Abstract
An affordable and fast liquid chromatography-tandem mass spectrometry (LC-MS/MS) method was developed for the accurate and precise determination of global DNA methylation levels in peripheral blood. Global DNA methylation extent was expressed as the ratio of methylated 2′-deoxycytidine (5MedC) to 2′-deoxyguanosine (dG), which were obtained after DNA extraction and hydrolysis and determined by positive electrospray LC–ESI-MS/MS. The cost-effective internal standards 15N3-dC and 15N5-dG were incorporated for the accurate quantification of 5MedC and dG, respectively. The desired nucleoside analytes were separated and eluted by LC within 2.5 min on a reverse phase column with a limit of detection of 1.4 femtomole on column for 5MedC. Sample preparation in 96-well format has significantly increased the assay throughput and filtration was found to be a necessary step to assure precision. Precision was performed with repeated analysis of four DNA QC sample over 12 days, with mean intra- and inter-day CVs of 6% and 11%, respectively. Accuracy was evaluated by comparison with a previously reported method showing a mean CV of 4% for 5 subjects analyzed. Furthermore, application of the assay using a benchtop orbitrap LCMS in exact mass full scan mode showed comparable sensitivity to tandem LCMS using multiple reaction monitoring.
Keywords: Global DNA methylation assay, Liquid chromatography mass spectrometry, 5-Methyl-2′-deoxycytidine, Internal standard, Orbitrap MS
1. Introduction
DNA methylation is an epigenetic regulation that controls normal organismal development and cellular differentiation in mammals [1]. Normal methylation of DNA involves the covalent addition of a methyl group to the 5-position of cytosine on the CpG dinucleotide at gene promoter regions and forms a 5-methyl-2′-deoxycytidine (5MedC) modified DNA [2]. Aberrant DNA methylation patterns are found in Wilms tumor, ovarian epithelial carcinoma and breast cancer cells [3–5] that are characterized by hypermethylation of CpG islands and global genomic hypomethylation [2]. Early detection of the alterations of DNA methylation is critical in understanding carcinogenesis and developing markers for diagnosis.
The current methods for assessing DNA methylation can be classified into gene-specific methods and non-specific (global) analyses [6]. The former relies on restriction enzymes in combination with Southern blot analyses or sodium bisulfite modifications, and map the DNA methylation pattern of specific genetic loci by integrating the methylation pattern with gene expression and transcription [6]. Alternatively, global DNA methylation assays have been developed to quantify the large-scale cellular DNA methylation levels. These assays are based on the identification and separation of methylated nucleosides following enzymatic hydrolysis of DNA, and therefore provide a quantitative and accurate tool for the evaluation of the relationship between genome-wide changes and cell development. Recent findings about global DNA methylation levels functioning as reliable cancer biomarkers have provoked great interest in developing highly sensitive and efficient analytical tools for the determination of methylated and unmethylated DNA.
Many analytical methods have been reported to quantify global DNA methylation levels including immunoassays for 5MedC [7] and chromatographic techniques such as HPLC in combination with UV [8], fluorescence [9] or mass spectrometric (MS) detection, and GC/MS [10]. Among these techniques, liquid chromatography (LC) in combination with MS detection provides a highly sensitive and accurate way for the quantification of global genomic methylation levels [11–14]. It measures 2′-deoxycytidine (dC) and also its 5-methyl adduct (5MedC) in the digested DNA samples after chromatographic separations, and the degree of methylation is expressed as the percentage of the methylated adduct to the sum of dC plus 5MedC [11]. One drawback of current LCMS methods, however, is the scarcity of suitable internal standards for dC and 5MedC to correct experimental and instrumental errors. Friso et al. have used (methyl-d3, ring-6-d1)-5MedC as an internal standard for 5MedC in their LCMS method [11], but this chemical requires custom synthesis and is extremely expensive. Quinlivan and Gregory have reported the biosynthesis of 15N3-5MedC and 15N3-dC [15] as internal standards for 5MedC and dC, respectively, but the preparation procedure is time-consuming and requires Escherichia coli culture that cannot be easily prepared in a clinical and analytical laboratory environment. We intended to establish a fast assay with readily available and affordable internal standards for the reliable and accurate determination of global methylation. Furthermore, we aimed to improve current LCMS methods suffering from long analysis time which prevents high throughput, an essential aspect in modern clinical laboratories. Finally, we evaluated whether recently introduced orbitrap mass spectrometers improve the global methylation assay in comparison to tandem mass spectrometers and whether internal standard corrected ratios of 5MedC/dG are equivalent to non-internal standard corrected ratios of 5MedC/(5MedC + dC).
