Abstract
Background
For many pathogenic microorganisms, iron acquisition represents a significant stress during the colonization of a mammalian host. Heme is the single most abundant source of soluble iron in this environment. While the importance of iron assimilation for nearly all organisms is clear, the mechanisms by which heme is acquired and utilized by many bacterial pathogens, even those most commonly found at sites of infection, remain poorly understood.
Methods
An alternative protocol for the production and purification of the outer membrane hemoglobin receptor (HmbR) from the pathogen Neisseria meningitidis has facilitated a biophysical characterization of this outer membrane transporter by electronic absorption, circular dichroism, electron paramagnetic resonance, and resonance Raman techniques.
Results
HmbR co-purifies with 5-coordinate high spin ferric heme bound. The heme binding site accommodates exogenous imidazole as a sixth ligand, which results in a 6-coordinate, low-spin ferric species. Both the 5- and 6-coordinate complexes are reduced by sodium hydrosulfite. Four HmbR variants with a modest decrease in binding efficiency for heme have been identified (H87C, H280A, Y282A, and Y456C). These findings are consistent with an emerging paradigm wherein the ferric iron center of bound heme is coordinated by a tyrosine ligand.
Conclusion
In summary, this study provides the first spectroscopic characterization for any heme or iron transporter in Neisseria meningitidis, and suggests a coordination environment heretofore unobserved in a TonB-dependent hemin transporter.
General Significance
A detailed understanding of the nutrient acquisition pathways in common pathogens such as N. meningitidis provides a foundation for new antimicrobial strategies.
Keywords: Heme transport, HmbR, Hemoglobin receptor, Septic shock, Pathogenesis, Ton-B dependent transport
1. Introduction
Iron is essential for many types of biochemical reactions throughout biology. One obstacle faced by any organism that infects a host is the acquisition of iron from their environment, which is often highly regulated by the host and tightly sequestered in heme and other iron binding proteins. Of interest to this work are gram-negative bacteria that have evolved rather sophisticated mechanisms to meet their iron requirements. These are classified into two broad categories, referred to as siderophore and receptor mediated transport [1,2]. In the former, a low molecular weight compound with a high affinity for iron, referred to as a siderophore, is synthesized and secreted by the bacteria into the extracellular milieu. Upon binding to ferric iron, the siderophore is recognized and transported into the cell by a TonB-dependent receptor. After subsequent entry into the cytoplasm via an ABC (ATP-binding cassette) transport system, the iron is reduced and released [3–5]. The second mechanism involves direct binding of a heme-containing host protein to a specific TonB-dependent receptor on the surface of a Gram-negative cell. The binding interaction excises the heme molecule from the native protein and transports it into the periplasm [6–8]. Both of these types of receptors require the proton motive force of the TonB accessory protein for proper transport, and thus are referred to as TonB dependent transporters (TBDTs) [9]. Once inside the periplasm, heme is further shuttled by a periplasmic binding protein to an ABC system on the inner membrane that transports it into the cytoplasm [10]. There it is processed by a heme oxygenase to yield free iron and products that are consistent with the organism’s physiology [11].
The bacterial pathogen Neisseria meningitidis is one of the leading causes of bacterial meningitis and meningococcal septicemia [12]. The increasing number of strains resistant to traditional antibiotics has driven research into developing novel drugs for treating meningococcal infections [13]. Since virulence of this pathogen is tightly coupled to the availability of iron in the local environment of the host, one antimicrobial strategy is to disrupt its iron uptake system. Although this approach has been well recognized for several years, to date, no reliable drug treatment, based on targeting this essential pathway, exists. This deficiency is due, in part, to a lack of fundamental understanding of how iron transport occurs in this, and other bacterial pathogens. Targeting the iron uptake system of Neisseria meningitidis could be of particular interest, since synthesis of siderophores has not been definitively established in this organism, strongly suggesting that specific receptors for iron- and heme-containing host proteins are the sole means by which iron requirements are fulfilled [14]. Indeed, several such transport proteins have been identified in N. meningitidis, including transferrin, lactoferrin, and hemoglobin receptors, as well as a dual specificity hemoglobin/haptoglobin receptor [8]. Disruption of any one of these transport systems should, in theory, decrease the iron available to the organism and cause a decrease in virulence. The hemoglobin receptor (HmbR) is one of the more promising targets due to its high prevalence in invasive strains, specificity for human hemoglobin, surface accessibility, and monomeric nature [15,16]. Additionally, the expression of HmbR significantly increases when hemoglobin is the sole source of iron, suggesting that it is a major iron delivery protein that contributes to the aggressive nature of meningococcal septicemia [17]. Although HmbR is a promising drug target, like other heme transporters, very little is known about its structure, and how it extricates and transports heme from hemoglobin.
In this study we have isolated HmbR and have obtained electronic absorption, circular dichroism (CD), electron paramagnetic resonance (EPR), and resonance Raman (rR) spectra on the purified protein. Consistent with the fold of homologous proteins, the secondary structure is dominated by β sheet structure and HmbR copurifies with a 5-coordinate, high-spin heme bound. This can be converted to a 6-coordinate low spin ferric state with the addition of imidazole, and both of these species are susceptible to reduction upon treatment with sodium hydrosulfite. We also identified residues involved in maintaining proper heme binding properties, and propose that a residue with an oxygen-centered partial negative charge, such as tyrosine, acts as an axial ligand in coordinating the heme, unlike a histidine residue observed in other TBDTs, such as HasA from Seratia marcesens [18] and ShuA from Shigella dysenteriae [7,19], and others [20]. As a whole, our investigation provides the first spectroscopic characterization of any heme or iron transporter in Neisseria meningitidis, and indicates that the cell-surface binding pocket of HmbR may provide a heme axial ligand environment distinct from the above TBDTs characterized to date.
2. Materials and methods
2.1. Creation of HmbR variants
The plasmids used in this study are listed in Table 1. Residues selected for mutational analysis were determined in two ways. Mutations H87C, H280A, Y282A, and Y497C were selected from regions on HmbR determined to be important in heme binding from a previous study [21]. Mutations Y456C and Y763C were selected from conserved residues identified from a ClustalW sequence alignment of HmbR with other outer membrane heme and iron transport proteins, specifically, BpR, HemR, HutA, ShuA, and YpR (accession numbers NP_879220, CAA48250, YP_002812154, AAC27809, and Q56989, respectively)[22]. Each mutation was engineered using Quickchange site-directed mutagenesis. In brief, complimentary mutagenic oligonucleotide primers were designed to contain the desired mutation flanked by a 15–20 base pair extension on each end homologous to the parent plasmid (DPB1809). Each PCR contained a total volume of 50 μL with 4 μM of each primer, 200 μM of each dNTP, and 10 μg of parent DNA, in ThermoPol reaction buffer (20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100). After preheating each condition at 95 °C for 5 min, 1.0 μL of VentR® DNA Polymerase (New England Biolabs; #M0254S) was added to each tube, and the PCRs were allowed to proceed (annealing: 55–58 °C, 1 min; extension: 68–72 °C, 20 min; denaturing: 95 °C, 1 min; 18 complete cycles). Following the PCR, 1.0 μL of DpnI (New England Biolabs; #R0176S) was added to each tube and allowed to react at 37 °C for 6 h. The suspensions were then transformed into competent DH5α E. coli, and identification of plasmids with the desired mutations was performed by subsequent isolation and sequencing of plasmid DNA from candidate colonies.
Table 1.
