Abstract
There are no minimally invasive diagnostic metrics for acute kidney transplant rejection (AR), especially in the setting of the common confounding diagnosis, acute dysfunction with no rejection (ADNR). Thus, though kidney transplant biopsies remain the gold standard, they are invasive, have substantial risks, sampling error issues and significant costs and are not suitable for serial monitoring. Global gene expression profiles of 148 peripheral blood samples from transplant patients with excellent function and normal histology (TX; n = 46), AR (n = 63) and ADNR (n = 39), from two independent cohorts were analyzed with DNA microarrays. We applied a new normalization tool, frozen robust multi-array analysis, particularly suitable for clinical diagnostics, multiple prediction tools to discover, refine and validate robust molecular classifiers and we tested a novel one-by-one analysis strategy to model the real clinical application of this test. Multiple three-way classifier tools identified 200 highest value probesets with sensitivity, specificity, positive predictive value, negative predictive value and area under the curve for the validation cohort ranging from 82% to 100%, 76% to 95%, 76% to 95%, 79% to 100%, 84% to 100% and 0.817 to 0.968, respectively. We conclude that peripheral blood gene expression profiling can be used as a minimally invasive tool to accurately reveal TX, AR and ADNR in the setting of acute kidney transplant dysfunction.
Keywords: Acute dysfunction with no rejection, acute kidney rejection, gene expression profiling, microarrays, molecular classifiers
Introduction
Improvements in kidney transplantation have resulted in significant reductions in clinical acute rejection (AR) (8–14%) (1). Unfortunately, histological AR without evidence of kidney dysfunction (i.e. subclinical AR) occurs in >15% of protocol biopsies done within the first year (2–4). Without a protocol biopsy, patients with subclinical AR would be treated as excellent functioning transplants (TX). Moreover, 10-year allograft loss rates remain unacceptably high, 57% with deceased donor kidneys (5) and biopsy studies document significant rates of a progressive interstitial fibrosis and tubular atrophy in >50% of protocol biopsies starting as early as 1 year posttransplant (6–8).
Two factors contribute to AR: the failure to optimize immunosuppression and individual patient nonadherence (9,10). Currently, there is no validated test to measure or monitor the adequacy of immunosuppression, the failure of which is often first manifested as an AR episode. Subsequently, inadequate immunosuppression results in chronic rejection and allograft failure. The current standards for monitoring kidney transplant function are serum creatinine and estimated GFRs. Unfortunately, serum creatinine and eGFR are relatively insensitive markers requiring significant global injury before changing (3,11,12) and are influenced by multiple nonimmunological factors.
The gold standard for AR remains a kidney biopsy. Performing routine protocol biopsies is one strategy to diagnose and treat AR prior to extensive injury. A study of 28 patients 1 week posttransplant with stable creatinines showed that 21% had unsuspected “borderline” AR and 25% had inflammatory tubulitis (13). Other studies reveal a 29% prevalence of subclinical rejection (14) and that subclinical rejection with chronic allograft nephropathy was a risk factor for late graft loss (3). A study of 517 renal transplants followed after protocol biopsies showed that finding subclinical rejection significantly increased the risk of chronic rejection (15).
Limitations of biopsies include sampling errors, significant costs and patient risks. AR is a dynamic process and predicting rejection and managing immunosuppression require serial monitoring not possible using biopsies. Moreover, many patients present with acute dysfunction but no rejection is documented by biopsy (ADNR). Thus, there is a pressing need to develop a minimally invasive, objective metric for the diagnosis of AR and the adequacy of immunosuppression that can also identify ADNR.
We originally reported a peripheral blood gene expression signature by DNA microarrays to diagnose AR (16). Subsequently, others have reported quantitative polymerase chain reaction (qPCR) signatures of AR in peripheral blood based on genes selected from the literature or using microarrays (17–22). As the biomarker field has evolved, validation requires independently collected sample cohorts and avoidance of over-training during classifier discovery (23,24). Another limitation is that the currently published biomarkers are designed for two-way classifications, AR versus TX, when many biopsies reveal ADNR and that demands three-way classifiers.
We prospectively followed over 1000 kidney transplants from five different clinical centers (Transplant Genomics Collaborative Group) to identify 148 cases of unequivocal biopsy-proven AR (n = 63), ADNR (n = 39) and TX (n = 46). Global gene expression profiling was done on peripheral blood using DNA microarrays and robust three-way class prediction tools (25–27). Classifiers comprising the 200 highest value probesets ranked by the prediction accuracies with each tool were created with three different classifier tools to insure that our results were not subject to bias introduced by a single statistical method. Importantly, even using three different tools, the 200 highest value probeset classifiers identified were essentially the same. These 200 classifiers had a sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and area under the curve (AUC) for the validation cohort depending on the three different prediction tools used ranging from 82% to 100%, 76% to 95%, 76% to 95%, 79% to 100%, 84% to 100% and 0.817 to 0.968, respectively. Next, the Harrell bootstrapping method (28) based on sampling with replacement was used to demonstrate that these results, regardless of the tool used, were not the consequence of statistical over-fitting. Finally, to model the use of our test in real clinical practice, we developed a novel one-by-one prediction strategy in which we created a large reference set of 118 samples and then randomly took 10 samples each from the AR, ADNR and TX cohorts in the validation set. These were then blinded to phenotype and each sample was tested by itself against the entire reference set to model practice in a real clinical situation where there is only a single new patient sample obtained at any given time.
Materials and Methods
Patient populations
We studied 46 kidney transplant patients with well-functioning grafts and biopsy-proven normal histology (TX; controls), 63 patients with biopsy-proven acute kidney rejection (AR) and 39 patients with acute kidney dysfunction without histological evidence of rejection (ADNR). Inclusion/exclusion criteria are presented in Table S2. Subjects were enrolled serially as biopsies were performed by five different clinical centers (Scripps Clinic, Cleveland Clinic, St. Vincent Medical Center, University of Colorado and Mayo Clinic Arizona). Human Subjects Research Protocols approved at each Center and by the Institutional Review Board of the Scripps Research Institute covered all studies.
