny
Abstract
Background. Developing a universal standardized microbial typing and nomenclature system that provides phylogenetic and epidemiological information in real time has never been as urgent in public health as it is today. We previously proposed to use genome similarity as the basis for immediate and precise typing and naming of individual organisms or viruses. In this study, we tested the validity of the proposed system and applied it to the epidemiology of infectious diseases using Ebola virus disease (EVD) outbreaks as the example.
Methods. One hundred twenty-eight publicly available ebolavirus genomes were compared with each other, and average nucleotide identity (ANI) was calculated. The ANI was then used to assign unique codes, hereafter referred to as Life Identification Numbers (LINs), to every viral isolate, whereby each LIN consisted of a series of positions reflecting increasing genome similarity. Congruence of LINs with phylogenetic and epidemiological relationships was then determined.
Results. Assigned LINs correlate with phylogeny at the species and infraspecies level and can even identify some individual transmission chains during the 2014–2015 EVD epidemic in West Africa.
Conclusions. Life Identification Numbers can provide a fast, automated, standardized, and scalable approach to precisely identify and name viral isolates upon genome sequence submission, facilitating unambiguous communication during disease epidemics among clinicians, epidemiologists, and governments.
Although naming of viral species is regulated by well established nomenclature rules described in the International Code of Virus Classification and Nomenclature [1], there are no general rules for classification below the species level, leaving it up to specialist groups to develop family-specific rules to name strains, variants, and isolates. In the case of the Filoviridae, which includes the genus Ebolavirus, a taxonomic revision was published in 2010 [2]. Based on this revision, the genus Ebolavirus contains 5 species: Bundibugyo ebolavirus, Reston ebolavirus, Sudan ebolavirus, Taï Forest ebolavirus, and Zaire ebolavirus, whereby a viral isolate is considered to be a member of a species if it is less than 30% different (based on its full-length genomic sequence) from the virus type of the species and more than 30% different from the type virus of the type species (ie, Z ebolavirus). Each species has 1 member virus, which is also the type of virus: Bundibugyo virus (BDBV), Reston virus (RESTV), Sudan virus (SUDV), Taï Forest virus (TAFV), and Ebola virus (EBOV), respectively. Each type virus has a type variant represented by a specific isolate [3, 4]. The isolate name is composed of the following: virus name, isolation host-suffix, country of sampling, year of sampling, genetic variant designation, and isolate designation [4].
Because of such complex nomenclature rules that are specific to every viral family and because of the frequency of taxonomic revisions, it can be very challenging for nonexperts to correctly classify and name a newly discovered virus that causes an emerging disease outbreak. This situation can lead to confusion about the identity of the pathogen, which can in turn delay an effective international response to contain such an outbreak before it turns into an epidemic.
Fortunately, because next-generation sequencing [5] is so affordable today, the opportunity exists to develop new classification and naming systems that can overcome the above limitations and that can provide approaches to specifically type and name any individual isolate, strain, or organism as soon as its genome sequence becomes available. We previously described such an approach and demonstrated its suitability for providing codes that largely correlate with phylogenetic and epidemiological relationships using the genomes of various bacteria, animals, humans, and foot and mouth disease virus (FMDV) [6].
In short, genome similarity-based codes (ie, Life Identification Numbers [LINs]) that we propose to assign to every genome-sequenced organism and virus consist in a series of positions, each of which reflects a different threshold of percentage of DNA identity, expressed as average nucleotide identity (ANI) [7]. The more similar the genomes of 2 organisms (or viral isolates) are, the more similar their LINs will be. Organisms with very different genomes will have LINs that are different at their left-most position, organisms with intermediate similarity will have identical symbols (any number or letter can be used) up to an intermediate position in their LINs, and almost identical organisms will have LINs that are identical almost to the right-most position. Only isolates with 100% DNA identity will have exactly the same LIN (Figure 1).
Figure 1.
Basic principal of Life Identification Number (LIN) assignment. Each LIN is composed of a series of positions corresponding to average nucleotide identity values increasing from left to right of the LIN. The actual symbol at each position can be any number or letter and does not reflect the degree of similarity between genomes. The information content in a LIN is its similarity to other LINs. For example, the first genome added to the database is assigned “0” in all positions (Example 1). A genome with relatively low genome similarity compared with Example 1 (74% for Example 2) will have a LIN that is the same up to the LIN position B corresponding to the 70% threshold (because 74 is higher than 70) but different at position C corresponding to the 80% threshold (because 74 is lower than 80). A genome more similar to Example 1 (99.4% for Example 3) has a LIN that is identical to Example 1 up to a position further to the right (position L), and almost identical genomes have LINs identical to each other nearly up to the right-most position (Examples 1 and 5). Identical genomes have identical LINs (Examples 3 and 4).
We propose to assign LINs sequentially as genomes become available, whereby the LIN of the organism with the most similar genome that already has an assigned LIN will be used as the basis for assigning the new LIN [6]. The only time a LIN of an isolate or organism should be adjusted is when a higher quality genome sequence becomes available. This approach will provide stability of LINs and minimize confusion inevitably associated with taxonomic revisions.
One inherent limitation of identifiers assigned based on overall genome similarity is as follows: in the case of organisms or viruses with very recent common ancestors—in which a single mutation may be the only indication of a new transmission chain during an outbreak—LINs’ ability to reflect phylogenetic and epidemiological relationships is limited [6]. However, we still need to establish exactly what that limit is.
In this study, we determined to what depth LINs can be informative of phylogenetic relationships among members of the genus Ebolavirus. Forty-seven publicly available ebolavirus genomes from previous outbreaks and 81 genomes of ebolavirus isolates from the 2014–2015 Ebola virus disease (EVD) epidemic [8, 9] were compared, provisional LINs were assigned, and these assigned LINs were compared with whole genome phylogenies. The results reveal that LINs reflect evolutionary relationships from the species level all the way down to single transmission chains identified by phylogenetic reconstruction. However, they do not reflect every node revealed by phylogeny, which needs to be considered when interpreting LINs in an epidemiological context. We finally propose that LINs should be assigned to every genome in GenBank (and other public databases) upon genome sequence submission, to provide the healthcare and research community with a system for fast and precise identification of, and clear communication about, viruses and other pathogens.