Plasmids used in this study.
| Plasmid | HmbR Variant |
|---|---|
| pDPB1809 | Wild-type |
| pDZMl | H87C |
| pDZM2 | H280A |
| pDZM3 | Y282A |
| pDZM4 | Y456C |
| pDZM5 | Y497C |
| pDZM6 | Y763C |
2.2. Purification of HmbR
Wild-type and single amino acid HmbR mutants were purified by identical methods. In general, competent BL21-CodonPlus (DE3)-RIPL E. coli (Stratagene) were transformed with the appropriate plasmid, and grown in 2-L Erlenmeyer flasks each containing 1 L of LB with 50μg/mL carbenicillin. When the cells reached A600 0.6–0.9, HmbR expression was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 2.5 h. Owing to low yields, approximately 24 L of cell pellets were utilized for the purification of each HmbR variant. Cell pellets were collected and resuspended in 225 mL of 10 mM potassium phosphate, pH 6.8 supplemented with 2 complete EDTA-free protease inhibitor tablets (Roche; #04693132001), and approximately 1 mg each of PMSF, trypsin inhibitor, aprotinin, leupeptin, chymostatin, pepstatin, and DNAse. The cell suspension was then subsequently passed twice through a French pressure cell, and then spiked with 1 mM of MgSO4, and cleared of large cellular debris by spinning at 6370 g for 15 min. The resulting supernatant was supplemented with hemin to a final concentration of 2.1 μM (using a 4 mM stock concentration containing 10 mM potassium hydroxide prepared in ethanol), and fractionated at 100,000 g for 1.25 h to recover membranes. The isolated membranes were washed once in 150 mL of 20 mM potassium phosphate, pH 6.8, 2% Triton X-100, recovered at 100,000 g for 1.25 h, and incubated overnight at 4 °C in 150 mL of 20 mM potassium phosphate, pH 7.4, 500 mM NaCl, and 1% n-octyl-β-D-glucopyranoside (OG). The unsolubilized material was recovered at 100,000 g for 1.25 h, and incubated overnight at 4 °C in 150 mL total volume of 20 mM potassium phosphate, pH 7.4, 500 mM NaCl, and 1% Anzergent™3-14 (Anatrace). Solubilized protein was recovered by centrifugation (100,000 g for 1.25 h) and then supplemented with 10 mM imidazole, and allowed to incubate with 20 mL of a Ni-NTA resin overnight. The suspension was then transferred to a gravity-flow column, washed with 90 mL of 20 mM potassium phosphate, pH 7.4, 500 mM NaCl, 10 mM imidazole, and 1% Anzergent™3-14, followed by elution with 20 mM potassium phosphate, pH 7.4, 500 mM NaCl, 500 mM imidazole, and 0.5% Anzergent™3-14. Elution fractions containing the highest protein concentrations were pooled into a centrifugal filter unit with a 50 kDa nominal molecular weight cut-off (NMWC) (Millipore; #4323), and concentrated to an approximate volume of 1.0 mL by 10-min centrifugation intervals at 1500 g. The imidazole was removed from these samples by two different methods. For hemochromogen assays (described below), 500 μL of each protein variant (concentrations ranging from 2 to 10 mg/mL) was transferred to a Slide-a-Lyzer cassette (Pierce, 7000 NMWC), and allowed to equilibrate in 1 L 20 mM potassium phosphate, pH 7.4, 500 mM NaCl, and 0.5% Anzergent™3-14 for 24 h. For all other applications, the imidazole was removed from the samples by iterative dilutions and concentrations in an Amicon concentrator (5000 NMWC) using 20 mM potassium phosphate, pH 7.4, 500 mM NaCl, and 0.5% Anzergent™3-14 until the concentration of imidazole was less than 5.0 mM. The final imidazole concentration was determined by calculating the dilution factor of the retentate after each spin.
2.3. Electronic absorption spectrum
Electronic absorption spectra of different HmbR species were recorded both aerobically on a nanodrop (ND-1000) spectrophotometer (400 μM/mL protein, 1.5 μL/sample) and anaerobically on a Shimadzu (UV-1601) spectrophotometer (66 μM/mL, 600 μL/sample) at room temperature. Each method evaluated purified HmbR with or without the presence of 500 mM imidazole, in both ferric and ferrous states. To record spectra under anaerobic conditions, samples and buffers were degassed using a vacuum manifold, and transferred to an anaerobic chamber. A 100 mM stock solution of sodium hydrosulfite was subsequently prepared in the appropriate buffers and titrated into samples at 0, 160, 330, and 500 μM intervals unless stated otherwise. Reduction of the bound heme was attempted with either Cleland’s reagent or sodium hydrosulfite. In both cases, the reduced sample was less stable and led to precipitation over time, however, reduction with sodium hydrosulfate occurred quickly and provided ample time to work with the sample. Electronic absorption spectra were obtained using samples that were anaerobically sealed in quartz cuvettes with a 1-cm path length. All plots were generated using Excel graphing software from data electronically extracted from the nanodrop (ND-1000) software program (version 3.3).
2.4. Determination of molar percent of heme bound to HmbR variants
In order to determine the molar percent of heme present within purified HmbR samples, we performed a pyridine hemochromogen assay as previously described [23]. In general, samples dialyzed using Slide-a-Lyzer cassettes as described above were diluted to 3.33 μM in dialysis buffer (20 mM potassium phosphate, pH 7.4, 500 mM NaCl, 0.5% Anzergent™3-14, and 5 mM imidazole) and further diluted to 2.50 μM by the addition of 250 mM NaOH and 25% pyridine. Half the volume of these samples was removed and reduced with sodium hydrosulfite, and the absorbance difference between the reduced and unreduced samples was determined at 530 nm. Heme concentrations were calculated using a millimolar extinction coefficient of 20.7 cm−1 mM−1. These were compared to protein concentrations previously determined through a detergent compatible protein assay (Bio-Rad DC). Each HmbR variant was evaluated by this method a minimum of three independent times.
2.5. Circular dichroism
A circular dichroism spectrum was recorded for purified HmbR obtained as described above in the ferric state. The sample was diluted to 3.9 μM/mL in 10 mM potassium phosphate, pH 7.4, 250 mM NaCl, and 0.25% Anzergent™, and placed in a 0.1 cm cuvette for data collection. A Jasco J-715 spectropolarimeter obtained measurements from 190 to 240 nm using a 10 nm bandwidth. Following data collection, the Y-axis was converted to molar ellipticity (Δε) and plotted as a function of wavelength utilizing software associated with the instrument.
2.6. Electron paramagnetic resonance
EPR spectra were recorded on samples containing approximately 200 μM of HmbR in sample buffer (20 mM potassium phosphate, pH 7.4, and Anzergent™3-14) with or without the addition of 500 mM imidazole using a Bruker ESP300D spectrometer equipped with an Oxford Instruments ESR-900 helium flow cryostat to maintain the temperature of the samples at 10 K. The modulation amplitude was 6.477 G with a modulation frequency of 100 kHz. The microwave frequency and power were 9.6 GHz and 5 mW, respectively.
2.7. Resonance Raman spectroscopy
Resonance Raman scattering was excited with 406.7-nm emission from a Kr+ laser using a power of 14 mW, in the 135° backscattering geometry. Samples were prepared in 0.05% Anzergent™, 300 mM NaCl, and 100 mM buffer (MES, Tris, or CAPS, pH 6.4, 7.8, and 9.7, respectively). Samples were contained in 5 mm NMR tubes and spun at 20 Hz at room temperature to avoid laser-induced degradation. Scattered light was collected, filtered to remove Rayleigh scattered light and focused onto the slit of a 0.67-m spectrograph. Spectra were recorded with a liquid nitrogen cooled CCD camera. Raman shift axes were calibrated against known Raman shifts for toluene and DMF. No spectral artifacts attributable to laser-induced sample damage were observed.
