Abstract
Different organisms have independently and recurrently evolved similar phenotypic traits at different points throughout history. This phenotypic convergence may be caused by genotypic convergence and constrained by historical contingency. To investigate how convergence may be driven by selection in a particular environment and constrained by history, we analyzed nine life-history traits and four metabolic traits during an experimental evolution of six yeast strains in four different environments. In each of the environments, the population converged towards a different life-history strategy. However, phenotypic convergence was partly associated with the selection of mutations in genes involved in the same pathway. In a fifth of our evolution experiments, mutations in the same gene, BMH1, were selected, in three out of the six ancestral genotypes. Two types of BMH1 mutation with opposite phenotypic effects on several traits were found. The evolution of most traits, as well as the occurrence of BMH1 mutations, was significantly influenced by the ancestral strain. However, this effect could not be easily predicted from ancestors’ phylogeny or past-selection. All together, our data demonstrate that phenotypic and its underlying genotypic convergence depends on a complex interplay between the evolutionary environment, pleiotropy and the ancestor genetic background but are not straightforwardly predicable.
Keywords: adaptive landscape, pleiotropy, Saccharomyces cerevisiae, 14-3-3 protein, BMH1
Introduction
Throughout the tree of life, evolutionarily divergent lineages have recurrently and independently evolved similar phenotypic traits. Such convergent evolution has been observed in numerous plant and animal species (Arendt and Reznick 2008; Manceau et al. 2010), but evidence for trait convergence is sparse in microorganisms, likely due to the paucity of visually observable phenotypes. Examples of convergent evolution in microbes include the independent evolution of fruiting bodies and thallus type in microscopic fungi (Plata and Lumbsch 2011), and fermentation in yeast species (Ostrowski et al. 2008).
Phenotypic convergence can be caused either by natural selection or by chance. Physical or biological constraints can restrict available phenotypes to a subset of phenotypic space (DePristo et al. 2005; Weinreich et al. 2006; Gompel and Prud’homme 2009; Chevin et al. 2010; Feldman et al. 2012), and in this restricted space, chance alone can cause phenotypic convergence. However, if convergent evolution occurred only by chance, it would not be driven by the ecological niche. By contrast, if natural selection drives convergent evolution, convergence would be expected in environments that are similar in some aspects. However, the extent to which the environment drives convergent evolution is unknown, as most studies have focused on either natural populations in uncontrolled environments (Arendt and Reznick 2008; Gompel and Prud’homme 2009; Elmer and Meyer 2011) or laboratory-evolved populations in a single well-controlled environment (Wichman et al. 1999). In addition, most studies have focused on a limited number of traits, and it is unclear how convergent evolution for a single phenotypic trait is constrained by changes in correlated traits. Co-variation of life-history traits (i.e. traits involved in the life-cycle of an organism) and/or morphological traits has been well described (Roff 2002), with the course of evolution shaped by natural selection (Schluter 1996; Arnold et al. 2001). However, how the convergence of one trait impacts the evolution of other traits remains to be studied empirically (Kolbe et al. 2011).
Phenotypic convergence can occur through mutations in different sets of genes that cause similar phenotypes in distinct lineages. Alternatively, phenotypic convergence may be caused by convergent or parallel evolution at the genotypic level (Wichman et al. 1999; Arendt and Reznick 2008; Remold et al. 2008; Gompel and Prud’homme 2009; Elmer and Meyer 2011; Feldman et al. 2012; Tenaillon et al. 2012). Parallel genotypic evolution arises when mutations occur in independent lineages that start from the same genotype, while convergence refers to mutations produced from different ancestral genotypes (Zhang and Kumar 1997). Parallel and convergent genotypic evolution may arise at several levels: the same nucleotide mutating independently several times (Wichman et al. 1999; Rozpedowska et al. 2011), different mutations in the same gene (Rosenblum et al. 2010) or in a multigene family (Christin et al. 2007; Srithayakumar et al. 2011), through mutations in different genes sharing the same function (Elias and Tawfik 2012) or in the same network (Lozovsky et al. 2009).
Phenotypic and/or genotypic evolution can be constrained by historical factors, which can produce different phenotypic and/or genotypic outcomes despite similar environmental conditions. This has been defined as historical contingency (Travisano et al 1995, Blount et al. 2008). Historical contingency arises because the effects of mutations are contingent on the alleles that have been retained from history through epistasis. This constrains mutation roads and as a consequence the evolution of phenotypes. Selection in similar environment is supposed to eliminate the effect of historical contingency. Indeed, selection is supposed to lead to the same phenotypic solution regardless of the genotypic background, as illustrated by the numerous examples of convergence. This has lead to the idea that the effect of historical contingency should be detected at the genomic level, but may be less frequently detected at the phenotypic level, especially for traits correlated to fitness (Teotonio et al. 2009, Joshi et al. 2033, Nguyen et al. 2011, Bedhomme et al. 2013). However, an extensive study including multiple traits in multiple environments is lacking to test for this hypothesis.
In this study, we analyzed the evolution of 13 metabolic and life-history traits across multiple environments and genotypes using the budding yeast Saccharomyces cerevisiae as a model system. Yeast is one of the few microorganisms for which life-history traits have been studied (Spor et al. 2008; Spor et al. 2009; Granek et al. 2011; Magwene et al. 2011; Wang et al. 2011), and yeast populations display different life-history strategies depending on their ecological niche of origin (Spor et al. 2009). Using experimental evolution, we ask how selection and historical contingency interplay on phenotypic and genotypic evolution. We found evidence of genotypic convergence underlying multiple trait convergence in specific environments, suggesting that mutational paths on the adaptive landscape are restricted by selection. We also found that the evolution of most traits, including fitness components, is constrained by history. By further investigating the convergence in one gene, we demonstrate that genotypic convergence underlying multi-trait convergence depends partly on the environment and on the ancestors’ genetic background, highlighting the role of pleiotropy and history in shaping rugged fitness landscape.
Results
We chose six diverse S. cerevisiae strains for which there exists whole-genome sequence, with the goal of maximizing habitat, genetic and phenotypic variability (Spor et al. 2008; Liti et al. 2009; Spor et al. 2009) (Table 1). Three replicates of each strain were propagated in serial batch cultures under four selection regimes (1%_48h, 1%_96h, 15%_48h and 15%_96h) differing by the glucose content (1% or 15%) and by the cycle length (48 hours or 96 hours). The 48 h and 96 h cycle length experiments were stopped after at least 325 or 165 generations, respectively. At the end of the experiment, we studied nine life-history traits (growth rates during fermentation (Rferm) and respiration (Rresp), the time to diauxic shift (Tshift), population sizes at Tshift and the serial transfer time points (Kferm, PopSize48h, PopSize96h) and cell sizes at Tshift and serial transfer time points (Sferm, S48h and S96h)), and four metabolic traits (the specific glucose consumption rate (Jspec), the biomass yield from fermentation (Yferm) and the quantity of ethanol produced and released in the medium at the two serial transfer time points (Eth48h and Eth96h)).
Table 1. Saccharomyces cerevisiae strains used in this study.
| Strains ID |
Lab ID |
Genetic Group |
Geographical Origin |
Habitat | Environmental Source |
|---|---|---|---|---|---|
| NCYC110 | 247 | WA | West Africa | Industrial | Ginger Beer from Z. officinale |
| YJM981 | 304 | W/E | Bergamo, Italy | Clinical Isolate | Vagina |
| YPS128 | 104 | NA | Pennsylvania, USA | Forest | Oak Exudate |
| UWOPS83-787.3 | 270 | Mosaic | Bahamas | Fruit | Opuntia stricta |
| S288C | 96 | Mosaic | California, USA | Laboratory | Rotting fig |
| Y55 | 97 | Mosaic / WA WA | France | Laboratory | Wine |
Genetic groups were obtained from inference of S. cerevisiae population structure using the program Structure (Liti et al. 2009). WA (West Africa), W/E (Wine/European), NA (North America), Mosaic / WA (Mosaic, mostly West African).
Phenotypic responses to selection
We analyzed the evolution of each trait relative to its ancestral population using mixed-effects analysis of variance (see Material and Methods). The selection regime effect, which measures the differences in phenotypic evolution between the four selection regimes, across all strains, was significant for all traits except Rresp, indicating that the strains evolved towards different phenotypes depending on the selection regime (Table 2). We summarize the response to selection in each selection regime using the average trait evolution across all strains (Table 3). A value of 0 indicates no detectable response to the selection on average, while a positive (negative) value indicates an increase (decrease) of the average value of the trait in the evolved populations compared to the ancestral strains. The strongest response to selection occurred in the selection regime with the strongest glucose starvation, i.e. the 1%_96h selection regime. The 1%_96h regime selected for a more extreme strategy (decreased cell size S and increased population size, PopSize), which tends to a previously described life-history strategy found among strains originating from soil and oak bark [14]. Populations selected under this regime also displayed an increase of Rferm and Jspec. The 1%_48h regime selected for an intermediate life-history strategy that is close to the average life-history strategy of the ancestral populations at 48h (PopSize48h and S48h are near zero), however the characteristics of the evolved population are different regarding the traits in fermentation (increased Rferm, Kferm, Jspec, and decreased Sferm, Table 2 and 3). Globally, both 1% regimes selected for a decreased cell size compared to the 15% regimes. The difference in cycle lengths (48h versus 96h) mainly led to the evolution of metabolic changes - both 96h cycle length regimes selected for populations that left more residual ethanol compared to the 48h cycle length regimes. In 15% glucose regimes, the 96h regime also selected for an increase in the amount of time spent in fermentation (Tshift) with a decrease in the glucose consumption rate (Jspec).
Table 2. Analysis of variance of the relative response to selection of each life-history and metabolic trait.