2. Materials and methods
2.1. Reagents and materials
All nucleoside standards, including dC, 5MedC, and 2′-deoxyguanosine (dG), nuclease P1, phosphodiesterase I, ammonium acetate, and ammonium bicarbonate were purchased from Sigma–Aldrich (St. Louis, MO). The internal standards, 15N3-2′-deoxycytidine (15N3-dC) and 15N5-2′-deoxyguanosine (15N5-dG) were purchased from Cambridge Isotope laboratories (Andover, MA). Alkaline phosphatase was purchased from Roche Applied Science (Indianapolis, IN). Ultracel-10 96-well filter plate with 10 kDa protein cutoff weight was purchased from Millipore (Billerica, MA). All solvents were of LCMS grade from Fisher Scientific (Pittsburg, PA).
2.2. DNA extraction and hydrolysis
Buffy coat was isolated from 4 mL of whole blood collected in a heparin-coated tube by centrifugation at 3000 × g for 20 min at 4 °C. DNA was extracted from the buffy coat with QIAamp mini-prep kit (Qiagen, Valencia, CA). Briefly, the buffy coat was incubated with protease, RNase and lysis buffer, and the DNA was precipitated out by adding ethanol. The crude DNA was then purified using a mini spin column provided from the kit, washed and eluted out with TE buffer. The purity of the extracted DNA was between 1.8 and 2.0 as determined by A260/A280 ratios. The DNA concentration was determined using a NanoDrop spectrophotometer and stored at −20 °C until analysis after adjusting the concentration to 100 ng μL−1 with TE buffer.
The hydrolysis was performed according to a previous report [16] with some modifications. We found that 1 μg of DNA is sufficient for the downstream LCMS analysis. But in our current application, we used 3 μg of DNA instead for easy aliquoting from upstream procedures. Briefly, 30 μL of genomic DNA (100 ng μL−1 in TE buffer) was first denatured at 100 °C for 3 min on a heating block followed by chilling immediately on ice. 3 μL of 0.1 M ammonium acetate (pH5.2) and 4 μL of nuclease P1 (3 mg mL−1 in 20 mM sodium acetate pH 5.2) were added. The mixture was incubated at 45 °C for 2 h. Subsequently, 3.7 μL of 1 M aq. ammonium bicarbonate and 4 μL of phosphodiesterase I (0.001 unit μL−1) were added and the resulting solution was incubated at 37 °C for 1 h. Next, 1 μL of alkaline phosphatase (1 unit μL−1) and 4.5 μL of alkaline phosphatase buffer (10×, Roche) were added, and the incubation was continued at 37 °C for 1 h. After this enzymatic hydrolysis, 84 μL internal standards (15N3-dC and 15N5-dG, 4.17 μg mL−1 in 20 mM sodium acetate buffer, pH 5.2) were added to the hydrolysates to make the final internal standard concentration of 2.6 μg mL−1 which is in the same range as the analytes. As an additional purification step we further cleaned this hydrolysate by filtration to remove large proteins. 40 μL of the DNA digest (total of 134 μL from 3 μg of DNA) was diluted with 160 μL of DI water and the mixture was filtered through an Ultracel-10 96-well plate with 10 kD cutoff weight by centrifugation at 3000 × g for 90 min to remove the proteins/enzymes added for DNA hydrolysis.
Assay throughput and efficiency were improved by applying a 96-well format during all sample preparation steps. DNA hydrolysis was performed using 96-well PCR plates which can be heated on a PCR heating block. The enzyme reagents were prepared as a master mixture and were aliquot to the reaction wells using a multi-channel pipette. After adding internal standards and filtration, the samples were transferred to a 96-well HPLC plate with a pierceable lid for autosampler injection.
2.3. LCMS procedure
The analysis was performed with a model Accela ultra-HPLC system connected to a TSQ Quantum Ultra triple-quadrupole mass spectrometer (Thermo Electron, Waltham, MA) using a HTC Pal autosampler (Leap technologies, Carrboro, NC). 10 μL of DNA digest containing the internal standard was injected into a Hypersil Gold C18 column (2.1 mm × 50 mm, 1.9 μm, Thermo) coupled with a pre-column filter (2.1 mm, 0.2 μm, Thermo), using a mobile phase consisting of 5% of 0.1% formic acid in methanol and 95% of 0.1% aq. formic acid at a flow rate of 350 μL min−1. The total run time using the isocratic elution was 2.5 min.
