Abstract
Background
While formal reporting, surveillance, and response structures remain essential to protecting public health, a new generation of freely accessible, online, and real-time informatics tools for disease tracking are expanding the ability to raise earlier public awareness of emerging disease threats. The rationale for this study is to test the hypothesis that the HealthMap informatics tools can complement epidemiological data captured by traditional surveillance monitoring systems for meningitis due to Neisseria meningitides (N. meningitides) by highlighting severe transmissible disease activity and outbreaks in the United States.
Methods
Annual analyses of N. meningitides disease alerts captured by HealthMap were compared to epidemiological data captured by the Centers for Disease Control’s Active Bacterial Core surveillance (ABCs) for N. meningitides. Morbidity and mortality case reports were measured annually from 2010 to 2013 (HealthMap) and 2005 to 2012 (ABCs).
Findings
HealthMap N. meningitides monitoring captured 80-90% of alerts as diagnosed N. meningitides, 5-20% of alerts as suspected cases, and 5-10% of alerts as related news articles. HealthMap disease alert activity for emerging disease threats related to N. meningitides were in agreement with patterns identified historically using traditional surveillance systems. HealthMap’s strength lies in its ability to provide a cumulative “snapshot” of weak signals that allows for rapid dissemination of knowledge and earlier public awareness of potential outbreak status while formal testing and confirmation for specific serotypes is ongoing by public health authorities.
Conclusions
The underreporting of disease cases in internet-based data streaming makes inadequate any comparison to epidemiological trends illustrated by the more comprehensive ABCs network published by the Centers for Disease Control. However, the expected delays in compiling confirmatory reports by traditional surveillance systems (at the time of writing, ABCs data for 2013 is listed as being provisional) emphasize the helpfulness of real-time internet-based data streaming to quickly fill gaps including the visualization of modes of disease transmission in outbreaks for better resource and action planning. HealthMap can also contribute as an internet-based monitoring system to provide real-time channel for patients to report intervention-related failures.
Introduction
Neisseria meningitidis (N. meningitidis) is a human pathogen that spreads between individuals through direct contact with respiratory secretions [1]. The bacteria can cause septicemia, pneumonia, and meningitis. N. meningitidis strains can be categorized, based on the composition of the bacteria’s capsule, into 13 different serogroups of which five are responsible for the great majority of invasive disease (serogroups A, B, C, W, and Y) [2]. Up to 14% of meningococcal infected individuals die [2,3] and among the survivors, up to 20% have serious sequelae including hearing loss, limb amputation, and neurologic disability [4]. Due to the disease’s rapid progression (< 24–48 hours), serious sequelae, and potential for person-to-person spread, a single case of meningococcal disease will trigger immediate and increasing media attention during which public health officials are working to avoid panic, to identify whether there is an outbreak, and to generate an effective response to treat disease and prevent further transmission.
Recently, two United States (US) universities have experienced serogroup B strain outbreaks that were widely reported by both traditional media and internet-based news reporting. The widespread adoption of more sophisticated information technology has increased public awareness of potentially serious diseases as seen during the emergence and global spread of the 2009 pandemic influenza A/H1N1 virus [5]. While formal reporting, surveillance, and response structures remain essential to protecting public health, a new generation of online, freely accessible, and real-time disease surveillance-monitoring tools have become available. These tools complement and expand existing public health professionals’ capacities to detect weak signals and to raise earlier concerns regarding emerging diseases [6–8]. In this report, we used the HealthMap informatics tool [9] to investigate the burden of “meningitis” that can be captured in the US. We aim to test the hypothesis that HealthMap complements data captured by traditional surveillance and provides insights related to the mode of disease transmission linked to emerging outbreaks.
Methods
HealthMap
HealthMap (http://www.healthmap.org/) is a multistream real-time surveillance platform that continually aggregates reports on new and ongoing infectious disease outbreaks. The system performs extraction, categorization, filtration, and integration of these reports, facilitating knowledge management and earlier awareness of potential outbreak status [10].
HealthMap system capabilities
The HealthMap automated text processing system proceeds in four stages: (1) Acquisition (2) Characterization (3) Filtering, and (4) Clustering. Trained public health analysts are employed to review content and correct misclassifications. Finally, the correctly classified content is used to improve the automated algorithms through a training feedback loop.
The first step is data acquisition. The system collects data via five main channels: news aggregators (such as Google News, Yahoo News, Moreover); specific Real-time Simple Syndication (RSS) feeds including WHO (World Health Organization), OIE (Office International des Epizooties/World Organisation for Animal Health), and others; e-mail subscriptions (GPHIN [Global Public Health Intelligence Network (developed by Health Camada)], ProMED, etc); custom-parsed HTML (HyperText Markup Language) scraping or CSV (Comma-separated Values)/XML (Extensible Markup Language) feeds; and user submitted reports. The framework allows one to add a new feed through a custom PHP (Personal Home Page Tools) Hypertext Preprocessor class where the feed-specific code fetches the content, parses it, and then extracts a set of common fields.
In the second step, the common outputs of the feed framework then pass to the characterization engine where we extract disease and location entities, flag the report as not-disease-related by rule (if applicable), and identify the source publication. This module loads large dictionaries of terms into memory and then matches them against the input text using a rapid matching algorithm. Algorithm performance is linear on the size of the input text and memory consumption is linear on the size of the dictionary. Under some conditions, the characterization module may make calls back to the acquisition module to request additional input text from the source. The dictionaries currently support identification of about 250 diseases. HealthMap has an internal location database of over 25,000 locations. The characterization engine generally assigns countries or first level administrative units; analysts generally change these to cities, hospitals, or schools, if that level of specificity is warranted.
Once disease and location are assigned, the document then passes to a document-level Bayesian filtering module (third step). The filter breaks the document into unigrams, digrams, trigrams (and tetragrams in some languages), and then computes a likelihood score based on training data from tens of thousands of previously recorded documents. It then assigns the document to one of five categories based on relevance; documents falling in the two most relevant categories, Breaking News and Warning, are posted to the map, whereas documents in the Old News, Context, or Not Disease Related categories are hidden from the map. These alerts are retained in the database for use in future research.
The last step in the automated pipeline is document clustering to group duplicate reports together. The clustering module compares the given document to each previously collected document over a fixed time window, and computes a similarity score based on the texts and classification outputs. If the maximum score is above a set threshold, the new document is assigned as a “child” of the previous document, and hidden from the map. The similarity calculation is based on six factors: (1) character-level headline comparison; (2) word-level headline comparison (fraction of words in common, with inverse term frequency taken into account); (3) disease and place comparison from the output of the named-entity extraction engine; (4) country-level comparison (i.e., if the two reports have different locations within the same country) boost similarity score somewhat; (5) timestamp (reports closer in time are more likely to be similar); and (6) filter category comparison based on the output of the Bayesian filtering module.