Source is the tested effect, df the degrees of freedom. MS the mean square and F Fisher’s F. The significant p-values after FDR correction (global level 0.05) are indicated in grey.
| K ferm | PopSize 48h | PopSize 96h | S ferm | S 48h | S 96h | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Source | df | F | P | F | P | F | P | F | P | F | P | F | P |
| Block | 5 | 5 | 5.10−04 | 4.7 | 8. 10−04 | 9.4 | 6.10−07 | 1.1 | 0.35 | 2.8 | 0.024 | 1 | 0.42 |
| eval medium | 1 | 259.6 | 7.10−26 | 19.9 | 3.10−05 | 0.003 | 0.95 | 128.7 | 6.10−18 | 116.4 | 6.10−17 | 1.3 | 0.26 |
| strain | 4 | 6.6 | 1.10−04 | 19.2 | 7.10−11 | 7.1 | 7.10−05 | 9.1 | 5.10−06 | 32.6 | 1.10−15 | 13.7 | 2.10−08 |
| selection regime | 3 | 3.6 | 0.017 | 8.1 | 9.10−05 | 10.6 | 7.10−06 | 8.9 | 4.10−05 | 40.2 | 1.10−15 | 20.9 | 6.10−10 |
| eval medium*strain | 4 | 13.1 | 4.10−08 | 5.5 | 6.10−04 | 24.1 | 8.10−13 | 37.2 | 4.10−17 | 27.8 | 4.10−14 | 13.2 | 3.10−08 |
| eval medium*selection regime | 3 | 0.84 | 0.48 | 1.5 | 0.216 | 7.7 | 1.10−04 | 4 | 0.01 | 14.4 | 2.10−07 | 2.2 | 0.1 |
| strain*selection regime | 12 | 0.94 | 0.52 | 1.9 | 0.05 | 3.5 | 4.10−04 | 4 | 10−04 | 2.95 | 0.002 | 2.3 | 0.017 |
|
| |||||||||||||
| Residuals means square | 0.1 | 0.027 | 0.046 | 0.002 | 0.001 | 0.002 | |||||||
| R ferm | R resp | T shift | J spec | Y ferm | Eth 48h | Eth 96h | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source | df | F | P | F | P | F | P | F | P | F | P | F | P | F | P |
| Block | 5 | 15.2 | 3.10−10 | 3.01 | 0.016 | 13.67 | 2.10−09 | 0.77 | 0.552 | 3.8 | 0.004 | 4.92 | 6.10−04 | 1.24 | 0.300 |
| eval medium | 1 | 18.4 | 5.10−05 | 0.75 | 0.39 | 56.28 | 1.10−10 | 2.23 | 0.141 | 236 | 2.10−24 | 177.65 | 2.10−21 | 37.27 | 4.10−08 |
| ancestor | 4 | 9.6 | 2.10−06 | 4.11 | 0.005 | 1.96 | 0.11 | 13.4 | 8.10−08 | 7.5 | 4.10−05 | 60.83 | 8.10−23 | 12.53 | 7.10−08 |
| selection regime | 3 | 3.4 | 0.02 | 0.51 | 0.68 | 5.51 | 0.002 | 3.8 | 0.014 | 3.2 | 0.027 | 4.87 | 0.004 | 12.98 | 6.10−07 |
| eval mediumstrain | 4 | 6.8 | 1.10−04 | 1.87 | 0.13 | 1.79 | 0.14 | 13.3 | 9.10−08 | 10 | 2.10−06 | 58.33 | 3.10−22 | 3.45 | 0.012 |
| eval medium*selection regime | 3 | 1.28 | 0.28 | 2.29 | 0.085 | 1.7 | 0.18 | 3.25 | 0.028 | 0.35 | 0.79 | 4.40 | 0.007 | 12.14 | 1.10−06 |
| ancestor*selection regime | 12 | 1.80° | 0.063 | 0.48 | 0.92 | 1.81° | 0.063 | 1.88° | 0.056 | 0.55 | 0.88 | 2.31 | 0.014 | 1.62 | 0.104 |
| Residuals | 0.02 | 0.05 | 0.01 | 3.59 | 0.09 | 1.92 | 0.05 | ||||||||
Table 3. Main effect of the selection regime on the relative response to selection of life-history and metabolic traits.
Mean relative response to selection is given for each trait in each selection regime. For each trait, significance of the differences was assessed by HSD Tukey tests. Levels that are not connected by a common letter (A, B or C) are significantly different.
|
Selection
regime |
K ferm | PopSize 48h | PopSize 96h | S ferm | S 48h | S 96h | R ferm | R resp | T shift | J spec | Eth 48h | Eth 96h | Y ferm |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1%_48h | 1.1 A | 0.088 B | 0.569 B | −0.137 BC | −0.070 B | −0.061 B | 0.083 A | NS | −0.030 B | 1.467 A | 0.73 A | −0.381 C | 0.67 A |
| 1%_96h | 1.08 AB | 0.247 A | 0.832 A | −0.160 C | −0.115 C | −0.098 C | 0.060 AB | 0.113 A | 0.013 B | 0.4 AB | 1.72 A | −0.208 B | 0.713 A |
| 15%_48h | 0.87 AB | −0.018 B | 0.499 B | −0.085 A | −0.003 A | −0.018 A | 0.021 AB | 0.085 A | −0.028 B | 1.61 AB | 1.61 A | −0.239 C | 0.514 A |
| 15%_96h | 0.82 B | 0.013 B | 0.461 B | −0.116 AB | −0.012 A | −0.008 A | −0.03 B | 0.069 A | 0.070 A | 0.155 B | 1.61 A | −0.004 A | 0.554 A |
Multiple traits convergence within selection regimes
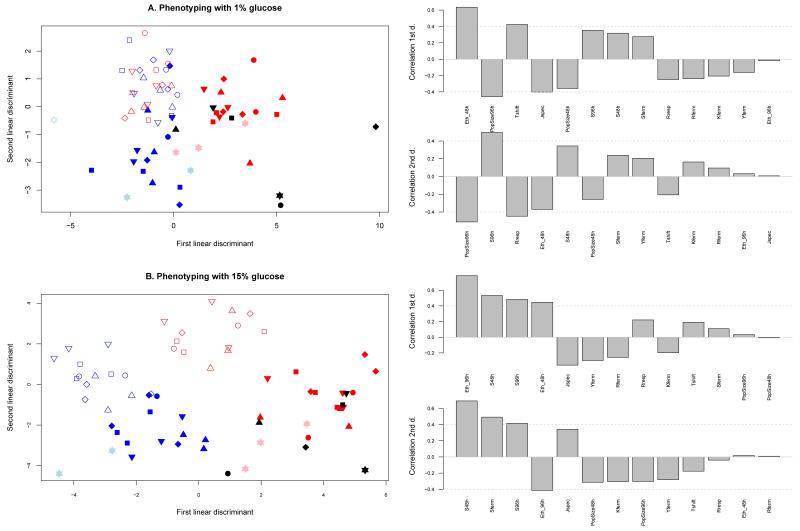
To determine which traits converged most within a selection regime and diverged most between selection regimes, we carried out one linear discriminant analysis (LDA) on all traits for each evaluation medium (1 % and 15 % glucose, respectively Figure 1A and 1B) setting the four selection regimes as four a priori categories. The LDA axes represent the combination of life-history trait values towards which directional selection has driven the populations in each selection regime, i.e. the multivariate phenotypic convergence landscape. The first two axes explained 89.1 % and 95.9 % of the variance in 1% glucose and 15% glucose evaluation media respectively. The probability of a population being correctly assigned to its selection regime was 0.99 in the 15% glucose evaluation medium, indicating convergence within selection regime and diversifying selection among selection regimes for several traits (see Figure 1B). In the 1% glucose evaluation medium, this probability was 0.88 in the 1% glucose evaluation medium as we were not able to discriminate between the 1%_48h regime and the 15%_48h regime (see Figure 1A). Among the 13 traits, cell size (S96h, S48h, Sferm), traits related to fitness during the respiration phase (PopSize96h, PopSize48h, Rresp) as well as metabolic traits (Jspec, Eth48h, Eth96h, Tshift) were significantly correlated to at least one of the LDA axes indicating that they were the most convergent traits within selection regime and divergent traits between selection regimes. By contrast, the traits related to fitness during fermentation (Rferm, Kferm, Yferm) did not significantly explain any of the axes of the phenotypic convergence landscape. Indeed, these traits evolved towards the same points in the different selection regimes.
Figure 1. Canonical plots of the linear discriminant analysis.
A. Phenotypic evaluation of ancestral strains and evolved populations in the 1 % glucose medium. B. Phenotypic evaluation of ancestral strains and evolved populations in the 15 % glucose medium. The correlations between traits and how they explain the linear discriminant axis are presented on the right. The colors of the symbols correspond to the different selection regimes (empty blue: 1%_48h, filled blue: 1%_96h, empty red: 15%_48h and filled red: 15%_96h). The black symbols correspond to the ancestral strains. The shapes of the symbols correspond to the evolved and ancestral state of a given strain (stars for S288c, down triangle for Y55, up triangle for YPS128, diamond for NCYC110, square for UWOPS83-787.3, and circle for YJM981).
Effect of historical contingency on phenotypic convergence
The effect of historical contingency was tested by a MANOVA performed on all traits of the evolved populations evaluated either in the 1% glucose or in the 15% glucose media. The ancestor effects were significant in both evaluation media (Pillai=1,5, P<0.001 in 1% glucose media; Pillai=1.9, P<0.001 in 15% glucose media) indicating that the genetic background constrained the phenotypic landscape. An ancestor effect was found separately for each trait except for Tshift and Eth48h in the 1% medium of evaluation and for Rferm. Rresp. Eth48h in the 15% medium of evaluation (Table S1). In 14 cases out of the 26 evaluated traits (13 traits in two evaluation media), an interaction effect between selection regime and ancestor effect was found indicating that the ancestor effect may depend on the environment (Table S1).