Pathology
All subjects had kidney biopsies (either protocol or “for cause”) graded for evidence of AR by the Banff’ 2007 criteria (29). All biopsies were read by local pathologists and then reviewed and graded in a blinded fashion by a single pathologist at an independent center (LG). The local and single pathologist readings were then reviewed by DRS to standardize and finalize the phenotypes prior to cohort construction and any diagnostic classification analysis. C4d staining was done per the judgment of the local clinicians and pathologists on 69 of the 148 samples (47%; Table 1). Positive was defined as linear, diffuse staining of peritubular capillaries. Donor-specific antibodies were not measured on these patients, and thus we cannot exclude the new concept of C4d negative antibody-mediated rejection (ABMR) (30,31).
Table 1.
Clinical characteristics for the 148 study samples
| All study samples | Multivariate analysis1 | |||||
|---|---|---|---|---|---|---|
| TX | AR | ADNR | Significance2 | Significance (phenotypes) |
Significance (phenotypes/cohorts) |
|
| Subject numbers | 46 | 63 | 39 | – | – | – |
| Recipient age ± SD (years) | 50.1 ± 14.5 | 44.9 ± 14.3 | 49.7 ± 14.6 | NS3 | NS | NS |
| % Female recipients | 34.8 | 23.8 | 20.5 | NS | NS | NS |
| % Recipient African American | 6.8 | 12.7 | 12.8 | NS | NS | NS |
| % Pre-TX Type II diabetes | 25.0 | 17.5 | 21.6 | NS | NS | NS |
| % PRA >20 | 29.4 | 11.3 | 11.5 | NS | NS | NS |
| HLA mismatch ± SD | 4.2 ± 2.1 | 4.3 ± 1.6 | 3.7 ± 2.1 | NS | NS | NS |
| % Deceased donor | 43.5 | 65.1 | 53.8 | NS | NS | NS |
| Donor age ± SD (years) | 40.3 ± 14.5 | 38.0 ± 14.3 | 46.5 ± 14.6 | NS | NS | NS |
| % Female donors | 37.0 | 50.8 | 46.2 | NS | NS | NS |
| % Donor African American | 3.2 | 4.9 | 13.3 | NS | NS | NS |
| % Delayed graft function | 19.0 | 34.4 | 29.0 | NS | NS | NS |
| % Induction | 63.0 | 84.1 | 82.1 | NS | NS | NS |
| Serum creatinine ± SD (mg/dL) | 1.5 ± 0.5 | 3.2 ± 2.8 | 2.7 ± 1.8 | TX vs. AR = 0.00001; TX vs. ADNR = 0.0002; AR vs. ADNR = NS | TX vs. AR = 0.04; TX vs. ADNR = 0.01 | TX vs. AR vs. ADNR = 0.00002; AR vs. ADNR = NS |
| Time to biopsy ± SD (days) | 512 ± 1359 | 751 ± 1127 | 760 ± 972 | NS | NS | NS |
| Biopsy ≤365 days (%) | 27 (54.2%) | 38 (49.0%) | 23 (52.4%) | NS | NS | NS |
| Biopsy >365 days (%) | 19 (45.8%) | 32 (51.0%) | 18 (47.6%) | NS | NS | NS |
| % Calcineurin inhibitors | 89.7 | 94.0 | 81.1 | NS | NS | NS |
| % Mycophenolic acid derivatives | 78.3 | 85.7 | 84.6 | NS | NS | NS |
| % Oral steroids | 26.1 | 65.1 | 74.4 | TX vs. AR = 0.001; TX vs. ADNR = 0.001 | TX vs. ADNR = 0.04 | NS |
| C4d positive staining (%)4 | 0/13 (0%) | 12/36 (33.3%) | 1/20 (5%) | NS | NS | NS |
ADNR, acute dysfunction with no rejection by biopsy histology; AR, acute rejection; TX, excellent functioning transplant.
A multivariate logistic regression model was used with a Wald test correction. In the first analysis (phenotypes), we used all 148 samples and in the second analysis (phenotypes/cohorts), we did the analysis for each randomized set of two cohorts (discovery and validation).
Significance for all comparisons were determined with paired Student’s t-test for pair-wise comparisons of data with standard deviations and for dichotomous data comparisons by chi-square.
NS, not significant (p ≥ 0.05).
Subjects with biopsy-positive staining for C4d and total number of subjects whose biopsies were stained for C4d with (%).
Gene expression profiling and statistical analysis
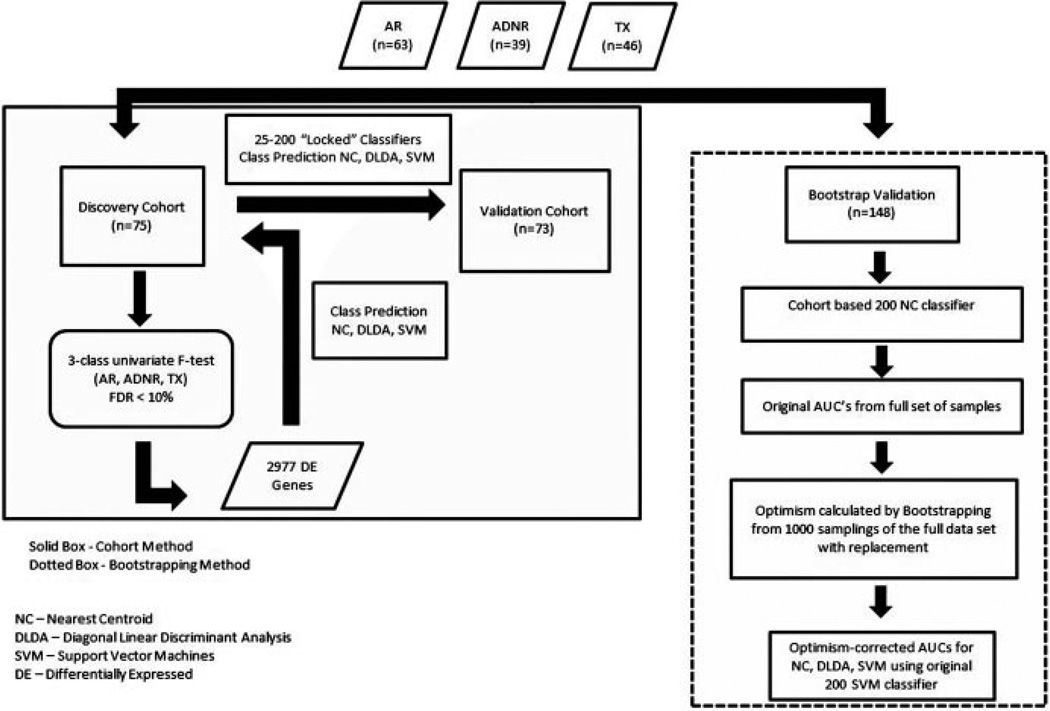
RNA was extracted from Paxgene tubes using the Paxgene Blood RNA system (PreAnalytiX GmbH, Hombrechtikon, Switzerland) and Ambion GLOBINclear (Life Technologies, Carlsbad, CA). Biotinylated cRNA was prepared with Ambion MessageAmp Biotin II kit (Ambion) and hybridized to Affymetrix Human Genome U133 Plus 2.0 GeneChips (Affymetrix Inc., Santa Clara, CA). Normalized signals were generated using frozen robust multi-array analysis (fRMA) in R (32,33). The complete strategy used to discover, refine and validate the biomarker panels is shown in Figure 1. Class predictions were performed with multiple tools: nearest centroids (NC), support vector machines (SVM) and diagonal linear discriminant analysis (DLDA). Predictive accuracy is calculated as true positives + true negatives/true positives + false positives + false negatives + true negatives. Other diagnostic metrics given are sensitivity, specificity, PPV, NPV and AUC. Receiver operating characteristic (ROC) curves were generated using pROC in R (34). Clinical study parameters were tested by multivariate logistic regression with an adjusted (Wald test) p-value and a local false discovery rate (FDR) calculation (q-value). Chi-square analysis was done using GraphPad (35). CEL files and normalized signal intensities are posted in NIH Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo (accession number GSE15296).