3. Results
3.1. Absorption spectra of purified HmbR
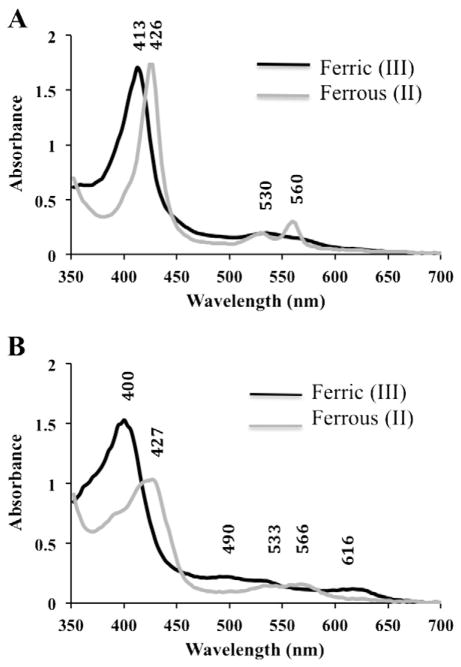
The protocol described above (Section 2.2) for the purification of HmbR yielded protein of the predicted size (90 KDa) with more than 90% purity as judged by SDS-PAGE densitometry analysis using Coomassie stain (Supplemental Fig. 1). The removal of imidazole from purified protein fractions resulted in a qualitative color change from bright red to rust brown. Based on this observation, we hypothesized that the imidazole could interact with the heme that was bound to HmbR during purification, consistent with the behavior reported for other outer membrane heme transporters [7,18,19]. In order to test this, we monitored the electronic absorption spectra of samples in the absence and presence of imidazole, in both the ferric and ferrous states (Fig. 1). The ferric HmbR sample, in the presence of 500 mM imidazole, exhibited a Soret maximum at 413 nm that shifted to 426 nm when reduced. In addition, this sample also had the corresponding Q-band maximum at 535 nm, which split into α and β bands at 560 nm and 530 nm, respectively (Fig. 1, Panel A). These spectral features are characteristic of hexacoordinate, low spin heme hemophores [24]. HmbR samples that were free of imidazole conveyed slightly different behavior in their absorption spectra. Specifically, the ferric sample had a Soret maximum at 400 nm that shifted to 427 nm when reduced, consistent with high-spin pentacoordinate hemes (Fig. 1, Panel B) [24]. This sample also had small α and β bands at 616 nm and 490 nm, respectively, which shifted to 566 nm and 533 nm when reduced. Reduction of HmbR under aerobic or anaerobic conditions was possible, however, the intensity of these bands was significantly depressed relative to the more prominent peaks observed in the presence of imidazole. Cleland’s reagent is a relatively mild reductant that can be used under aerobic conditions. In contrast, and in order to confirm that the observed reduction in the intensity of absorption spectra peaks was not a result of spurious artifacts resulting from the re-oxidation of the aerobically-reduced HmbR during the course of the experiment, we also performed a titration experiment under anaerobic conditions using limiting quantities of sodium hydrosulfite, and acquired electronic absorption spectra from these samples. No change in the overall spectra was seen when compared to the spectra obtained when measurements were recorded aerobically. In fact, attempts to reoxidize the sample were unsuccessful, leading to protein precipitation. While the redox state of the heme molecule may be important in the context of the transport mechanism and handoff to a periplasmic transporter, at this time, we can only qualitatively conclude that reduction appears to result in a less soluble/stable protein. Hence, the scan of the reduced sample devoid of imidazole must be interpreted with caution, as some of the peaks could be attributed to a mix of non-native protein configurations in the conditions tested.
Fig. 1.
Electronic Absorption Spectra of HmbR. Purified HmbR in sample buffer (20 mM phosphate, pH 7.4, 500 mM NaCl) was subjected to UV-visible absorption scans either A) as purified (with 500 mM imidazole) or B) after removal of imidazole. Black lines represent the spectra of samples in the ferric state, and gray lines represent the spectra of samples reduced to the ferrous state by the addition of 5 mM sodium hydrosulfite to the sample. Each spectrum was obtained using 1.5 μL of protein at approximately 35 mg/mL on a nanodrop (ND-1000) spectrophotometer. Soret, α, and β peaks are indicated by their respective wavelengths within the graphs.
3.2. Point mutations in conserved residues of HmbR do not abolish heme binding
Our purification strategy utilizes detergent micelles in order to solubilize HmbR, which complicates determining the absolute stoichiometry for heme binding to the receptor. However, the intensities of our absorption spectra were less than would be anticipated for a 1:1 ratio of heme:HmbR. We therefore quantified the molar percent of heme to protein present using an established pyridine hemochromogen assay and determined that approximately 30% of the HmbR present was saturated with heme (Table 2). In order to test the heme binding mechanism, and to potentially obtain protein with an increased affinity for heme binding for facilitating spectroscopic studies, we created a number of point mutations either targeting tyrosines in highly conserved regions of HmbR compared to other proteins (Y456C and Y763C), or residues within regions previously determined to be functionally significant for heme binding within HmbR itself (H87C, H280A, Y282A, and Y497C) [21]. Of the six variants evaluated, four showed a statistically significant decrease in the amount of residual heme that co-purified with HmbR (17%–20%) relative to wild-type (30%). These mutations were H87C (17.6%), H280A (18.2%), Y282A (20.5%), and Y456C (17.6%) (Table 2). The remaining two (Y497C and Y763C) had a diminished effect, but not pronounced enough to be considered statistically significant. These results are consistent with the previous deletion study of HmbR, which showed that partial heme transport capacity was retained, even when large stretches of amino acids were deleted from the transporter [21] (see discussion).
Table 2.
Molar percent of heme to protein of HmbR variants.
| HmbR Variant | Molar Percent of Heme to Protein | P-value1 |
|---|---|---|
| Wt | 30.4 ± 0.4 | * |
| H87C | 17.6 ± 0.4 | .007 |
| H280A | 18.2 ± 0.1 | .009 |
| Y282A | 20.5 ± 0.1 | .019 |
| Y456C | 17.6 ± 0.2 | .009 |
Relative to wild-type.
3.3. Circular dichroism of wild type HmbR
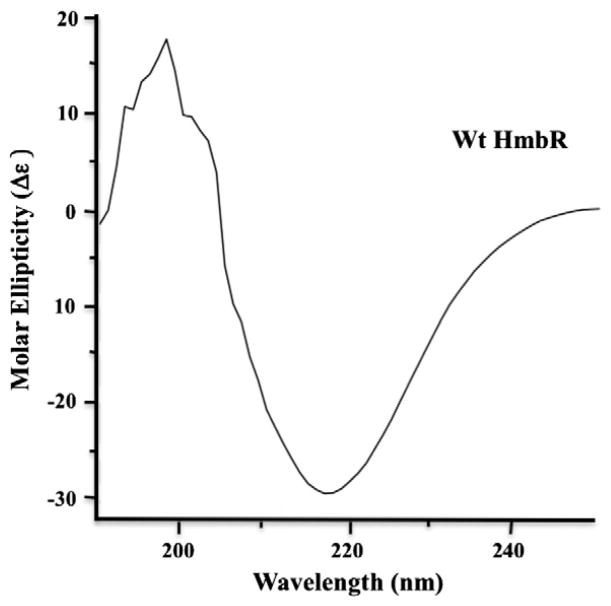
We considered the possibility that the low molar binding ratio of heme to HmbR may be due to mild protein denaturation leading to a loss in affinity for heme. This would also explain why we were unable to identify mutants with enhanced heme binding. While no crystal structure has been reported for HmbR, it does perform a similar biological function to a number of TonB-dependent heme transporters with known structures, as well as some sequence identity (27% and 21% for HasR and ShuA, respectively), with higher (up to 30%) sequence similarity (SIAS server), [7,18,22]. Predictive modeling of HmbR suggests a tertiary structure containing a classical 22-stranded β-barrel [21]. Bearing this in mind, we obtained a circular dichroism spectrum on the wild-type ferric imidazole-free sample in order to establish whether the overall native structure was intact. The spectrum is consistent with a secondary structure composition dominated by β-sheets and similar to the CD spectrum of the functionally related TBDT, ShuA (Fig. 2) [7, 19]. Although we cannot rule out the possibility that a critical residue was oxidized during purification, or that a conformational change occurred which was not detected by CD, the spectrum does not exhibit a significant contribution of the random coil signature, consistent with the protein being essentially completely folded and is further addressed in the discussion. The irregular appearance of the curve between 195 and 205 nm is likely attributable to a combination of residual oxygen in the solution and efficient off-axis scattering of the UV light by the detergent micelles in which the protein was solubilized.
Fig. 2.