We then asked whether ancestral strains that were phenotypically more similar evolved more similar phenotypes. We measured the distances between pairwise ancestral strains and the distances between the derived pairwise evolved populations in each selection regime on the phenotypic landscape. On the LDA axis, we did not find any significant correlation between the phenotypic distances of evolved populations and their ancestors ones. When analyzing each trait separately, only five of the 52 correlation tests (13 traits × 4 selection regimes) were significant indicating that phenotypic evolution cannot be predicted easily from the ancestors’ phenotype. Significant correlations were found for Sferm in the 15%_96h selection regime (r=0.7, p<0.001) as well as Kferm (r=0.76, p<0.001), Popsize96h (r=0.62, p=0.002), Yferm (r=0.59, p=0.003) and Eth96h (r=0.69, p<0.001) in the 1%_96H selection regime. We did not find any significant correlations between the phenotypic distances of evolved populations and the phylogenetic relationships of their ancestral strains.
Genotypic response to selection
To determine if genotypic convergence was underlying the observed phenotypic convergence, we sequenced the genomes of the ancestral strains Y55, YPS128, UWOPS83-787.3 and YJM981, and of clones that were evolved in the 1%_48h and 15%_48h conditions, for a total of eight ancestral-evolved pairs. We identified 27 SNPs and indels across the eight evolved clones relative to their respective ancestor (Table 4).
Table 4. Characteristics of the mutations discovered in the evolved populations.
| Strain : Selection regime | Chromosome | Position | Parent genotype |
Evolved genotype |
Systematic name | Gene name |
Mutation effect |
Functional effect |
|---|---|---|---|---|---|---|---|---|
| Y55 : 1%_48h | 5 | 546253 | C/C | C/G | YER177W | BMH1 | Y 216 * | nonsense |
| 13 | 470745 | G/G | G/C | YMR102C | S 536 * | nonsense | ||
| Y55 : 15%_48h | 1 | 126548 | C/C | C/T | YAL016W | TPD3 | Q 557 * | nonsense |
| 2 | 9561 | C/C | C/T | 5′ of tL(UAA)B1; 3′ of YBLWdelta3 |
intergenic | |||
| 4 | 1015447 | T/T | T/A | YDR277C | MTH1 | I 85 F | disruptive | |
| 6 | 201121 | G/G | G/T | YFR023W | PES4 | silent | synonymous | |
| 12 | 21757 | G/G | G/A | YLL060C | GTT2 | P 28 L | disruptive | |
| 14 | 451643 | G/G | G/T | YNL092W | M 257 I | tolerated | ||
| 16 | 488074 | G/G | G/A | YPL033C | SRL4 | silent | synonymous | |
| YPS128 : 1%_48h | 14 | 220425 | G/G | G/A | 5′ of URE2; 3′ of JJJ1 |
intergenic | ||
| 15 | 998511 | G/G | A/A | YOR353C | SOG2 | T 772 I | disruptive | |
| YPS128 : 15%_48h | 4 | 1333026 | C/C | T/T | YDR435C | PPM1 | E 313 K | disruptive |
| 11 | 604204 | C/C | C/G | YKR088C | TVP38 | R 286 T | tolerated | |
| 12 | 229054 | G/G | G/T | YLR040C | A 177 D | tolerated | ||
| 15 | 496908 | T/T | A/A | YOR092W | ECM3 | L 594 * | nonsense | |
| 15 | 497041 | C/C | A/A | 5′ of ECM3; 3′ of YOR093C |
intergenic | |||
| UWOPS83-787.3 : 1%_48h |
5 | 546126 | G/G | T/T | YER177W | BMH1 | G 174 V | disruptive |
| 7 | 965094 | */* | +T/+T | YGR237C | protein truncation |
nonsense | ||
| 14 | 682330 | */* | +T/+T | YNR031C | SSK2 | protein truncation |
nonsense | |
| UWOPS83-787.3 : 15%_48h |
5 | 546138 | A/A | G/G | YER177W | BMH1 | N 178 S | disruptive |
| 15 | 824405 | */* | −G/−G | YOR267C | HRK1 | protein truncation |
nonsense | |
| YJM981 : 1%_48h | 4 | 1433639 | */* | */+G | YDR490C | PKH1 | protein truncation |
nonsense |
| 5 | 546294 | T/T | T/G | YER177W | BMH1 | L 230 * | nonsense | |
| 10 | 430543 | T/T | T/A | YJL005W | CYR1 | silent | synonymous | |
| 12 | 849668 | C/C | C/A | YLR361C-A | S 4 I | NA | ||
| 13 | 409669 | G/G | G/T | YMR070W | MOT3 | A 173 S | tolerated | |
| YJM981 : 15%_48h | 12 | 753687 | C/C | C/T | YLR310C | CDC25 | W 1103 * | nonsense |
The vast majority of SNPs were non-synonymous changes (21/27). Of the remaining mutations, three were synonymous changes in protein sequences, and three were located in intergenic regions. Of the 21 non-synonymous variants, ten were nonsense mutations and eleven were missense mutations. Unlike nonsense mutations, which typically result in loss of protein or domain function, missense mutations may cause loss of function, gain of function, or no functional change. Of the eleven missense mutations, SIFT (Kumar et al. 2009) predicted that six would disrupt protein function, four would be tolerated, while one had insufficient homology data for an accurate prediction. Assuming that protein-modifying mutations are most likely to influence phenotype, these data suggest that 59% of all found variants (16/27) most likely cause the evolved phenotypes, with at least one protein-modifying mutation per evolved clone.
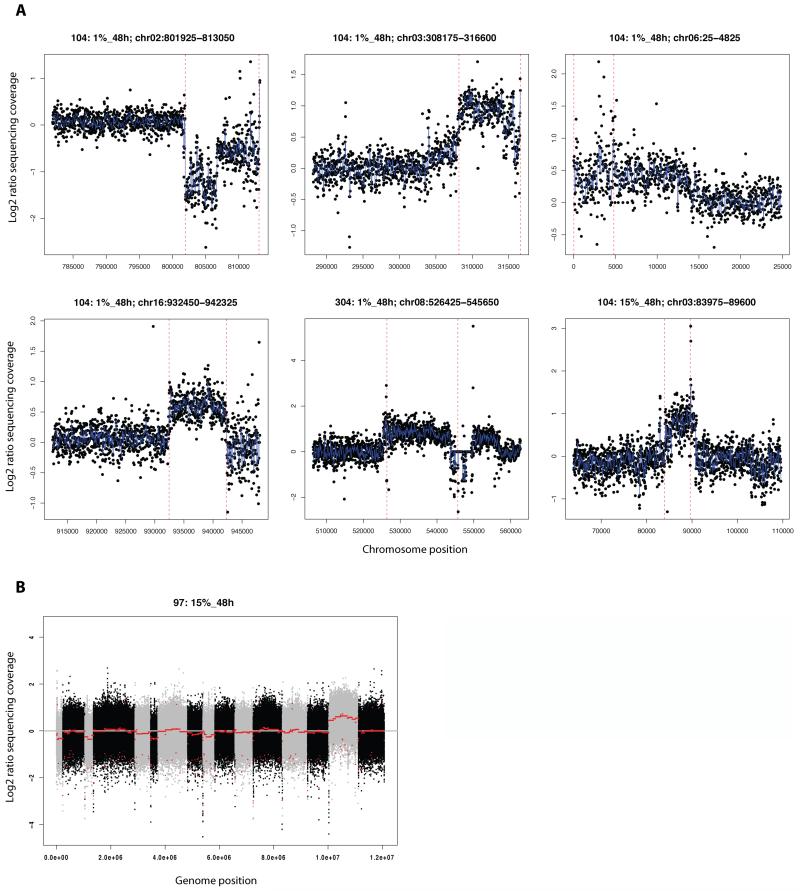
We found evidence of seven copy number variants (CNVs) in the evolved clones relative to their ancestral strain (Figure 2), with four in YPS128 evolved in 1%_48h strain alone. Three of these four increase copy number, most likely resulting from a heterozygous duplication event, while the other CNV is likely a heterozygous deletion. All four CNVs are in telomeric or subtelomeric regions. The three remaining CNVs each occur in one strain apiece. Lastly, chromosome 15 appears to be a homozygously duplicated, creating two extra copies, in Y55 evolved in 15%_48h.
Figure 2. Copy Number Variants (CNVs) identified by sequencing coverage of the evolved strain versus the ancestral strain.
A) Zoomed-in coverage plots of each sub-chromosomal CNV. Red-dotted lines delimit the regions identified as CNVs by DNAcopy. Blue line is a running median of log2 ratios. B) Whole-genome coverage plot for the 97: 15%_48h evolved strain showing duplication of chromosome 15. Alternating grey/black colors are chromosomes. Red line is a running median of log2 ratios.
Convergence on genes involved in same function or pathway
We determined Gene Ontology (GO) enrichments (Boyle et al. 2004) for the genes mutated across all strains (Table 5). GO enrichments shared by both the 1%_48h and 15%_48h conditions include general signal transduction as well as Ras protein signal transduction indicating genetic convergence on genes involved in these pathway.
Table 5. GO terms significantly enriched in the mutated gene set.
| Selection regime | GO terms | GO ID | P-value | Expected FP | Gene(s) annotated to the term |
|---|---|---|---|---|---|
| All | Signal transduction | 7165 | 0.00023 | 0 | MTH1, PKH1, BMH1, CYR1, CDC25, SSK2, SOG2 |
| Intracellular signal transduction | 35556 | 0.00296 | 0.04 | PKH1, BMH1, CYR1, CDC25, SSK2 | |
| Ras protein signal transduction | 7265 | 0.02499 | 0.34 | BMH1, CYR1, CDC25 | |
|
| |||||
| 1%_48h | Signal transduction | 7165 | 0.0002 | 0 | PKH1, BMH1, CYR1, SSK2, SOG2 |
| Intracellular signal transduction | 35556 | 0.0007 | 0 | PKH1, BMH1, CYR1, SSK2 | |
| MAPKKK cascade | 165 | 0.02237 | 0.1 | PKH1, SSH2 | |
| Cellular response to stimulus | 51716 | 0.04528 | 0.16 | PKH1, BMH1, CYR1, SSK2, SOG2 | |
|
| |||||
| 15%_48h (protein modifying mutations only) |
Mitotic cell cycle | 278 | 0.04208 | 0.24 | TPD3, BMH1, TVP38, CDC25 |
The 1%_48h condition was enriched for the GO terms signal transduction, MAPKKK cascade and cellular response to stimulus (Table 5). The drivers of these enrichments were BMH1, PKH1, SSK2 and SOG2. Of these, only BMH1 obviously functions in nutrient signaling (Burbelo and Hall 1995; Bertram et al. 1998). The others are involved in cell wall integrity and osmolarity sensing through the HOG pathway.