MS measurements were carried out with positive electrospray ionization (ESI). Tandem MS was performed using a spray voltage of 4500 V, heated capillary temperature of 300 °C, nitrogen as sheath gas (pressure 30 units) and auxiliary gas (pressure 5 units) and argon as the collision gas at 1.0 Torr. The scan time for each mass was set at 0.1 s and scan width at 0.7 unit. The skimmer offset was set at 5 V to reduce sodium adducts and dimer formation. To minimize the loss of sensitivity due to contamination on the sample cone, the divert valve was set to detector position from 0.25 to 2.2 min, and the ion transfer tube was replaced when the fore pressure was below 0.8 unit. Data acquisition and analysis were performed using Thermo’s Xcalibur software.
The MS data were also obtained on an Orbitrap MS (model Exactive, Thermo) in full scan mode. MS detection was conducted under positive ESI mode same as triple quadrupole MS. The in source CID was set at 5 eV to dissociate dimers and adducts. Maximum injection time was set at 250 ms and the scan range is 100–300. No higher energy collisional dissociation (HCD) was applied. Data acquisition and analysis were performed using Thermo’s Xcalibur software. Detection of the analytes was set within 10 ppm of the calculated mass.
2.4. Standard preparation
Stock solutions of the nucleosides were initially prepared gravimetrically by dissolving in methanol and diluted to a final concentration of 500 μg mL−1 (dC and dG) and 100 μg mL−1 (5MedC) in methanol as the working stock. The molar extinction coefficients (ε in M−1 cm−1) were determined using methanol diluted stock solution at 5 μg mL−1 as follows: dG (λ = 255 nm, ε = 14,949), dC (λ = 274 nm, ε = 11,348), and 5MedC (λ = 280 nm, ε = 10,313), and we used these ε-values for the concentration determination of stock solutions in all subsequent experiments. Six calibration standards containing a mixture of 5MedC/dC/dG were freshly prepared daily by serial dilution of the stock solutions in 20 mM sodium acetate (pH 5) buffer to the following concentrations (ng mL−1): 125/2500/2500, 250/5000/5000, 375/7500/7500, 500/10,000/10,000, 750/15,000/15,000, and 1000/20,000/20,000. 15N3-dC was used as the internal standard for 5MedC and dC, whereas 15N5-dG was applied as internal standard for dG (final concentration of internal standard: 2.6 μg mL−1). The calibration curve covered the expected DNA concentrations in unknown samples, and was plotted by the peak ratio of analytes/internal standard versus analyte concentrations. The calibration curve was linear (R2 > 0.99) in this range and was plotted for every batch. The standards were treated in the very same way as DNA samples including enzyme incubation.
2.5. Method validation
The method was validated based upon linearity, accuracy and consistency. Linearity was measured using a six-point calibration curve. Four sets of quality control DNA samples (QC 1–4) were prepared from the buffy coats of four individuals. The coefficient of variation (CV) was evaluated using a total of four QCs in duplicate or quadruplicate that were inserted randomly to a 70-DNA-sample batch and analyzed in every batch for 12 batches. Our method was further validated by comparison of 5 individual DNA methylation results (5MedC/dG) using a reported method that applied isotope-labeled internal standards [11], and their CVs were calculated.