Following automated processing, HealthMap alerts are posted directly to the public map, partner pages, and RSS feeds. However, at the same time, a team of trained analysts will review all incoming data and correct misclassifications and assign additional higher resolution metadata classifications. For these curation tasks, a feature-rich analyst dashboard allows users to search, review and annotate the alerts. For example, analysts can highlight text in a report and assign a named entity category such as executing a geocoding query with the highlighted text and bringing up a map for use in assigning precise location to a report. The analyst corrections feed back into the machine learning algorithms and improve the automated content analysis. Case counts data is entered and additional tags are assigned when relevant. The current list of tags includes such diverse topics as schools, healthcare facilities, sports-associated, recall (of a product), and cruise.
HealthMap data sources
HealthMap uses a large variety of inputs to detect outbreak news displayed on the main web page, to produce password protected sites for partners with sensitive data, and to do research on novel disease surveillance methods.
The standard data inputs for the main webpage are: news aggregators (such as Google News, Yahoo News, Baidu), specific RSS feeds (WHO, OIE, FAO), constructed datasets of language-specific RSS feeds, email subscriptions (GPHIN, ProMED, etc), custom-parsed HTML scraping or CSV/XML feeds, and user submitted reports. HealthMap queries news aggregators at regular intervals with a list of queries designed to return outbreak news stories with excellent sensitivity while avoiding poor specificity. Post-collection processing ensures non-outbreak related articles are not displayed on the map. Specific RSS feeds and e-mail subscriptions allow the system to receive high quality data from known providers. Custom-parsed scraping programs allow the system to retrieve data that is otherwise difficult to access.
User-submitted reports represent a diverse group of resources. Users may submit URLs or eyewitness descriptions of outbreaks to HealthMap via the website, e-mail, telephone, text message or the Outbreaks Near Me mobile app for Android and iPhone. These reports are processed through various means depending on the method of delivery.
Social media sources are also included in HealthMap. Twitter and Facebook data are collected and processed by the parser. The short nature of tweets does present challenges to automated categorization, but enhanced dictionaries have helped to address that issue.
HealthMap subject matter domains
Although HealthMap was developed specifically to provide a comprehensive real time display of infectious disease outbreak knowledge, the system has proven flexible and robust enough to support many other projects. HealthMap has been used in diverse government-sponsored and academic research-oriented projects including monitoring violence against civilians in the Syrian conflict, and enhancing situation awareness for response efforts for a natural disaster. The core architecture and processing capabilities are language and subject matter independent. HealthMap is constantly expanding its language capabilities. The core HealthMap languages have extensive language support: English, Spanish, Portuguese, French, Russian, Chinese, and Arabic. More recently added language capabilities are German, Korean, Japanese, Vietnamese, Malay and Indonesian. HealthMap also processes alerts in Italian and Thai for specific projects. HealthMap aims to continue to adding language capabilities to further expand its global coverage of hyper-local outbreaks. The relatively straightforward process for adding languages demonstrate the flexibility of the underlying automated parser.
Study design and population
HealthMap data capture and analyses
HealthMap became operational for N. meningitidis disease capture in 2010 and was used for this study to detect cases of meningococcal disease and potential outbreaks in the US from 2010 to 2013. Heat maps were generated using the following search conditions: meningitis-Neisseria, United States, new & ongoing outbreaks, and yearly intervals. Alerts reflect online information from news, expert-curated discussions, and validated official reports (e.g., World Health Organization, ProMED Mail, and Google News feeds that were active during this time interval). Alerts were individually crosschecked manually to ensure they reflected confirmed cases of meningococcal disease with citation of the appropriate state health departments or institutions. Accuracy of HealthMap capture was calculated based on downloading all the citations and individually counting the alerts to verify those capturing diagnoses, suspected cases, or general news.
An “alert” in HealthMap is defined as any unique data source, typically a unique URL link. If the data source (i.e. Associated Press or Reuters) writes a story and 30 newspapers pick it up and write slightly different versions, then the system receives all of those URLs as distinct alerts, but the artificial intelligence of the system will group them together as having identical information despite existing in different locations in the Internet. The term alert is used sometimes interchangeably as the presence of a case, but occasionally it could also be a warning.
Centers for Disease Control data capture and analyses
Traditional surveillance data were obtained from the Centers for Disease Control (CDC) Active Bacterial Core surveillance (ABCs) Emerging Infections Program Network [11] for N. meningitidis from 2005 (to include assessment of the impact of vaccine introduction on disease incidence) to 2012 (most recent finalized report available online). Non-B serogroup and serogroup B case rates per 100,000 population reflects ABCs figures in ten US states representing 39 to 43 million inhabitants over eight years. Total cases in the US reflect CDC estimates based on applying ABC case rate to US population census.
Results
The HealthMap meningitis system—increasing visibility of meningococcal cases
Geographic distribution of HealthMap alerts in the US
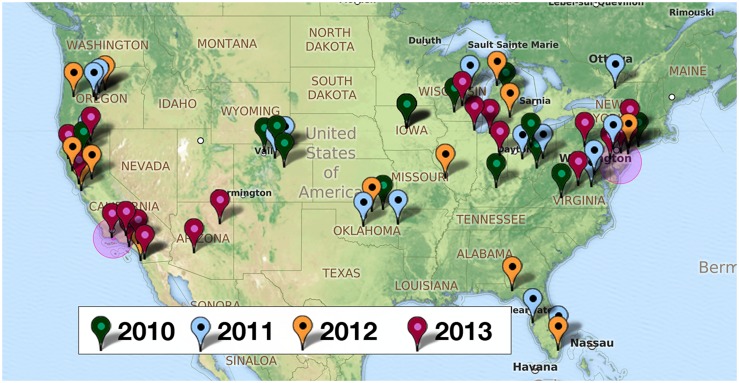
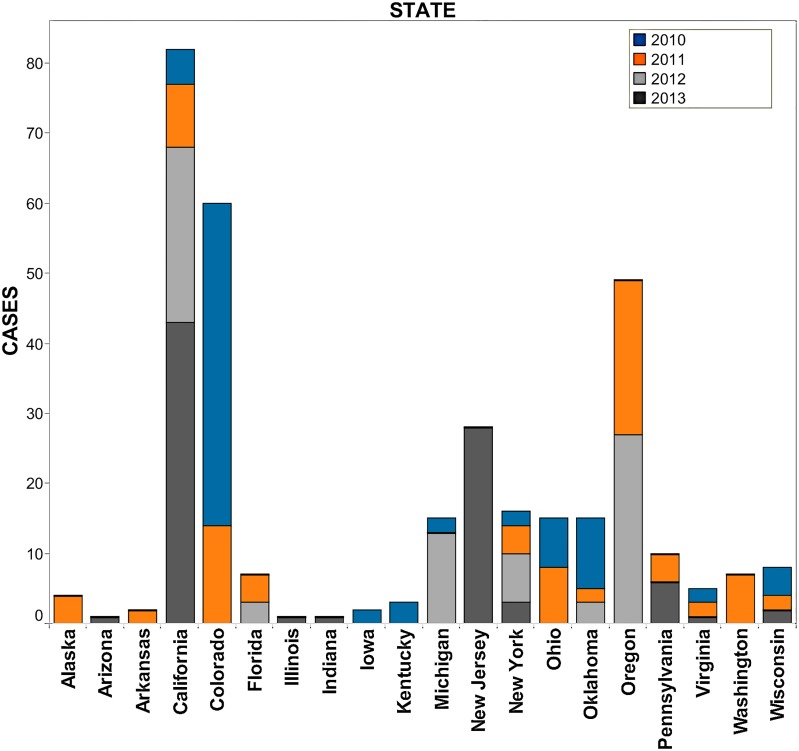
Fig 1 illustrates the distribution of HealthMap meningococcal alerts from 2010–2013 that are color-coded by year. The two pink shaded circles represent country levels alerts indicated by HealthMap for 2013 due to the serogroup B outbreaks at Princeton University and University of California Santa Barbara. Compared to a retrospective surveillance study reviewing outbreaks from July 1994 to June 2002 [12], there is a similar geographic distribution of N. meningitidis disease activity in HealthMap. The distribution of cases captured for the school setting (57.8%) and the community setting (42.2%) by HealthMap alerts were similar to that reported previously in the setting of N. meningitidis outbreaks (44.9% and 36.2%, respectively) [12]. Fig 2 is a breakdown of state-specific HealthMap case alerts for N. meningitidis annually from beginning of 2010 to end of 2013 (2010 = blue, 2011 = orange, 2012 = light grey, and 2013 = dark grey). From this breakdown, it shows that 20 states represented the case alert activity captured by HealthMap during this four-year period. The top-two states, by year, comprising the majority of internet-based case alerts were Oklahoma/Colorado (12.0%/55.4%) in 2010, Colorado/Oregon (16.7%/26.2%) in 2011, Oregon/California (34.6%/32.1%) in 2012, and California/New Jersey (50.0%/32.6%) in 2013.
