Abstract
Diverse innate lymphoid cell (ILC) subtypes have been defined, based on effector function and transcription factor expression. ILCs derive from common lymphoid progenitors, although the transcriptional pathways leading to ILC lineage specification remain poorly characterized. Here we demonstrate that transcriptional regulator TOX is required for the in vivo differentiation of common lymphoid progenitors to ILC lineage-restricted cells. In vitro modeling demonstrates that TOX deficiency results in early defects in progenitor cell survival or expansion as well as later stage ILC differentiation. In addition, comparative transcriptome analysis of bone marrow progenitors reveals that TOX-deficient cells fail to upregulate many aspects of the ILC gene program, including Notch gene targets, implicating TOX as a key determinant of early ILC lineage specification.
INTRODUCTION
Recently identified types of immune effector cells, termed innate lymphoid cells (ILCs), have been found in mouse and human tissues, including lung, gut, skin, and adipose tissue (reviewed in ref.1). Despite lacking antigen receptors, these cells nevertheless display a wide range of effector functions, in many cases mirroring those seen in T helper cell subsets. ILCs likely provide a more rapid response to certain pathogens than provided by the adaptive immune system, as well as playing a role in modulating subsequent innate and adaptive immune responses1. In addition, ILCs can play a reparative role in response to tissue injury, where cytokine secretion by infected or damaged tissue, rather than foreign antigen production, is the activating signal2.
Like T helper cell subsets, ILCs are classified based on their effector cytokine secretion profile and development of each subset is associated with key transcriptional regulators. T-bet-dependent group 1 ILCs (ILC1s) are IL-12 responsive, secrete IFN-γ and TNF, and are involved in controlling intracellular infections3. Group 2 ILCs (ILC2s) secrete IL-5 and IL-13 upon stimulation with IL-33 and, like TH2, are GATA-3 dependent4. However, GATA-3 also plays an obligatory role in development of other ILC lineages5. In addition, ILC2 development is dependent on transcriptional regulators RORα and TCF-16,7. Activation of ILC2s can in turn regulate eosinophils8, alternatively activated macrophages9, as well as TH2 cells in the context of allergen-induced airway inflammation10. RORγt-dependent group 3 ILCs (ILC3) include fetal lymphoid tissue inducer cells (LTi), which are required for lymph node organogenesis11, and CD4+ LTi-like cells found in the adult12. Other ILC3s express the natural cytotoxicity receptor (NKp46+)13, are dependent on TCF-1 for development14 and are involved in maintaining intestinal homeostasis15. ILC3s secrete IL-22 and IL-17A when activated with IL-2316 and granulocyte-macrophage colony-stimulating factor in response to IL-1β production by macrophages15. Splenic ILC3s have been identified in both human and mouse, and provide marginal zone B cell help through T cell-independent mechanisms17.
All ILCs arise from common lymphoid progenitors (CLP) in BM and fetal liver through a Notch-6,18–20 and Id2-dependent process21,22. PLZF, a transcriptional regulator also implicated in NKT cell function23, marks a subset of α4β7+ ILC lineage-specific progenitors that can give rise to all ILCs, except LTi and cNK24. These data suggested the presence of an earlier common ILC progenitor. Indeed, Id2-reporter mice were used to identify a cell population termed the common progenitor to all helper-like ILCs (CHILP), which give rise to multiple ILC lineages, including LTi, and contain a subpopulation of PLZFhi cells3. Neither the PLZFhi nor CHILP populations can differentiate into the cNK lineage. The basic leucine zipper transcription factor NFIL3 was shown to be required for the development of cNK, ILC1s, ILC2s and ILC3s25–27, and in its absence, the Lin−α4β7+CD127+c-KitloSca-1loFlt3− progenitor population, including a minor subset of CXCR6+ cells, failed to develop 27,28. However, the relationship between these cells and CHILP is unclear because Id2 was not used as an identifying marker for the CXCR6+ cell population27. More restricted ILC1 (ILC1p) and ILC2 (ILC2p) precursors in the BM have also been identified4.