Figure 1.
Flow chart describing the cohort and bootstrapping strategies for biomarker discovery and validation.
Results
Patient population
Subjects were consented and biopsied in a random and prospective fashion at five centers (n = 148; Table 1). Blood was collected at the time of biopsy. TX represented protocol biopsies of transplants with excellent, stable graft function and normal histology (n = 46). AR patients were biopsied “for cause” based on elevated serum creatinine (n = 63). We excluded subjects with recurrent kidney disease, BK virus (BKV) or other infections. ADNRs were biopsied “for cause” based on suspicion of AR but had no AR by histology (n = 39). Differences in steroid use, less in TX, reflect more protocol biopsies done at a steroid-free center. As expected, creatinines were higher in AR and ADNR than in TX. Creatinine was the only significant variable by multivariable logistic regression by either phenotype or cohort. C4d staining, when done, was negative in TX and ADNR. C4d staining was done in 56% of AR subjects by the judgment of the pathologists and was positive in 12/36 (33%) of this selected group.
Three-way predictions
We randomly split the data from 148 samples into two cohorts, discovery and validation (Figure 1). Discovery was 32 AR, 20 ADNR and 23 TX, and validation was 32 AR, 19 ADNR and 22 TX. Normalization used fRMA (32,33). Probesets with median Log2 signals less than 5.20 in ≥70% of samples were eliminated. A three-class univariate F-test was done on the discovery cohort (1000 random permutations, FDR <10%; BRB ArrayTools) yielding 2977 differentially expressed probesets (Table S1). The NC algorithm (25) was used to create a three-way classifier for AR, ADNR and TX in the discovery cohort revealing 200 high-value probesets defined by having the lowest class predictive error rates (Table 2; see also Supplemental Statistical Methods).
Table 2.
Diagnostic metrics for the three-way nearest centroid classifiers for AR, ADNR and TX in discovery and validation cohorts
| Method | Classifiers | % Predictive accuracy (discovery cohort) |
% Predictive accuracy (validation cohort) |
Sensitivity (%) |
Specificity (%) |
Positive predictive value (%) |
Negative predictive value (%) |
AUC | Sensitivity (%) |
Specificity (%) |
Positive predictive value (%) |
Negative predictive value (%) |
AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 200 | TX vs. AR | 92 | 83 | 87 | 96 | 95 | 89 | 0.917 | 73 | 92 | 89 | 79 | 0.837 |
| Classifiers | TX vs. ADNR | 91 | 82 | 95 | 90 | 91 | 95 | 0.913 | 89 | 76 | 76 | 89 | 0.817 |
| AR vs. ADNR | 92 | 90 | 87 | 100 | 100 | 86 | 0.933 | 89 | 92 | 89 | 92 | 0.893 | |
| 100 | TX vs. AR | 91 | 83 | 87 | 93 | 91 | 90 | 0.903 | 76 | 88 | 84 | 82 | 0.825 |
| Classifiers | TX vs. ADNR | 98 | 81 | 95 | 100 | 100 | 95 | 0.975 | 84 | 79 | 80 | 83 | 0.814 |
| AR vs. ADNR | 98 | 90 | 95 | 100 | 100 | 97 | 0.980 | 88 | 92 | 88 | 92 | 0.900 | |
| 50 | TX vs. AR | 92 | 94 | 88 | 96 | 95 | 90 | 0.923 | 88 | 91 | 89 | 89 | 0.891 |
| Classifiers | TX vs. ADNR | 94 | 95 | 92 | 98 | 98 | 90 | 0.944 | 92 | 90 | 88 | 89 | 0.897 |
| AR vs. ADNR | 97 | 93 | 95 | 97 | 100 | 97 | 0.969 | 89 | 91 | 89 | 89 | 0.893 | |
| 25 | TX vs. AR | 89 | 92 | 81 | 96 | 95 | 84 | 0.890 | 88 | 90 | 90 | 89 | 0.894 |
| Classifiers | TX vs. ADNR | 95 | 95 | 95 | 95 | 95 | 95 | 0.948 | 92 | 92 | 89 | 89 | 0.898 |
| AR vs. ADNR | 96 | 91 | 95 | 96 | 95 | 96 | 0.955 | 85 | 90 | 89 | 88 | 0.882 |
ADNR, acute dysfunction with no rejection by biopsy histology; AR, acute rejection; AUC, area under the curve; TX, excellent functioning transplant.