Circular dichroism of ferric HmbR in sample buffer (10 mM potassium phosphate, pH 7.4, 250 mM NaCl, and 0.25% Anzergent™) at approximately 0.34 mg/mL protein concentration. Data was collected using a Jasco J-715 spectropolarimeter and a 0.1 cm cuvette containing the sample. Measurements were taken from 190 to 250 nm using a 10 nm bandwidth. Data were plotted as a function of molar ellipticity (Δε) utilizing software associated with the instrument. The irregularity of the curve between 195 and 205 nm is likely attributable to a combination of residual oxygen in the solution and efficient off-axis scattering of the UV light by the detergent micelles in which the protein was solubilized.
3.4. Electron paramagnetic resonance of HmbR
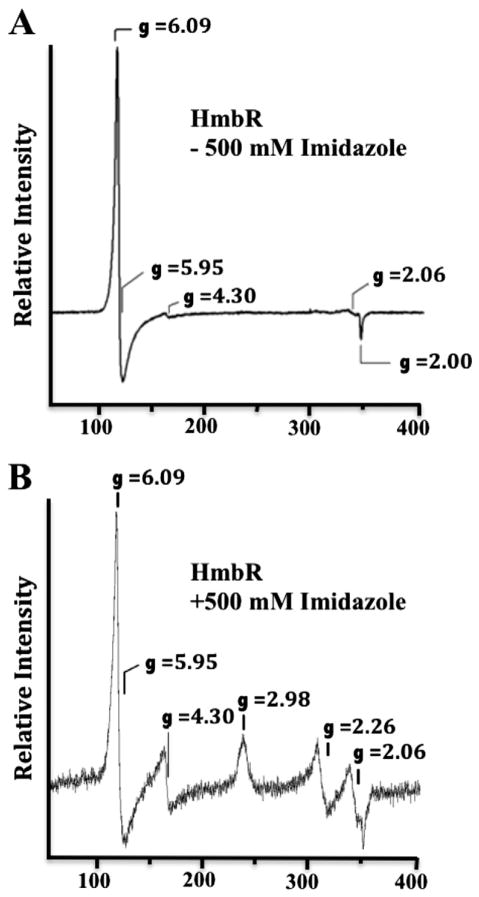
The electronic absorption spectra of HmbR indicate a hexacoordinate heme in the presence of imidazole, and a pentacoordinate in the absence of imidazole. To augment these findings, and to elucidate details on the spin states and coordination environment of the heme, we performed EPR on ferric HmbR with and without 500 mM imidazole. In the absence of imidazole, the EPR spectrum reveals a classic 5-coordinate high spin axial ferric heme signal characterized by g-values of 6.09 and 2.00 (Fig. 3, Panel A). In the presence of imidazole, the EPR spectrum reveals significant new features at gz = 2.98 and gy = 2.26 (Fig. 3 Panel B). Due possibly to g strain, the gx feature was not observed in this spectrum. Spectra of hexacoordinate LS hemes having low-field g values between 2.9 and 3.6 typically arise from complexes in which two axial heterocyclic ligand rings, such as imidazole, are orientated orthogonal to one another [25]. However, the axial ligand planes in complexes that exhibit an intermediate g value (i.e. gy) >2.0 are typically parallel. We did not quantify the high spin to low spin signals due to the high microwave power and low temperature involved, factors that can distort making accurate quantifications. In an initial effort to potentially stabilize the detergent solubilized HmbR with ferrous heme bound, samples were treated with Cleland’s reagent (dithiothreitol) or sodium hydrosulfite. Both compounds appeared to reduce the bound heme, although sodium hydrosulfite was much more effective and both appeared to decrease the stability of the sample. Additionally, we observed that the heme can accommodate cyanide. However, the addition of these reducing agents or cyanide dramatically altered the stability of the protein, which complicates performing reliable EPR studies. For this reason, we do not elaborate on these results here. In any case, the EPR spectra are consistent with a change from pentacoordinate to hexacoordinate ligation upon addition of exogenous imidazole to ferric holoHmbR. Alternatively, this is consistent with the formation of a 6-coordinate low spin ferric heme in which the two axial ImH planes are approximately parallel and oriented along a vector connecting opposing meso carbon atoms of the porphyrin macrocycle [26]. Finally, the excreted bacterial hemophores, HasAs, comprise His/Tyr axial ligand sets [27,28] and exhibit EPR features at gz = 2.8, gy = 2.2 and gx =1.7, [27,29] similar to those observed here for ferric HmbR in the presence of imidazole. Thus, the EPR is perhaps most consistent with the increasingly common axial Tyr ligand in bacterial hemin transport proteins [30,31].
Fig. 3.
Electron paramagnetic resonance spectra of HmbR. EPR spectra were recorded on samples containing approximately 200 μM of HmbR in sample buffer (20 mM phosphate, pH 7.4, and Anzergent™3-14) either without imidazole (Panel A), or in the presence of 500 mM imidazole (Panel B). All data were collected using a Bruker ESP300D spectrometer equipped with an Oxford Instruments ESR-900 helium flow cryostat to maintain the temperature of the samples at 10 K. The modulation amplitude was 6.477 G with a modulation frequency of 100 kHz. The microwave frequency and power were 9.6 GHz and 5 mW, respectively. The signal at g = 4.3 is attributed to minute contamination of the protein sample by rhombic iron(III). The feature at g = 2.06 is attributed to copper contamination of the EPR cavity.
3.5. Resonance Raman spectroscopy of HmbR
The ferric state of HmbR was examined by resonance Raman (rR) spectroscopy with the goal of identifying the native axial ligand or ligands in the absence of exogenous imidazole. Fig. 4 shows the Soret-excited rR spectra of this species at pH 6.4, 7.8, and 9.7. The high-frequency signature of the heme in Fig. 4A reveals marker bands ν2, ν3, and ν4 at 1570, 1489, and 1370 cm−1, respectively, all of which fall within the frequency ranges of five-coordinate high spin (5cHS) ferric heme. The low frequency spectra in Fig. 4B contain a combination of in-plane, out of plane and peripheral substituent bands typical of ferric hemes. The band at 518 cm−1 has been observed in a number of heme proteins having proximal Tyr ligands and is consistent with the axial ligand being coordinated through an oxygen atom with at least a partial negative charge [26,32–35]. The persistence of this band at pH 6.4, the I(ν3):I(ν4) ratio of ~0.7 (vide infra), and the invariance of the overall rR signature with changing pH suggest that the axial ligand is not hydroxide. This rR fingerprint and its insensitivity to pH are strongly reminiscent of rR spectra recorded from ShuT, a pentacoordinate periplasmic heme binding protein in which the proximal hemin ligand is the phenol side chain of a Tyr residue [26] (Table 3).
Fig. 4.
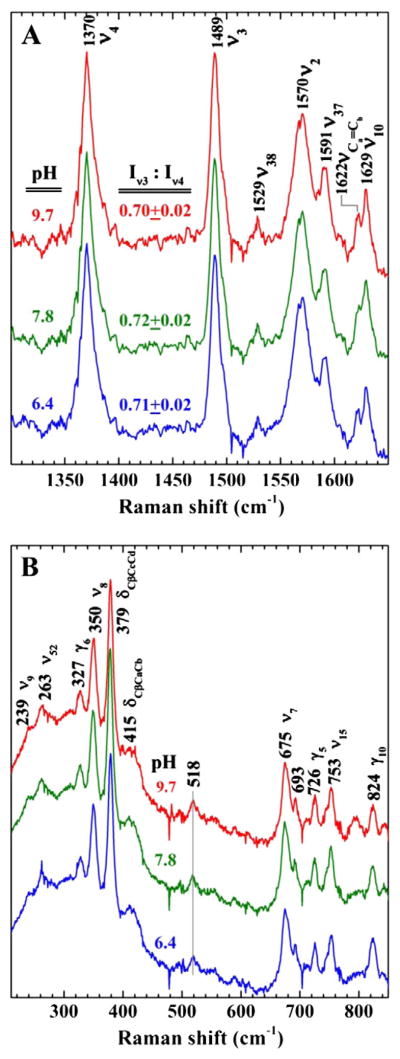
Resonance Raman spectrum of HmbR. Resonance Raman of HmbR showing the A) high frequency and B) low frequency spectra. Each panel has three spectra (bottom, middle, and top) corresponding to different buffers (MES, Tris, or CAPS) and pH conditions (6.4, 7.8, and 9.7), respectively. Samples were prepared in 0.05% Anzergent™, 300 mM NaCl, and 100 mM of the indicated buffer. Data were obtained using a 406.7 nm emission from a Kr+ laser using a power of 14 mW, a 135° backscattering geometry, and spun at 20 Hz in 5 mM NMR tubes at room temperature to avoid laser induced degradation. The laser bean was calibrated against Raman frequencies of toluene and DMF, and focused to a line on the spinning NMR tubes. The identity of certain peaks is indicated in each graph.