METHODS
One hundred twenty-eight ebolavirus genomes were downloaded from the National Center for Biotechnology Information (NCBI) nucleotide database on September 3, 2014, and pairwise ANI was calculated using our previously described custom pipeline [6] based on jSpecies [10]. Calculations of ANI were performed in the order of the dates associated with each ebolavirus genome sequence in GenBank (which is either the date the genome sequence was submitted or the date it was last updated). In this process, every genome was only compared to those genomes that would have been available on those dates (Table 1). When 2 or more genomes had the same associated date, accession numbers were used to establish the order of comparison. Life Identification Numbers were then assigned to each genome sequentially based on the ANI with the most similar genome to which a LIN had already been assigned (ie, all genomes in rows above the genome in question in Table 1) using ANI thresholds shown in Figure 1.
Table 1.
ANI Values and LINs Assigned to Each EBOV Genomea
| Shortened GenBank Definition | Date | Order | Most Similar Genome | ANI | LIN |
|---|---|---|---|---|---|
| AF522874.1 Reston_strain_Pennsylvania_1989-1990 | 09/04/02 | 1 | AF522874.1 | 100 | 0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| AY354458.1 Zaire_strain_Zaire_1995 | 02/06/04 | 2 | AF522874.1 | 69.44778 | 0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| AY729654.1 Sudan_strain_Gulu_2000 | 10/07/05 | 3 | AY354458.1 | 70.22714 | 0.1.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| EU338380.1 Sudan_EBOV-S-2004 | 01/23/08 | 4 | AY729654.1 | 94.95444 | 0.1.1.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| AB050936.1 Reston_ebolavirus | 06/25/08 | 5 | AF522874.1 | 98.74722 | 0.0.0.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| FJ217161.1 Bundibugyo_ebolavirus | 11/21/08 | 6 | AY354458.1 | 72.39182 | 0.1.2.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| FJ217162.1 Tai_Forest_Cote_d'Ivoire_1994 | 11/21/08 | 7 | FJ217161.1 | 75.35462 | 0.1.3.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| FJ968794.1 Sudan_UGA_strain_Boniface_2007 | 05/29/09 | 8 | EU338380.1 | 99.63444 | 0.1.1.0.0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| FJ621583.1 Reston_Reston_strain_Reston_2008-A | 07/14/09 | 9 | AF522874.1 | 97.69167 | 0.0.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| FJ621584.1 Reston_Reston_strain_Reston_2008-C | 07/14/09 | 10 | AF522874.1 | 96.98167 | 0.0.0.0.0.0.2.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| FJ621585.1 Reston_strain_Reston_2008-E | 07/14/09 | 11 | AB050936.1 | 98.68222 | 0.0.0.0.0.0.0.2.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| HQ613402.1 Zaire_034-KS_2008 | 11/07/11 | 12 | AY354458.1 | 97.84833 | 0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| HQ613403.1 Zaire_DRC_2007_M-M | 11/07/11 | 13 | HQ613402.1 | 99.93889 | 0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.1.0.0.0.0.0.0.0 |
| JN638998.1 Sudan_2011_Nakisamata | 08/06/12 | 14 | AY729654.1 | 99.39556 | 0.1.1.0.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| JQ352763.1 Zaire_1995_strain_Kikwit | 01/23/13 | 15 | AY354458.1 | 99.98333 | 0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.1.0.0 |
| KC242784.1 Zaire_EBOV_COD-2007-9_Luebo | 03/08/13 | 16 | HQ613403.1 | 99.98333 | 0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.1.0.0.0.0.1.0.0 |
| KC242785.1 Zaire_EBOV_COD-2007-0_Luebo | 03/08/13 | 17 | HQ613403.1 | 99.97222 | 0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.1.0.0.0.1.0.0.0 |
| KC242786.1 Zaire_EBOV_COD-2007-1_Luebo | 03/08/13 | 18 | HQ613403.1 | 99.98889 | 0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.1.0.0.0.0.2.0.0 |
| KC242787.1 Zaire_EBOV_COD-2007-23_Luebo | 03/08/13 | 19 | HQ613403.1 | 99.98889 | 0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.1.0.0.0.0.3.0.0 |
| KC242788.1 Zaire_EBOV_COD-2007-43_Luebo | 03/08/13 | 20 | KC242787.1 | 99.95611 | 0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.1.0.1.0.0.0.0.0 |
| KC242789.1 Zaire_EBOV_COD-2007-4_Luebo | 03/08/13 | 21 | KC242786.1 | 99.98889 | 0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.1.0.0.0.0.4.0.0 |
| KC242790.1 Zaire_EBOV_COD-2007-5_Luebo | 03/08/13 | 22 | HQ613403.1 | 99.96667 | 0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.1.0.0.1.0.0.0.0 |
| KC242791.1 Zaire_EBOV_COD-1977-Bonduni | 03/08/13 | 23 | AY354458.1 | 98.86611 | 0.1.0.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KC242792.1 Zaire_EBOV_GAB-1994-Gabon | 03/08/13 | 24 | AY354458.1 | 99.065 | 0.1.0.0.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KC242796.1 Zaire_EBOV_COD-1995-13625_Kikwit | 03/08/13 | 25 | AY354458.1 | 99.945 | 0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.1.0.0.0.0.0.0 |
| KC242797.1 Zaire_EBOV_GAB-1996-1Oba | 03/08/13 | 26 | KC242792.1 | 99.81833 | 0.1.0.0.0.0.0.0.1.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0 |
| KC242798.1 Zaire_EBOV_GAB-1996-1Ikot | 03/08/13 | 27 | KC242792.1 | 99.65722 | 0.1.0.0.0.0.0.0.1.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KC242799.1 Zaire_EBOV_COD-1995-13709_Kikwit | 03/08/13 | 28 | KC242796.1 | 99.99444 | 0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.1.0.0.0.0.1.0 |
| KC242800.1 Zaire_EBOV_GAB-1996-Ilembe | 03/08/13 | 29 | KC242786.1 | 97.88 | 0.1.0.0.0.0.2.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KC242801.1 Zaire_EBOV_COD-1976-deRoover | 03/08/13 | 30 | KC242791.1 | 99.98889 | 0.1.0.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.1.0.0 |