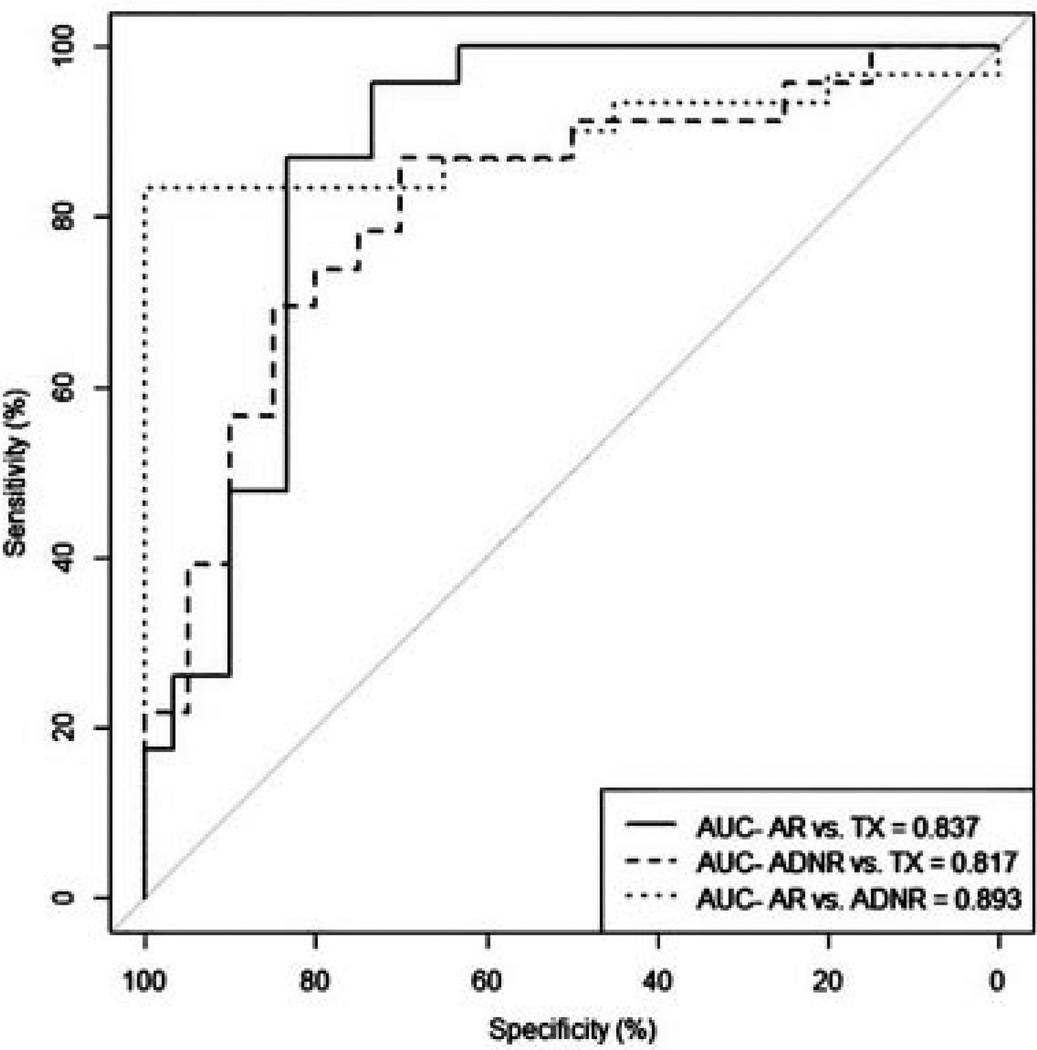
Thus, testing our locked classifier in the validation cohort demonstrated predictive accuracies of 83%, 82% and 90% for the TX versus AR, TX versus ADNR and AR versus ADNR, respectively (Table 2). The AUCs for the TX versus AR, the TX versus ADNR and the AR versus ADNR comparisons were 0.837, 0.817 and 0.893, respectively (Figure 2). The sensitivity, specificity, PPV and NPV for the three comparisons were in similar ranges and are shown in Table 2. To determine a possible minimum classifier set, we ranked the 200 probesets by p-values and tested the top 25, 50, 100 and 200 (Table 2). The conclusion is that given the highest value classifiers discovered using unbiased whole genome profiling, the total number of classifiers necessary for testing can be as little as 25. However, below that number the performance of our three-way classifier falls off dramatically to about 50% AUC at 10 or lower (data not shown).
Figure 2.
Performance of the 200-probeset nearest centroids (NC) classifier discovered and locked in the discovery cohort tested on the validation cohort based on area under the curve (AUC).
Alternative prediction tools
Robust molecular diagnostic strategies should work using multiple tools. Therefore, we repeated the entire three-way locked discovery and validation process using DLDA and SVM (Table S3). All the tools perform nearly equally well with 100–200 classifiers, though small differences were observed.
It is also important to test whether a new classifier is subject to statistical over-fitting that would inflate the claimed predictive results. This testing can be done with the method of Harrell et al using bootstrapping where the original data set is sampled 1000 times with replacement and the AUCs are calculated for each (28). The original AUCs minus the calculated AUCs for each tool create the corrections in the AUCs for “optimism” in the original predictions that adjust for potential over-fitting (Table S4). Therefore, we combined the discovery and validation cohorts and performed a three-class univariate F-test on the whole data set of 148 samples (1000 random permutations, FDR <10%; BRB ArrayTools). This yielded 2666 significantly expressed genes from which we selected the top 200 by p-values. Results using NC, SVM and DLDA with these 200 probesets are shown in Table S4. Optimism-corrected AUCs from 0.823 to 0.843 were obtained for the 200-probeset classifier discovered with the two cohort-based strategy. Results for the 200-classifier set obtained from the full study sample set of 148 were 0.851–0.866. These results demonstrate that over-fitting is not a major problem as would be expected from a robust set of classifiers (Figure S1). These results translate to sensitivity, specificity, PPV and NPV of 81%, 93%, 92% and 84% for AR versus TX; 90%, 85%, 86% and 90% for ADNR versus TX and 85%, 96%, 95% and 87% for AR versus ADNR.
