Abstract
Objectives
Differences in gut bacteria have been described in several autoimmune disorders. In this exploratory pilot study, we compared gut bacteria in multiple sclerosis patients and healthy controls and evaluated the influence of glatiramer acetate and vitamin D treatment on the microbiota.
Methods
Subjects were otherwise healthy white women with or without relapsing-remitting multiple sclerosis who were vitamin D insufficient. Multiple sclerosis patients were untreated or were receiving glatiramer acetate. Subjects collected stool at baseline and after 90 days of vitamin D3 (5,000 IU/day) supplementation. The abundance of operational taxonomic units was evaluated by hybridization of 16S rRNA to a DNA microarray.
Results
While there was overlap of gut bacterial communities, the abundance of some operational taxonomic units, including Faecalibacterium, was lower in multiple sclerosis patients. Glatiramer acetate-treated MS subjects showed differences in community composition compared to untreated subjects, including Bacteroidaceae, Faecalibacterium, Ruminococcus, Lactobacillaceae, Clostridium, and Other Clostridiales. Compared to the other groups, untreated multiple sclerosis subjects had an increase in the Akkermansia, Faecalibacterium, and Coprococcus genera after vitamin D supplementation.
Conclusions
While overall bacterial communities were similar, specific operational taxonomic units differed between healthy and multiple sclerosis subjects. Glatiramer acetate and vitamin D supplementation were associated with differences or changes in the microbiota. This study was exploratory, and larger studies are needed to confirm these preliminary results.
Keywords: Multiple sclerosis, gut microbiome, vitamin D, glatiramer acetate, autoimmunity
INTRODUCTION
Multiple sclerosis (MS), an autoimmune neurologic disease caused by a combination of genetic and environmental exposures, has increased in incidence in the past several decades [1], suggesting changing environmental risk factors.
The gut microbiota is symbiotic with the human and is crucial to normal immune function [2]. Differences in gut bacteria exist in those with autoimmune disorders (e.g. inflammatory bowel disease)[2] compared with healthy individuals, and antibiotic and probiotic regimens are being assessed as potential therapies. Altering gut bacteria influences the development and severity of experimental autoimmune encephalomyelitis (EAE), a mouse model of MS [3]. A more recent study has shown that unaltered commensal bacteria can trigger, after exposure to myelin oligodendrocyte glycoprotein, a spontaneous form of EAE [4]. No study has assessed gut bacteria in patients with MS. We sought to evaluate the gut microbiota in healthy controls (HC) and those with MS and to explore differences associated with MS treatment and supplementation with vitamin D, both of which are used commonly in MS clinical practice.
MATERIALS AND METHODS
We capitalized on a vitamin D3 supplementation study (NCT01667796) to conduct this sub-study. All subjects provided written informed consent, and the study was approved by the UCSF Institutional Review Board and thus is in compliance with the Declaration of Helsinki. Since the parent study was a small pilot study, strict inclusion and exclusion criteria were used to reduce heterogeneity that might otherwise have impacted the results. Subjects were white, non-Hispanic females aged 18 to 60 years with screening body mass index of 18-30 kg/m2 and screening 25-hydroxyvitamin D level ≤ 30 ng/mL. Subjects had to be willing to stop taking additional multivitamins or cod liver oil and to avoid tanning beds for the duration of the study. Subjects could not be pregnant or nursing and had to be willing to avoid pregnancy during the study. Those with known gastrointestinal disorders were excluded from the overall study because such diseases could impact the absorption of vitamin D; for similar reasons, subjects could not be on a no-fat diet. Those with known renal or liver disease, history of nephrolithiasis, history of hypercalcemia or hypercalciuria, screening serum calcium >10 mg/dL, anemia (hemoglobin <11 g/dL) hyperthyroidism, infection with Mycobacterium species, sarcoidosis or other serious chronic illness (including cancer, cardiac, HIV) were excluded. Further, those who were taking thiazide diuretics, digoxin, diltiazem, verapamil, cimetidine, heparin, low-molecular weight heparin, or medication associated with malabsorption were excluded. Finally, subjects were excluded if in the month prior to screening, they had smoked cigarettes, used illicit drugs, or had taken non-topical steroids. MS patients had relapsing-remitting disease by McDonald criteria with Expanded Disability Status Scale scores ≤ 3.0 and without major heat sensitivity, criteria that were chosen to minimize differences in sun exposure behaviors. Also, patients were taking either no MS medication or glatiramer acetate (GA) in order to minimize heterogeneity associated with medications, although this criterion was broadened after the current sub-study was completed due to slow enrollment.