Table 3.
Resonance Raman frequencies of tyrosinate coordinated heme binding proteins.
4. Discussion
The acquisition, transport, and degradation of heme by a pathogenic organism represents a crucial pathway for iron and/or heme acquisition during host infection when critical nutrients/cofactors may be limiting, qualities that make proteins involved in this process good drug targets [1–8,10,11]. Ideally, the best targets are those that are involved in the primary stages of heme or iron extrication with exposure to the extra-cellular environment to facilitate drug accessibility. However, the very nature of these qualities mandates that the protein be fully or partially embedded in the membrane, which complicates structural and functional characterization studies. Nonetheless, in this work, we present the first isolation and spectroscopic characterization of an outer membrane heme transporter in Neisseria meningitidis. Electronic absorption, EPR, and resonance Raman data support the conclusion that a 5-coordinate high spin ferric heme is bound to HmbR in the “as isolated” state. We can also conclude that the heme is bound in such a way that the heme-binding pocket is accessible to solution. This is supported by the conversion of some of the axial high-spin Fe(III) EPR signal into that of a 6-coordinate low spin ferric species, upon the addition of imidazole (Fig. 3). This behavior would not be expected for a heme sequestered by hexacoordination in a closed protein conformation.
Interestingly, the hemin bound protein was substoichiometric at only 30% (Table 2). This partial loading was not a result of a lack of porphyrin in the cells, as addition of d-aminolevulinic acid (ALA) to cell cultures did not increase HmbR heme saturation (data not shown), and even routine addition of excess exogenous hemin to crude cell lysates during protein preparation did not confer higher heme binding ratios. Hemin was not added to purified HmbR directly because it resulted in irreversible protein precipitation. In a protein that harbors heme or hemin as a static cofactor, such low loading could be taken as an indication of low hemin affinity and could even call into question whether the binding of hemin is physiologically relevant. However, TonB-dependent heme transporters participate in active translocation of hemin into the periplasm [1]. As such, it is difficult to assign importance of high affinity to function because the stability of any particular hemin-bound state of the protein is likely to depend upon the status of its interactions with the cognate hemin donor and acceptor proteins as well as the energy transducing machinery (i.e. the TonB complex), which directs the free energy necessary for active transport across the outer membrane. Given that none of those native components are present in the heterologous expression system used, it is perhaps not surprising that the receptor is isolated with substoichiometric hemin. This is actually quite a common phenomenon when dealing with heme containing proteins and transporters produced in such systems. For example, utilizing the same established hemochromogen assay that we used here, a previous study established that the purified C. elegans Dual Oxidase 1 peroxidase domain from insect cells contained only 50% heme incorporation into the protein [36]. Even endogenous expression of the cytochrome c synthetase (CcsBA) from E. coli results in purified protein loaded with as little as 25% heme [37], and even lower heme loading ratios have been reported, as in the case of the Serratia marcescens HasR heme receptor, which contained only 10% heme incorporation, three times lower than our reported value [38]. One potential concern is that the detergents used to solubilize HmbR induce a non-native conformation with consequent loss of hemin during purification. While we have observed variable heme:protein stoichiometries ranging from 30% to 50% using different detergents and buffers to purify wild-type HmbR, we achieved the most stable and consistent results using the current protocol, which was the result of a solubility and stability screen involving 17 different conditions, modified from a previous study (data not shown), [39]. Although our stabilized protein results in a sub-stoichiometric heme:protein content, we highly caution addition of endogenous heme after protein purification to any heme binding protein (putative or established) to achieve what might be considered a desired binding ratio, because the heme can be promiscuously bound and/or interfere with the native structure of the protein, which in turn can alter the heme coordination environment and interpretation of results. Here, we ascertain that the heme binding domain in HmbR is natively folded, as deemed by the following: 1) The circular dichroism spectrum in Fig. 2 reveals that HmbR possesses an overall native β-barrel fold after purification, 2) the spectral signature of this spectrum is consistent with previous reports of the behavior of other TonB-dependent hemin transport proteins in the detergents we have used [7,40,41], and 3) the electronic absorption, 4) EPR, and 5) rR analysis all show conformational heterogeneity of the heme that co-purified with HmbR. Taken together, we deem that the heme environment analyzed in this study is biologically relevant, and is not due to promiscuous interactions with HmbR, even if other regions of the protein outside the heme binding domain were altered.
The HmbR residues that were mutated where selected because their side chains are candidate ligands for hemin. Based largely on sequence homology with other TBDT’s for hemin, all of the mutated residues are either on the extracellular loop analogous to that shown to harbor a coordinating His residue in HasR, or on the apex of the plug domain that resides inside the β-barrel structure [18]. Thus, all of these candidate ligands are likely to be in or near the cell-surface binding site of the receptor. All of the HmbR variants investigated here have affinity for hemin, as judged by their having been isolated and purified with some hemin bound, and are therefore not obligatory for hemin binding (Table 1). This observation further supports the idea that multiple ligands can participate in compensatory hemin binding, and that complete loss of affinity for hemin might only be observed when two or more mutations are made in crucial heme ligands involved in the complete transport mechanism. These alternative ligation states are consistent with considerable flexibility in the binding site, an essential quality of any membrane transport protein, and a finding that is consistent with a previous study that also failed to identify an HmbR mutant with complete loss in heme binding capacity [21]. In that study, nineteen deletions and site directed HmbR mutants were generated. Two internal deletions in the putative extracellular L2 and L3 loops did demonstrate a decrease in heme binding, but did not occlude the presence of heme altogether [21], similar to what we observed with the site mutants generated in the current study. Although all of the variants in this study clearly have some affinity for hemin, it is not possible to conclude, based only on fractional loading of the variants, whether all of these hemin-bound states are physiologically relevant to transport, as the reduced heme loading ratios could be a consequence of tertiary structural changes induced by the mutations themselves, or due to uncontrollable variables as applied to otherwise routine protein preparations. More importantly, in the absence of heme dissociation constants, it cannot be assumed that reduced heme loading is a result of reduced heme affinity. It is, nevertheless, interesting to consider that there may be multiple ligation states that are relevant to transport. For example, it is conceivable that, in contrast to the proposed mechanism for hemin acquisition via HasR (PDB IDs 3CSL, 3CSN, and 3DDR), there are sequential ligation states involving multiple Tyr residues on the extracellular loop that are populated during the transfer of hemin from host Hb to the cell surface binding site of HmbR. Pursuit of this question will require in vivo growth experiments with the HmbR mutants constructed here, among others.