3. Results
3.1. LC-MS/MS
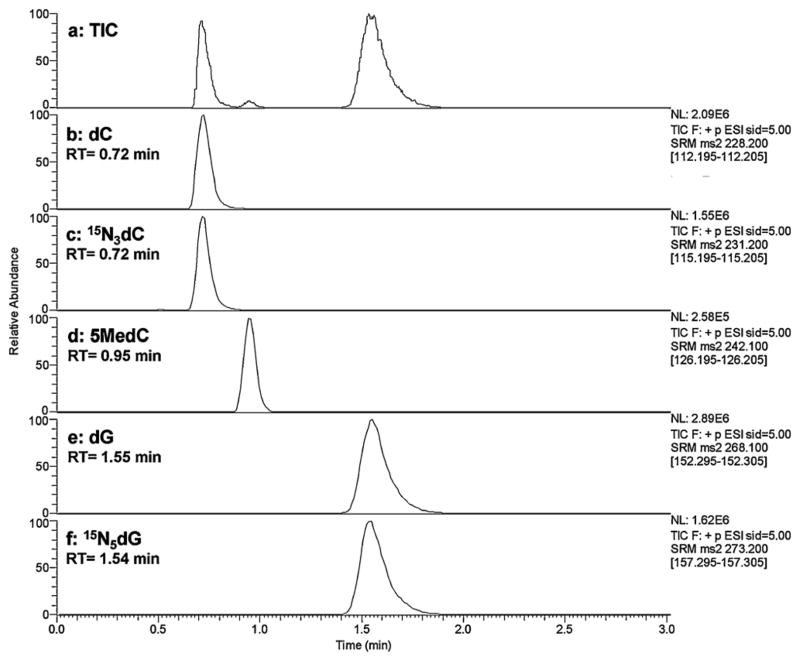
Baseline separation of all nucleosides of interest (5MedC, dC and dG) and two internal standards (15N3-dC and 15N5-dG) was achieved within 2.5 min (Fig. 1) using a Hypersil Gold C18 reverse phase column with an isocratic mobile phase consisting of 0.1% formic acid and 5% methanol in water (flow rate of 350 μL min−1, back pressure below 3500 psi). ESI tandem mass spectrometry was optimized with direct infusion of 1 μg mL−1 5MedC in 0.1% formic acid in methanol. Analyses of nucleosides were conducted in positive electrospray ionization mode, and quantification was accomplished in multiple reaction monitoring (MRM) mode by monitoring the [M + H]+ parent ion to product ion (generated by the loss of deoxyribose moiety) transitions: 5MedC m/z 242.1/126, dC m/z 228.2/112.2, dG m/z 268.1/152.3, 15N3-dC m/z 231.2/115.2, and 15N5-dG m/z 273.2/157.3. The collision energy was set at 14 V for all the analytes, and scan time is 100 ms for each pair. The calibration curve was linear from 50 fmol to 200 pmol, and the limit of detection (LOD) of 5MedC was 1.4 fmol on column based on signal to noise ratio (S/N = 3).
Fig. 1.
Typical LCMS chromatogram of QC DNA hydrolysates showing separation of Total Ion Current (a), dC (b), 15N3-dC (c), 5MedC (d), dG (e) and 15N5-dG (f) with a TSQ Quantum Ultra triple-quadrupole mass spectrometer. Numbers give retention times in minutes.
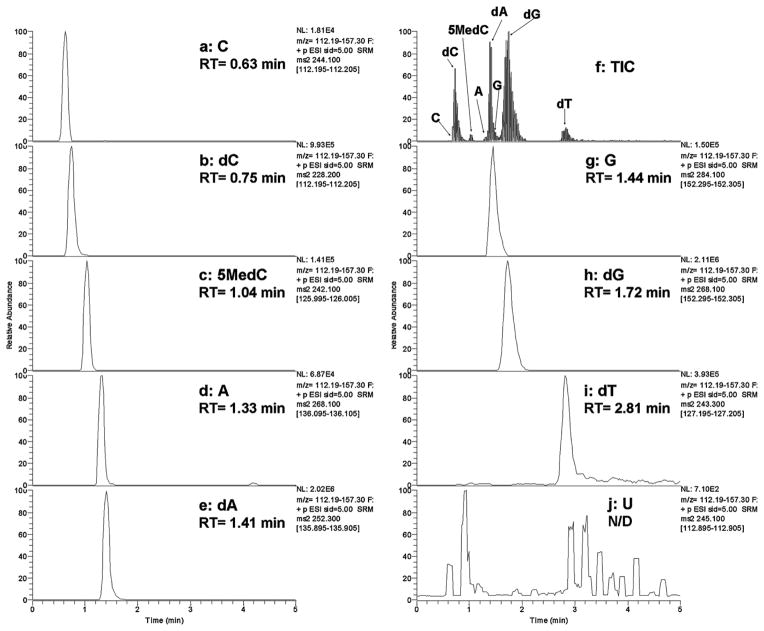
In addition, in order to identify potential RNA contamination and find out its effects on DNA nucleoside ionization, we separated the RNA and DNA nucleosides using the same analytical column and isocratic mobile phases as detailed in the experimental session. MRM scan data of the five deoxyribonucleosides (5MedC, dC, dG, dA and dT) and the four ribonucleosides (C, G, A and U) were acquired. The transitions pairs of m/z 242.1/126.3, 228.2/112.2, 268.1/152.3, 252.3/135.9, 243.3/127.2, 244.1/112.2, 284.1/152.3, 268.1/136.1, and 245.1/112.9 were selected for the detection of 5MedC, dC, dG, dA, dT, C, G, A and U, respectively. Fig. 2 shows the chromatogram of the nine nucleosides mentioned above in QC DNA sample. As shown in Fig. 2, RNA nucleosides were separated from DNA nucleosides with slight overlapping between C/dC and A/dA. However, the intensity of RNA nucleosides (C, G and A) is less than 4% of their counter parts in DNA nucleosides (dC, dG and dA, respectively), suggesting that the extracted DNA is pure and their negative ionization effects on DNA nucleosides would be negligible. The detection of urindine (U) was less sensitive than other nucleosides possibly was attributed to its weak proton affinity.