Validation in one-by-one predictions
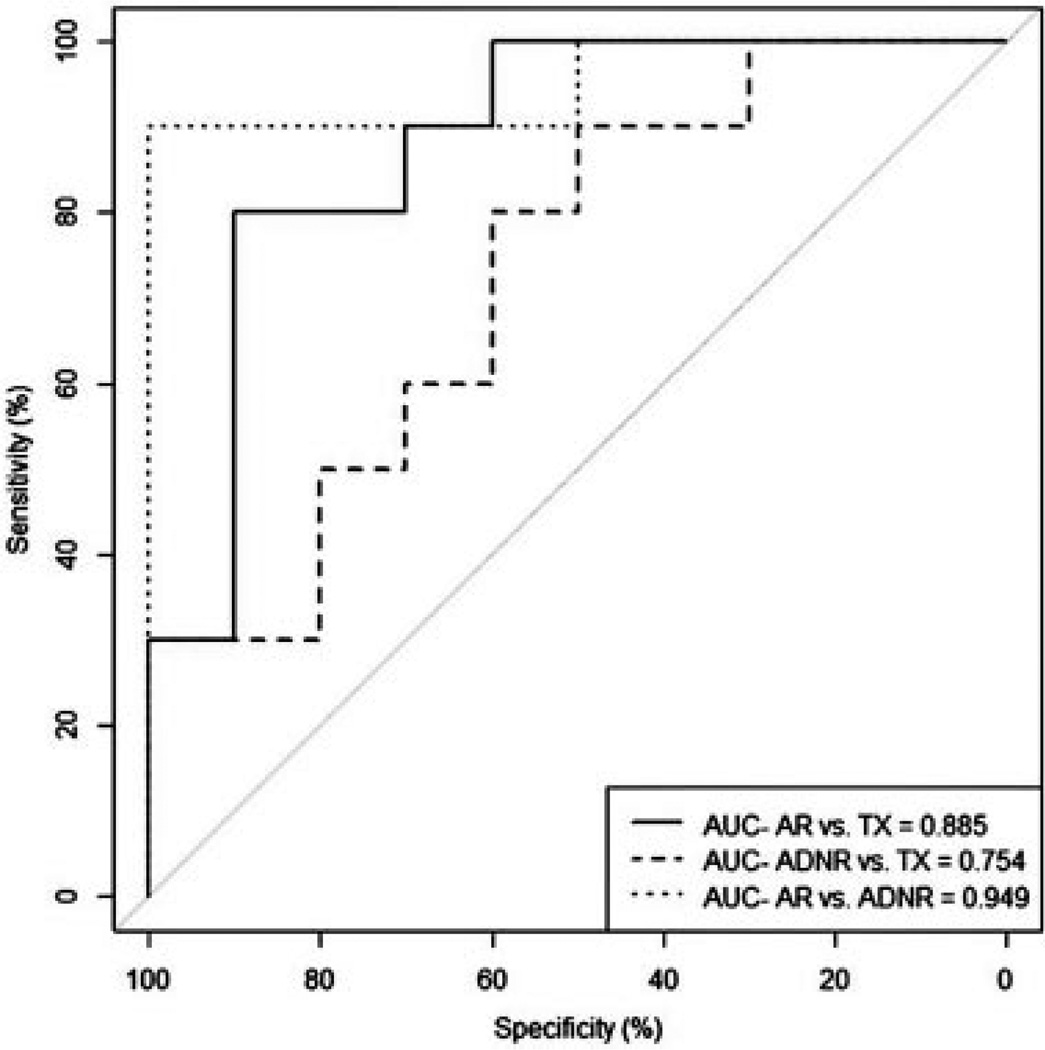
In clinical practice, the diagnostic value of a biomarker is challenged each time a single patient sample is acquired and analyzed. Thus, prediction strategies based on large cohorts of known clinical classifications do not address the performance of biomarkers in their intended application. Two problems exist with cohort-based analysis. First, signal normalization is typically done on the entire cohort, which is not the case in a clinical setting for one patient. Quantile normalization is a robust method but has two drawbacks: it cannot be used in clinical settings where samples must be processed individually or in small batches and data sets normalized separately are not comparable. fRMA overcomes these limitations by normalization of individual arrays to large publicly available microarray databases, allowing for estimates of probe-specific effects and variances to be precomputed and “frozen” (32,33). The second problem with cohort analysis is that all the clinical phenotypes are already known and classification is done on the entire cohort. To address these challenges, we removed 30 random samples from the validation cohort (10 AR, 10 ADNR, 10 TX), blinded their classifications and left a reference cohort of 118 samples with known phenotypes. Classification was done by adding one blinded sample at a time to the reference cohort. Using the 200 genes, three-way classifier derived in NC, we demonstrated an overall predictive accuracy of 80% and individual accuracies of 80% AR, 90% ADNR and 70% TX and AUCs of 0.885, 0.754 and 0.949 for the AR versus TX, the ADNR versus TX and the AR versus ADNR comparisons, respectively (Figure 3).
Figure 3.
Performance of the 200-probeset nearest centroids (NC) classifier discovered and locked in the discovery cohort using a one-by-one validation on 30 randomly selected samples (10 AR, 10 ADNR and 10 TX) from the validation cohort based on area under the curve (AUC). ADNR, acute dysfunction with no rejection by biopsy histology; AR, acute rejection; TX, excellent functioning transplant.
Discussion
After 50 years of kidney transplantation, there is no validated molecular diagnostic for AR to obviate the necessity of a transplant biopsy in the face of acute transplant dysfunction. Ideally, molecular markers will serve as early warnings for immune-mediated injury, before renal function deteriorates, and also permit optimization of immunosuppression. We studied a total of 148 subjects with biopsy-proven phenotypes identified in five different clinical centers by following over 1000 transplant patients. Global RNA expression of peripheral blood was used to profile 63 patients with biopsy-proven AR, 39 patients with ADNR and 46 patients with excellent function and normal histology (TX).