| JX477165.1 Reston_RESTV-PHL-2009-09A_Farm_A | 03/08/13 | 31 | FJ621583.1 | 99.91667 | 0.0.0.0.0.0.1.0.0.0.0.0.0.0.1.0.0.0.0.0.0.0.0.0 |
| JX477166.1 Reston_RESTV-PHL-USA-1996-Ferlite,Philippines-Alice,TX | 03/08/13 | 32 | AB050936.1 | 99.88389 | 0.0.0.0.0.0.0.1.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0 |
| KC242783.2 Sudan_SUDV_SSD-1979-Maleo | 03/14/13 | 33 | FJ968794.1 | 99.77556 | 0.1.1.0.0.1.0.0.0.0.1.1.0.0.0.0.0.0.0.0.0.0.0.0 |
| KC589025.1 Sudan_EboSud-639_2012 | 06/24/13 | 34 | AY729654.1 | 99.19889 | 0.1.1.0.0.0.0.0.2.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KC545389.1 Sudan_EboSud-602_2012 | 06/24/13 | 35 | AY729654.1 | 99.29278 | 0.1.1.0.0.0.0.0.3.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KC545390.1 Sudan_EboSud-603_2012 | 06/24/13 | 36 | KC545389.1 | 99.99444 | 0.1.1.0.0.0.0.0.3.0.0.0.0.0.0.0.0.0.0.0.0.0.1.0 |
| KC545391.1 Sudan_EboSud-609_2012 | 06/24/13 | 37 | KC545389.1 | 99.98889 | 0.1.1.0.0.0.0.0.3.0.0.0.0.0.0.0.0.0.0.0.0.1.0.0 |
| KC545392.1 Sudan_EboSud-682_2012 | 06/24/13 | 38 | KC545389.1 | 99.99444 | 0.1.1.0.0.0.0.0.3.0.0.0.0.0.0.0.0.0.0.0.0.0.2.0 |
| KC545393.1 Bundibugyo_EboBund-112_2012 | 06/24/13 | 39 | FJ217161.1 | 98.71333 | 0.1.2.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KC545394.1 Bundibugyo_EboBund-120_2012 | 06/24/13 | 40 | KC545393.1 | 99.97222 | 0.1.2.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.1.0.0.0 |
| KC545395.1 Bundibugyo_EboBund-122_2012 | 06/24/13 | 41 | KC545393.1 | 99.97778 | 0.1.2.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.2.0.0.0 |
| KC545396.1 Bundibugyo_EboBund-14_2012 | 06/24/13 | 42 | KC545393.1 | 99.96111 | 0.1.2.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.1.0.0.0.0 |
| KM034550.1 Zaire_SLE-2014-Makona-EM095 | 08/08/14 | 43 | KC242786.1 | 97.11944 | 0.1.0.0.0.0.3.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KM034551.1 Zaire_SLE-2014-Makona-EM096 | 08/08/14 | 44 | KM034550.1 | 99.85222 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0 |
| KM034552.1 Zaire_SLE-2014-Makona-EM098 | 08/08/14 | 45 | KM034551.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM034553.1 Zaire_SLE-2014-Makona-G3670.1 | 08/08/14 | 46 | KM034552.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.2.0.0 |
| KM034554.1 Zaire_SLE-2014-Makona-G3676.1 | 08/08/14 | 47 | KM034552.1 | 99.97722 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.1.0.0.0 |
| KM034556.1 Zaire_SLE-2014-Makona-G3677.1 | 08/08/14 | 48 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM034558.1 Zaire_SLE-2014-Makona-G3679.1 | 08/08/14 | 49 | KM034551.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0 |
| KM034559.1 Zaire_SLE-2014-Makona-G3680.1 | 08/08/14 | 50 | KM034554.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.1.0.1.0 |
| KM034560.1 Zaire_SLE-2014-Makona-G3682.1 | 08/08/14 | 51 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM034561.1 Zaire_SLE-2014-Makona-G3683.1 | 08/08/14 | 52 | KM034554.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.1.0.2.0 |
| KM034562.1 Zaire_SLE-2014-Makona-G3686.1 | 08/08/14 | 53 | KM034554.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.1.1.0.0 |
| KM034563.1 Zaire_SLE-2014-Makona-G3687.1 | 08/08/14 | 54 | KM034550.1 | 99.11222 | 0.1.0.0.0.0.3.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KM233035.1 Zaire_SLE-2014-Makona-EM104 | 08/08/14 | 55 | KM034552.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.3.0.0 |
| KM233036.1 Zaire_SLE-2014-Makona-EM106 | 08/08/14 | 56 | KM034552.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233037.1 Zaire_SLE-2014-Makona-EM110 | 08/08/14 | 57 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233038.1 Zaire_SLE-2014-Makona-EM111 | 08/08/14 | 58 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233039.1 Zaire_SLE-2014-Makona-EM112 | 08/08/14 | 59 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233040.1 Zaire_SLE-2014-Makona-EM113 | 08/08/14 | 60 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233041.1 Zaire_SLE-2014-Makona-EM115 | 08/08/14 | 61 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233042.1 Zaire_SLE-2014-Makona-EM119 | 08/08/14 | 62 | KM233036.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.4.0.0 |
| KM233043.1 Zaire_SLE-2014-Makona-EM120 | 08/08/14 | 63 | KM034552.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.2.0 |
| KM233044.1 Zaire_SLE-2014-Makona-EM121 | 08/08/14 | 64 | KM034552.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.3.0 |
| KM233045.1 Zaire_SLE-2014-Makona-EM124.1 | 08/08/14 | 65 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233049.1 Zaire_SLE-2014-Makona-G3707 | 08/08/14 | 66 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233050.1 Zaire_SLE-2014-Makona-G3713.2 | 08/08/14 | 67 | KM233036.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.4.0 |
| KM233053.1 Zaire_SLE-2014-Makona-G3724 | 08/08/14 | 68 | KM233036.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.5.0 |
| KM233054.1 Zaire_SLE-2014-Makona-G3729 | 08/08/14 | 69 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233055.1 Zaire_SLE-2014-Makona-G3734.1 | 08/08/14 | 70 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233056.1 Zaire_SLE-2014-Makona-G3735.1 | 08/08/14 | 71 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233058.1 Zaire_SLE-2014-Makona-G3750.1 | 08/08/14 | 72 | KM233036.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.6.0 |