We asked subjects enrolled by March, 2011 to participate in this sub-study. 25-hydroxyvitamin D levels (referred to herein as “vitamin D levels” were obtained at the screening visit and, in all but one participant, after the study was completed. Participants were asked to collect their first morning stool at baseline and after 90 days of oral vitamin D3 5,000 IU/day supplementation. Samples were shipped overnight on ice packs to the processing facility, where they were immediately stored at −80° C. The UltraClean Fecal DNA Isolation Kit (MoBio, Carlsbad) was used to batch-isolate total DNA, which was then amplified by PCR with bacterial 16S rRNA gene degenerate forward primer: 27F.1 5’-AGRGTTTGATCMTGGCTCAG-3’ and a non-degenerate reverse primer: 1492R.jgi 5’-GGTTACCTTGTTACGACTT-3’. Amplified products were concentrated with a solid-phase reversible immobilization method and quantified with an Agilent 2100 Bioanalyzer®. PhyloChip Control Mix™ was added, followed by thirty-five cycles of bacterial 16S rRNA gene PCR amplification. The products were fragmented, labeled with biotin, and hybridized to the PhyloChip™ Array (version G3) [5]. The array contains representative sequences from operational taxonomic units (OTUs), analogous to a bacterial strain, which were clustered at 0.5% divergence using the Greengenes rRNA database [6]. A GeneArray® scanner (Affymetrix) was used to wash, stain, and scan the PhyloChip arrays. Standard Affymetrix software was used to capture the scans. Hybridization values, the fluorescence intensity, for each taxon were calculated as a trimmed average; maximum and minimum values were removed prior to averaging. After maximum and minimum values were removed, the mean fluorescence intensity was log2 transformed before multiplying by 1000; doubling of the fluorescence intensity is thus indicated when the score changes by 1000.
Statistical analyses
Based on the abundances of the OTUs determined by array hybridization, gut communities were compared by calculation of dissimilarities with weighted Unifrac. Dendrograms were generated by clustering the samples from the dissimilarity matrix hierarchically using the average-neighbor method. Two-dimensional ordination plotting by principal coordinates analysis was used to visualize inter-sample relationships by using the dissimilarity values to position the points relative to each other. The Prediction Analysis for Microarrays (PAM), using a nearest shrunken centroid method, was used to generate lists of significant taxa whose abundance characterizes each class [7]. The Adonis test was used to determine the significance of microbial differences associated with variables of interest.
To assess changes in operational taxonomic unit (OTU) abundances associated with vitamin D supplementation, the within-person change in abundance was calculated for each OTU by OTUpost-OTUpre. Distributions of OTU changes in each group were compared using a Wilcoxon rank sum test with a Bonferroni correction, which adjusts for multiple comparisons.
RESULTS
We collected fecal samples from 15 subjects. A flow diagram depicting the sample sources is presented in Figure 1.The median (range) age of MS patients was 42 years (30, 48) and of healthy controls was 38 years (29, 51) (p=0.95 [Wilcoxon rank-sum test]). The mean 25-hydroxyvitamin D levels (measured in one batch by liquid chromatography-mass spectrometry at Heartland Assays [Ames, IA]), were similar between the two groups at baseline (25.9 ± 4.4 ng/mL in the MS subjects versus 23.2 ± 5.7 ng/mL in the healthy controls (p=0.31 [t-test]) and after 90 days of supplementation (55.6 ± 17.0 ng/mL in the MS subjects versus 59.8 ± 11.7 ng/mL in the healthy controls, p=0.58 [t-test]). When a Wilcoxon rank-sum test was employed, the differences also didn't appear to be significant. Among MS patients (n=7; 5 GA-treated), one submitted the initial stool sample late (excluded from pre-/post-vitamin D analyses), while two gave specimens only after supplementation. Thus, four MS subjects (2 GA-treated) had samples available before and after supplementation.