The 15%_48h condition did not have significant GO enrichments when all mutated genes were considered, but when only genes with protein-modifying mutations were used, “mitotic cell cycle” was enriched (Table 5). This enrichment was driven by TPD3, BMH1, TVP38 and CDC25. Tpd3p is a regulatory subunit of protein phosphatase 2A (PP2A) and is required for transcription by RNA pol III (van Zyl et al. 1992), which when mutated results in larger cell size (Jorgensen et al. 2002), as well as a general decrease in resistance to stress (Auesukaree et al. 2009). We observed a nonsense mutation in the clone evolved from Y55 in the 15%_48h condition, which may cause the larger cell size characteristic of the 15%_48h phenotype. Cdc25 activates Ras by phosphorylation of GDP, leading to increased signaling through the cAMP/PKA pathway, which stimulates progression through the cell cycle (Broek et al. 1987). The CDC25 mutation in the evolved clone of YJM981 under the 15%_48h condition is a nonsense mutation, likely resulting in reduced Ras activity and decreased signaling through the cAMP/PKA pathway.
Genotypic convergence at the BMH1 locus
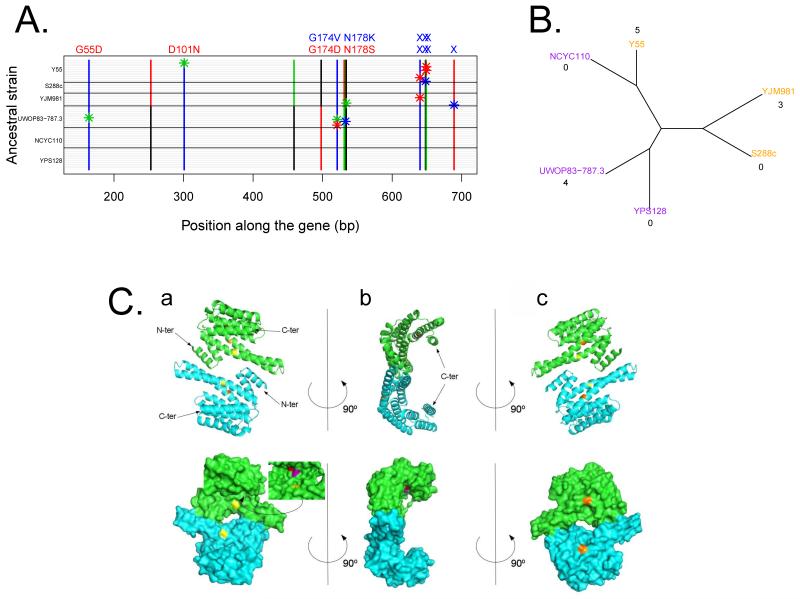
Because four out of the eight sequenced clones had BMH1 mutations (Table 4), we determined whether there were BMH1 mutations in the other evolved populations, and also sequenced BMH2, which encodes a paralogous protein of the 14-3-3 protein family. No mutations were detected in BMH2, but 12 out of the 60 clones had BMH1 mutations (Figure 3A), with eight being heterozygous for the mutant allele and four being homozygous. The 12 mutations were distributed across nine sites and can be grouped in two classes: six mutations are STOP mutations that occurred within the 642-690 bp positions from the ATG and six mutations are non synonymous mutations that mostly occurred at 522-534 bp from the ATG (Figure 3A). At one site, the exact same mutation occurred twice independently.
Figure 3. Polymorphism in the BMH1 gene and encoded protein.
A) Each grey horizontal line represents the allele sequence of a clone isolated from a population after evolution. The ancestral strain is shown on the left. Polymorphic sites are shown with vertical lines. Nucleotides are indicated with color (T in red, A in green, C in blue, G in black). Bars represent the ancestral allele and stars show mutations that had occurred along the evolutionary course. Corresponding changes in the protein are indicated on top of the nucleotide mutations. Crosses are stop mutations. Amino-acid substitutions are indicated by their position in the protein. Mutations that have occurred in 1% selection regimes are indicated in blue, mutations that have occurred in 15% selection regimes in red. B) Neighbor-joining tree based on the proportion of different nucleotides between genomes of the six ancestral strains. The BMH1 allele is indicated by the color of the strain’s name (orange or purple). The number of mutations that has occurred in BMH1 during the course of evolution is indicated below the name of each strain. C) Model of dimeric Bmh1p (region 1 to 235). A: front view, B: side view, C: back view, top row: cartoon representation, bottom row : surface representation. The two chains composing the dimer are shown in different colors (cyan and green). The positions affected by the mutation are highlighted in colors: position 55 in yellow, position 101 in orange, position 174 in red and position 178 in purple.
Using a mathematical model, we checked whether the number of BMH1 mutations observed at the end of the experimental evolution could be found under the hypotheses that there is no selection. By simulating 107 experimental evolutions of M1=32 populations (respectively M2=28 populations) with 34 bottlenecks (96h selection regime) or 69 bottlenecks (48h selection regime), we estimated that the probabilities of observing 4 mutants after evolution in the 96h selection regime or 8 mutants after evolution in the 48h selection regime under the null hypotheses that there is no selection were below 10−7.
BMH1 mutations were found in Y55, YJM981, UWOPS83-787.3 but not in the other ancestor strains leading to a significant ancestor effect on convergence (Figure 3A and 3B, Fisher’s Exact Test, P=0.005). The occurrence of mutations in BMH1 was not related to the ancestral BMH1 allele (Table 6) and does not appear to be related to the phylogenetic distance between the ancestral strains (Figure 3B).
Table 6. Distribution of the 13 Bmh1p mutations.
There are 2 different ancestral sequences of Bmh1p (W1 and W2), one in S288C, Y55, YJM981 strains, the other in YPS128, NCYC110, UWOP83-787.3 strains.
| Ancestor strain | Selection regimes | |||
|---|---|---|---|---|
| 1%_48h | 1%_96h | 15%_48h | 15%_96h | |
|
| ||||
| S288c | ||||
| Y55 | E214stop Y216stop |
2 × K217stop | D101N | |
| YJM981 | L230stop N178K |
E214stop | ||
|
| ||||
| YPS128 | ||||
| NCYC110 | ||||
| UWOP83-787.3 | G174V |
G174D
N178S G55D |
||
Phenotypic effect of BMH1 mutations
The distribution of the two classes of BMH1 mutations (truncation versus amino-acid changes) was different between the two media although not statistically significant (Fisher’s Exact Test, P=0.06). STOP mutations, resulting in truncated proteins, emerged only in the 1 % glucose medium (Table 6). From the predicted 3D protein structure, these truncated proteins will lack the final helix of the C-terminus as well as a glutamine repeat known to be involved in protein/protein interactions (Figure 3C). Non synonymous mutations lead to amino-acid changes at positions 55, 101, 174, 178 of the protein (Table 6) and were found in strains evolved in both glucose concentrations. Amino-acid changes at 55, 174, and 178 are located inside the groove where Bmh1p is expected to interact with other proteins as predicted by docking with arbitrary peptides and with two known Bmh1 protein partners (Serine/threonine-protein phosphatase PP1-2 and Heat shock protein Ssb1).