| KM233061.1 Zaire_SLE-2014-Makona-G3752 | 08/08/14 | 73 | KM233036.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.7.0 |
| KM233062.1 Zaire_SLE-2014-Makona-G3758 | 08/08/14 | 74 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233063.1 Zaire_SLE-2014-Makona-G3764 | 08/08/14 | 75 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233064.1 Zaire_SLE-2014-Makona-G3765.2 | 08/08/14 | 76 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233065.1 Zaire_SLE-2014-Makona-G3769.1 | 08/08/14 | 77 | KM034552.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.5.0.0 |
| KM233069.1 Zaire_SLE-2014-Makona-G3770.1 | 08/08/14 | 78 | KM233042.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.4.0.0 |
| KM233071.1 Zaire_SLE-2014-Makona-G3771 | 08/08/14 | 79 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233072.1 Zaire_SLE-2014-Makona-G3782 | 08/08/14 | 80 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233073.1 Zaire_SLE-2014-Makona-G3786 | 08/08/14 | 81 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233074.1 Zaire_SLE-2014-Makona-G3787 | 08/08/14 | 82 | KM034552.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.6.0.0 |
| KM233075.1 Zaire_SLE-2014-Makona-G3788 | 08/08/14 | 83 | KM034556.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.8.0 |
| KM233076.1 Zaire_SLE-2014-Makona-G3789.1 | 08/08/14 | 84 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233077.1 Zaire_SLE-2014-Makona-G3795 | 08/08/14 | 85 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233078.1 Zaire_SLE-2014-Makona-G3796 | 08/08/14 | 86 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233079.1 Zaire_SLE-2014-Makona-G3798 | 08/08/14 | 87 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233080.1 Zaire_SLE-2014-Makona-G3799 | 08/08/14 | 88 | KM034552.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.9.0 |
| KM233081.1 Zaire_SLE-2014-Makona-G3800 | 08/08/14 | 89 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233082.1 Zaire_SLE-2014-Makona-G3805.1 | 08/08/14 | 90 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233084.1 Zaire_SLE-2014-Makona-G3807 | 08/08/14 | 91 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233085.1 Zaire_SLE-2014-Makona-G3808 | 08/08/14 | 92 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233086.1 Zaire_SLE-2014-Makona-G3809 | 08/08/14 | 93 | KM233036.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.10.0 |
| KM233087.1 Zaire_SLE-2014-Makona-G3810.1 | 08/08/14 | 94 | KM034552.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.7.0.0 |
| KM233089.1 Zaire_SLE-2014-Makona-G3814 | 08/08/14 | 95 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233090.1 Zaire_SLE-2014-Makona-G3816 | 08/08/14 | 96 | KM233086.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.10.0 |
| KM233091.1 Zaire_SLE-2014-Makona-G3817 | 08/08/14 | 97 | KM233036.1 | 99.98333 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.8.0.0 |
| KM233092.1 Zaire_SLE-2014-Makona-G3818 | 08/08/14 | 98 | KM233036.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.9.0.0 |
| KM233093.1 Zaire_SLE-2014-Makona-G3819 | 08/08/14 | 99 | KM233036.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.11.0 |
| KM233094.1 Zaire_SLE-2014-Makona-G3820 | 08/08/14 | 100 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233095.1 Zaire_SLE-2014-Makona-G3821 | 08/08/14 | 101 | KM233086.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.10.0 |
| KM233096.1 Zaire_SLE-2014-Makona-G3822 | 08/08/14 | 102 | KM233086.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.10.0 |
| KM233097.1 Zaire_SLE-2014-Makona-G3823 | 08/08/14 | 103 | KM034552.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.12.0 |
| KM233098.1 Zaire_SLE-2014-Makona-G3825.1 | 08/08/14 | 104 | KM233061.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.7.0 |
| KM233100.1 Zaire_SLE-2014-Makona-G3826 | 08/08/14 | 105 | KM233036.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.13.0 |
| KM233101.1 Zaire_SLE-2014-Makona-G3827 | 08/08/14 | 106 | KM233100.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.13.0 |
| KM233102.1 Zaire_SLE-2014-Makona-G3829 | 08/08/14 | 107 | KM233061.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.14.0 |
| KM233103.1 Zaire_SLE-2014-Makona-G3831 | 08/08/14 | 108 | KM233074.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.6.0.0 |
| KM233104.1 Zaire_SLE-2014-Makona-G3834 | 08/08/14 | 109 | KM233061.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.7.0 |
| KM233105.1 Zaire_SLE-2014-Makona-G3838 | 08/08/14 | 110 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233106.1 Zaire_SLE-2014-Makona-G3840 | 08/08/14 | 111 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233107.1 Zaire_SLE-2014-Makona-G3841 | 08/08/14 | 112 | KM034552.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.0.0 |
| KM233108.1 Zaire_SLE-2014-Makona-G3845 | 08/08/14 | 113 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233109.1 Zaire_SLE-2014-Makona-G3846 | 08/08/14 | 114 | KM233036.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.10.0.0 |
| KM233110.1 Zaire_SLE-2014-Makona-G3848 | 08/08/14 | 115 | KM233061.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.7.0 |
| KM233111.1 Zaire_SLE-2014-Makona-G3850 | 08/08/14 | 116 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KM233112.1 Zaire_SLE-2014-Makona-G3851 | 08/08/14 | 117 | KM233061.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.7.0 |