TOX (thymocyte selection-associated high-mobility group box protein) is a member of the HMG-box superfamily of DNA binding factors29,30 and is required for development of T cell subsets including CD4+ T, regulatory T and natural killer T (NKT) cells, as well as cNK and fetal LTi cells31–33. As a consequence of the loss of LTi, TOX-deficient (Tox−/−) mice lack lymph nodes. Based on some phenotypic overlap between Id2 and TOX deficiency, we investigated whether TOX, like Id2, might also play a broader role in ILC development. Here we show coincident expression of TOX and Id2 in CHILP. Moreover, intrinsic TOX deficiency resulted in low ILC1, ILC2, and splenic ILC3 numbers, due to the inhibition of Lin−α4β7+CD127+CD25−Flt3− progenitor development. Whole transcriptome analysis suggests loss of TOX leads to a post CLP and pre CHILP block at a potential transitional stage of ILC progenitor cell development. Moreover, induction of key ILC transcriptional regulators and cell survival mediators was diminished in the absence of TOX, and evidence suggests a failure to fully activate a Notch-mediated gene program. In vitro modeling demonstrated an early cell-intrinsic defect not only in expansion and/or survival of progenitors in the absence of TOX, but also failure to upregulate a number of key factors for ILC development. Together, these data support a role for TOX as an essential factor in ILC lineage specification.
RESULTS
CHILP co-express Tox and Id2
To assess expression of TOX in the development of ILCs, we generated a tandem dimer Tomato (TOM) reporter strain of mouse, ToxTOM (Fig. 1a). As all ILC development is Id2-dependent21, we additionally bred ToxTOM mice to Id2GFP reporter animals34. Tox is upregulated during the NK cell precursor (NKp) to immature NK cell (iNK) transition in BM, with subsequent down regulation in mature NK cells (mNK)32. ToxTOMId2GFP mice recapitulated this expression pattern (Fig. 1b). Fewer mNK cells expressed TOM, consistent with Tox mRNA in this cell population32, while GFP remained high (Fig. 1b). Together, these data support the utility of the Tox reporter strain to study ILC development.
Figure 1.
TOX is expressed in ILC progenitors and mature ILC lineages. (a) Deletion of the neomycin cassette was accomplished by breeding to FLPase recombinase expressing mice. FLPase was removed in subsequent breeding to generate ToxTOM reporter mice. Restriction enzyme sites (P=PstI, S=StuI, R=EcoRI) are shown in surrounding non-coding sequences only. ToxTOM mice were also bred to Id2GFP mice to generate a dual reporter strain used in many experiments. (b) Flow cytometry of Lin−CD122+ cells from BM and spleen for TOM and GFP in NKp, iNK, and mNK populations (n=3). Numbers refer to frequency of gated population here and in subsequent figures. (c) Expression of TOM and GFP in BM CLP reporter mice as indicated (n=3). (d) TOM and GFP expression in BM Lin−α4β7+CD127+ cells subgated by CD25 (n=3). (e) Lung ILC2s from WT and ToxTOMId2GFP mice were analyzed for reporter expression (n=3). (f) Expression of Tox and Gata3 by qRT-PCR in IL-33-expanded ILC2s, in vitro generated TH2 cells, and CD19+B220+ splenic B cells (n=2–3). (g, h) Expression of TOM in WT and ToxTOM lamina propria cells gated as shown (n=3). (i, j) Expression of TOM in splenic subpopulations (n=3) as in (g). *P <0.05, NS= non-significant, P >0.05 (Student’s t-test). Data (± s.d.) are representative of three (b–e, g–j) or two (f) independent experiments.
Adult ILC subsets develop from CLP6,19,28 (cell population definitions as in Supplementary Table 1). There was little reporter expression in ToxTOMId2GFP CLP (Fig. 1c). Among Lin−α4β7+ CD127+CD25− cells, ~80% were GFP+, fitting the phenotypic definition of CHILP (Fig. 1d). These cells were also TOM+ (Fig. 1d). However, a subpopulation of this α4β7+ subset did not express Tox or Id2 reporters. Lin−α4β7+ CD127+CD25+ cells, containing ILC2p4, also expressed GFP and TOM (Fig. 1d).
Mature lung ILC2s (defined as in Supplementary Fig. 1a) expressed TOM in a heterogeneous pattern (Supplementary Fig. 1b, c), as well as Tox mRNA (Fig. 1f). A proportion of NKT cells (Supplementary Fig. 1d) also expressed TOM, consistent with the known role for TOX in NKT cell development31, although with differing pattern of reporter expression when compared to ILC2s (Supplementary Fig. 1b). Unlike Gata3, expressed by ILC2s and TH2 cells4,35, Tox was solely expressed in ILC2s (Fig. 1f). Neither Gata3 nor Tox was expressed by B cells (Fig. 1f), as expected31.
The small intestine lamina propria (LP) contains multiple ILC subsets1. All ILC subtypes in LP expressed TOM (Fig. 1g, h). ILC3s are also present in spleen17 and TOM was expressed in a proportion of all splenic ILC3 subtypes (Fig 1i, j). Together with previous results, these data confirm expression of Tox in all known ILC subsets.