We addressed several important and often overlooked aspects of biomarker discovery. To avoid over-training, we used a discovery cohort to establish the predictive equation and its corresponding classifiers, then locked these down and allowed no further modification. We then tested the diagnostic on our validation cohort. To demonstrate the robustness of our approach, we used multiple, publically available prediction tools to establish that our results are not simply tool-dependent artifacts. We used the bootstrapping method of Harrell to calculate optimism-corrected AUCs and demonstrated that our predictive accuracies are not inflated by over-fitting. We also modeled the actual clinical application of this diagnostic, with a new strategy optimized to normalizing individual samples by fRMA. We then used 30 blinded samples from the validation cohort and tested them one by one. Finally, we calculated the statistical power of our analysis (36) and determined that we have greater than 90% power at a significance level of p < 0.001. We conclude that peripheral blood gene expression profiling can be used to diagnose AR and ADNR in patients with acute kidney transplant dysfunction. An interesting finding is that we got the same results using the classic two-cohort strategy (discovery vs. validation) as we did using the entire sample set and creating our classifiers with the same tools but using the Harrell bootstrapping method to control for over-fitting. Thus, the current thinking that all biomarker signatures require independent validation cohorts may need to be reconsidered.
In the setting of acute kidney transplant dysfunction, we are the first to address the common clinical challenge of distinguishing AR from ADNR by using three-way instead of two-way classification algorithms. In the study of Li et al (18), a five-gene test was devised by qPCR from peripheral blood that effectively classified AR from a stable group (similar to our TX group). This two-way classifier was shown to have good specificity for AR against a set of non-AR biopsy-documented phenotypes comprising 12 borderline AR, 37 chronic allograft nephropathy, 16 calcineurin inhibitor (CNI) toxicities and 7 “other pathology.” Only their 16 CNI toxicities would be equivalent to our ADNR group, but if validated by testing on an independent cohort, this is an alternative strategy to use a two-way classifier to account for ADNR.
Two recent publications have described new urine signatures for AR. The first study with over 4000 urine specimens from 220 total patients describes a three-gene signature by qPCR that discriminated between biopsy specimens showing AR versus TX with AUCs of 0.83 in the validation cohort of 67 patients (37). Samples equivalent to our ADNR population were not tested but the signature was tested in cases with urinary tract, cytomegalovirus (CMV) and BKV infection. The second study involved urine profiling of six genes by qPCR and detection of CXCL9 and CXCL10 by protein ELISA in 280 adult and pediatric subjects as a two-way classifier of AR versus TX (38). Again, no clear ADNR group equivalent was studied but urinary detection of CXCL9 was revealed to have significant predictive accuracy. The strength of both these new studies was serial samples revealing the potential of these signatures to predict the future risk of presenting with clinical AR. We are currently doing a prospective serial monitoring study of blood and biopsies to validate the three-way classifiers presented here in 300 kidney transplants followed for 2 years.
The use of the Affymetrix microarray platform represents a well-established commercial technology, FDA-approved for diagnostic testing. New instrumentation enables automated, high-throughput analysis at the rate of >800 samples per week per instrument and a total undiscounted cost of approximately $275 per sample plus labor and indirect costs. Sampling of several million lymphocytes in the blood in each assay also eliminates the kind of sampling errors that are common to single biopsy cores.
We acknowledge limitations of this work that must be addressed in the next study. This is not a prospective, blinded study, and ultimately, validation of biomarkers for clinical use requires such a design. We had insufficient numbers of samples to test whether we could classify the different subtypes of T cell–mediated, histologically defined AR. Thus, we can diagnose AR, but biopsies will still be indicated when more detailed histological phenotyping is necessary. The majority of our patients were Caucasian. All the patients in this study were treated with CNIs and mycophenolic acid derivatives but the addition of steroids had no impact on the results (data not shown). Full confidence in our biomarkers will require validations in multiple new clinical centers to establish any race- and/or therapy-dependent differences, the impact of bacterial and viral infections and to remove any concern for the study of highly selected and phenotyped subjects.
Our population had no cases of pure ABMR. However, we had 12 mixed ABMR/T cell–mediated rejection cases but only 1 of the 12 was misclassified for AR. About 30% of our AR subjects had biopsies with positive C4d staining. However, supervised clustering to detect outliers did not indicate that our signatures were influenced by C4d status. At the time this study was done it was not common practice to measure donor-specific antibodies. However, we note the lack of correlation with C4d status for our data, recent data that ABMR can be present without C4d staining detected and the fact that the presence of DSA antibodies is not diagnostic of ABMR (30,31).
An open question remains what is the mechanism of ADNR since these patients were biopsied based on clinical judgments of suspected AR after efforts to exclude common causes of acute transplant dysfunction. While our results do not address this question, it is evident that renal transplant dysfunction is common to both AR and ADNR. The key point is that the levels of kidney dysfunction based on serum creatinines were not significantly different between AR and ADNR subjects. Thus, these gene expression differences are not based simply on renal function or renal injury. The second point is that our review of the biopsy histology for the ADNR patients simply revealed nonspecific and only focal tubular necrosis, interstitial edema, scattered foci of inflammatory cells that did not rise to even borderline AR and nonspecific arteriolar changes consistent but not diagnostic of CNI toxicity.
Finally, biopsy-based diagnosis is subject to the challenge of sampling errors and differences between the interpretations of individual pathologists (39). It is a fair question to ask whether the biopsy diagnoses are always correct and this goes to the underlying assumption in the field that the biopsy is the “gold standard.” To mitigate this limitation, we used the Banff schema classification and an independent central biopsy review of all samples to establish the phenotypes. Another question is how these signatures would reflect known causes of acute kidney transplant dysfunction (e.g. urinary tract infection, CMV and BK nephropathy). Our view is that there are already well-established, clinically validated and highly sensitive tests available to diagnose each of these. Thus, implementation and interpretation of our molecular diagnostic for AR and ADNR assume that clinicians would do this kind of laboratory testing in parallel. In complicated cases, a biopsy will still be required, though we note that a biopsy is also not definitive for sorting out AR versus BK.
There are several questions to answer in the next clinical study. First, can we use our molecular diagnostic to predict outcomes such as AR, especially diagnose subclinical AR, prior to enough tissue injury to result in kidney transplant dysfunction? Second, can we measure and ultimately optimize the adequacy of long-term immunosuppression by serial monitoring of blood gene expression? The design of the present study involved blood samples collected at the time of biopsies. Thus, we cannot claim at this point that our expression signatures are predictive of AR or ADNR. However, we would suggest that the absence of an AR gene profile in a patient sample would be a first measure of adequate immunosuppression and could be integrated into a serial blood monitoring protocol. Demonstrating the diagnosis of subclinical AR and the predictive capability of our classifiers would create the first objective measures of adequate immunosuppression. One potential value of our approach using global gene expression signatures developed by DNA microarrays rather than highly reduced qPCR signatures is that these more complicated predictive and immunosuppression adequacy signatures can be derived later from prospective studies like CTOT08. In turn, an objective metric for the real-time efficacy of immunosuppression will allow the individualization of drug therapy and enable the long-term serial monitoring necessary to optimize graft survival and minimize drug toxicity.