| KM233113.1 Zaire_SLE-2014-Makona-G3856.1 | 08/08/14 | 118 | KM233036.1 | 99.99444 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.15.0 |
| KM233115.1 Zaire_SLE-2014-Makona-G3857 | 08/08/14 | 119 | KM233061.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.7.0 |
| KM233116.1 Zaire_SLE-2014-Makona-NM042.1 | 08/08/14 | 120 | KM233036.1 | 100 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.0.1.1.0 |
| KJ660346.2 Zaire_GIN-2014-Kissidougou-C15 | 08/26/14 | 121 | KM034554.1 | 99.98889 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.1.2.0.0 |
| KJ660347.2 Zaire_GIN-2014-Gueckedou-C07 | 08/26/14 | 122 | KJ660346.2 | 99.98333 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.1.3.0.0 |
| KJ660348.2 Zaire_GIN-2014-Gueckedou-C05 | 08/26/14 | 123 | KJ660347.2 | 99.97778 | 0.1.0.0.0.0.3.0.0.0.0.0.1.0.0.0.0.0.0.0.2.0.0.0 |
| NC_002549.1 Zaire_COD-1976_Yambuku-Mayinga | 08/27/14 | 124 | KC242791.1 | 99.98889 | 0.1.0.0.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0.2.0.0 |
| NC_004161.1 Reston_USA-1989-Philippines89-Pennsylvania | 08/27/14 | 125 | AF522874.1 | 100 | 0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KC242793.1 Zaire_EBOV_GAB-1996-1Eko | 09/12/14 | 126 | KC242797.1 | 100 | 0.1.0.0.0.0.0.0.1.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0 |
| KC242794.1 Zaire_EBOV_GAB-1996-2Nza | 09/12/14 | 127 | KC242798.1 | 100 | 0.1.0.0.0.0.0.0.1.0.1.0.0.0.0.0.0.0.0.0.0.0.0.0 |
| KC242795.1 Zaire_EBOV_GAB-1996-1Mbie | 09/12/14 | 128 | KC242797.1 | 100 | 0.1.0.0.0.0.0.0.1.0.0.0.1.0.0.0.0.0.0.0.0.0.0.0 |
Abbreviations: ANI, average nucleotide identity; EBOV, Ebola virus; LINs, Life Identification Numbers; NCBI, National Center of Biotechnology Information.
a Each genome was analyzed in the order it was submitted to NCBI GenBank (or the date it was most recently revised). For each genome the following is shown in the table: date submitted to NCBI (or date the sequence was last revised), the order in which the genome was analyzed, the most similar genome out of all the genomes submitted to GenBank on an earlier date, the corresponding ANI, and the assigned LIN using the ANI cutoffs described in Figure 1 (instead of labeling each position with a subscript LIN, positions are simply separated by a period).
For phylogenetic reconstruction, the genomes were aligned using MAFFT [11, 12]. A maximum-likelihood (ML) tree was constructed using RAxML [13] with 20 ML search replicates and 1200 nonparametric bootstrap replicates under the GTRGAMMA model. Nonparametric bootstrap branch support values were mapped onto the best log-likelihood ML tree, and clades with less than 50% bootstrap support were collapsed into polytomies using TreeCollapseCL4 [14].
RESULTS
Sequential Calculation of Average Nucleotide Identity and Identification of Most Similar Genomes
All results of sequential ANI calculation are listed in Table 1. However, for the purpose of clarity, examples of individual results are described here in the order in which they were obtained: the ebolavirus genome sequence in our dataset with the earliest associated date (September 2002) had accession number AF522874.1. It was not compared with any other genome, because all other ebolavirus genome sequences were either submitted to GenBank or updated in GenBank after September 2002. The genome with accession number AY35458.1 was the second genome in our dataset (because the associated date, February 2004, was the second genome submitted to GenBank, or updated, in temporal order). Thus, this genome was compared only with genome AF522874.1, and the ANI between the 2 genomes was determined to be 69.44778%. The third genome submitted to GenBank, ie, the genome with accession number AY729654.1 (submitted in October 2005), was then compared to the first 2 genomes. Genome AY729654.1 was found to be more similar to genome AY35458.1 than to genome AF522874.1. Therefore, Table 1 lists genome AY729654.1 as most similar to genome AY35458.1 with the corresponding ANI of 70.22714%. This type of comparison was repeated 125 times, with the 128th ebolavirus genome, genome KC242795.1, being compared with the first 127 ebolavirus genomes, whereby genome KC242797.1 was identified to be the most similar genome to genome KC242795.1 with an ANI value of 100%, indicating identical sequences.
Assignment of Life Identification Numbers and Their Correlation With Membership in Ebolavirus Species and Their Association With Separate Ebola Virus Disease Outbreaks
Life Identification Numbers were assigned based on the sequentially calculated ANI values and the sequentially identified most similar genomes reported in Table 1. The same thresholds as those in reference [6] were used at each LIN position. The LIN assigned to the first genome has zeroes at all positions, but any other number or letter could have been used. Zero is simply the default symbol we use to start the LIN assignment, but the information provided by LINs is not represented by the symbol themselves but by the presence of identical symbols at the same position in different isolates. For example, the LIN assigned to the fourth isolate in Table 1 is identical to the third isolate up to position E, because the 2 isolates have genomes that are 94.95444% identical to each other, which is higher than 90% (the threshold corresponding to position E; also see Figure 1). However, they are different at position F because 94.95444% is lower then 95% (threshold corresponding to position F; also see Figure 1).