Fig 1. HealthMap data for Neisseria meningitidis alerts from 2010 to 2013.
Markers indicate state, province, or local alerts. The two pink-shaded circles represent country-level alerts for 2013 due to the serogroup B outbreaks at University of California Santa Barbara and Princeton University. Image generated using ZeeMaps. Reprinted under a CC BY license with permission from ZeeMaps.
Fig 2. Number of HealthMap meningococcal case alerts in twenty US states reporting disease activity from 2010–2013.
The majority of internet-based case alerts derived from meningococcal disease activity in Oklahoma/Colorado (12.0% and 55.4% in 2010), Colorado/Oregon (16.7%/26.2% in 2011), Oregon/California (34.6% and 32.1% in 2012), and California/New Jersey (50.0% and 32.6% in 2013). Bars indicate actual number of alerts. Blue color indicates case alerts in 2010, orange color indicates case alerts in 2011, light grey color indicates case alerts in 2012, and dark grey color indicates case alerts in 2013.
Accuracy of HealthMap capture of meningococcal disease
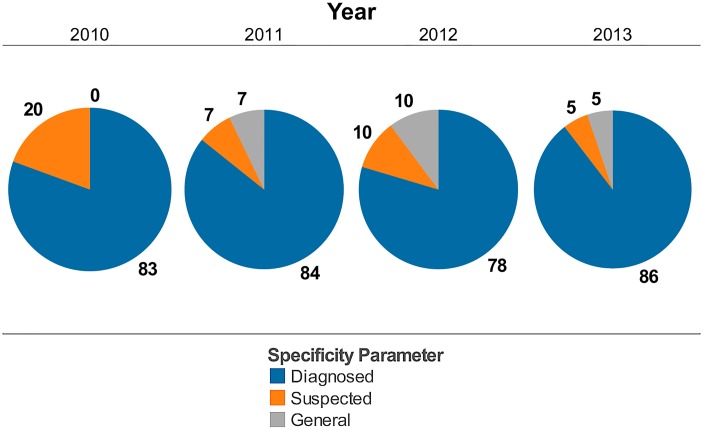
Fig 3 includes information on the specificity of the disease cases captured by the HealthMap automated search algorithm. On average, 83.3% of reported cases identified by HealthMap represented diagnosed cases (blue color), 10.5% represented suspected cases (orange color), and 5.6% were related news but not actual cases of N. meningitidis (light grey color). Table 1 indicates the total cases captured through internet data sources utilized by HealthMap for 2010–2013 (30, 28, 22, and 40 cases, respectively) and illustrates that serogroup specific information for these cases was not available in 53.3%, 82.1%, 68.1%, and 52.5% of alerts.
Fig 3. Specificity of HealthMap case alerts for actual Neisseria meningitidis disease activity from 2010–2013.
The specificity, on average, for HealthMap alerts during 2010 to 2013 were as follows: 83.3% were diagnosed cases, 10.5% were suspected cases, and 5.6% were related news but not actual cases of Neisseria meningitidis. Pie graphs illustrate percentages for alerts per year with numerical values indicating actual number of alerts. Blue color indicates diagnosed Neisseria meningitidis, orange color indicates suspected Neisseria meningitidis, and light grey color indicates general information associated with meningococcal disease (related news articles but not actual cases).
Table 1. Meningococcal disease cases identified by HealthMap alerts.
| Year | School (# of cases) | Community (# of cases) | Alive / Dead | Distribution |
|---|---|---|---|---|
| 2010 | Red Bluff, CALIFORNIA (1) | Red Bluff/Tehama County, | 16/14 | Total = 30 cases |
| 2010 | Denver/Fort Collins, COLORADO | CALIFORNIA (1B+1) | B = 23% | |
| (3C+1) | Larimer County, COLORADO | C = 23% | ||
| 2010 | Clare County, MICHIGAN (1) | (4C+1B) | Y = 0% | |
| 2010 | Long Island, NEW YORK (1) | Bullitt County, KENTUCKY (1) | ? = 54% | |
| 2010 | Athens/Franklin County, OHIO (4B) | Franklin County, OHIO (1B) | ||
| 2010 | Oologah, OKLAHOMA (7) | |||
| 2010 | Roanoke College, VIRGINIA (1) | |||
| 2010 | Madison, WISCONSIN (2) | |||
| 2011 | Shasta County, CALIFORNIA (1) | Craig, ALASKA (1) | 19/9 | Total = 28 cases |
| 2011 | Boulder, COLORADO (1) | Fort Smith, ARKANSAS (1) | B = 4% | |
| 2011 | Washington, DISTRICT OF | Los Angeles County, | C = 7% | |
| COLUMBIA (1) | CALIFORNIA (7) | Y = 7% | ||
| 2011 | Canton, NEW YORK (1) | Larimer County, COLORADO | ? = 82% | |
| 2011 | Columbus, OHIO (2) | (1Y) | ||
| 2011 | Norman, OKLAHOMA (1) | Hillsborough/Broward | ||
| 2011 | Prinville, OREGON (1C) | County, FLORIDA (2) | ||
| 2011 | Bethlehem, PENNSYLVANIA (1) | Morgan County, OHIO (1) | ||
| 2011 | Caroline County, VIRGINIA (1) | Bend/Prinville/Deschutes | ||
| 2011 | Greenbay, WISCONSIN (1) | County, OREGON | ||
| (1C+1B+1Y+1) | ||||
| 2012 | Sacramento/San Francisco/ | Clearlake/Ukiah, CALIFORNIA | 14/8 | Total = 22 cases |
| San Diego, CALIFORNIA (3+1B) | (2) | B = 5% | ||
| 2012 | Tallahassee/Miami, FLORIDA (2) | New York, NEW YORK (4C) | C = 27% | |
| 2012 | East Lansing/Traverse City, | Pawnee County, OKLAHOMA | Y = 0% | |
| MICHIGAN (2) | (1) | ? = 68% | ||
| 2012 | St. Louis, MISSOURI (1) | Prinville/Eugene, OREGON | ||
| 2012 | Prinville/Eugene, OREGON | (1C+2) | ||
| (1C+2) | ||||
| 2013 | Maricopa County, ARIZONA (1B) | Navajo/Maricopa County | 34/6 | Total = 40 cases |
| 2013 | Petaluma/San Diego/Shasta | ARIZONA (2B) | B = 43% | |
| County/Santa Barbara, | Huntington Beach/San | C = 5% | ||
| CALIFORNIA (3+5B) | Diego/West Hollywood/ | Y = 0% | ||
| 2013 | Chicago, ILLINOIS (1C) | Willits, CALIFORNIA | ? = 52% | |
| 2013 | South Bend/Muncie, INDIANA (2) | (6+1C) | ||
| 2013 | Princeton/West Long Branch, | New York, NEW YORK (4) | ||
| NEW JERSEY (8B+1) | ||||
| 2013 | Wester Chester/Reading/ | |||
| University Park, | ||||
| PENNSYLVANIA (4) | ||||
| 2013 | Charlottesville, VIRGINIA (1B) | |||
| 2013 | Madison, WISCONSIN (1) |
Example of community-identified and reported meningitis outbreaks in the United States in a four-year period. Based on the source of information and the stage of the outbreak, HealthMap is able to provide geographical and numerical information on cases and also displays information related to disease-causing serotype (when available).