Supplementary Material
Acknowledgments
The authors would like to thank Conrad H. Lu, Tina Tan and Donald L. Tschirhart, St. Vincent Medical Center; Frank Wesley Hall, David Bylund, Edward Kane Jr. and Michael Quigley, Scripps Clinic; Jonathan L. Myles, Cleveland Clinic Foundation for local pathology reviews from the participating clinical centers. The authors also acknowledge Joanna Sung and Deandra Holland, Scripps Clinic; Barbara Mastroianni and Kathy Savas, Cleveland Clinic; Anjela Tsirunyan and Caron Hutchinson, St. Vincent Medical Center; Rebecca Rush and Anna Gordon, Mayo Clinic Scottsdale and Janis Cicerchi, Andrea Singleton and Lois Clegg, University of Colorado Hospital for the clinical coordination of this research study. Funding was provided by NIH grant U19 AI52349, U01 AI084146, the Molly Baber Research Fund and the Verna Harrah Research Fund.
Abbreviations
- ABMR
antibody-mediated rejection
- ADNR
acute dysfunction with no rejection by biopsy histology
- AR
acute rejection
- AUC
area under the curve
- BKV
BK virus
- CMV
cytomegalovirus
- CNI
calcineurin inhibitor
- DLDA
diagonal linear discriminant analysis
- eGFR
estimated GFR
- FDR
false discovery rate
- NC
nearest centroids
- NPV
negative predictive value
- PPV
positive predictive value
- qPCR
quantitative polymerase chain reaction
- ROC
receiver operating characteristic
- SVM
support vector machines
- TX
excellent functioning transplant
Footnotes
Disclosure
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. SRH, DRS, SMK and MMA are founding scientists and have ownership stock in Transplant Genomics, Inc.
Additional Supporting Information may be found in the online version of this article.
Supplemental Methods
Figure S1: The individual AUCs derived with the entire 148 sample set using a three-way NC classifier for AR versus ADNR versus TX with the Harrell method adjustment for over-fitting.
Table S1: List of 2977 differentially expressed probesets obtained by a three-class univariate F-test on the discovery cohort (1000 random permutations, FDR < 10%; BRB ArrayTools)
Table S2: List of study subject inclusion/exclusion criteria
Table S3: Diagnostic metrics for the three-way DLDA and SVM classifiers for AR, ADNR and TX in discovery and validation cohorts
Table S4: Optimism-corrected area under the curves (AUCs) for the three-way AR, ADNR and TX classifier done using all three predictive tools (NC, DLDA and SVM) with Harrell bootstrapping of all 148 samples 1000 times with replacement to create optimism-corrected AUCs. The individual AUCs for each comparison using only the NC tool are shown in Figure S1
References
- 1.Meier-Kriesche HU, Schold JD, Srinivas TR, Kaplan B. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am J Transplant. 2004;4:378–383. doi: 10.1111/j.1600-6143.2004.00332.x. [DOI] [PubMed] [Google Scholar]
- 2.Rush D, Somorjai R, Deslauriers R, Shaw A, Jeffery J, Nickerson P. Subclinical rejection—A potential surrogate marker for chronic rejection—May be diagnosed by protocol biopsy or urine spectroscopy. Ann Transplant. 2000;5:44–49. [PubMed] [Google Scholar]
- 3.Moreso F, Ibernon M, Goma M, et al. Subclinical rejection associated with chronic allograft nephropathy in protocol biopsies as a risk factor for late graft loss. Am J Transplant. 2006;6:747–752. doi: 10.1111/j.1600-6143.2005.01230.x. [DOI] [PubMed] [Google Scholar]
- 4.Nankivell BJ. Subclinical renal allograft rejection and protocol biopsies: Quo vadis? Nat Clin Pract Nephrol. 2008;4:134–135. doi: 10.1038/ncpneph0701. [DOI] [PubMed] [Google Scholar]
- 5.US Department of Health and Human Services. OPTN/SRTR Annual Report. [Accessed February 12, 2013]; Available at: http://www.ustransplant.org/annual_reports/current/509a_ki.htm. [Google Scholar]
- 6.Nankivell BJ, Borrows RJ, Fung CL, O’Connell PJ, Allen RD, Chapman JR. The natural history of chronic allograft nephropathy. N Engl J Med. 2003;349:2326–2333. doi: 10.1056/NEJMoa020009. [DOI] [PubMed] [Google Scholar]
- 7.Chapman JR. Longitudinal analysis of chronic allograft nephropathy: Clinicopathologic correlations. Kidney Int Suppl. 2005:S108–S112. doi: 10.1111/j.1523-1755.2005.09920.x. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz A, Mengel M, Gwinner W, et al. Risk factors for chronic allograft nephropathy after renal transplantation: A protocol biopsy study. Kidney Int. 2005;67:341–348. doi: 10.1111/j.1523-1755.2005.00087.x. [DOI] [PubMed] [Google Scholar]
- 9.Lerut E, Kuypers DR, Verbeken E, et al. Acute rejection in non-compliant renal allograft recipients: A distinct morphology. Clin Transplant. 2007;21:344–351. doi: 10.1111/j.1399-0012.2007.00647.x. [DOI] [PubMed] [Google Scholar]
- 10.Morrissey PE, Reinert S, Yango A, Gautam A, Monaco A, Gohh R. Factors contributing to acute rejection in renal transplantation: The role of noncompliance. Transplant Proc. 2005;37:2044–2047. doi: 10.1016/j.transproceed.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Mengel M, Gwinner W, Schwarz A, et al. Infiltrates in protocol biopsies from renal allografts. Am J Transplant. 2007;7:356–365. doi: 10.1111/j.1600-6143.2006.01635.x. [DOI] [PubMed] [Google Scholar]
- 12.Yilmaz S, Isik I, Afrouzian M, et al. Evaluating the accuracy of functional biomarkers for detecting histological changes in chronic allograft nephropathy. Transpl Int. 2007;20:608–615. doi: 10.1111/j.1432-2277.2007.00494.x. [DOI] [PubMed] [Google Scholar]