Figure 1.
These samples were not available (n=2) or not used (n=1; sample given late) for the pre- vs post-vitamin D supplementation analysis.
MS= Multiple sclerosis
HC= Healthy control
GA= glatiramer acetate
*One GA-treated subject (who didn't contribute a pre-vitamin D supplementation sample) took antibiotics for an infection prior to the study (azithromycin for sinus infection), and ** one baseline sample was inadvertently stored at −20° C for several days before being moved to −80° C. There was no apparent impact on results of either affected sample (i.e. they did not appear to be outliers).
Overall comparison of MS subjects versus healthy controls
There was no apparent whole community compositional shift in MS (Figure 2; p= 0.15 when all samples were included; p=0.74 when including only pre-vitamin D supplementation samples; p=0.21 when including only post-vitamin D supplementation samples). Prior to supplementation, there were significantly fewer OTUs classified as Bacteroidaceae (37 OTUs) and Faecalibacterium (57 OTUs), and a higher number of Ruminococcus OTUs (38 OTUs) in MS compared to HCs (Supplementary Table 1). Using PAM to identify a small subset of taxa with contrasting hybridization scores between the groups, OTUs within RikenellaceaeII, Lachnospiraceae, Porphyromonadaceae, Bacteroides and Oscillibacter (Supplementary Table 2) collectively may predict whether samples prior to vitamin D supplementation came from MS patients or healthy controls.
Figure 2.

PCoA by phenotype and supplementation. Principal component analysis is a transformation of Weighted Unifrac distance, a pair-wise distance between samples based the calculation of the shared branches a the phylogenetic tree of the representative rRNA genes from 19757 operational taxonomic units present in at least one sample, weighted by abundance. HC, healthy control; MS, affected; pre/post, refers to supplementation with Vitamin D. Axis 1: 33% of variation explained. Axis 2: 21% of variation explained. All samples were used to generate the plots; for plot clarity, the excluded sample (late sample from one MS subject) was removed from this figure (see Supplementary Figure 1 for unaltered plot).
Overall differences between treated versus untreated MS subjects
Whole community differences were observed when comparing GA-treated and untreated MS subjects using an Adonis Test (p= 0.007) when all samples were considered (p=0.66 when comparing only pre-vitamin D supplementation samples; p=0.048 when comparing only post-vitamin D supplementation samples) (Figure 3). OTUs showing statistically significant different abundance between these groups (Supplementary Table 3) include Bacteroidaceae (500 OTUs), Faecalibacterium (186 OTUs), Ruminococcus (104 OTUs), Lactobacillaceae (77 OTUs), Clostridium (49 OTUs) and Other Clostridiales (201 OTUs).
Figure 3.

Differential clustering based on glatiramer acetate treatment. Principal component analysis is a transformation of Weighted Unifrac distance, a pair-wise distance between samples based the calculation of the shared branches a the phylogenetic tree of the representative rRNA genes from 19757 operational taxonomic units present in at least one sample, weighted by abundance. MS, affected where patients are either on GA or untreated. Axis 1: 33% of variation explained. Axis 2: 21% of variation explained. All samples were used to generate the plots; for plot clarity, the excluded samples (HCs) were removed from this figure (see Supplementary Figure 2 for unaltered plot).
Effects of vitamin D supplementation
For the overall group, vitamin D supplementation was associated with a trend for a whole community shift (p=0.062), although the within-group p values were attenuated (p=0.58 for the MS patients; p=0.20 for HCs). Compared to baseline, in samples post-supplementation, the abundance of Faecalibacterium (81 OTUs) and Enterobacteriaceae (244 OTUs) increased while Ruminococcus decreased (53 OTUs) after vitamin D supplementation (Supplementary Table 4). At the individual level, when restricting analyses to only the OTUs found in both pre- and post-supplementation samples, most OTUs increased slightly in the HC samples following vitamin D supplementation (Figure 4). In contrast, while the MS patients who were treated with GA showed few changes in OTU abundances, with the majority of OTUs remaining consistent in their abundance (median change near 0), untreated MS subjects showed a slight decrease of abundance for the majority of OTUs by Wilcoxon Rank Sum (p < 0.001) (Figure 4).