Fig. 2.
Typical LCMS chromatogram of QC DNA hydrolysates showing separation of cytidine (a), dC (b), 5MedC (c), adenosine (d), dA (e), total ion current (f), guanosine (g), dG (h), 2-deoxythymidine (i), and uridine (j) with a TSQ Quantum Ultra triple-quadrupole mass spectrometer. Numbers give retention times in minutes. Uridine was not detected in the analysis timeframe between 0 and 5 min (signals shown are of very low intensity and show as large peaks due to the >500-fold magnified scale relative to other panels).
3.2. Orbitrap MS
Filtered DNA digests were also analyzed by orbitrap MS under full scan mode and quantified using exact masses without fragmentation (Table 1). The limit of detection was found to be 3–4 fmol on column, 2-fold less sensitive than tandem MS (model TSQ, Thermo), and the linearity for the expected DNA range was excellent between the calibration range (100–500 fmol, R2 > 0.99). Overall, orbitrap based results showed excellent quantitation linearity and LODs for the determination of global methylation levels. Table 1 shows the detailed comparison of the instrument parameters and performances.
Table 1.
Comparison of triple-quadrupole (model TSQ Quantum) and orbitrap (model Exactive) mass spectrometers for global DNA methylation analysis.
| TSQ triple-quadrupole | Exactive orbitrap | |
|---|---|---|
| LC | Isocratic: 0.1% formic acid in methanol/0.1% aq. formic acid 5/95 (v/v) | |
| Column | Hypersil Gold C18 50 mm × 2.1 mm | |
| Injection volume | 10 μL | |
| Source | (+) ESI, skimmer offset 5 V | |
| MS detector | Triple quadrupole | Orbitrap |
| Scan type | MRM | Full scan |
| Collision energy | 14 eV | N/A |
| Quantification | Transition pairs | Parent ions (exact masses, ±5 ppm) |
| 5MedC (m/z 242.1/126.3) | 5MedC (m/z 242.11353) | |
| dC (m/z 228.2/112.2) | dC (m/z 228.09788) | |
| dG (m/z 268.1/152.3) | dG (m/z 268.10403) | |
| 15N3-dC (m/z 231.2/115.2) | 15N3-dC (m/z 231.08839) | |
| 15N5-dG (m/z 273.2/157.3) | 15N5-dG (m/z 273.08844) | |
| Calibration range | 50 fmol to 200 pmol (R2 > 0.99) | 100 fmol to 500 fmol (R2 > 0.99) |
| Limit of detection (LOD)a | 1.4 fmol on column | 3.3 fmol on column |
| DNA methylation range 5MedC/dG (800 DNA samples) | 2–6% | 2–6% |
| Intra-day CV | 3–17% (mean 6%) (14 days) | 3–13% (3 days) |
| Inter-day CV | 9–13% (mean 11%) (14 days) | 9% (3 days) |
At signal to noise ratio of 3.
3.3. Expression of DNA methylation level
The DNA methylation level was expressed as either [5MedC]/[dG] with adjustment to the respective 15N labeled internal standards or as [5MedC]/([5MedC] + [dC]) without internal standard adjustments. These two ratios showed an excellent correlation with a Pearson correlation of 0.94, and good linearity of r = 0.87 for a total of 27 QC samples analyzed, with the 5MedC/dG ratios showing a better intra- and inter-day CVs than 5MedC/(5MedC + dC) ratios (intra-day: 6% vs. 9%; inter-day: 2.6% vs. 4%, respectively). In addition, we found that internal standards are necessary for measuring [5MedC]/[dG] ratios accurately, since the [5MedC]/[dG] ratio without internal standard adjustment deviates significantly from the above two calculated ratios (r = 0.20), possibly due to the different ion suppression effects on the fast (5MedC) and slow eluting (dG) compounds.