Epidemiology of vaccine-preventable serogroup ACWY versus serogroup B
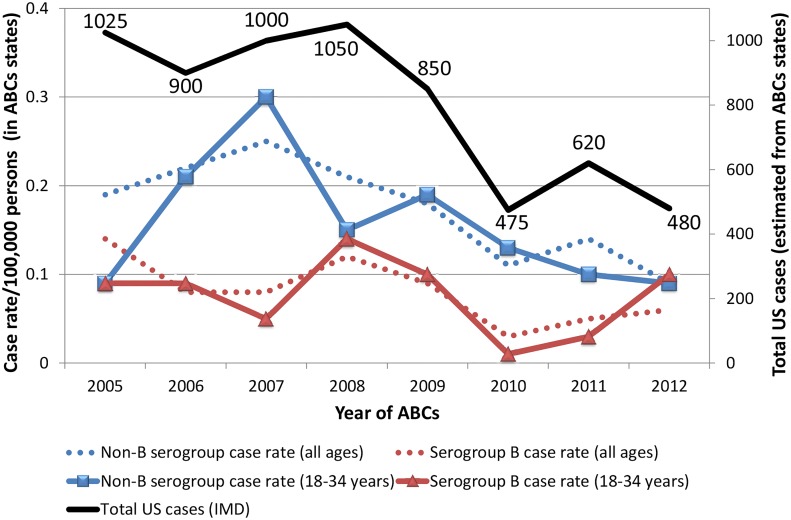
Fig 4 illustrates the trend in meningococcal cases from 2005 to 2012 using ABCs surveillance data obtained from the CDC surveillance for N. meningitidis [11]. ABCs surveillance reflects ten US states representing 39 to 43 million inhabitants over eight years (Fig 5). There is a decline of total cases (Fig 4, solid black line) in the ABCs states from 1,025 cases in 2005 to what appears to be a plateau ranging from 475 to 620 cases during 2010 to 2012. The dotted blue line indicates the decreasing trend in case rates per 100,000 persons (all age groups) for non-B serogroups from 2007 to 2012. The dotted red line indicates a more variable trend in case rates per 100,000 persons (all age groups) for the serogroup B that appears to show an increasing trend since 2010. This variability in case rate for serogroup B is expected since the MenACWY vaccine introduced in 2005 would not be protective against serogroup B. No deviation from these trends occur when analyzing specifically case rates in 18–34 year old individuals (university age group) as indicated for non-B serogroups (solid blue line) and serogroup B (solid red line). Case rates from serogroup B meningococcus in children < 1 year of age were not included but are oscillating at a much higher case rate than all other age groups with values between 1.0 to 2.9 cases per 100,000 population compared to non-B serogroups that range from 0.6 to 1.2 cases per 100,000 population.
Fig 4. Invasive meningococcal disease (IMD) reported over the past eight years by Active Bacterial Core Surveillance.
Data plotted from Active Bacterial Core Surveillance (ABCs) Emerging Infections Program Network (states highlighted in white). There is a decline of total cases (solid black line) in the ABCs states from 1,025 cases in 2005 to what appears to be a plateau ranging from 475 to 620 cases during 2010 to 2012. The dotted blue line indicates the decreasing trend in case rates per 100,000 persons (all age groups) for non-B serogroups from 2007 to 2012. The dotted red line indicates a more variable trend in case rates per 100,000 persons (all age groups) for the serogroup B that appears to show an increasing trend since 2010. This variability in case rate for serogroup B is expected since the MenACWY vaccine introduced in 2005 would not be protective against serogroup B. No deviation from these trends occur when analyzing specifically case rates in 18–34 year old individuals (university age group) as indicated for non-B serogroups (solid blue line) and serogroup B (solid red line). Case rates for serogroup B meningococcus in children < 1 year of age are not shown but are oscillating at a much higher case rate than all other age groups with values between 1.0 to 2.9 cases per 100,000 population compared to non-B serogroups that range from 0.6 to 1.2 cases per 100,000 population.
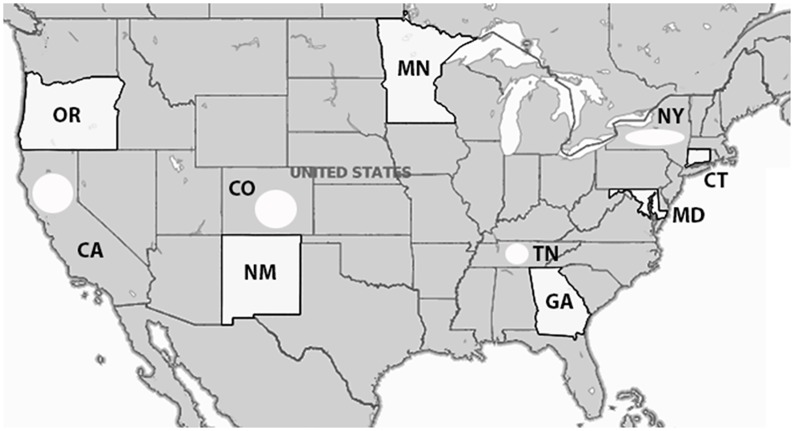
Fig 5. Population under suveillance by ABC for Neisseria meningitidis (adapted from CDC website).