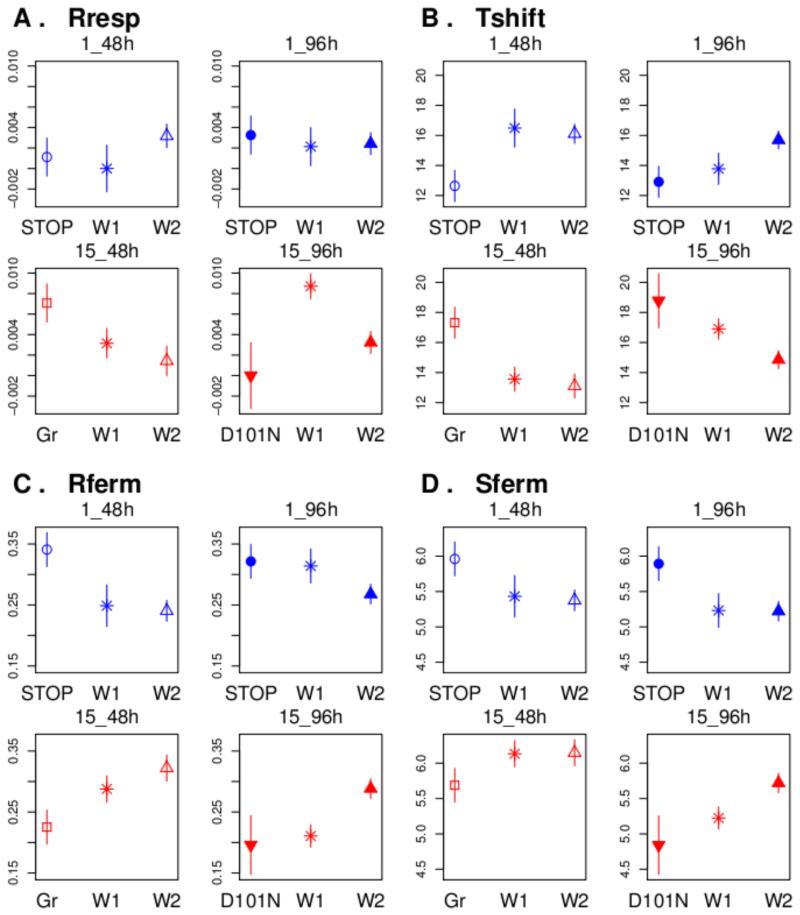
We tested the effect of BMH1 mutations on the value of each of the 13 traits separately. We distinguished five classes of Bmh1 protein: the ancestral sequences of strains S288c, Y55, YJM981, the ancestral sequences of strains of YPS128, NCYC110, UWOP83-787.3, the truncated proteins by stop mutations, the proteins having amino-acid substitutions pointing toward the groove, and the protein having the unique amino-acid substitution pointing toward the back inside. The last three classes of BMH1 mutations changed the phenotypic value of a similar set of traits: Rferm, Tshift, Sferm and Rresp (Figure 4, Table 7). Interestingly, knowing the class of BMH1 mutation provides as good or better prediction of the variation of these traits than knowing the ancestral strain (ANOVA’s R2 comparison, Table 7). Globally, the mutation effects were clearer in 15 % evaluation medium than in 1 %. In 15 % glucose medium, substitutions G55D, G174D and N178S, which are located in the Bmh1p groove of strain UWOP83-787.3, decreased average Rferm (P=0.026), increased Tshift (P=0.01) and Rresp (P=0.047), when compared to the ancestral protein sequence W2. Interestingly, the substitution D101N located in the back of the protein in strain Y55 had also an effect on Tshift and Rferm (decreased Rferm P=0.05, increased Tshift, P=0.019). The effects of stop mutations were opposite to the effects of amino-acid substitutions: they increased Rferm (P=0.013 for populations evolved in 1%_48h, P=0.168 for populations evolved in1%96h), decreased Tshift (P=0.02 for populations evolved in 1%_48h, P=0.056 for populations evolved in 1%_96h) and increased Sferm (P=0.086 for populations evolved in 1%_48h, P=0.047 for populations evolved in 1%_96h). Because stop mutations only occurred in the 1 % selection regimes, we also analyzed whether the effect of stop mutations changed with the media of evaluation. We found that stop mutations had antagonistic effects in 15 % and 1 % glucose media: while stop mutations increased Rferm and decreases Tshift in 15 % glucose media, it decreased Rferm and increased Tshift in 1 % glucose media. Note that stop mutations were selected in the media where their effect on the reproduction rate was negative, demonstrating that reproduction rate is not always the best proxy for fitness.
Figure 4. Effect of Bmh1p mutations in each selection regime where the mutations had occurred as evaluated in 15% glucose medium.
W indicates evolved populations that have kept their ancestral protein sequences (W1 or W2), Gr indicates evolved populations that have had a substitution pointing on the groove of the protein, STOP indicates evolved populations having a truncated protein.
Table 7. Testing the effect of Bmh1p mutations.
Two ANOVA models were carried out, the first one with ancestor strains as genetic information, the other one with Bmh1p class of sequences as genetic information. The first column presents the R2 of the first model. The other columns show the R2 of the second model as well as the P-values associated with the Bmh1p main and interactions effects. Significant effects after FDR correction are highlighted in grey.
| R2 | R2 | Pvalue | Pvalue | Pvalue | Pvalue | |
|---|---|---|---|---|---|---|
| Traits | Strains model | Bmh1p model | Bbmh1p |
Bmh1p
selection regime |
Bmh1p
eval medium |
Bmh1p
evalmedium selection regime |
| PopSize48h | 0.607 | 0.295 | 0.979 | 0.967 | 0.853 | 0.769 |
| PopSize96h | 0.572 | 0.194 | 0.624 | 0.683 | 0.232 | 0.707 |
| Kferm | 0.543 | 0.350 | 0.704 | 0.541 | 0.067 | 0.976 |
| Sferm | 0.783 | 0.292 | 0.156 | 0.038 | 0.365 | 0.979 |
| S48h | 0.853 | 0.535 | 0.722 | 0.138 | 0.611 | 0.755 |
| S96h | 0.664 | 0.317 | 0.178 | 0.015 | 0.650 | 0.681 |
| Rferm | 0.324 | 0.266 | 0.522 | 0.551 | 0.132 | 0.034 |
| Rresp | 0.173 | 0.180 | 0.054 | 0.476 | 0.015 | 0.863 |
| Tshift | 0.486 | 0.464 | 0.125 | 0.062 | 0.048 | 0.022 |
| Jspec | 0.789 | 0.712 | 0.194 | 0.886 | 0.741 | 0.778 |
| Yferm:1% | 0.349 | 0.130 | NA | NA | 0.053 | 0.428 |
| Yferm:15% | 0.587 | 0.222 | NA | NA | 0.632 | 0.005 |
| Eth_48h:1% | 0.448 | 0.393 | NA | NA | 0.403 | 0.328 |
| Eth_48h:15% | 0.107 | 0.069 | NA | NA | 0.709 | 0.120 |
| Eth_96h:1% | −0.020 | −0.188 | NA | NA | 0.912 | 0.912 |
| Eth_96h:15% | 0.763 | 0.633 | NA | NA | 0.002 | 0.939 |
Discussion
By varying both the environment and genetic background during experimental evolution, we found direct evidence of multiple traits divergence between experimental conditions, as well as extensive convergent evolution of diverse genetic backgrounds to common environments at the phenotypic and genetic levels.
Divergent evolution between environments
Each environment selected for a different combination of phenotypic values, indicating that diversifying selection had occurred between the four selection regimes, and that divergence was due to various life-history and metabolic traits rather than a single fitness component. Results from numerous experiments have shown that environmental variation in space and time indeed leads to ecologically divergent populations (Rainey et al. 2000; Jessup et al. 2004; Kassen and Rainey 2004; MacLean 2005; Habets et al. 2006, Kolbe et al. 2012). In yeast, it is known that populations from natural habitats such as oak and soil have a small cell size, large population and high growth rate, while yeast populations from domesticated habitats (beer, wine, bread) have a bigger cell size, a higher survival rate but a lower growth rate and population size (Spor et al, 2008, 2009, Albertin et al. 2011). Here, we show in the lab that decreasing the amount of glucose resource and increasing the time of glucose starvation (1% _96h selection regime) indeed select for yeast populations of small cell size but high growth rate and high population size in comparison to environments with high sugar availability. By analyzing evolution in action under differing controlled environments, we provide direct evidence that differences in sugar availability, as found between forest and domesticated environments, can explain the difference of life-history strategies in the yeast ecological niches.
Convergent evolution in phenotype and genotype
Phenotypic convergent evolution is a well-documented phenomenon that shapes the adaptation of diverged organisms evolving in a common niche. Most studies investigating phenotypic convergence have focused on a single trait and/or a single environment. Here, we provide direct experimental support for multi-trait convergence in response to selection in four different environments. We chose ancestral strains that were widely distributed on the S. cerevisiae phylogenomic tree (Liti et al. 2009) that were also phenotypically divergent for their life-history and metabolic strategies (Spor et al. 2009). Despite this diversity, strains evolved towards a common phenotype in each selection regime. In Escherichia coli, Fong et al demonstrated parallel genotypic evolution and convergent growth phenotypes after evolving nine replicates of a single strain in two environmental conditions (Fong et al. 2005). Our study extends these findings to show that phenotypic convergence occurred between different genetic backgrounds.
After sequencing the genomes in a subset of our evolved strains, we find evidence that the same pathways, genes and even nucleotides are recurrently mutated. The Ras/cAMP/PKA pathway and protein phosphatase 2A (PP2A) complex are repeated targets, with mutations in the genes BMH1, CDC25, CYR1 and TPD3, PPM1, respectively. The gene BMH1 is mutated in at least 20% of the populations, and four sites were mutated independently twice. These repeated mutational events strongly suggest that phenotypic convergence can be caused by convergence at the genotypic and pathway level. Mutations in the Ras/cAMP pathway had already been found in many other evolution experiments performed in chemostats (Kao and Sherlock 2008; Kvitek and Sherlock 2011; Wenger et al. 2011), indicating that this pathway is a frequent target of adaptive mutation both in continuous and batch culture.
Finding recurrent mutations in BMH1 was unexpected, and to our knowledge no previous experimental evolution studies have discovered adaptive mutations in this gene. BMH1 is highly pleiotropic, and have been shown to bind at least 271 proteins (Kakiuchi et al. 2007). Molecular analyses have implicated it in many functions such as chaperone-like protein, protein tethers, cell signaling, cell cycle control, transcription regulation, post-transcriptional regulation, life-span and apoptosis (Aitken 2006; van Heusden and Steensma 2006; Bruckmann et al. 2007; Paul and van Heusden 2009; Wang et al. 2009; Clapp et al. 2012; Veisova et al. 2012). Here, we showed that Bmh1 has an impact on the reproduction rate in fermentation and respiration, the length of fermentation (until the diauxic shift) and on cell size. Which traits controlled by Bmh1p have been selected for in our experiment remains to be studied.
We also see evidence for phenotypic convergence caused by mutations in disparate pathways in the 15%_48h condition. Mutations in genes within the PP2A complex (TPD3, PPM1), Ras pathway/network (CDC25, BMH1), glucose sensing pathway (MTH1), and membrane potential/cytoplasmic pH regulation (HRK1) are all likely causative mutations. However, each mutation affects a different pathway, with multiple pathways often being mutated in individual clones, suggesting that disparate genetic changes can converge on a common phenotype. Fong et al. (2005) demonstrated that the transcriptional states of their evolved populations were very different from each other, despite similarity in endpoint growth phenotypes. They also showed that the evolutionary response involved an initial widespread expression shift followed by a large number of compensatory gene expression changes. Recently, other studies have also highlighted that phenotypic convergence could be achieved through diverse genetic mechanisms (Steiner et al. 2009; Chou and Marx 2012).