Based largely on our Raman spectra in Fig. 4, we propose that an endogenous axial ligand coordinates to Fe(III) through an oxygen atom that has a partial negative charge, as deemed by the following evidence. Firstly, we report a I(ν3)/I(ν4) ratio of 0.7. Such high I(ν3)/I(ν4) ratios (i.e. between 2/3 and 1.0) are a common Soret-excited rR signature of 5c HS hemes having an axial anionic ligand bound through an oxygen atom. Similar spectra have been reported for other bacterial heme trafficking proteins, including the periplasmic hemin binding protein, ShuT, from S. dysenteriae [26] and the hemophore mutant, HasA(H32A), from S. marcesense [56]. Additionally, this rR signature has been reported for the heme-containing Asperillus niger catalase [42], the myoglobin mutant Mb(H93Y) [43], and the heme chaperone, CcmE, [44]. Although high I(ν3)/I(ν4) ratios are not diagnostic for proximal Tyr coordination, together with the insensitivity of the rR spectral signature to pH, it constitutes strong evidence for axial Tyr ligation of the heme. Secondly, and most convincingly, the current literature reports a range of νFe − Tyr modes for tyrosinate ligated hemes with a frequency range between 502 and 613 cm−1 (Table 2) [26,31–35]. The low-frequency rR spectra in Fig. 4B reveal a band at 518 cm−1, which falls within this range. Much like the rR spectra of heme catalases [45], this band is insensitive to changes in pH from 6.4 to 9.7, indicating that it is not susceptible to axial ligand substitution by solvent and does not participate directly in acid–base reactions over this range. As in the case of ShuT [26], this behavior argues that the rR signature is not attributable to a hydroxide complex and that the 518-cm−1 band does not originate from an Fe–OH stretching mode. Interestingly, our value of the νFe−Tyr frequency of 518 cm−1 for holo HmbR is in the lower part of the published range of 502–613 cm−1, and actually represents the second lowest frequency reported thus far for any tyrosinate ligated heme, with the lowest being 502 cm−1 for the ferric heme of Chlamydomonas chloroplast hemoglobin [32]. In that study, it was postulated that the low frequency was due to the 6-coordinate low-spin configuration of the heme group, with one mutant displaying a vibrational mode as low as 498 cm−1. Although the precise reason for why tyrosinate ligated hemes present specific νFe − Tyr modes is beyond the scope of this paper, the lower value we observed could be due to the microenvironment of the Fe–O–Ph bond, driven by factors such as Fe–O bond strengths and angles, the orientation of the tyrosinate ring relative to the heme plane, and the side chains that interact with the axial heme ligands, among others [26,35]. It is also plausible, if not likely, that the lower νFe–Tyr frequency reports a state of pentacoordinate hemin:HmbR in which the Fe–Tyr bond is allosterically strained. Weakening of the Fe–Tyr bond is a foregone conclusion of the progression of hemin to its next binding site along the transport coordinate. That we observe a weak Fe–Tyr bond in purified hemin:HmbR suggests that the observed hemin-bound state is activated toward the axial ligand exchange necessary for hemin transport across the outer cell membrane. In other words, the aforementioned weak bond is intrinsic to the complex and does not require interaction with TonB. Future studies should be aimed at monitoring the heme iron during active transport to test this hypothesis.
Our results contribute to an emerging theme among extracellular and cell surface hemin binding sites in hemin acquisition systems of both Gram-negative and Gram-positive pathogens [26,31–35] (Table 3). Specifically, all of the secreted hemophores, from both Gram-negative and Gram-positive pathogens, for which hemin-bound structures have been determined exhibit H-bond-stabilized axial Tyr coordination to the Fe(III) center of hemin. In the HasAs from S. marcesens (1DK0), P. aerugenosa (3MOL), and Y. pestis (4JET) the H-bonding partner for the proximal Tyr is the ImH side chain of a His residue [46–48]. Moreover, in the Y. pestis protein, crystal structure reveals Tyr to be the only endogenous ligand [48]. In the heme-binding NEAT domains of the IsdX1 (3SIK) and IsdX2 (4H8P) hemophores from B. anthracis, the bound hemin is also pentacoordinate, and the H-bond partner of the proximal Tyr is the phenol side chain of another Tyr residue in a YXXXY hemin binding motif [49–51]. Additionally, the hemin-binding NEAT domains of the cell-wall-tethered hemin receptors, IsdB (3RTL), IsdA (2ITF) and IsdC (2O6P) from S. aureus also exhibit H-bond-stabilized Tyr bonding within the YXXXY motif [52–55]. All of the holohemophores and Gram-positive cell wall receptors, 5- and 6-coordinate alike, appear to have large formation constants. Thus, it appears as though H-bond-assisted Tyr coordination is a widely used and very stable coordination environment for hemin acquisition and transport. By abolishing the H-bonding interaction of the coordinated Try75 in holoHasASM(H83A) 13.8 kJ/mol of stability is lost (255-fold decrease in Kf) relative to WT [56–58]. Thus, it is reasonable to hypothesize that loss of stability in these H-bond stabilized Tyr complexes is activated by a reorganization of the protein structure that disrupts the proximal H-bonding interaction. This mechanism is biologically sound, as HmbR has to extract the heme from hemoglobin, yet must be capable of releasing it to a peri-plasmic transporter through interaction with TonB. We suggest that the pentacoordinate O-bound axial ligand in the pH-insensitive, pentacoordinate holoHmbR is a Tyr residue whose coordination to the acquired hemin is stabilized by H-bonding with either a nearby His or Tyr residue. This would account for the driving force necessary to acquire hemin from host Hb with the installation of a means to trigger release of the Tyr-ligated hemin through a conformational change, driven by interaction with TonB, that breaks the stabilizing H-bond. Such a release of hemin from the cell-surface binding site would be crucial to the non-equilibrium protein dynamics that translocate hemin across the outer membrane, a process that remains poorly understood at this time.
HmbR-mediated transport is likely the primary means of iron acquisition during meningococcal septicemia, and positively correlated with the virulence of the pathogen [17]. This necessarily requires a source of hemoglobin, which is likely derived from hemolysis resulting from the secretion of specific lytic factors produced by the bacteria [59,60]. Previous reports have demonstrated that the release of free oxyhemoglobin from intravascular erythrocyte lysis quickly undergoes autoxidation to yield methemoglobin [61]. Furthermore, we have observed a unique electronic absorption peak when HmbR was titrated with as little as 0.25 molar equivalents of methemoglobin in its tetramer form, and this signature was different from HmbR titrated with hemin, or hemoglobin alone (data not shown). Hence, our identification of a ferric heme in HmbR is consistent with the suggestion that methemoglobin is the biologically relevant substrate. Future investigations evaluating the degree of HmbR specificity for methemoglobin over hemin should be compared with other forms of hemoglobin utilizing more analytically sensitive techniques. Insight into the structural basis for excision of hemin from methemoglobin and transfer to HmbR remains difficult to obtain without a high-resolution structure or kinetic data. To date, only one structure for a TonB-dependent heme tranporter has been determined, which is the ShuA heme transport protein [19] that serves the same function as HmbR in the pathogen Shigella dysenteriae. Despite their functional similarity, differences between the two proteins raise challenges to their direct comparison. In particular, they only share approximately 15% sequence homology [22]. Second, the structure of ShuA identifies two candidate histidine residues involved in coordinating the heme, whereas our data indicate one residue with a Raman signature that supports an oxygen ligand. Finally, the extracellular regions of ShuA and HmbR, like all TBDTs, are highly variable. Thus, the involvements of those structures in stabilizing the hemin-bound states can neither be compared nor delineated without high-resolution structural information and/or kinetic data for their heme transfer reactions. Hence, the design of drugs targeting heme transport proteins will likely need to take into account the differences between receptors of different organisms, even if they have identical substrates. Toward this end, crystallographic and kinetic studies on HmbR are under way to elucidate these important structural details.
Acknowledgments
We are grateful to Dr. Donna Perkins-Balding for supplying the parent HmbR expression plasmid used in these studies (DPB1809), and members of the Lanzilotta laboratory for critical discussions and technical assistance. This work was supported in part by NSF grant MCB 0835432 to W.N.L. and NIH grant AI072719 to K.R.R. and NIH grant GM094039 to G.S.L.R.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagen.2014.06.009.