Non-B serogroup and serogroup B case rates reflects ABCs figures in ten US states (white shading of entire state indicates that total state population under surveillance while white shaded circle indicates that population of certain counties within the state are under surveillance). Numbers of inhabitants being followed per state are as follows: CT = 3,590,347; GA = 9,919,945; MD = 5,884,563; MN = 5,379,139; NM = 2,085,538; OR = 3,899,353; TN = 3,872,994 (20 urban counties); CA = 3,460,180 (3 county Bay area); CO = 2,532,982 (5 county Denver area); NY = 2,178,020 (7 county Rochester area and 8 county Albany area). Total population under surveillance by ABCs is 42,803,061.
Detecting HealthMap signals across national borders for emerging N. meningitidis threats
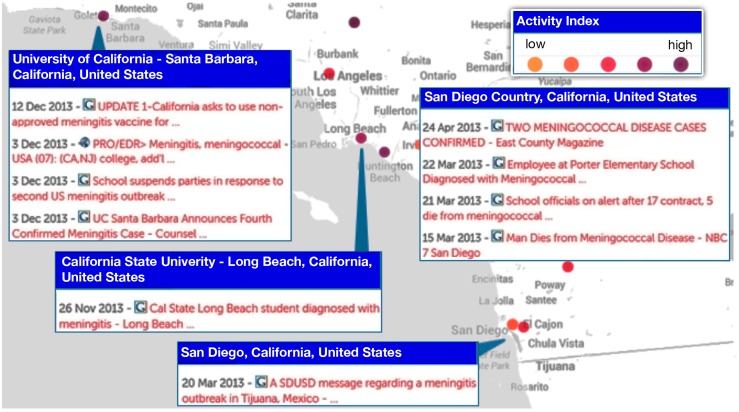
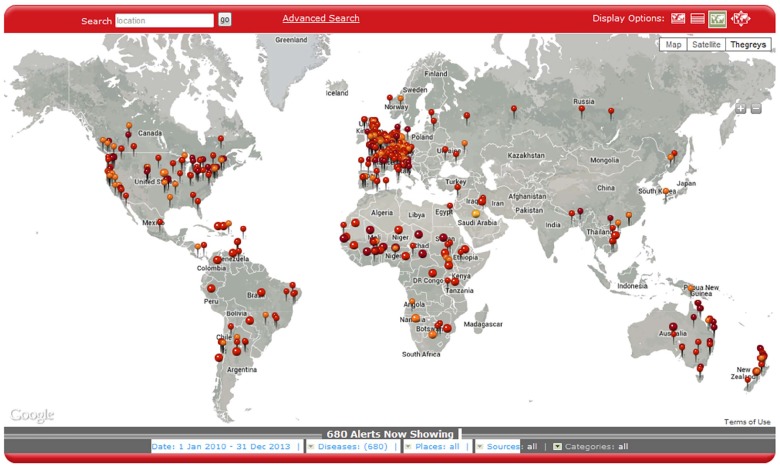
Fig 6 is an illustration of a close-up view from HealthMap illustrating the multiple N. meningitidis alerts for Southern California for 2013 including one in San Diego referring to a meningitis outbreak in Tijuana, Mexico [13]. What becomes readily apparent from the image is the proximity of Tijuana to the southern border of the United States. In the first three months of 2013, there had been 17 cases (including six deaths) in Tijuana [13]. If the Tijuana outbreak cases were counted as US cases, this would increase the total number of cases for 2013 from 40 to 57 patients (Table 1). In Fig 7, HealthMap is illustrating N. meningitidis disease alerts globally and illustrates N. meningitidis disease activity near the northern border of the United States with Canada (involving cities in British Columbia and Ontario). Of the eight cases captured by HealthMap in Canada between 2010 to 2013, the bordering cities reported one case in a student in Western Ontario (2010), one death in a student in Hamilton (2010), and one death in a student in Vancouver Island (2012).
Fig 6. Local view of HealthMap data for Neisseria meningitidis alerts from 2010 to 2013.
Multiple alerts in Southern California including one in San Diego refering to a meningitis outbreak across the border in Tijuana, Mexico. Marker color (light orange to darker purple) reflects increasing significance assigned by HealthMap users or news volume associated with the alert. Each marker may represent more than one Neisseria meningitidis alert (as seen in the break-out box). What becomes readily apparent from the image is the proximity of Tijuana to the southern border of the United States. In the first three months of 2013, there had been 17 cases (including six deaths) in Tijuana. Individuals today are more mobile than before which underscores the relevance of disease activity awareness in cities bordering countries.
Fig 7. Global view of HealthMap data for Neisseria meningitidis alerts from 2010 to 2013.
Multiple alerts globally including Neisseria meningococcus activity near the northern border of the US with Canada (cities in British Columbia and Ontario). Marker color (light orange to darker red) indicates increasing newsworthiness. Smaller marker size reflects local alert for city, state, or province while larger marker size reflects a country-level alert. Of the eight cases captured by HealthMap in Canada between 2010 to 2013, the bordering cities reported one case in a student in Western Ontario (2010), one death in a student in Hamilton (2010), and one death in a student in Vancouver Island (2012). HealthMap disease activity across borders could contribute to understanding disease transmission and subsequent outbreaks.
“Connecting the dots” to disease transmission using HealthMap
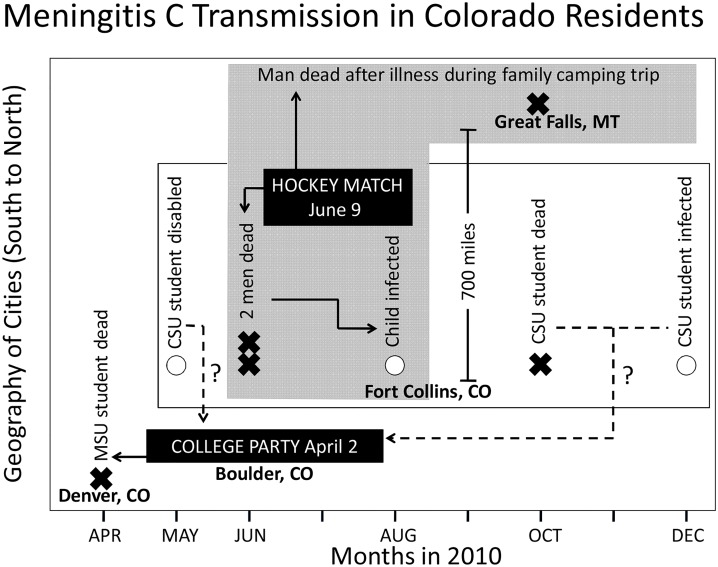
Fig 8 is an example of how real-time informatics can be used to connect what appear to be unrelated meningococcal disease cases (due to serogroup C) that differ by state, city, site, and time. The data indicate one death in April (Metropolitan State University student in Denver, Colorado), two survivors and one death between May and December (Colorado State University students at Fort Collins, Colorado), two deaths and one survivor between June and August (two men and one child in Fort Collins, Colorado), and one death in October (a man in Great Falls, Montana). Using the news data captured with HealthMap, what becomes apparent is that the disease transmission may have started at a college party in Boulder, Colorado attended by the student from Denver who died. This party was also attended by Colorado State University (CSU) students from Fort Collins. While it is not clear whether the three CSU students infected with meningococcus serogroup C attended the party in Boulder, the infection in one of the three students predated the infection resulting in the death of two men and the hospitalization of one child in Fort Collins. The infected child was related to one of the two men who died. These two men were players in a hockey match in June (which CSU students attended), and the man dying four months later in Great Falls, Montana was also a player in this hockey match.