The cause of convergence
It is a common debate whether convergent evolution occurs because of natural selection toward a common adaptive phenotype, or for reasons unrelated to adaptation such as random evolutionary changes or constraints inherent to the biological system (DePristo et al. 2005; Weinreich et al. 2006; Gompel and Prud’homme 2009; Feldman et al. 2012). Here, we provide strong evidence of convergent phenotypic and genotypic evolution occurring because of adaptation.
We show that mutations in a single gene converge specifically in one environment and have antagonistic effects in other environments, which is strong evidence that convergence occurred due to selection. Other mutations in the same gene, even at the same site, occurred and were selected for in different genetic backgrounds and in different environments. In addition, we show that positions of the mutations in BMH1 appear to be nonrandom. None of the changes are located in the N-terminus of the protein, which is involved in dimerization. Instead, observed missense and nonsense mutations are predicted to be located in regions where protein-protein interactions occur and may change the capacity for interaction of Bmh1p with its potential partners. Finally, by simulating our experimental evolution under the neutral hypothesis, we show that such genetic convergence is unlikely without selection.
Genotypic convergence is thought to occur because of variation in adaptive potential between loci: variation in mutation rate, variation in the magnitude of mutation effect, variation in the number and type of traits controlled directly or indirectly by the locus (pleiotropy, epistasy). Theoretical studies (Chevin et al. 2010) have shown that the probability of parallel evolution is increased when pleiotropy increased. Here we found that among 8 genomes, the only gene that has been mutated independently several times is highly pleiotropic.
The effect of historical contingency
We showed that phenotypic and genotypic evolution depended on the ancestor. This was true for all but two life-history traits (Rresp and Rferm in 15% evaluation medium), for all but two metabolic traits (Tshift in 1% evaluation medium and Eth48h, Table S1) as well as for the occurrence of selected mutations in BMH1 (Table 6). The influence of the initial genotypes remains apparent even if adaptive convergence within a selection regime occurred; in fact, the phenotypic evolution was even better explained by initial genotype than by selection regime in most traits (Table S1). However, predicting the phenotypic evolution based on phenotypic or genetic distances between ancestors appear not to be feasible. Results from previous studies have led to the idea that the influence of historical contingency is higher for traits that have less impact on fitness (Travisano et al. 2005, Joshi et al. 2003). Here, we present the most complete study thus far for testing this idea by including the analysis of many different cellular levels: genomic, metabolic, morphological and reproduction traits. By contrast to previous studies, we found that the ancestors effect persists for all traits’ and has not disappear for traits that are strongly selected for. As illustrated by convergence in BMH1, we show that this can be partly explained by the fact that genetic convergence that is contingent on an ancestor’s genetic background occur in genes that are highly pleiotropic and that determine variation in many different types of traits (metabolic, morphological, reproduction traits). Finally, we found that the effect of historical contingency depends on the environment indicating that trait plasticity should also be considered when analyzing the effect of history. All together our data highlight the role of epistasis, pleiotropy and environment into life-history evolution.
Fitness landscape in microbes
The fitness landscape topology found here correspond to a rugged landscape with various connected peaks organized around a convergent point. Whether the top of the peaks is reached in our experiment remains to be studied. Ruggedness has been related to a high level of epistasis (Kvitek and Sherlock 2011). We show here that it is also related to constraints emerging from pleiotropy. By and large, researchers have used the non-competitive reproduction rate (also called growth rate) as a proxy for evolutionary fitness in microbes (Orr 2009). It remains debatable how accurate of a proxy this is, since adaptive strategies can impact traits other than reproduction rate, such as the combination of traits representing fitness, often referred to as life-history traits (Roff 2002). For example, it has been shown that a simple increase in reproduction rate does not have to reflect the advantage of diploids over haploids in experimental evolution (Gerstein and Otto 2011). Similarly, in our case, the 15%_96h selection regime did not select for a higher reproduction rate during fermentation (Rferm). However, it selected for a decreased population size, increased cell size and increase in specific glucose consumption rate, phenotypes that suggest the cells are not increasing reproduction rate but instead are metabolizing the abundant glucose to create biomass. Thus, phenotypic adaptation of multiple traits should be considered when investigating the fitness landscape of evolved microbial populations.
In conclusion, by analyzing the evolution in action and its genotypic cause, we found direct evidence of both convergent evolution in a particular environment and divergent selection among contrasted environments. In addition we highlight the need to analyze multiple fitness components for a better understanding of adaptation. We demonstrated examples of distinct genetic changes from disparate genetic backgrounds converging on a common phenotype, as well as these disparate genetic backgrounds finding convergent genotypic solutions. Finally we show that genotypic convergence underlying multiple traits evolution is constrained by historical contingency and depends on the environment.
Materials & Methods
Yeast Strains
Six strains were chosen from the Saccharomyces Genome Re-sequencing Project (Table 1) that are distributed on the S. cerevisiae phylogenomic tree (Liti et al. 2009), and differ for several life-history and metabolic traits (Spor et al. 2008; Spor et al. 2009). They are all homothallic autodiploids, except S288c, which is haploid. For each phenotypic evaluation, a single new colony was isolated from the −80°C stock.
Experimental Evolution
Three replicates of each of the six strains were serially propagated (2 mL into 40 mL of fresh liquid medium) under four selection regimes over 5 months. Two different glucose conditions (1 % and 15 % glucose in 3 % YNB with amino acids) and two different “winter” lengths (48 h and 96 h) were chosen. The cultures were incubated at 30 °C at 200 rpm. In 1% glucose, the yeast populations first grow fermentatively and then switch to respirative growth after the diauxic shift. Cycles of 48 h have a very short period of respiration, unlike the cycles of 96 h. In 15 % glucose, the yeast populations grow only fermentatively but are subject to osmotic stress. Cycles of 48 h are composed of a long period of growth followed by a short period of stationary phase whereas cycles of 96 h have a long stationary phase. The four selection regimes are denoted 1%_48h, 1%_96h, 15%_48h and 15%_96h. All six strains were propagated in these four conditions in triplicate. Over 5 months, the populations transferred every 48 hours were transferred 74 times (about 325 generations), while the populations transferred every 96h were transferred 37 times. At each transfer, the populations were diluted 20 fold leading to an effective population size ranging from 106 to 107 depending on the strain and the selection regime. Out of the 72 evolved populations, 12 were discarded due to cross-contamination, as detected using microsatellite analysis. The 60 remaining evolved population included a duplicate of evolution for all ancestors strains in all four selection regimes, except for strain S288c (the haploid strain) in the 1_48h and 15_48h selection regimes and for strain YJM981 in the 1_96h selection regime.
Phenotyping Ancestral Strains and Evolved Populations
Phenotyping of two clones from each of the six ancestral strains and a single end-clone from each of the 60 evolved populations was performed in both 1 % and 15 % glucose as described in Spor et al. (2009), leading to a total of 144 sets of phenotyping kinetics. For each kinetic, nine life-history (reproduction rate in fermentation Rferm, time to diauxic shift Tshift, reproduction rate in respiration Rresp, carrying capacity Kferm and cell size Sferm at the diauxic shift time point, population sizes PopSize48h, PopSize96h, and cell sizes S48h,S96h measured at the serial transfer time points) and four metabolic traits (specific glucose consumption rate denoted Jspec (g × min−1 × cell−1), ethanol amount denoted Eth48h and Eth96h expressed in g × L−1 and yield Yferm, measured as the ratio of the biomass to the quantity of glucose consumed) were quantified.
Statistical Analysis
Response to selection
The 144 kinetics were randomly distributed into five blocks of experiments. For each trait Z, the phenotypic evolution was calculated as follows:
where the value of the ancestral strain is averaged over the replicates.
Variation of each variable ΔZ among media and strains was first analyzed using the following mixed model of analysis of variance (ANOVA):
where ΔZ is the variable, Block is the random block effect (experimental repetition, m = 1, 2, 3, 4, 5), eval medium is the evaluation condition effect (l = 1, 2), strain is the diploid strain effect (i = 1, 2, 3, 4, 5), selection regime is the selection regime effect (j = 1, 2, 3, 4), eval medium*strain (fixed), eval medium*selection regime (fixed) and strain*selection regime (fixed) are interaction effects and ε is the residual error. The interaction effects were chosen to study the interactions between the historical contingency (ancestors’ effect) and selection effects. To account for multiple testing, the significance of the different effects was assessed using the false discovery rate method (ref).
For each trait, normality and homogeneity of residual distributions were checked. Least square means and REML variance estimates were obtained using the JMP® Software. The significance of differences between selection regime means was assessed using the Tukey HSD method.
Identifying convergence and divergence
To identify whether the selection regime and the genetic background of the ancestor had an impact on the traits we carried out a MANOVA on the 13 traits. Those analyses were conducted on the phenotypic values of each trait for each replicate of evolution from each diploid ancestor in each selection regime. However, prior to those analyses, we corrected the data for a block effect estimated by a simple regression model performed trait by trait and for each evaluation medium separately. Missing values were replaced by the mean over replicates of evolution. The PopSize48h,PopSize96h, and Kferm were log10 transformed. We analyzed separately the data obtained with the two evaluation medium. We discarded the S288c strain from those analyses as it was the single haploid strain.
To study further the phenotypic landscape of our evolved strains, we performed linear discriminant analyses (LDA) of those data for each evaluation medium. The four selection regimes defined the four categories used a priori to compute the discriminant functions. The a posteriori assignment probabilities to a priori categories indicate if the objects can be properly assigned to an a priori group. After using the data of diploid evolved strains to build the axes of the LDA, the ancestral strains as well as the evolved strains (including the discared xx strain) were projected on those axes. The output of the analyses in each evaluation medium (l =1, 2) were the coordinates of each strain evolved in each selection regime (j = 1, 2, 3, 4) as well as the coordinates of each ancestor strain. The haploid S288c evolved populations were excluded from the LDA analysis but projected afterwards on the phenotypic landscape in the cases of evolved populations with no missing data.