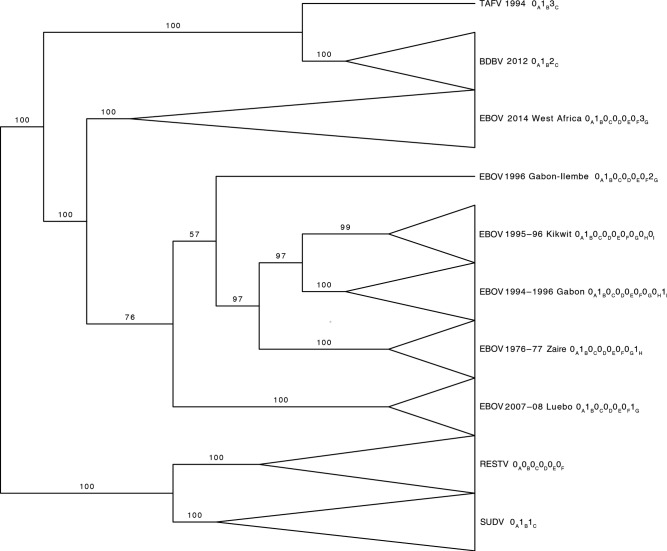
Table 1 and Figure 2 show that all ebolavirus isolates share the symbol 0 in position A (0A) because their genomes are all over 60% identical to each other. Isolates of RESTV are the only isolates identified by the symbol 0 in position B. Thus, Life Identification Number 0A0B is sufficient to distinguish RESTV isolates from all other ebolavirus isolates. Members of the other 4 species of Ebolavirus are instead uniquely identified by a specific symbol at LIN position C: all EBOV isolates can be uniquely identified as 0A1B0C; all SUDV isolates can be uniquely identified as 0A1B1C; all BDBV isolates can be uniquely identified as 0A1B2C; and all TAFV isolates can be uniquely identified as 0A1B3C. Therefore, the first 3 LIN positions are sufficient to identify ebolavirus isolates as members of each species of Ebolavirus (as classified by Kuhn et al [2]).
Figure 2.
Life Identification Numbers (LINs) assigned to ebolavirus isolates are informative of species membership and the outbreak during which they were isolated. A maximum likelihood tree was constructed and midpoint rooting was applied. Nonparametric bootstrap support values as a percentage of 1200 bootstrap replicates are shown above branches. Clades are labeled using virus abbreviations described in reference [2]. For Ebola virus (EBOV), the geographic location of outbreaks is also listed. Clades corresponding to individual species and to individual outbreaks are displayed as collapsed branches. Only those LIN positions that distinguish species and outbreaks from each other are shown. Abbreviations: BDBV, Bundibugyo virus; RESTV, Reston virus; SUDV, Sudan virus.
Although different outbreaks are caused by different variants of EBOV, Figure 2 shows that the isolates from these outbreaks share the same LIN up to position F: 0A1B0C0D0E0F. Isolates from the first outbreak of EBOV in Zaire in 1976 are then uniquely identified by LIN positions G and H (0G1H), and the isolates from the Luebo outbreak in 2007 by position G (1G). The isolates of the Gabon outbreak in 1996 share the same LIN with the isolates from the Kikwit outbreak up to position H (0A1B0C0D0E0F0G0H). This is consistent with a recent common ancestor of the viruses that caused these 2 outbreaks, which is confirmed by phylogenetic reconstruction (see the tree shown in Figure 2 and previous publications [8, 9]). Consequently, the isolates of these 2 latter outbreaks are only differentiated at the position with the next higher similarity threshold, ie, position I, at which the isolates from the Kikwit outbreak have a 0 (0I) and the isolates from the Gabon outbreak have a 1 (1I). A single isolate from Gabon from an outbreak in 1996 was previously shown to represent a separate genetic lineage compared with the other EBOV isolates from the earlier Gabon outbreak in 1994 [8], and this is clearly identified with a 2 in position G (2G).
Finally, isolates from the 2014 EBOV epidemic are different from all other EBOV isolates at position G. These isolates share a 3 at that position (3G) and then zeroes at all following positions up to position T. Only isolates G3687.1 and EM095 are exceptions, having a different symbol at positions I and M, respectively, possibly because of sequencing errors due to low median genome coverage: 20× for G3687.1 and 16x for EM0957.
Therefore, the provisional LINs assigned here immediately reveal that the 2014 EVD epidemic in West Africa is caused by a variant that is distinct from all other EBOV variants that have caused outbreaks of EVD in the Democratic Republic of Congo, Uganda, or Gabon in the past (because of the distinctive LIN at position G: 3G). In addition, the LINs immediately reveal that all isolates from the current EVD outbreak are extremely similar to each other (because of the conserved LIN at position L: 0L).
It is important to note that the LINs assigned here are provisional and are based only on comparisons among ebolavirus genomes. We do not propose to actually use these provisional LINs. Instead, we propose that permanent LINs should be assigned in the future by NCBI or an independent LIN database.