Figure 4.

Distribution of change of overall microbial abundance after vitamin D supplementation. Change in relative microbial OTU abundance in fecal samples between pre- and post- vitamin D supplementation samples in each subject group using only OTUs present in both samples. The “*” signifies a significant change in the distribution between HC and untreated.
While overall abundances did not markedly change in ways that distinguished the subject groups, a few specific OTUs distinguished each MS subgroup from the other two groups. Compared to HC and GA-treated MS subjects, untreated MS subjects had an increase in the Akkermansia, Faecalibacterium, and Coprococcus genera after vitamin D supplementation (Figure 5A). Compared to healthy participants and untreated MS subjects, those treated with GA had increases in Janthinobacterium and decreases in Eubacterium and Ruminococcus after high-dose vitamin D supplementation (Figure 5B).
Figure 5.
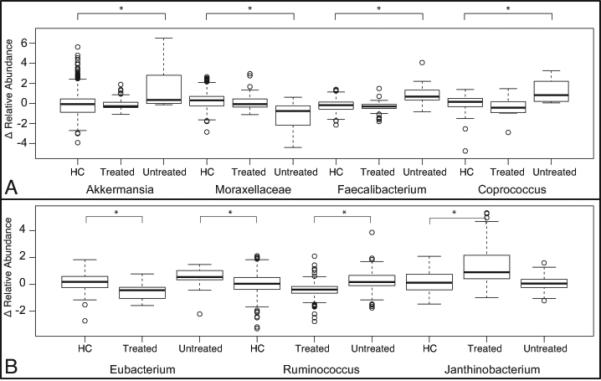
Distribution of statistically significant changes of OTU abundance in taxa. Change in relative abundance of taxa between the pre- and post- Vitamin D supplement time points where statistically significant changes between (A) untreated MS and healthy control or treated MS and (B) treated MS and untreated MS or healthy control (HC). The “*” signifies a significant change in the distribution between HC and untreated.
DISCUSSION
In this first exploratory study of gut bacteria in MS, the most important finding is that the gut bacterial communities differed in treated versus untreated MS patients. While we utilized all stool samples for the overall analysis due to the small sample size, which may lead to inter-subject variability accounting for apparent differences, the difference remained statistically significant when considering only samples from MS subjects given after vitamin D supplementation. That the differences were not statistically significant before vitamin D supplementation probably is related to the small number of available samples from that time point, although we can't exclude a true lack of difference between GA treated and untreated at that time. That MS patients receiving GA appear to have differences in gut bacteria compared to untreated patients is intriguing with respect to possible mechanisms of action of GA, although it clearly requires further investigation in a much larger cohort in order to establish the finding is replicable and to ascertain if the association is one of cause and effect. It is certainly plausible that GA may influence gut microbial populations. GA has been shown to modulate the immune response in the gut in animal models of inflammatory bowel disease (which is known to be associated with gut microbial alterations), reducing the lymphocytic response to colonic extract in mesenteric lymph nodes.[8] The investigators of this study speculated that GA may locally displace microbial or self-antigen from antigen-presenting cells, thus reducing T-cell activation. GA has also been shown to improve a mouse model of inflammatory bowel disease by inducing CD8+ T regulatory cells.[9] Regardless of the putative mechanism or direction of the relationship, the finding that the gut microbial profile may differ in treated versus untreated MS patients has immediate implications for design of studies of the gut microbiota in MS: investigators should carefully consider the whether patients are treated or not and may even consider that different MS therapies could differentially impact gut microflora, in order to avoid heterogeneity and confounding.