3.4. Method validation
QC DNA samples were rigorously analyzed for method validation purposes, see details of the experimental part in Section 2.5. Our results indicated that our developed method is consistent with the global DNA methylation level expressed as 5MedC/dG and adjusted with 15N3-dC and 15N5-dG. The intra-day CVs of QC 1–4 range from 3–17% with a mean of 6%, and the inter-day CV range 9–13% with a mean of 11% based on four sets of QC samples included in each of 12 batches (Table 2). In addition, we further validated our method accuracy by comparing the DNA methylation levels (5MedC/(5MedC + dC)) from 5 individuals to Friso and Choi et al.’s method which calculates ([5MedC]/([dC] + [5MedC]) ratios using (methyl-d3, ring-6-d1)-5MedC and 15N3-dC as internal standard for 5MedC and dC, respectively [11]. A total of 25 DNA samples from 5 subjects were compared by the two methods and the global DNA methylation levels range from 4.6% to 5.1% with a CV range from 1% to 7% (mean of 4%) between the two assays. See supporting information for additional sensitivity parameters of the mass spectrometric detection of the assay, including the calibration equations and R2 values, as well as limits of detection and detailed method validation data.
Table 2.
Global DNA methylation assay method validation based on 12 batches (n = 12).
| QC1 (%) | QC2 (%) | QC3 (%) | QC4 (%) | |
|---|---|---|---|---|
| 5MedC/dG mean | 3.51 | 3.69 | 3.69 | 3.80 |
| Intra-day CV range | 1–15 | 0–18 | 1–16 | 3–17 |
| Intra-day CV mean | 5 | 5 | 8 | 6 |
| Inter-day CV mean | 12 | 9 | 13 | 12 |
4. Discussion
4.1. LCMS
We described here an efficient high throughput LCMS method for the determination of global DNA methylation levels. All the interested nucleoside analytes were separated within 2.5 min on a sub-2 μm 50 mm reverse phase column, which is four-times faster than previously reported methods [11,12]. It is important to note that due to the usage of a highly aqueous (95% water) mobile phase [17] for the water soluble nucleosides elution, the column gradually lost its retention capacity after repeated injections. As a result, all the peaks were shifted to earlier retention times and dC, and the 15N3-dC and 5MedC peaks partially overlapped. The original retention could be restored by conditioning the column with the mobile phase containing 85% of methanol for 15 min, and this step was therefore repeated at the end of the batch after eighty injections to keep dC and 5MedC baseline separated. The loss of retention could also be avoided using a gradient elution using a higher content of methanol in the middle of the run (methanol/water with 0.1% formic acid): 0–1.5 min, 5/95; 1.6–4 min, 80/20; 4.1–10 min, 5/95. However, this gradient elution led to a longer analysis time (10 min) as well as high intra-day CVs of the same sample injected 24 h apart (CV > 30%), possibly due to the changes of mass spectrometer performance over time. Therefore, we applied the shorter isocratic runs (2.5 min) for our analysis.
4.2. Comparison with benchtop Orbitrap MS
The global methylation assay was previously evaluated on ion-trap [11] and triple-quadrupole [12,13,15] mass spectrometers, and showed good linearity of 5MedC from 40 fmol to 200 pmol on column. Recently, benchtop orbitrap mass spectrometers have been developed allowing high resolution, fast scanning and accurate mass. One major advantage of the orbitrap spectrometer is that it obtains full scan mass data besides the target analytes and is very useful for re-interrogation of data once interest in new analytes emerges, or for metabolomics research, i.e. RNA ribonucleosides data for contamination evaluation. Therefore, we implemented the developed assay on orbitrap MS (model Exactive, Thermo) and found excellent quantitation linearity and detection limit, indicating that orbitrap MS is an excellent alternative for assaying global DNA methylation. Both orbitrap and triple quadrupole MS showed similar LOD and linear range, see details of the comparison in Table 1. Importantly, although the advantages of retrospective analysis of data for other components are beyond the scope of this investigation, orbitrap MS will show more rewards when a large number of analytes are included, for example, when RNA data are also needed.