The axes of the LDA represent a linear combination of the traits that discriminate the most selection regime. Hence, the traits that correlate with the axes are those for which convergence was observed within a selection regime and divergence was observed between selection regimes. To account for multiple testing, the significance of correlations was assessed using the false discovery rate method.
Determining the genotype of evolved clones
Whole-genome sequencing and SNP/indel detection
Genomic DNA was extracted from final clones either by spooling (Treco 1987), or using Qiagen G/100 Genomic Tips. Single-end Illumina sequencing libraries were generated using the Illumina Genomic DNA Sample Prep Kit starting with 5 μg of genomic DNA, and sequencing flow cells were prepared using the Illumina Standard Cluster generation Kit. Samples were sequenced on the Illumina Genome Analyzer II, generating 36 bp reads. The data were mapped to the S288c reference (from SGD, Dec 3, 2008) and to additional contigs generated from other S. cerevisiae strains (Dunn et al. 2012) using BWA v0.5.8 (Li and Durbin 2009) with default parameters.
SNP calling was performed on uniquely mapping reads only using the Genome Analysis Toolkit v1.0.4905 (McKenna et al. 2011) on all strains simultaneously with ad hoc quality filtering (see Supp. Material and Methods). SNPs were validated using Sanger sequencing.
Indels were called using Samtools v0.1.7 (Li et al. 2009) with default settings; the false positive rate was decreased using an in-house developed method (see Supp. Material and methods). Indels were validated using Sanger sequencing.
Copy-number variation (CNV) detection
Sequence coverage was used to identify CNVs between each evolved and ancestral strain pair as in Araya et al. (2010). Briefly, raw sequencing coverage was averaged over 25 bp segments across the genomes of each evolved clone and ancestor and log2(evolved/ancestor) ratios were calculated. These log2 ratios were then adjusted by either the genome-wide log2 mean to identify whole-chromosome CNVs, or each chromosome’s log2 mean to identify intrachromosomal CNVs. Genome segments were identified using the R package DNAcopy v1.22.1 (Venkatraman and Olshen 2007) with default parameters except: smooth.CNA(smooth.region=5); segment(min.width=5, undo.splits=”sdundo”, undo.SD=4). Results of DNAcopy were plotted and regions of varying copy number were identified visually. Putative CNVs occupying partial chromosomes were tested using real-time quantitative PCR of genomic DNA as described (Hoebeeck et al. 2007), except that each putative CNV was normalized to a single non-varying locus located on the same chromosome as the putative CNV. The whole-chromosome 15 CNV in Y55:15%_48h was normalized to a single non-varying locus on a different chromosome.
Gene Ontology (GO) enrichment analysis
GO biological process enrichments were determined using GO::TermFinder (Boyle et al. 2004) at SGD (http://www.yeastgenome.org) using default parameters, except dubious ORFs were omitted from the analysis, with a False Discovery Rate cutoff of 0.05. All genes containing at least one mutation within its ORF were included.
Convergence on BMH1
Testing the presence of selection on BMH1
We built a theoretical model (see Supp. Material and methods) of our experimental evolution and computed the probability of observing recurrent BMH1 mutations among M evolved clones at the end of our experimental evolution under the hypotheses that there is no selection.
Evaluating the impact of BMH1 mutations
We tested for the effect of BMH1 mutations on Z the value of a single trait in an evolved population. We used the following linear model:
where Yijlb is Z, eval medium is the effect of the culture medium (l = 1,2), selection regime (j = 1,2,3,4) is the effect of the selection regime, Bmh1p the effect of mutations in Bmh1p (b=1,2,3,4,5), (Bmh1p*eval medium)lb, (Bmh1p*selection regime)jb, (eval medium*selection regime)jl, (Bmh1p*eval medium*selection regime]ljb interaction effects, and εijlb the residual error. The effects of the Bmh1p protein changes (Bmh1p effect) were tested from the ANOVA using contrast. FDR corrections were carried out using a value of 15%. For three traits (Yferm, Eth_48h, Eth_96h), the ANOVA was carried out separately for each evaluation medium because of residuals heterocedasticity.
This ANOVA model using BMH1 as sole genetic information was compared to the following model using the genetic differences between ancestral strains:
Similar R2 between the two models would indicate that genetic variation in the gene where genetic convergence occurred explained as much traits variation than genome differences between evolved strains.
BMH1: structure and docking predictions
We generated a structural model of yeast dimeric Bmh1p to locate the amino-acid changes and truncation on the 3D protein structure. We used its homology with the human protein 2BR9, whose structure has been resolved by crystallography (Yang et al. 2006, see Supp. Material and methods). Arbitrary and specific docking predictions were then carried out (see Supp. Material and methods).
Supplementary Material
Acknowledgements
The authors would like to thank Gianni Liti for kindly providing strains. We are also grateful to Mickael Brockhurst, the GQF team and Frank Rosenzweig for fruitful discussions and critical reading of the manuscript.
Footnotes
Data archive will be located in the supplementary information
Accession Numbers
The Illumina sequence data are available at the NCBI Sequence Read Archive under accession SRA029322.1.
Literature Cited
- Aitken A. 14-3-3 proteins: A historic overview. Seminars in Cancer Biology. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Albertin W, Marullo P, Aigle M, Dillmann C, de Vienne D, Bely M, Sicard D. Population Size Drives Industrial Saccharomyces cerevisiae Alcoholic Fermentation and Is under Genetic Control. Applied and Environmental Microbiology. 2011;77:2772–2784. doi: 10.1128/AEM.02547-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya C, Payen C, Dunham M, Fields S. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC genomics. 2010;11:88. doi: 10.1186/1471-2164-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt J, Reznick D. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends in Ecology & Evolution. 2008;23:26–32. doi: 10.1016/j.tree.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Pfrender ME, Jones AG. The adaptive landscape as a conceptual bridge between micro-and macroevolution. Genetica. 2001;112:9–32. [PubMed] [Google Scholar]
- Auesukaree C, Damnernsawad A, Kruatrachue M, Pokethitiyook P, Boonchird C, Kaneko Y, Harashima S. Genome-wide identification of genes involved in tolerance to various environmental stresses inSaccharomyces cerevisiae. Journal of applied genetics. 2009;50:301–310. doi: 10.1007/BF03195688. [DOI] [PubMed] [Google Scholar]
- Bedhomme S, Lafforgue G, Elena SF. Genotypic but not phenotypic historical contingency revealed by viral experimental evolution. BMC evolutionary biology. 2013;13:1–13. doi: 10.1186/1471-2148-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram PG, Zeng C, Thorson J, Shaw AS, Zheng XF. The 14-3-3 proteins positively regulate rapamycin-sensitive signaling. Current biology. 1998;8:1259–S1. doi: 10.1016/s0960-9822(07)00535-0. [DOI] [PubMed] [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proceedings of the National Academy of Sciences. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004a;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004b;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D, Toda T, Michaeli T, Levin L, Birchmeier C, Zoller M, Powers S, Wigler M. The S. cerevisiae< i> CDC25</i> gene product regulates the< i> RAS</i>/adenylate cyclase pathway. Cell. 1987;48:789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Bruckmann A, Hensbergen PJ, Balog CIA, Deelder AM, Steensma HY, van Heusden GPH. Post-Transcriptional Control of the Saccharomyces cerevisiae Proteome by 14-3-3 Proteins. Journal of Proteome Research. 2007;6:1689–1699. doi: 10.1021/pr0605522. [DOI] [PubMed] [Google Scholar]
- Burbelo PD, Hall A. 14-3-3 proteins: hot numbers in signal transduction. Current Biology. 1995;5:95–96. doi: 10.1016/s0960-9822(95)00022-4. [DOI] [PubMed] [Google Scholar]
- Chevin L-M, Martin G, Lenormand T. FISHER’S MODEL AND THE GENOMICS OF ADAPTATION: RESTRICTED PLEIOTROPY, HETEROGENOUS MUTATION, AND PARALLEL EVOLUTION. Evolution. 2010;64:3213–3231. doi: 10.1111/j.1558-5646.2010.01058.x. [DOI] [PubMed] [Google Scholar]
- Chou H-H, Marx CJ. Optimization of Gene Expression through Divergent Mutational Paths. Cell Reports. 2012;1:133–140. doi: 10.1016/j.celrep.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Salamin N, Savolainen V, Duvall MR, Besnard G. C4 Photosynthesis Evolved in Grasses via Parallel Adaptive Genetic Changes. Current Biology. 2007;17:1241–1247. doi: 10.1016/j.cub.2007.06.036. [DOI] [PubMed] [Google Scholar]
- Clapp C, Portt L, Khoury C, Sheibani S, Norman G, Ebner P, Eid R, Vali H, Mandato CA, Madeo F, Greenwood MT. 14-3-3 Protects against stress-induced apoptosis. Cell Death and Disease. 2012;3:e348. doi: 10.1038/cddis.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nature Reviews Genetics. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- Dunn B, Richter C, Kvitek DJ, Pugh T, Sherlock G. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Research. 2012;22:908–924. doi: 10.1101/gr.130310.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M, Tawfik DS. Divergence and Convergence in Enzyme Evolution: Parallel Evolution of Paraoxonases from Quorum-quenching Lactonases. Journal of Biological Chemistry. 2011;287:11–20. doi: 10.1074/jbc.R111.257329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer KR, Meyer A. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends in Ecology & Evolution. 2011;26:298–306. doi: 10.1016/j.tree.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED, Brodie ED, Pfrender ME. From the Cover: Constraint shapes convergence in tetrodotoxin-resistant sodium channels of snakes. Proceedings of the National Academy of Sciences. 2012a;109:4556–4561. doi: 10.1073/pnas.1113468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED, Brodie ED, Pfrender ME. From the Cover: Constraint shapes convergence in tetrodotoxin-resistant sodium channels of snakes. Proceedings of the National Academy of Sciences. 2012b;109:4556–4561. doi: 10.1073/pnas.1113468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong SS, Joyce AR, Palsson BØ. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome research. 2005;15:1365–1372. doi: 10.1101/gr.3832305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Otto SP. Cryptic Fitness Advantage: Diploids Invade Haploid Populations Despite Lacking Any Apparent Advantage as Measured by Standard Fitness Assays. PLoS ONE. 2011;6:e26599. doi: 10.1371/journal.pone.0026599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B. The causes of repeated genetic evolution. Developmental Biology. 2009;332:36–47. doi: 10.1016/j.ydbio.2009.04.040. [DOI] [PubMed] [Google Scholar]