In addition to noting the whole community differences in GA-treated versus untreated MS patients, we distinguished that among MS patients, vitamin D supplementation altered the gut microbiota differently depending on GA treatment status. Of great interest is that compared to healthy and GA-treated MS subjects, there was increased abundance of Faecalibacterium after vitamin D supplementation in MS subjects not on GA. Faecalibacterium is associated with a reduced inflammatory state due to its role in butyrate production,[10] and strategies to boost the abundance of Faecalibacterium have been hypothesized to be potentially therapeutic for patients with ulcerative colitis.[11] Several Coprococcus strains are also butyrate producers and may thus be anti-inflammatory.[12] Akkermansia, which also increased in the untreated MS patients after vitamin D supplementation, is a mucin-degrading bacteria that is thought to be involved in immune tolerance toward commensal gut microbes, as evidenced by changes in gene expression of multiple pathways involving the immune response after germ-free mice were colonized by Akkermansia in one study [13]. In a mouse model of inflammatory bowel disease, Akkermansia-derived extracellular vesicles protected against the disease.[14] Little is known about the roles of Eubacteria (genera), Ruminococcus and, Janthinobacterium, all of which changed in abundance after vitamin D supplementation in treated MS subjects. The effect of vitamin D on the gut microbiota has been investigated in some mouse models of inflammatory bowel disease. Since bacteria do not possess vitamin D receptors, it is thought that any effects of vitamin D therapy are mediated by effects on the host that lead to secondary effects on gut bacteria.[15] In a mouse model of inflammatory bowel disease, knockout of Cyp27B1, the enzyme that converts 25-hydroxyvitamin D to the active 1, 25-dihydroxyvitamin D, is associated with elevated levels of pro-inflammatory Helicobacteraceae, which can induce IBD symptoms.[16] The authors of the study suggest that disruption of vitamin D activity (through knockout models) increases the inflammatory milieu in the gut, leading to dysbiosis in which disease-inducing pathogens are able to outcompete commensal bacteria. While data regarding the effect of vitamin D in humans are sparse, our study suggests that further investigations of how vitamin D supplementation impacts gut microbial populations in MS patients are warranted.
This study also identified specific OTUs in bacterial genera that exhibited abundance differences in MS subjects compared to HCs, some of which were also identified in subjects with inflammatory bowel disorders [11, 17]. Particularly of note the anti-inflammatory Faecalibacterium appeared to be less abundant in MS patients overall. This finding is consistent with the observation that Faecalibacterium is less abundant in patients with newly-diagnosed, untreated Crohn's disease than in healthy controls.[18] Patients with Type 1 diabetes have been shown to have reduced butyrate-producing and mucin-degrading bacteria compared to controls such that if confirmed, the results herein may imply similarities in the mechanisms by which gut bacteria influence the development of autoimmune disease.[19] On the other hand, we were unable to define an overall MS-specific bacterial community type. This finding could be viewed as consistent with studies of other autoimmune disease and supports the concept that instead of a major shift, an “unhealthy” community is relative to one's “healthy” community of bacteria. However, it is still possible that the small sample size may have precluded detection of a specific overall MS community type.
The study has several limitations. The small sample size may preclude detecting other important differences, but the use of a conservative Bonferroni correction in this study limits concerns that significant results are due to chance. That only one sample was available per subject before and after supplementation may have introduced outliers; future studies should acquire more than one sample to represent each time point of interest. Whether the results would have differed if the patients had not had vitamin D insufficiency at baseline is unclear. Self-reported race and ethnicity were used, rather than genetic ancestry markers, to attempt to minimize heterogeneity in skin pigmentation that would have potentially confounded the results of the parent study. Larger studies may want to consider using such markers in the screening or analysis process to account for differences in host genetics that may influence a person's microbial colonization but may also need to consider self-reported cultural groupings so as to account for differences in behaviors that may influence the microbiota.[20] Finally, the study would be strengthened by the use of metagenomic approaches. However, the study also has several strengths including the relative homogeneity of MS cases and the exclusion of conditions that might affect the gut microbiota.