4.3. Additional sample clean-up by filtration
Although filtering the DNA digest to remove excess proteins before LCMS analysis is not a routine procedure in reported methods, we found that this step is necessary to assure assay precision. Without the filtering step the sensitivities of early eluting peaks including dC, 15N3-dC (RT = 0.7 min) and 5MedC (RT = 0.95 min) were significantly reduced after 24 h continuous mass analysis, whereas dG and 15N5-dG (RT = 1.55 min) responses were only slightly affected over that period. In addition, the ion source fore pressure was decreased after repeated injections indicating a possible clogging on the ion transfer tube. We reasoned that the complex matrix of the DNA digest may contribute to the ion suppression to dC, 15N3-dC, and 5MedC [18]. To overcome this problem, we diluted the samples with water and filtered it through a membrane with 10 kDa cutoff weight to remove large proteins in order to purify the samples for HPLC injection. We found that the sensitivity for filtered samples did not decrease for over 400 injections and that no significant changes occurred in the ion source fore pressure over that period of time. Furthermore, by applying the filtration step, the intra- and inter-day CVs have been significantly improved, with the intra-day CVs of QCs less than 10% and the inter-day CV less than 13% for all four QC samples over 12 days, compared to a ~20% intra-day CV and >30% inter-day CV without filtration (Table 1). Therefore, the filtration step to remove proteins is critical for accurate measurement of DNA methylation.
4.4. Importance of internal standards
The DNA global methylation level was traditionally reported as the ratio of 5MedC to total dC levels ([5MedC]/([dC] + [5MedC]) [10,11,15], whereas each analyte was adjusted with their corresponding isotope-labeled internal standards for accurate measurement by LCMS. As we stated earlier, these internal standards for 5MedC, namely deuterium 2H-((methyl-d3, ring-6- d1)-5MedC) and 15N-labeled (15N3-5MedC), are either extremely expensive [11] or require laborious biosyntheses [15]. Although the initial high investment of internal standards is by and large compensated when large sample numbers need to be analyzed, however, this is not the case if only few samples on an irregular basis need to be analyzed. On the other hand, Song et al. [12] reported expressing the DNA methylation levels by 5MedC to dG ratio ([5MedC]/[dG]) based on the assumption that [dG] = [5MedC] + [dC], without adjusting each analyte with isotope-labeled internal standards. We assume that using the [5MedC]/[dG] ratio is a good and cost-effective alternative since this ratio does not require adjustments due to loss of material during work-up and analysis since nominator and denominator values would be affected in the identical fashion and cancel each other out, and the high cost of 5MedC isotope internal standards can be avoided. Nevertheless, while calculating [5MedC]/[dG] ratios, we found the ion suppression affect the fast eluting compound (5MedC and dC) more than the slow eluting compound (dG) and the [5MedC]/[dG] ratios of QCs without isotope internal standards varied significantly (CV > 20%). On the other hand, considering the very similar chemical structures and minute differences in retention times (0.25 min) and m/z values (14 Da) between 5MedC and dC, we utilized the inexpensive and commercially readily available 15N3 isotope of dC as the internal standard for 5MedC. Due to our validation results we concluded that 5MedC and 15N3-dC will experience very similar ion suppression or other unwanted effects and ultimately their ratio (analyte versus internal standard) will lead to consistent values. Moreover, the linearity of the calibration standards containing an enzyme matrix identical to that in DNA extracts over a wide concentration range also indicates that ion suppression was present at the same extent for 5MedC and 15N3-dC. For dG we used its 15N5 isotope as internal standard. The correctness of our approach was evidenced by our internal standard adjusted results of 5MedC/dG values which showed good consistency for the QC samples with an intra-day mean CV of 6% and inter-day mean CV of 11%, whereas 5MedC/dG values without any internal standard adjustment showed CVs of over 20%. Furthermore, 5Med/dG ratios with internal standard adjustments showed smaller intra-and inter-day CVs than 5MedC/(5MedC + dC) ratios without the use of internal standards indicating that the former approach is more consistent to express DNA methylation levels. It is important to point out that our CV values are based on most vigorous analyses of four QC samples in duplicate or quadruplicate that are randomly inserted in a 80 DNA sample sequence for a period of 12 days. This highlights the repeatability and consistency of our assay. Furthermore, we also found good precision for QC samples and excellent correlation with previously reported methods [11].