Fig 8. Understanding disease transmission from disease alerts in HealthMap.
The figure illustrates on the y-axis the geographic location from south to north and on the x-axis, monthly intervals between April and December of 2010. The circles indicate survivors of meningococcal infection while the cross-marks indicate deaths. Data captured from real-time informatics trace eight subjects infected with Neisseria meningococcus back to possible disease initiation at a college party in Boulder, Colorado, with subsequent amplification at a hockey match in Fort Collins, Colorado, two months later.
Discussion
Real-Time “snapshot” of meningococcal disease activity by HealthMap
Surveillance monitoring systems based on Internet news can provide new dynamics to public health data analysis and, as illustrated in the 2009 A/H1N1 influenza pandemic, help to overcome certain limitations of existing public health surveillance systems including reporting delays, inconsistent population coverage, and poor accuracy in detecting emerging diseases. The current university-linked serogroup B outbreaks in 2013–2014 and the serogroup B outbreaks described previously in Oregon from 1994–1996 and in Ohio in 2010 [14] highlight the unpredictable nature of N. meningitidis outbreaks and the challenges facing public officials who need to rapidly identify whether an outbreak is occurring and trigger the necessary intervention to treat and prevent disease transmission. Given the historical data available for comparison and the recent advancement in the capabilities of informatics, we used HealthMap, an automated text processing system that collects data from diverse online sources including media reports and health department press releases, to detect cases of meningococcal disease and potential outbreaks in the US from 2010 to 2013. HealthMap application to emerging diseases was previously proven during the global surveillance monitoring of H1N1 influenza pandemic in 2009 [5]. This real-time informatics system captured a national distribution of meningitis cases (Fig 1) and increased prevalence for school-based settings (Table 1) similar to what has been published in an older US study using traditional and more comprehensive surveillance [12]. While the Internet-based data streaming approach used by HealthMap underestimates US disease burden (Table 1) when compared to estimates from ABCs (Fig 4), it is a very rapid method for acquiring a snapshot of meningococcal disease visibility as a function of time (Figs 2 and 3). Predicting when a sporadic case will evolve into an outbreak of the proportions seen at Princeton, New Jersey, and Santa Barbara, California, remains problematic with traditional surveillance and could be bridged by the application of real-time informatics such as HealthMap.
Detailed epidemiology captured by traditional surveillance systems
The real-time informatics approach used by HealthMap has been active since 2006 and its capacities have evolved significantly for disease monitoring [15–22] with the addition of data capture for N. meningitidis being fully operational as of 2010. However, internet-based data streaming cannot yet provide the level of epidemiological detail captured by traditional surveillance approaches which have also investigated patterns in meningococcal disease historically. Traditional surveillance has demonstrated that, since 1991, the frequency of local meningococcal outbreaks has increased with the majority of cases being due to serogroup C [23]. Since 1997, local outbreaks due to serogroups Y and B have also been reported [23]. A study published in 2006 characterized outbreak-associated and sporadic cases in the US during an eight-year period from 1994 through 2002 and identified 69 outbreaks (3–14 outbreaks per year) involving 229 patients from 30 states [12]. For all outbreaks, serogroup C was predominant (66.4%) followed by B (23.6%), Y (10.0%), and W (0%). During the same period, sporadic cases were predominantly caused by serogroup B (36.7%), followed by Y (32.5%), C (28.6%), and W (2.2%) [12]. This 2006 publication [12] concluded that 1) outbreak-associated cases were more likely to die compared to sporadic disease cases, 2) outbreak-associated cases most often involved serogroup C, and 3) institutional outbreaks generally last less than two weeks.
However, in the case of the 2013 outbreaks involving Princeton University and University of California Santa Barbara, the serogroup involved was meningococcus B and these institutional outbreaks persisted for months as captured by HealthMap data-streaming. These recent outbreaks suggests a change in meningococcal serogroup distribution which could be expected due to the introduction of vaccines protective against A, C, W, and Y (decreasing incidence of non-serogroup B meningococcus captured by ABCs in Fig 4). Since the peak of meningococcal disease in the 1990s in the US, disease incidence has decreased to 0.2 cases per 100,000 population in all age groups in 2011 [24]. The burden of disease is highest among infants younger than 1 year of age (2.6 cases per 100,000 persons), adolescents 16 to 21 years of age (0.4 cases per 100,000 persons), and adults older than 65 years of age (0.3 cases per 100,000 persons) [24]. While serogroups B, C, and Y are each responsible for approximately one third of meningococcal cases in the US, the proportion caused by each serogroup varies by age group. For example, 60% of disease in children younger than 60 months of age is caused by serogroup B. However, 73% of disease in persons aged ≥ 11 years is caused by serogroup C, Y, or W. While no serogroup B vaccine is licensed in the US for children younger than 60 months of age, quadrivalent meningococcal glycoconjugate vaccines protective against serogroups A, C, W, and Y (MenACWY) have been licensed in the US since 2005 [24]. MenACWY vaccine coverage has increased to over 70% by 2011 in adolescents 13 to 17 years of age [24] with the estimated number of annual cases from serogroups C and Y decreasing by 75% in individuals 11 to 14 years of age from 2000–2004 to 2005–2009 [23]. However, for individuals 15 to 18 years of age, serogroup C and Y cases have decreased only by 27% and a peak in meningococcal disease has persisted among persons aged 18 to 21 years [23].
Using informatics to put into context emerging N. meningitidis disease threats and related public health interventions
The ability of informatics-based approaches to filter through local, national, and global disease activity in real-time provides another feature not readily apparent from traditional surveillance approaches—modes of disease transmission. Individuals today are more mobile than before which underscores the relevance of disease activity awareness in cities bordering countries. From Fig 7, it becomes apparent that HealthMap disease activity across borders could contribute to understanding disease transmission and subsequent outbreaks. As an example, the HealthMap alerts for Southern California in 2013 brought to attention a meningitis outbreak in Tijuana, Mexico (Fig 6). According to the San Diego Deputy Public Health director, the outbreak in Tijuana in 2013 was three times the incidence seen in Tijuana annually [13]. Tijuana is the largest city in the Mexican state of Baja California and is situated on the US-Mexico border adjacent to San Diego, California. This is the most traversed international border in the world [25] with the two border crossing stations in Tijuana accounting for an estimated 300,000 border crossings each day. Notably, a subsequent epidemiological study on meningococcal disease in Latin America from 1945 to 2010 demonstrated that serogroup A has virtually disappeared, and that serogroups B and C are responsible for most cases [26]. The disease activity captured by HealthMap for Canadian cities (Fig 7) near the US border reflected only two cases in 2010 and one case in 2012 that may be reflecting the timely incorporation of vaccines into the national schedules by Health Canada. The quadrivalent conjugated vaccine protective for A,C,W,Y was licensed in Canada in 2007 [27] and, more recently, a recombinant serogroup B vaccine was approved on January 2014 [28].