Despite the study's small size, our results have important implications. First, that certain OTUs may differ between healthy and MS subjects, and in particular that differences in some of these OTUs have also been reported in inflammatory bowel diseases or are associated with immune function, lends support for further evaluations of the gut microbiota in MS. Second, that MS therapies and vitamin D supplementation (widely used in current MS practice) may influence gut bacterial populations suggests that studies of the gut microbiota in MS should take these immunomodulatory therapies into account in their design. This study provides only preliminary data about the gut microbiota in MS, but unraveling the relationships of therapies to microbial populations in the gut and elsewhere in the body will be important to understanding the independent effects of these factors in MS.
Supplementary Material
ACKNOWLEDGEMENTS
This study was funded by NIH K23 NS067055 and an Irene Perstein award (EMM). Dr. Mowry is currently receiving free glatiramer acetate in support of the conduct of a clinical trial, and some of the authors (CC, JK, TZD, and JW) are employees of Second Genome, Inc.
Footnotes
There are no conflicts of interest.
REFERENCES
- 1.Granieri E, Casetta I, Govoni V, et al. The increasing incidence and prevalence of MS in a Sardinian province. Neurology. 2000;55:842–848. doi: 10.1212/wnl.55.6.842. [DOI] [PubMed] [Google Scholar]
- 2.Stanghellini V, Barbara G, Cremon C, et al. Gut microbiota and related diseases: clinical features. Intern Emerg Med. 2010;5:S57–S63. doi: 10.1007/s11739-010-0451-0. [DOI] [PubMed] [Google Scholar]
- 3.Ochoa-Reparaz J, Mielcarz DW, Begum-Haque S, Kasper LH. Gut, bugs, and brain: role of commensal bacteria in the control of central nervous system disease. Ann Neurol. 2011;69:240–247. doi: 10.1002/ana.22344. [DOI] [PubMed] [Google Scholar]
- 4.Berer K, Mues M, Koutrolos M, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 5.Hazen TC, Dubinsky EA, DeSantis TZ, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 6.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aharoni R, Kayhan B, Brenner O, et al. Immunomodulatory therapeutic effect of glatiramer acetate on several murine models of inflammatory bowel disease. J Pharmacol Exp Ther. 2006;318:68–78. doi: 10.1124/jpet.106.103192. [DOI] [PubMed] [Google Scholar]
- 9.Yao Y, Han W, Liang J, et al. Glatiramer acetate ameliorates inflammatory bowel disease in mice through the induction of Qa-1-restricted CD8(+) regulatory cells. Eur J Immunol. 2013;43:125–136. doi: 10.1002/eji.201242758. [DOI] [PubMed] [Google Scholar]
- 10.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siaw YH, Hart A. Commentary: is Faecalibacterium prausnitzii a potential treatment for maintaining remission in ulcerative colitis? Aliment Pharmacol Ther. 2013;38:551. doi: 10.1111/apt.12404. [DOI] [PubMed] [Google Scholar]
- 12.Pryde SE, Duncan SH, Hold GL, et al. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 13.Derrien M, van Baarlen P, Hooiveld G, et al. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol. 2011;2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang CS, Ban M, Choi EJ, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila,, protect the progression of dextran sulfate sodium-induced colitis. PLoS One. 2013;8:e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantorna MT, McDaniel K, Bora S, et al. Vitamin D, immune regulation,the microbiota, and inflammatory bowel disease. Exp Biol Med. 2014 doi: 10.1177/1535370214523890. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ooi JH, Li Y, Rogers CJ, et al. Vitamin D regulates the gut microbiome and protects mice from dextran sodium sulfate-induced colitis. J Nutr. 2013;143:1679–1686. doi: 10.3945/jn.113.180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker AW, Sanderson JD, Churcher C, et al. High throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorkildsen LT, Nwosu FC, Avershina E, et al. Dominant fecal microbiota in newly diagnosed untreated inflammatory bowel disease. Gastroenterol Res Pract. 2013:636785. doi: 10.1155/2013/636785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown CT, Davis-Richardson AG, Giongo A, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortenberry JD. The uses of race and ethnicity in human microbiome research. Trends Microbiol. 2013;21:165–166. doi: 10.1016/j.tim.2013.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