Correlation Between Life Identification Numbers and Phylogenetic and Epidemiological Relationships of Isolates From the Ebola Virus Disease Epidemic in West Africa
The 2014–2015 EVD epidemic probably started with a single zoonotic event in Guinea and spread to Sierra Leone and Liberia [9]. Gire et al [9] concluded from genome sequences of 78 isolates from Sierra Leone that the epidemic in Sierra Leone started with the introduction of 2 separate viral lineages from Guinea. In fact, among the first patients in Sierra Leone, 2 divergent groups of EBOV, called SL1 and SL2, could be distinguished, and the most recent ancestor of these 2 groups was inferred to have existed before the Sierra Leone outbreak started. Life Identification Number position U correlates with these 2 groups: all but 2 SL1 isolates are identified by LIN 1U, and all SL2 isolates are identified by LIN 0U (Table 1, Figure 3, and Supplementary Figure 1). The 2 SL1 isolates that do not have LIN 1U are G3687.1 and EM095, mentioned previously, because they have a much lower ANI compared with all other EBOV isolates from the 2014 epidemic and consequently different symbols at LIN positions I and M, respectively (possibly because of sequencing errors due to low coverage). Two of the 3 Guinea isolates share LIN 1U with the SL1 group, suggesting that these 2 isolates and SL1 have a recent common ancestor. The third Guinea isolate is the only sequenced isolate with LIN 2U.
Figure 3.
Maximum likelihood phylogeny of ebolavirus genomes from the 2014 Ebola virus disease outbreak in West Africa with identifying Life Identification Number (LIN) positions mapped to taxa. Branch labels are nonparametric bootstrap support values as a percentage of 1200 bootstrap replicates. Branches with less than 50% bootstrap support were collapsed into polytomies. Branches are not to scale. All 2014 ebolavirus genomes shared all LIN positions up to position T (indicated in gray below the tree and by gray brackets) except for isolates G3687.1 and EM095, which were different at positions I and M, respectively. Group SL1 isolates are green, SL2 isolates are red, SL3 isolates are purple, and Guinean isolates are blue. All isolates are of variant “Makona” [15] and labels are based on isolate names in reference [9]. The labels “Kissidougou” and “Gueckedou” are the location of isolation in Guinea based on reference [8].
Gire et al [9] further identified a third viral group in Sierra Leone named SL3, which is a subgroup of SL2, with the only difference between SL3 and SL2 isolates being a single mutation in position 10,218. SL3 corresponds to the large clade in the upper portion of the phylogenetic tree in Figure 3. Because only a single mutation distinguishes SL2 from SL3, it is not surprising that SL3 and SL2 do not correlate with a conserved difference at any LIN position. Because LINs are assigned based on percentage of overall DNA identity between genomes, the mutations that distinguish isolates within SL2 and SL3 have a larger effect on LINs than the single mutation distinguishing SL2 from SL3.
In addition, within SL2 and SL3, there are 7 small clades that have bootstrap values higher than 60 and that probably correspond to small individual transmission chains (clades labeled I–VII in Figure 3). Five of these clades (II, III, IV, V, and VII) contain isolates that have exactly the same LIN in all positions. In clade I, isolate G3829 has a LIN ending in 14W0X, whereas the other 6 isolates in the same clade end with LIN 7W0X. In this case, isolate G3829 mutated to a point that it became too different to be grouped with the other isolates at LIN position W, and the phylogenetic signal that groups isolate G3829 with the rest of the clade was lost.
Finally, clade VI is the only clade in which isolates have the same identical LIN (ending in 1V0W0X) as isolates outside of the clade. More importantly, this result is simply due to the fact that draft genomes of slightly different length were used in tree construction, which influenced the location of these isolates in the tree. However, these isolates are actually 100% identical to each other in the part of the draft genome sequence that they share and which was considered in LIN assignment.
DISCUSSION
In today's interconnected world, emerging infectious diseases can spread globally within weeks, requiring international coordination in disease control. Therefore, it is important that communication about pathogens is not hindered by confusion about their identity because different names for the same pathogen, or the same name for different pathogens, are used by different researchers in different countries. In this study, we have shown that unique identifiers, such as LINs, can be assigned to individual ebolavirus isolates based on a simple measure of ANI to address this problem. The assigned LINs are not only informative of the species and the outbreak for which they are associated, but they even reflect some individual transmission chains.
Average nucleotide identity was originally developed to replace the experimental measurement of DNA-DNA hybridization (DDH) to determine whether 2 bacteria belong to the same species [7, 16, 17]. A 95% ANI was determined to correspond to approximately 70% DDH [18], which had been chosen years earlier as the minimum similarity between 2 bacteria to belong to the same species [19].
In viral taxonomy, a percentage of pairwise sequence identity has been used to demarcate taxa within viral families using computational approaches such as PASC [20] or DEmARC [21], both of which have been applied to the family of Filoviridae [22, 23]. Going 1 step further, we previously proposed that ANI could be used beyond assigning viruses to traditional taxa, such as species or genera [6]. We showed that ANI could be used to assign unique genome similarity-based codes to individual viral isolates, whereby we found that codes (which we now call LINs) assigned to individual isolates of FMDV collected during the 2001 United Kingdom FMDV outbreak largely reflected epidemiological relationships. This result was obtained by comparing assigned LINs with the results of an earlier molecular epidemiological investigation of that same outbreak [24].
In this study, we showed that ANI can be used to provide LINs for individual ebolavirus isolates and that (1) assigned LINs are informative of phylogenetic relationships at the species level, (2) LINs clearly correlate with separate EVD outbreaks, and (3) in many cases, LINs even reflect phylogenetic clades that correspond to likely individual transmission chains during the epidemic in West Africa in 2014. Thus, LINs are exceptionally informative of deep phylogenetic relationships among ebolavirus isolates.
However, there are challenges when comparing LINs with very deep phylogenetic relationships when isolates are associated with recent epidemics. First, neither LINs nor phylogeny may represent true evolutionary and epidemiological relationships, because not enough mutations were accumulated to create strongly supported phylogenetic clades or clearly distinguishable LIN classes. Second, some clades in the Maximum Likelihood and Bayesian trees provided in reference [9] for the current EVD epidemic have low statistical support and, not surprisingly, do not correspond to conserved LIN positions. However, even a single-nucleotide polymorphism (SNP) can potentially distinguish transmission chains from each other. In fact, a single SNP at position 10,218 was used to divide Sierra Leone viruses into groups SL2 and SL3 [9]. As a result, one question remains: how important would it be for isolate identifiers to reflect the distinction of 2 groups based on a single SNP? In the case of SL2 and SL3, viruses were sometimes isolated from the same patients [9]. Therefore, they do not represent separate transmission chains. Finally, we can assume that during additional transmission events, these viruses will accumulate several more mutations so that viruses after additional transmission events will become different enough at the whole genome level to be identified by different LINs.