References
- 1.Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 2.Genco CA, Dixon DW. Emerging strategies in microbial haem capture. Mol Microbiol. 2001;39:1–11. doi: 10.1046/j.1365-2958.2001.02231.x. [DOI] [PubMed] [Google Scholar]
- 3.Wyckoff EE, Mey AR, Payne SM. Iron acquisition in Vibrio cholerae. Biometals. 2007;20:405–416. doi: 10.1007/s10534-006-9073-4. [DOI] [PubMed] [Google Scholar]
- 4.Fetherston JD, Kirillina O, Bobrov AG, Paulley JT, Perry RD. The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague. Infect Immun. 2010;78:2045–2052. doi: 10.1128/IAI.01236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poole K, McKay GA. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front Biosci. 2003;1:d661–d686. doi: 10.2741/1051. [DOI] [PubMed] [Google Scholar]
- 6.Schryvers AB, Gray-Owen S. Iron acquisition in Haemophilus influenzae: receptors for human transferrin. J Infect Dis. 1992;165:S103–S104. doi: 10.1093/infdis/165-supplement_1-s103. (Supplemental) [DOI] [PubMed] [Google Scholar]
- 7.Burkhard K, Wilks A. Characterization of the outer membrane receptor ShuA from the heme uptake system of Shigella dysenteriae. J Biol Chem. 2007;282:15126–15136. doi: 10.1074/jbc.M611121200. [DOI] [PubMed] [Google Scholar]
- 8.Perkins-Balding D, Ratliff-Griffin M, Stojiljkovic I. Iron transport systems in Neisseria meningitidis. Microbiol Mol Biol Rev. 2004;68:154–171. doi: 10.1128/MMBR.68.1.154-171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson AD, Deisenhofer J. TonB-dependent receptors—structural perspectives. Biochim Biophys Acta. 2002;1565:318–332. doi: 10.1016/s0005-2736(02)00578-3. [DOI] [PubMed] [Google Scholar]
- 10.Krewulak KD, Vogel HJ. Structural biology of bacterial iron uptake. Biochim Biophys Acta. 2008;1778:1781–1804. doi: 10.1016/j.bbamem.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 11.Wilks A, Ikeda-Saito M. Heme utilization by pathogenic bacteria: not all pathways lead to biliverdin. Acc Chem Res. doi: 10.1021/ar500028n. (Article ASAP) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens DS. Biology and pathogenesis of the evolutionarily successful, obligate human bacterium Neisseria meningitidis. Vaccine. 2009;27:B71–B77. doi: 10.1016/j.vaccine.2009.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin SH, Kim KS. Treatment of bacterial meningitis: an update. Expert Opin Pharmacother. 2012;13(15):2189–2206. doi: 10.1517/14656566.2012.724399. [DOI] [PubMed] [Google Scholar]
- 14.West SE, Sparling PF. Response of Neisseria gonorrhoeae to iron limitation: alterations in expression of membrane proteins without apparent siderophore production. Infect Immun. 1985;47:388–394. doi: 10.1128/iai.47.2.388-394.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison OB, Evans NJ, Blair JM, Grimes HS, Tinsley CR, Nassif X, Kriz P, Ure R, Gray SJ, Derrick JP, Maiden MCJ, Feavers IM. Epidemiological evidence for the role of the haemoglobin receptor, HmbR, in meningococcal virulence. J Infect Dis. 2009;200:94–98. doi: 10.1086/599377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stojiljkovic I, Larson J, Hwa V, Anic S, So M. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. Am Soc Microbiol. 1996;178:4670–4678. doi: 10.1128/jb.178.15.4670-4678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stojiljkovic I, Hwa V, de Saint Martin L, O’Gaora P, Nassif X, Heffron F, So M. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 18.Krieg S, Huche F, Diederichs K, Izadi-Pruneyre N, Lecroisey A, Wandersman C, Delepelaire P, Welte W. Heme uptake across the outer membrane as revealed by crystal structures of the receptor-hemophore complex. Proc Natl Acad Sci U S A. 2009;106:1045–1050. doi: 10.1073/pnas.0809406106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobessi D, Meksem A, Brillet K. Structure of the heme/hemoglobin outer membrane receptor ShuA from Shigella dysenteriae: heme binding by an induced fit mechanism. Proteins. 2010;78:286–294. doi: 10.1002/prot.22539. [DOI] [PubMed] [Google Scholar]
- 20.Bracken CS, Baer MT, Abdur-Rashid A, Helms W, Stojiljkovic I. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J Bacteriol. 1999;181:6063–6072. doi: 10.1128/jb.181.19.6063-6072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkins-Balding D, Baer M, Stojiljkovic T. Identification of functionally important regions of a haemoglobin receptor from neisseria meningitidis. Microbiology. 2003;149:3423–3435. doi: 10.1099/mic.0.26448-0. [DOI] [PubMed] [Google Scholar]
- 22.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dailey HA, Fleming JE. Bovine ferrochelatase. Kinetic analysis of inhibition by N-methyprotoporphyrin, manganese, and heme. J Biol Chem. 1983;258:11453–11459. [PubMed] [Google Scholar]
- 24.Nygaard TK, Liu M, McClure MJ, Lei B. Identification and characterization of the heme-binding proteins SeShp and SeHts of Streptococcus equi subspecies equi. BMC Microbiol. 2006;6 doi: 10.1186/1471-2180-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoppellaro G, Bren KL, Ensign AA, Harbitz E, Kaur R, Hersleth HP, Ryde U, Hederstedt L, Andersson KK. Studies of ferric heme proteins with high anisitropic/ highly axial low spin (S = 1/2) electron paramagnetic resonance signals with bis-histidine and histidine–methionine axial iron coordination. Biopolymers. 2009;91(12):1064–1082. doi: 10.1002/bip.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eakanunkul S, Lukat-Rodgers GS, Sumithran S, Ghosh A, Rodgers KR, Dawson JH, Wilks A. Characterization of the periplasmic heme-binding protein ShuT from the heme uptake system of Shigella dysenteriae. Biochemistry. 2005;44:13179–13191. doi: 10.1021/bi050422r. [DOI] [PubMed] [Google Scholar]
- 27.Yukl ET, Jepkorir G, Alontaga AY, Pautsch L, Rodriguez JC, Rivera M, Moënne-Loccoz P. Kinetic and spectroscopic studies of hemin acquisition in the hemophore HasAp from Pseudomonas aeruginosa. Biochemistry. 2010;49(31):6646–6654. doi: 10.1021/bi100692f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnoux P, Haser R, Izadi N, Lecroisey A, Delepierre M, Wandersman C, Czjzek M. The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat Struct Biol. 1999;6(6):516–520. doi: 10.1038/9281. [DOI] [PubMed] [Google Scholar]
- 29.Izadi N, Henry Y, Haladjian J, Goldberg ME, Wandersman C, Delepierre M, Lecroisey A. Purification and characterization of an extracellular heme-binding protein, HasA, involved in heme iron acquisition. Biochemistry. 1997;36(23):7050–7057. doi: 10.1021/bi962577s. [DOI] [PubMed] [Google Scholar]
- 30.Rodgers KR, Lukat-Rodgers GS. Handbook of porphyrin science. 2011;30 ch.8. [Google Scholar]
- 31.Ferreira GC, Kadish KM, Smith KM, Guilard R, editors. World Scientific. 2013. [Google Scholar]
- 32.Das TK, Coulture M, Lee HC, Peisach J, Rousseau DL, Wittenberg BA, Wittenberg JB, Guertin M. Identification of the ligands to the ferric heme of Chlamydomonas chloroplast hemoglobin: evidence for the ligation of tyrosine-63 (B10) to the heme. Biochemistry. 1999;38:15360–15368. doi: 10.1021/bi991237e. [DOI] [PubMed] [Google Scholar]
- 33.Nagai M, Yoneyama Y, Kitagawa T. Characteristics in tyrosine coordinations of four hemoglobins M probed by resonance Raman spectroscopy. Biochemistry. 1989;28:2418–2422. doi: 10.1021/bi00432a012. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Moënne-Loccuz P, Hildebrand DP, Wilks A, Loehr TM, Mauk AG, Ortiz de Montellano PR. Replacement of the proximal histidine iron ligand by a cysteine or tyrosine converts heme oxygenase to an oxidase. Biochemistry. 1999;38:3733–3743. doi: 10.1021/bi982707s. [DOI] [PubMed] [Google Scholar]