- 13.Shapiro R, Randhawa P, Jordan ML, et al. An analysis of early renal transplant protocol biopsies—The high incidence of subclinical tubulitis. Am J Transplant. 2001;1:47–50. doi: 10.1034/j.1600-6143.2001.010109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hymes LC, Warshaw BL, Hennigar RA, Amaral SG, Greenbaum LA. Prevalence of clinical rejection after surveillance biopsies in pediatric renal transplants: Does early subclinical rejection predispose to subsequent rejection episodes? Pediatr Transplant. 2009;13:823–826. doi: 10.1111/j.1399-3046.2009.01200.x. [DOI] [PubMed] [Google Scholar]
- 15.Moreso F, Carrera M, Goma M, et al. Early subclinical rejection as a risk factor for late chronic humoral rejection. Transplantation. 2012;93:41–46. doi: 10.1097/TP.0b013e31823bb647. [DOI] [PubMed] [Google Scholar]
- 16.Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbs PJ, Tan LC, Sadek SA, Howell WM. Quantitative detection of changes in cytokine gene expression in peripheral blood mononuclear cells correlates with and precedes acute rejection in renal transplant recipients. Transpl Immunol. 2005;14:99–108. doi: 10.1016/j.trim.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Khatri P, Sigdel TK, et al. A peripheral blood diagnostic test for acute rejection in renal transplantation. Am J Transplant. 2012;12:2710–2718. doi: 10.1111/j.1600-6143.2012.04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabek O, Dorak MT, Kotb M, Gaber AO, Gaber L. Quantitative detection of T-cell activation markers by real-time PCR in renal transplant rejection and correlation with histopathologic evaluation. Transplantation. 2002;74:701–707. doi: 10.1097/00007890-200209150-00019. [DOI] [PubMed] [Google Scholar]
- 20.Sarwal M, Chua MS, Kambham N, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 21.Simon T, Opelz G, Wiesel M, Ott RC, Susal C. Serial peripheral blood perforin and granzymeB gene expression measurements for prediction of acute rejection in kidney graft recipients. Am J Transplant. 2003;3:1121–1127. doi: 10.1034/j.1600-6143.2003.00187.x. [DOI] [PubMed] [Google Scholar]
- 22.Vasconcellos LM, Schachter AD, Zheng XX, et al. Cytotoxic lymphocyte gene expression in peripheral blood leukocytes correlates with rejecting renal allografts. Transplantation. 1998;66:562–566. doi: 10.1097/00007890-199809150-00002. [DOI] [PubMed] [Google Scholar]
- 23.Lee JW, Devanarayan V, Barrett YC, et al. Fit-for-purpose method development and validation for successful biomarker measurement. Pharm Res. 2006;23:312–328. doi: 10.1007/s11095-005-9045-3. [DOI] [PubMed] [Google Scholar]
- 24.Chau CH, Rixe O, McLeod H, Figg WD. Validation of analytic methods for biomarkers used in drug development. Clin Cancer Res. 2008;14:5967–5976. doi: 10.1158/1078-0432.CCR-07-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dabney AR. Classification of microarrays to nearest centroids. Bioinformatics. 2005;21:4148–4154. doi: 10.1093/bioinformatics/bti681. [DOI] [PubMed] [Google Scholar]
- 26.Shen R, Ghosh D, Chinnaiyan A, Meng Z. Eigengene-based linear discriminant model for tumor classification using gene expression microarray data. Bioinformatics. 2006;22:2635–2642. doi: 10.1093/bioinformatics/btl442. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Y, Shen X, Pan W. Network-based support vector machine for classification of microarray samples. BMC Bioinformatics. 2009;10(Suppl 1):S21. doi: 10.1186/1471-2105-10-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao YM, Cenzer IS, Kirby KA, Boscardin JW. SAS Global Forum. San Francisco: 2013. [Accessed October 28, 2013]. Estimating Harrell’s optimism on predictive indices using bootstrap samples. Available at: http://support.sas.com/resources/papers/proceedings13/504-2013.pdf. [Google Scholar]
- 29.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: Updates and future directions. Am J Transplant. 2008;8:753–760. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- 30.Sis B, Jhangri GS, Bunnag S, Allanach K, Kaplan B, Halloran PF. Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am J Transplant. 2009;9:2312–2323. doi: 10.1111/j.1600-6143.2009.02761.x. [DOI] [PubMed] [Google Scholar]
- 31.Wiebe C, Gibson IW, Blydt-Hansen TD, et al. Evolution and clinical pathologic correlations of de novo donor-specific HLA antibody post kidney transplant. Am J Transplant. 2012;12:1157–1167. doi: 10.1111/j.1600-6143.2012.04013.x. [DOI] [PubMed] [Google Scholar]
- 32.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCall MN, Irizarry RA. Thawing frozen robust multi-array analysis (fRMA) BMC Bioinformatics. 2011;12:369. doi: 10.1186/1471-2105-12-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robin X, Turck N, Hainard A, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GP GraphPad QuickCalcs: Free statistical calculators. [Accessed October 26, 2012]; Available at: http://www.graphpad.com/quickcalcs/index.cfm. [Google Scholar]
- 36.PA. Power Atlas. [Accessed October 26, 2012]; Available at: http://www.poweratlas.org/. [Google Scholar]
- 37.Suthanthiran M, Schwartz JE, Ding R, et al. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. NEngl J Med. 2013;369:20–31. doi: 10.1056/NEJMoa1215555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hricik DE, Nickerson P, Formica RN, et al. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am J Transplant. 2013;13:2634–2644. doi: 10.1111/ajt.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mengel M, Sis B, Halloran PF. SWOT analysis of Banff: Strengths, weaknesses, opportunities and threats of the international Banff consensus process and classification system for renal allograft pathology. Am J Transplant. 2007;7:2221–2226. doi: 10.1111/j.1600-6143.2007.01924.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.