N. meningitidis outbreaks: potential of internet-based data streaming to de-convolute disease transmission or capture intervention-related signals
The unpredictable nature of meningococcal outbreaks and the protracted timeline during which enough cases accumulate to trigger declaration of an outbreak were recently highlighted by the serogroup B meningococcus university-linked outbreaks that occurred on both the east and west coasts of the US in 2013. More recently, on March 10, 2014, a student at Drexel University died from serogroup B meningococcal disease. CDC laboratory analysis, based on genetic fingerprinting, demonstrated that the strain infecting this student matched that in the Princeton University outbreak. This student had been in close contact with students from Princeton University about a week before illness, and the laboratory analysis indicated that the outbreak strain may still be present in the Princeton community and require vigilance for additional cases [29]. These university outbreaks highlight the limitations of antibiotic chemoprophylaxis interventions that require the identification and treatment of all individuals at risk in order to avoid subsequent disease transmission by untreated carriers of subclinical disease. This was evidenced during a serogroup B outbreak in a hotel [30] and another in a middle school [31] in 1995. However, real-time capture of cases by internet-based informatics can complement traditional surveillance monitoring approaches leading to increasing confidence and resourcing in outbreak settings by decreasing time to outbreak declaration and medical handling. As illustrated in Fig 8, modes of disease transmission could be traced to ensure targeted administration of antibiotic therapy or prophylactic interventions to the relevant at-risk population. Finally, internet-based monitoring systems like HealthMap can provide a real-time channel for patients to report intervention-related failures or can be applied to safety data mining by scanning social media channels like Twitter and Facebook on a macro-level for patient-reported safety insights. This could include the direct reporting to regulatory authorities of medication or vaccine-associated adverse events identified through social media and mobile phone apps. These widely accessible and user-friendly computer-based options can support the tracking of subject safety and could facilitate decision-making related to the use of unlicensed therapies or vaccines in emergency situations. One such emergency situation was the Princeton University serogroup B meningococcal disease outbreak that began in the US in March 2013. An emergency vaccination campaign with an imported vaccine (Bexsero, approved in Australia, Europe, and Canada but not in the US) was able to be implemented in the US in December 2013 (approximately nine months later) only under special regulatory procedures after the CDC filed an Investigational New Drug application with the Food and Drug Administration requesting special permission for use [32].
With regards to invasive meningococcal disease (IMD), a potential limitation of using HealthMap may be related to the inability to identify and report the serotypes specifically linked to the disease outbreak. Traditionally, all US states have mandated reporting of IMD [33] and require health care providers to immediately report suspected cases to assure rapid identification of close contacts and administration of antibiotic prophylaxis [23]. However, despite the swift execution of the protocol established to confirm these reports, there is always a window of delay secondary to the specialized public health molecular and microbiological laboratory testing necessary to confirm cases in establish IMD clusters. Usually, during an outbreak, the high case rates increase awareness in health authorities, but options for implementing the best tailored, prophylactic measure requires understanding the extent and mode of transmission involved in that particular outbreak. A national network of state epidemiologists and CDC maintain regular communication on the outbreak status through electronic alerts over the secure CDC Epi-X system and direct communication between states when out-of-state exposures occur.
Conclusions
International events with the potential to cross borders (e.g., 2009 H1N1 pandemic influenza) are probably the best example of the application of HealthMap and highlight its potential use for other emerging worldwide threats (e.g., SARS, avian influenza, or Ebola virus). The underreporting of disease cases by internet-based data streaming in specific N. meningitidis outbreak modes makes inadequate any comparison to epidemiological data captured by more comprehensive traditional N. meningitidis surveillance such as the ABCs network published by the CDC. However, the natural delays in compiling reports from traditional surveillance (at the time of writing, ABCs N. meningitidis data for 2013 is listed as being provisional) emphasize the utility of real-time internet-based data streaming for providing a real-time “snapshot” of meningococcal disease. In this investigation on surveillance of N. meningitidis disease activity and transmission using the HealthMap information tool, what becomes apparent is that this information technology can complement epidemiological data captured by traditional surveillance monitoring systems by quickly filling gaps (including the visualization of modes of meningococcal disease transmission in outbreaks) allowing for better resource and action planning. For the general public and the public health community, HealthMap surveillance of meningococcal disease activity can increase disease awareness by collecting an immense amount of disease/outbreak reports and putting them in a widely accessible and user-friendly platform. This user-friendly platform of HealthMap has the potential to additionally complement traditional surveillance monitoring by serving as an internet-based real-time channel for patients infected with N. meningitidis to report intervention-related failures.
Acknowledgments
The authors wish to acknowledge Mariagrazia Pizza and Rino Rappuoli for their critical review of the article and are grateful to Anna Tomasulo for her assistance with data preparation and Georgio Corsi for his assistance with artwork related to the figures. The authors affirm that all individuals who have contributed significantly to the article have been listed.
Data Availability
All relevant data are within the paper.
Funding Statement
Drs. Brownstein, Mekaru, and Freifeld are supported by grant 5 R01 LM010812-04 from the National Library of Medicine. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No other sources of funding have supported this work. Novartis provided support in the form of salaries for authors (SSA, EOO, GT), but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the “author contributions” section.