- Granek JA, Kayıkçı Ö, Magwene PM. Pleiotropic signaling pathways orchestrate yeast development. Current Opinion in Microbiology. 2011;14:676–681. doi: 10.1016/j.mib.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebeeck J, Speleman F, Vandesompele J. Real-time quantitative PCR as an alternative to Southern blot or fluorescence in situ hybridization for detection of gene copy number changes. Methods Mol Biol. 2007;353:205–226. doi: 10.1385/1-59745-229-7:205. [DOI] [PubMed] [Google Scholar]
- Jorgensen P. Systematic Identification of Pathways That Couple Cell Growth and Division in Yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Joshi A, Castillo RB, Mueller LD. The contribution of ancestry, chance, and past and ongoing selection to adaptive evolution. Journal of genetics. 2003;82:147–162. doi: 10.1007/BF02715815. [DOI] [PubMed] [Google Scholar]
- Kakiuchi K, Yamauchi Y, Taoka M, Iwago M, Fujita T, Ito T, Song S-Y, Sakai A, Isobe T, Ichimura T. Proteomic Analysis of in Vivo 14-3-3 Interactions in the Yeast Saccharomyces cerevisiae †. Biochemistry. 2007;46:7781–7792. doi: 10.1021/bi700501t. [DOI] [PubMed] [Google Scholar]
- Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nature Genetics. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe JJ, Revell LJ, Szekely B, Brodie ED, III, Losos JB. CONVERGENT EVOLUTION OF PHENOTYPIC INTEGRATION AND ITS ALIGNMENT WITH MORPHOLOGICAL DIVERSIFICATION IN CARIBBEAN ANOLIS ECOMORPHS. Evolution. 2011;65:3608–3624. doi: 10.1111/j.1558-5646.2011.01416.x. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Kvitek DJ, Sherlock G. Reciprocal Sign Epistasis between Frequently Experimentally Evolved Adaptive Mutations Causes a Rugged Fitness Landscape. PLoS Genetics. 2011;7:e1002056. doi: 10.1371/journal.pgen.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O’Kelly MJT, van Oudenaarden A, Barton DBH, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozovsky ER, Chookajorn T, Brown KM, Imwong M, Shaw PJ, Kamchonwongpaisan S, Neafsey DE, Weinreich DM, Hartl DL. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proceedings of the National Academy of Sciences. 2009;106:12025–12030. doi: 10.1073/pnas.0905922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwene PM, Kayikci O, Granek JA, Reininga JM, Scholl Z, Murray D. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences. 2011;108:1987–1992. doi: 10.1073/pnas.1012544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE. Convergence in pigmentation at multiple levels: mutations, genes and function. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010a;365:2439–2450. doi: 10.1098/rstb.2010.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE. Convergence in pigmentation at multiple levels: mutations, genes and function. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010b;365:2439–2450. doi: 10.1098/rstb.2010.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AH, Molineux IJ, Springman R, Bull JJ. MULTIPLE GENETIC PATHWAYS TO SIMILAR FITNESS LIMITS DURING VIRAL ADAPTATION TO A NEW HOST. Evolution. 2012;66:363–374. doi: 10.1111/j.1558-5646.2011.01433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. Fitness and its role in evolutionary genetics. Nature Reviews Genetics. 2009;10:531–539. doi: 10.1038/nrg2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski EA, Woods RJ, Lenski RE. The genetic basis of parallel and divergent phenotypic responses in evolving populations of Escherichia coli. Proceedings of the Royal Society B: Biological Sciences. 2008;275:277–284. doi: 10.1098/rspb.2007.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remold SK, Rambaut A, Turner PE. Evolutionary Genomics of Host Adaptation in Vesicular Stomatitis Virus. Molecular Biology and Evolution. 2008;25:1138–1147. doi: 10.1093/molbev/msn059. [DOI] [PubMed] [Google Scholar]
- Rivas Plata E, Lumbsch HT. Parallel evolution and phenotypic divergence in lichenized fungi: A case study in the lichen-forming fungal family Graphidaceae (Ascomycota: Lecanoromycetes: Ostropales) Molecular Phylogenetics and Evolution. 2011;61:45–63. doi: 10.1016/j.ympev.2011.04.025. [DOI] [PubMed] [Google Scholar]
- Roff DA. Life History Evolution. Sinauer Associates; MA: n.d. [Google Scholar]
- Rosenblum EB, Rompler H, Schoneberg T, Hoekstra HE. From the Cover: Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proceedings of the National Academy of Sciences. 2009;107:2113–2117. doi: 10.1073/pnas.0911042107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozpędowska E, Hellborg L, Ishchuk OP, Orhan F, Galafassi S, Merico A, Woolfit M, Compagno C, Piškur J. Parallel evolution of the make–accumulate–consume strategy in Saccharomyces and Dekkera yeasts. Nature Communications. 2011;2:302. doi: 10.1038/ncomms1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. Adaptive radiation along genetic lines of least resistance. Evolution. 1996;50:1766–1774. doi: 10.1111/j.1558-5646.1996.tb03563.x. [DOI] [PubMed] [Google Scholar]
- Spor A, Nidelet T, Simon J, Bourgais A, de Vienne D, Sicard D. Niche-driven evolution of metabolic and life-history strategies in natural and domesticated populations of Saccharomyces cerevisiae. BMC Evolutionary Biology. 2009;9:296. doi: 10.1186/1471-2148-9-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Wang S, Dillmann C, de Vienne D, Sicard D. “Ant” and “Grasshopper” Life-History Strategies in Saccharomyces cerevisiae. PLoS ONE. 2008;3:e1579. doi: 10.1371/journal.pone.0001579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srithayakumar V, Castillo S, Mainguy J, Kyle CJ. Evidence for evolutionary convergence at MHC in two broadly distributed mesocarnivores. Immunogenetics. 2011;64:289–301. doi: 10.1007/s00251-011-0588-7. [DOI] [PubMed] [Google Scholar]
- Steiner CC, Rompler H, Boettger LM, Schoneberg T, Hoekstra HE. The Genetic Basis of Phenotypic Convergence in Beach Mice: Similar Pigment Patterns but Different Genes. Molecular Biology and Evolution. 2008;26:35–45. doi: 10.1093/molbev/msn218. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. The Molecular Diversity of Adaptive Convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- Teotónio H, Chelo IM, Bradić M, Rose MR, Long AD. Experimental evolution reveals natural selection on standing genetic variation. Nature Genetics. 2009;41:251–257. doi: 10.1038/ng.289. [DOI] [PubMed] [Google Scholar]
- Travisano M, Mongold JA, Bennett AF, Lenski RE. Experimental tests of the roles of adaptation, chance, and history in evolution. Science. 267:87–90. doi: 10.1126/science.7809610. n.d. [DOI] [PubMed] [Google Scholar]
- Treco, Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. n.d. [Google Scholar]
- Van Heusden GPH. 14-3-3 proteins: Insights from genome-wide studies in yeast. Genomics. 2009;94:287–293. doi: 10.1016/j.ygeno.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Van Heusden GPH, Yde Steensma H. Yeast 14-3-3 proteins. Yeast. 2006;23:159–171. doi: 10.1002/yea.1338. [DOI] [PubMed] [Google Scholar]
- Van Zyl W, Huang W, Sneddon AA, Stark M, Camier S, Werner M, Marck C, Sentenac A, Broach JR. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Molecular and cellular biology. 1992;12:4946–4959. doi: 10.1128/mcb.12.11.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veisova D, Macakova E, Rezabkova L, Sulc M, Vacha P, Sychrova H, Obsil T, Obsilova V. Role of individual phosphorylation sites for the 14-3-3-protein-dependent activation of yeast neutral trehalase Nth1. Biochemical Journal. 2012;443:663–670. doi: 10.1042/BJ20111615. [DOI] [PubMed] [Google Scholar]
- Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- Wang C, Skinner C, Easlon E, Lin S-J. Deleting the 14-3-3 Protein Bmh1 Extends Life Span in Saccharomyces cerevisiae by Increasing Stress Response. Genetics. 2009;183:1373–1384. doi: 10.1534/genetics.109.107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Spor A, Nidelet T, Montalent P, Dillmann C, de Vienne D, Sicard D. Switch between Life History Strategies Due to Changes in Glycolytic Enzyme Gene Dosage in Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2010;77:452–459. doi: 10.1128/AEM.00808-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM. Darwinian Evolution Can Follow Only Very Few Mutational Paths to Fitter Proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Wenger JW, Piotrowski J, Nagarajan S, Chiotti K, Sherlock G, Rosenzweig F. Hunger Artists: Yeast Adapted to Carbon Limitation Show Trade-Offs under Carbon Sufficiency. PLoS Genetics. 2011;7:e1002202. doi: 10.1371/journal.pgen.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman HA. Different Trajectories of Parallel Evolution During Viral Adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- Yang X, Lee WH, Sobott F, Papagrigoriou E, Robinson C, Grossmann GJ, Sundström M, Doyle D, Elkin J. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc Natl Acad Sci U.S.A. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kumar S. Detection of convergent and parallel evolution at the amino acid sequence level. Molecular biology and evolution. 1997;14:527–536. doi: 10.1093/oxfordjournals.molbev.a025789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.