- 35.Adachi S, Nagano S, Ishimori K, Watanabe Y, Morishima I. Roles of proximal ligand in heme proteins: replacement of proximal histidine in human myoglobin with cysteine and tyrosine by site-directed mutagenesis as models for P-450, chloroperoxidase, and catalase. Biochemistry. 1993;32:241–252. doi: 10.1021/bi00052a031. [DOI] [PubMed] [Google Scholar]
- 36.Meitzler JL, Brandman R, Montellano PRO. Perturbed heme binding is responsible for the blistering phenotype associated with mutations in the Caenorhabditis elegans dual oxidase 1 (DUOX1) peroxidase domain. J Biol Chem. 2010;285(52):40991–41000. doi: 10.1074/jbc.M110.170902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frawley ER, Kranz RG. CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc Natl Acad Sci. 2009;106(25):10201–10206. doi: 10.1073/pnas.0903132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Létoffé S, Weckner K, Delepierre M, Delepelaire P, Wandersman C. Activities of the Serratia marcescens heme receptor HasR and isolated plug and beta-barrel domains: the beta-barrel forms a heme-specific channel. J Bacteriol. 2005;187(13):4637–4645. doi: 10.1128/JB.187.13.4637-4645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jancarik J, Pufan R, Hong C, Kim SH, Kim R. Optimum solubility (OS) screening: an efficient method to optimize buffer conditions for homogeneity and crystallization of proteins. Acta Crystallogr D. 2004;60(Pt9):1670–1673. doi: 10.1107/S0907444904010972. [DOI] [PubMed] [Google Scholar]
- 40.Smith AD, Wilks A. Extracellular heme uptake and the challenges of bacterial cell membranes. Curr Top Membr. 2012;69:359–392. doi: 10.1016/B978-0-12-394390-3.00013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukat-Rodgers GS, Rodgers KR, Caillet-Saguy C, Izadi-Pruneyre N, Lecroisey A. Novel heme ligand displacement by CO in the soluble hemophore HasA and its proximal ligand mutants: implications for heme uptake and release. Biochemistry. 2008;47:2087–2098. doi: 10.1021/bi7019518. [DOI] [PubMed] [Google Scholar]
- 42.Sharma KD, Andersson LA, Loehr TM. Comparative spectral analysis of mammalian, fungal, and bacterial catalases. 1989;264:12772–12779. [PubMed] [Google Scholar]
- 43.Egeberg KD, Springer BA, Martinis SA, Sligar SG, Morikis D, Champion PM. Alteration of sperm whale myoglobin heme axial ligation by site-directed mutagenesis. Biochemistry. 1990;29:9783–9791. doi: 10.1021/bi00494a004. [DOI] [PubMed] [Google Scholar]
- 44.Uchida T, Stevens JM, Daltrop O, Harvat EM, Hong L, Ferguson SJ, Kitagawa T. The interaction of covalently bound heme with the cytochrome c maturation protein CcmE. J Biol Chem. 2004;279(50):51981–51988. doi: 10.1074/jbc.M408963200. [DOI] [PubMed] [Google Scholar]
- 45.Chuang WJ, Johnson S, Van Wart HE. Resonance Raman spectra of bovine liver catalase: enhancement of proximal tyrosinate vibrations. J Inorg Biochem. 1988;34(3):201–219. doi: 10.1016/0162-0134(88)85030-x. [DOI] [PubMed] [Google Scholar]
- 46.Arnoux P, Haser R, Izadi-Pruneyre N, Lecroisey A, Czjzek M. Functional aspects of the heme bound hemophore HasA by structural analysis of various crystal forms. Proteins. 2000;41:202–210. doi: 10.1002/1097-0134(20001101)41:2<202::aid-prot50>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 47.Jepkorir G, Rodriguez JC, Rui H, Im W, Lovell S, Battaile KP, Alontaga AY, Yukl ET, Moenne-Loccoz P, Rivera M. Structural, NMR spectroscopic, and computational investigation of hemin loading in the hemophore HasAp from Pseudomonas aeruginosa. J Am Chem Soc. 2010;132:9857–9872. doi: 10.1021/ja103498z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar R, Lovell S, Matsumura H, Battaile K, Moënne-Loccoz P, Rivera M. The hemophore HasA from Yersinia pestis (HasAyp) coordinates hemin with a single residue, Tyr75, and with minimal conformational change. Biochemistry. 2013;52(16):2705–2707. doi: 10.1021/bi400280z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ekworomadu MT, Poor CB, Owens CP, Balderas MA, Fabian M, Olson JS, Murphy F, Balkabasi E, Honsa ES, He C, Goulding CW, Maresso AW. Differential function of lip residues in the mechanism and biology of an anthrax hemophore. PLoS Pathog. 2012;8:e1002559. doi: 10.1371/journal.ppat.1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Honsa ES, Owens CP, Goulding CW, Maresso AW. The near-iron transporter (NEAT) domains of the anthrax hemophore IsdX2 require a critical glutamine to extract heme from methemoglobin. J Biol Chem. 2013;288:8479–8490. doi: 10.1074/jbc.M112.430009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Honsa ES, Maresso AW. Mechanisms of iron import in anthrax. Biometals. 2011;24:533–545. doi: 10.1007/s10534-011-9413-x. [DOI] [PubMed] [Google Scholar]
- 52.Gaudin CF, Grigg JC, Arrieta AL, Murphy ME. Unique heme-iron coordination by the hemoglobin receptor IsdB of Staphylococcus aureus. Biochemistry. 2011;50:5443–5452. doi: 10.1021/bi200369p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grigg JC, Ukpabi G, Gaudin CF, Murphy ME. Structural biology of heme binding in the Staphylococcus aureus Isd system. J Inorg Biochem. 2010;104:341–348. doi: 10.1016/j.jinorgbio.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 54.Sharp KH, Schneider S, Cockayne A, Paoli M. Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus. J Biol Chem. 2007;282:10625–10631. doi: 10.1074/jbc.M700234200. [DOI] [PubMed] [Google Scholar]
- 55.Maresso AW, Schneewind O. Iron acquisition and transport in Staphylococcus aureus. Biometals. 2006;19:193–203. doi: 10.1007/s10534-005-4863-7. [DOI] [PubMed] [Google Scholar]
- 56.Caillet-Saguy C, Piccioli M, Turano P, Lukat-Rodgers G, Wolff N, Rodgers KR, Izadi-Pruneyre N, Delepierre M, Lecroisey A. Role of the iron axial ligands of heme carrier HasA in heme uptake and release. J Biol Chem. 2012;287:26932–26943. doi: 10.1074/jbc.M112.366385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caillet-Saguy C, Turano P, Piccioli M, Lukat-Rodgers GS, Czjzek M, Guigliarelli B, Izadi-Pruneyre N, Rodgers KR, Delepierre M, Lecroisey A. Deciphering the structural role of histidine 83 for heme binding in hemophore HasA. J Biol Chem. 2008;283:5960–5970. doi: 10.1074/jbc.M703795200. [DOI] [PubMed] [Google Scholar]
- 58.Izadi-Pruneyre N, Huché F, Lukat-Rodgers GS, Lecroisey A, Gilli R, Rodgers KR, Wandersman C, Delepelaire P. The heme transfer from soluble HasA hemophore to its membrane-bound receptor HasR is driven by protein-protein interaction from a high to a lower affinity binding site. J Biol Chem. 2006;281(35):25541–25550. doi: 10.1074/jbc.M603698200. [DOI] [PubMed] [Google Scholar]
- 59.Kamal N, Shafer WM. Biologic activities of the TolC-like protein of Neisseria meningitidis as assessed by functional complementation in Escherichia coli antimicrobial agents and chemotherapy. 2010;54:506–508. doi: 10.1128/AAC.01168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ulmer JB, Burke CJ, Shi C, Friedman A, Donnelly JJ, Liu MA. Pore formation and mitogenicity in blood cells by the class 2 protein of Neisseria meningitidis. J Biol Chem. 1992;267:19266–19271. [PubMed] [Google Scholar]
- 61.Hargrove MS, Whitaker T, Olson JS, Vali RJ, Mathews AJ. Quaternary structure regulates hemin dissociation from human hemoglobin. J Biol Chem. 1997;272:17385–17389. doi: 10.1074/jbc.272.28.17385. [DOI] [PubMed] [Google Scholar]