References
- 1. Bona G, Guidi C. Meningococcal vaccine evolution. J Prev Med Hyg 2012. September;53(3):131–5. [PubMed] [Google Scholar]
- 2. Bilukha O, Messonnier N, Fischer M. Use of meningococcal vaccines in the United States. Pediatr Infect Dis J 2007. May;26(5):371–6. [DOI] [PubMed] [Google Scholar]
- 3. Cohn AC, MacNeil JR, Harrison LH, Hatcher C, Theodore J, Schmidt M et al. Changes in Neisseria meningitidis disease epidemiology in the United States, 1998–2007: implications for prevention of meningococcal disease. Clin Infect Dis 2010. January 15;50(2):184–91. 10.1086/649209 [DOI] [PubMed] [Google Scholar]
- 4. Erickson LJ, De WP, McMahon J, Heim S. Complications of meningococcal disease in college students. Clin Infect Dis 2001. September 1;33(5):737–9. [DOI] [PubMed] [Google Scholar]
- 5. Brownstein JS, Freifeld CC, Madoff LC. Influenza A (H1N1) virus, 2009—online monitoring. N Engl J Med 2009. May 21;360(21):2156 10.1056/NEJMp0904012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brownstein JS, Freifeld CC, Madoff LC. Digital disease detection—harnessing the Web for public health surveillance. N Engl J Med 2009. May 21;360(21):2153–5, 2157. 10.1056/NEJMp0900702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hartley D, Nelson N, Walters R, Arthur R, Yangarber R, Madoff L et al. Landscape of international event-based biosurveillance. Emerg Health Threats J 2010;3:e3 10.3134/ehtj.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heymann DL, Rodier GR. Hot spots in a wired world: WHO surveillance of emerging and re-emerging infectious diseases. Lancet Infect Dis 2001. December;1(5):345–53. [DOI] [PubMed] [Google Scholar]
- 9. Brownstein JS, Freifeld CC, Reis BY, Mandl KD. Surveillance Sans Frontieres: Internet-based emerging infectious disease intelligence and the HealthMap project. PLoS Med 2008. July 8;5(7):e151 10.1371/journal.pmed.0050151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brownstein JS, Freifeld CC, Reis BY, Mandl KD. HealthMap: Internet-based emerging infectious disease intelligence In: Lemon SM, Hamburg MA, Sparling PF, Choffnes ER, Marck A, editors. Global Infectious Disease Surveillance and Detection: Assessing the Challenges—Finding Solutions.Washington, D.C.: Institute of Medicine of the National Academies; 2007. 10.1371/journal.pone.0006954 [DOI] [Google Scholar]
- 11.Centers for Disease Control and Prevention. Active Bacterial Core Surveillance Report, Emerging Infections Program Network, Neisseria meningitidis, 2003–2012. 2013. Available: http://www.cdc.gov/abcs/reports-findings/surv-reports.html.
- 12. Brooks R, Woods CW, Benjamin DK Jr, Rosenstein NE. Increased case-fatality rate associated with outbreaks of Neisseria meningitidis infection, compared with sporadic meningococcal disease, in the United States, 1994–2002. Clin Infect Dis 2006. July 1;43(1):49–54. [DOI] [PubMed] [Google Scholar]
- 13. Magee M. Meningitis warning in schools: outbreak results in six deaths in Tijuana and one in San Diego County. San Diego Union-Tribune 2013. March 21. [Google Scholar]
- 14. Mandal S, Wu HM, MacNeil JR, Machesky K, Garcia J, Plikaytis BD et al. Prolonged university outbreak of meningococcal disease associated with a serogroup B strain rarely seen in the United States. Clin Infect Dis 2013. August;57(3):344–8. 10.1093/cid/cit243 [DOI] [PubMed] [Google Scholar]
- 15. Brownstein JS, Freifeld CC. HealthMap: the development of automated real-time internet surveillance for epidemic intelligence. Euro Surveill 2007. November;12(11):E071129 [DOI] [PubMed] [Google Scholar]
- 16. Brownstein JS, Mandl KD. Reengineering real time outbreak detection systems for influenza epidemic monitoring. AMIA Annu Symp Proc 2006;866 [PMC free article] [PubMed] [Google Scholar]
- 17. Chunara R, Freifeld CC, Brownstein JS. New technologies for reporting real-time emergent infections. Parasitology 2012. December;139(14):1843–51. 10.1017/S0031182012000923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Freifeld CC, Brownstein JS, Menone CM, Bao W, Filice R, Kass-Hout T et al. Digital drug safety surveillance: monitoring pharmaceutical products in twitter. Drug Saf 2014. May;37(5):343–50. 10.1007/s40264-014-0155-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Freifeld CC, Chunara R, Mekaru SR, Chan EH, Kass-Hout T, Ayala IA et al. Participatory epidemiology: use of mobile phones for community-based health reporting. PLoS Med 2010;7(12):e1000376 10.1371/journal.pmed.1000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Freifeld CC, Mandl KD, Reis BY, Brownstein JS. HealthMap: global infectious disease monitoring through automated classification and visualization of Internet media reports. J Am Med Inform Assoc 2008. March;15(2):150–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scales D, Zelenev A, Brownstein JS. Quantifying the effect of media limitations on outbreak data in a global online web-crawling epidemic intelligence system, 2008–2011. Emerg Health Threats J 2013;6:21621 10.3402/ehtj.v6i0.21621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilson K, Brownstein JS. Early detection of disease outbreaks using the Internet. CMAJ 2009. April 14;180(8):829–31. 10.1503/cmaj.090215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Centers for Disease Control and Prevention. Meningococcal Disease: Epidemiology and prevention of vaccine-preventable diseases. 12 ed 2012. p. 193–204. [Google Scholar]
- 24. Cohn AC, MacNeil JR, Clark TA, Ortega-Sanchez IR, Briere EZ, Meissner HC et al. Prevention and control of meningococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 2013. March 22;62(RR-2):1–28. [PubMed] [Google Scholar]
- 25. Chacon-Cruz E, Sugerman DE, Ginsberg MM, Hopkins J, Hurtado-Montalvo JA, Lopez-Viera JL et al. Surveillance for invasive meningococcal disease in children, US-Mexico border, 2005–2008. Emerg Infect Dis 2011. March;17(3):543–6. 10.3201/eid1703.101254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Safadi MA, Gonzalez-Ayala S, Jakel A, Wieffer H, Moreno C, Vyse A. The epidemiology of meningococcal disease in Latin America 1945–2010: an unpredictable and changing landscape. Epidemiol Infect 2012. August 9;1–12. 10.1017/S0950268812002245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salvadori M, Bortolussi R. Meningococcal vaccines in Canada: An update. Paediatr Child Health 2011. October;16(8):485–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Office of Regulatory Affairs BaGTD. Bexsero. www hc-sc gc ca 2014 April 10. Available: http://www.hc-sc.gc.ca/dhp-mps/prodpharma/sbd-smd/drug-med/sbd_smd_2014_bexsero_147275-eng.php?utm_source=rss&utm_medium=rss&utm_campaign=summary-basis-of-decision-sbd-bexsero-2014.
- 29. Centers for Disease Control and Prevention. Meningococcal Disease Update. 2014. March 18 Available: http://www.cdc.gov/meningococcal/outbreaks/princeton.html. [Google Scholar]
- 30. Centers for Disease Control and Prevention. Outbreaks of group B meningococcal disease—Florida, 1995 and 1997. MMWR Morb Mortal Wkly Rep 1998. October 9;47(39):833–7. [PubMed] [Google Scholar]
- 31. Jackson LA, Alexander ER, DeBolt CA, Swenson PD, Boase J, McDowell MG et al. Evaluation of the use of mass chemoprophylaxis during a school outbreak of enzyme type 5 serogroup B meningococcal disease. Pediatr Infect Dis J 1996. November;15(11):992–8. [DOI] [PubMed] [Google Scholar]
- 32. Centers for Disease Control and Prevention. Princeton University meningococcal disease outbreak and University of California, Santa Barbara meningococcal disease outbreak. 2013. December 31 Available: http://www.cdc.gov/meningococcal/outbreaks/princeton.html. [Google Scholar]
- 33. Adams DA, Gallagher KM, Jajosky RA, Kriseman J, Sharp P, Anderson WJ et al. Summary of Notifiable Diseases—United States, 2011. MMWR Morb Mortal Wkly Rep 2013. July 5;60(53):1–117. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.