However, thresholds used at the different LIN positions could be further refined to improve correlation with phylogenetic relationships. For example, the isolate with accession number KM233102 was assigned LIN 14W, whereas the other isolates belonging to the same clade were assigned LIN 7W. KM233102 was assigned this LIN because the calculated ANI compared with the other isolates was calculated to be 99.9944%, which is less than the 99.999% threshold corresponding to LIN position W. Therefore, if an additional LIN position were to be introduced to the left of position W with a 99.994% threshold, KM233102 would still be in a group with the other isolates at position W and separate from them at the next position with threshold 99.999%. However, we believe that only after assigning LINs to many more viruses for many more disease outbreaks and comparing LINs with phylogeny each time should LIN position thresholds be optimized to best reflect phylogeny for assignment of permanent LINs. Finally, we emphasize that we do not propose to replace current viral taxonomy with LINs. Current viral taxonomy is very useful and highly informative in many aspects, and it is tailored to the evolutionary mechanisms at the base of viral diversity found in different viral families. Therefore, LINs should complement but not replace current viral taxonomy.
CONCLUSIONS
In summary, we have shown here that, in the case of ebolavirus, LINs in their current implementation are highly informative of similarity and relationships from the species level all the way to some individual transmission chains. Taken together with our previous results [6], these new results suggest that after further optimization, LINs have the potential to provide unique and stable identifiers for individual viral and bacterial isolates and eukaryotic organisms that reflect deep phylogenetic relationships. As such, we propose that LINs should be assigned in the future to every newly sequenced genome either by NCBI (or a database specifically developed for LIN assignment) to provide the healthcare and research community with a typing scheme of unprecedented precision and with identifiers for unambiguous communication about pathogen isolates and other organisms as soon as genomes sequences become available.
Supplementary Material
Supplementary material is available online at Open Forum Infectious Diseases (http://OpenForumInfectiousDiseases.oxfordjournals.org/).
Acknowledgments
We thank Dr. Caroline L. Monteil for critically reading the manuscript and for constructive criticism, all of which improved the final draft.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by funding from the National Science Foundation (grant IOS-1354215 to B. A. V. and grant DBI-1062472 to L. S. H.). H. M. was funded through an assistantship from the Egyptian government.
Potential conflicts of interest. Virginia Tech submitted a patent application (Serial No. 14/199,441) to protect the concept of genome similarity-based codes. B. A. V. and L. S. H. report that they are founders of This Genomic Life Inc. (Blacksburg, Virginia), which plans to license the above patent from Virginia Tech.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.International Committee on Taxonomy of Viruses. The International Code of Virus Classification and Nomenclature February 2013. Available at: http://ictvonline.org/codeOfVirusClassification_2012.asp. Accessed 22 September 2014.
- 2.Kuhn J, Becker S, Ebihara H, et al. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol. 2010;155:2083–103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn J, Andersen K, Bào Y, et al. Filovirus RefSeq Entries: Evaluation and selection of filovirus type variants, type sequences, and names. Viruses. 2014;6:3663–82. doi: 10.3390/v6093663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhn J, Bao Y, Bavari S, et al. Virus nomenclature below the species level: a standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch Virol. 2013;158:301–11. doi: 10.1007/s00705-012-1454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Dijk EL, Auger H, Jaszczyszyn Y, et al. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–26. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Marakeby H, Badr E, Torkey H, et al. A system to automatically classify and name any individual genome-sequenced organism independently of current biological classification and nomenclature. PLoS One. 2014;9:e89142. doi: 10.1371/journal.pone.0089142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 2005;102:2567–72. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baize S, Pannetier D, Oestereich L, et al. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med. 2014;371:1418–25. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 9.Gire SK, Goba A, Andersen KG, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–72. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–31. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katoh K, Misawa L, Kuma K, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodcroft E. TreeCollapserCL 4. Available at: http://emmahodcroft.com/TreeCollapseCL.html. Accessed 12 September 2014.
- 15.Kuhn J, Andersen K, Baize S, et al. Nomenclature- and database-compatible names for the two Ebola virus variants that emerged in Guinea and the Democratic Republic of the Congo in 2014. Viruses. 2014;6:4760–99. doi: 10.3390/v6114760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci. 2006;361:1929–40. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–64. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goris J, Konstantinidis KT, Klappenbach JA, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 19.Wayne LG, Brenner DJ, Colwell RR, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–4. [Google Scholar]
- 20.Bao Y, Kapustin Y, Tatusova T. Virus classification by pairwise sequence comparison (PASC) Encyclopedia of Virology (BWJ Mahy and MHV Van Regenmortel, Editors) 2008;5:342–8. [Google Scholar]
- 21.Lauber C, Gorbalenya AE. Partitioning the genetic diversity of a virus family: approach and evaluation through a case study of picornaviruses. J Virol. 2012;86:3890–904. doi: 10.1128/JVI.07173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao Y, Chetvernin V, Tatusova T. PAirwise sequence comparison (PASC) and its application in the classification of filoviruses. Viruses. 2012;4:1318–27. doi: 10.3390/v4081318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauber C, Gorbalenya AE. Genetics-based classification of filoviruses calls for expanded sampling of genomic sequences. Viruses. 2012;4:1425–37. doi: 10.3390/v4091425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cottam EM, Haydon DT, Paton DJ, et al. Molecular epidemiology of the foot-and-mouth disease virus outbreak in the United Kingdom in 2001. J Virol. 2006;80:11274–82. doi: 10.1128/JVI.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.