5. Conclusion
In conclusion, we present a fast, cost-effective, sensitive, consistent, validated, precise, and accurate LCMS assay for global DNA methylation analysis. The methylation levels were expressed as [5MedC]/[dG] ratios after adjustment of responses to inexpensive internal standards for nominator and denominator analytes (15N3-dC and 15N5-dG, respectively). We observed that using a [5MedC]/([dC] + [5MedC]) ratio for global DNA methylation without any internal standard adjustment leads to increased CVs. Applying a short sub-2 μm sized column the turn-around has been significantly improved with runs completed within 2.5 min, and RNA contamination was determined to be less than 4% and therefore had no appreciable effects on 5MedC ionization. Importantly, filtration was found to be a critical step for accurate measurements and to maintain the needed precision of the assay for complex biological samples. Finally, we compared the established methylation assay using a new benchtop orbitrap mass spectrometer (model Exactive, Thermo Scientific) to routinely used tandem mass spectrometry and found both MS alternatives to result in similar sensitivity and linearity, with the limit of detection of 5MedC 1.4 fmol on column by triple quadrupole MS and 3.3 fmol by orbitrap MS. Orbitrap MS might have more advantages when larger numbers of analytes are to be analyzed and/or when reinterrogation of data is desired.
Supplementary Material
Acknowledgments
We thank Dr. Sang-Woon Choi (Tufts University, Boston) for analyzing QC samples for method validation purposes. We acknowledge support of this study by NIH grants P30-CA71789 and S10-RR020890.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.aca.2011.07.014.
References
- 1.Jaenisch R, Bird A. Nat Genet. 2003;33:245. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA. Cancer Res. 1986;46:461. [PubMed] [Google Scholar]
- 3.Qu GZ, Grundy PE, Narayan A, Ehrlich M. Cancer Genet Cytogenet. 1999;109:34. doi: 10.1016/s0165-4608(98)00143-5. [DOI] [PubMed] [Google Scholar]
- 4.Bernardino J, Roux C, Almeida A, Vogt N, Gibaud A, Gerbault-Seureau M, Magdelenat H, Bourgeois CA, Malfoy B, Dutrillaux B. Cancer Genet Cytogenet. 1997;97:83. doi: 10.1016/s0165-4608(96)00385-8. [DOI] [PubMed] [Google Scholar]
- 5.Choi JY, James SR, Link PA, McCann SE, Hong CC, Davis W, Nesline MK, Ambrosone CB, Karpf AR. Carcinogenesis. 2009;30:1889. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oakeley EJ. Pharmacol Ther. 1999;84:389. doi: 10.1016/s0163-7258(99)00043-1. [DOI] [PubMed] [Google Scholar]
- 7.Oakeley EJ, Podesta A, Jost JP. Proc Natl Acad Sci U S A. 1997;94:11721. doi: 10.1073/pnas.94.21.11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magana AA, Wrobel K, Caudillo YA, Zaina S, Lund G, Wrobel K. Anal Biochem. 2008;374:378. doi: 10.1016/j.ab.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 9.Sonoki S, Lin J, Hisamatsu S. Anal Chim Acta. 1998;365:213. [Google Scholar]
- 10.Rossella F, Polledri E, Bollati V, Baccarelli A, Fustinoni S. Rapid Commun Mass Spectrom. 2009;23:2637. doi: 10.1002/rcm.4166. [DOI] [PubMed] [Google Scholar]
- 11.Friso S, Choi SW, Dolnikowski GG, Selhub J. Anal Chem. 2002;74:4526. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- 12.Song L, James SR, Kazim L, Karpf AR. Anal Chem. 2005;77:504. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Liu S, Xie Z, Blum W, Perrotti D, Paschka P, Klisovic R, Byrd J, Chan KK, Marcucci G. Nucleic Acids Res. 2007;35:e31/1. doi: 10.1093/nar/gkl1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humeny A, Beck C, Becker CM, Jeltsch A. Anal Biochem. 2003;313:160. doi: 10.1016/s0003-2697(02)00568-7. [DOI] [PubMed] [Google Scholar]
- 15.Quinlivan EP, Gregory JF., III Nucleic Acids Res. 2008;36:e119/1. doi: 10.1093/nar/gkn534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crain PF. Methods Enzymol. 1990;193:782. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 17.Walter TH, Iraneta P, Capparella M. J Chromatogr A. 2005;1075:177. doi: 10.1016/j.chroma.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 18.Annesley TM. Clin Chem (Washington DC, US) 2003;49:1041. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.