Common ILC progenitor development requires TOX
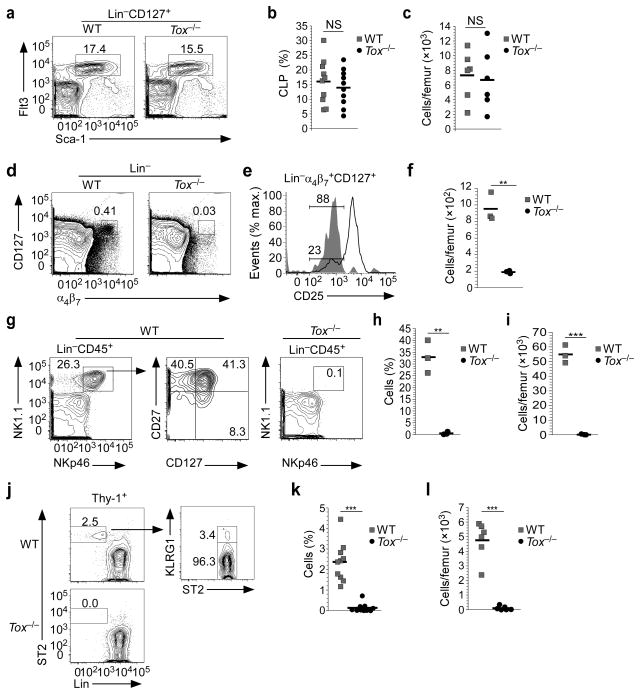
Tox−/− animals had normal numbers of CLP as compared to WT littermates (Fig. 2a–c). In contrast, there was a severe loss of Lin−α4β7+CD127+Flt3− cells in the absence of TOX (Supplementary Fig. 2a–c). In WT, a majority of Lin−α4β7+CD127+ cells expressed CD25 and these were largely absent in Tox−/− mice (Fig. 2e). Strikingly, the number of Lin−α4β7+CD127+CD25− cells, the majority of which are CHILP (Fig. 1d)3,28, was reduced to approximately 20% of their WT counterparts in Tox−/− animals (Fig. 2d–f). A heterogeneous population of Lin−CD45+NK1.1+NKp46+ cells consisting of cNK (CD27−CD127−), ILC1s (CD27+CD127+), ILC3s (CD27−CD127+), and ILC1 restricted progenitors (ILC1p) can be found in BM3. In the absence of TOX, this cell population was lost (Fig. 2g, h, i).
Figure 2.
TOX is required for the development of ILC progenitors. (a) Staining for BM CLP from WT and Tox−/− mice (n=6). (b, c) Compiled data of the frequency (b) and number (c) of Flt3+Sca-1int cells within the Lin−CD45+ cell population from WT and Tox−/− BM. (d) Lin−α4β7+CD127+ cells from BM of WT and Tox−/− animals (n=3). (e) Gated Lin−α4β7+CD127+ cells from WT and Tox−/− animals were analyzed for CD25. Shown is the frequency of CD25− cells (n=3). (f) Number of Lin−α4β7+CD127+CD25− cells in WT and Tox−/− animals. (g) Lin−CD45+NK1.1+NKp46+ cells were analyzed for cNK (CD27+CD127−) and ILC1s (CD27+CD127+) from WT and Tox−/− mice in BM. The lack of Lin−CD45+NK1.1+NKp46+ cells from Tox−/− mice precluded any additional subset analysis. (h, i) Compiled data of the frequency (h) and number (i) of NK1.1+NKp46+ cells within the Lin−CD45+ cell population from BM. (j) Staining for BM ILC2p from WT and Tox−/− mice. (k, l) Compiled frequency (k) and absolute numbers (l) of BM ILC2p. **P <0.01, ***P <0.001, NS= non-significant, P >0.05 (Student’s t-test). Symbols (b, c, f, h, I, k, l) represent individual mice analyzed independently; horizontal lines denote the mean.
ILC2s develop from CLP thorough a lineage-committed progenitor (ILC2p) 4. ILC2p also expressed TOM (Fig. 1d) and were absent from Tox−/− BM (Fig. 2j–l). ILC2p are IL-33R+ and can be expanded in vivo upon injection of IL-334. IL-33 led to a modest increase in ILC2p in the BM of Tox−/− animals, but not to the level seen in WT mice (Supplementary Fig. 2d–f).
TOX-dependent development of ILC-committed progenitors would predict failure to develop mature ILC in Tox−/− mice. Both cNK cells and ILC1s were present in lung of WT but not Tox−/− mice (Fig 3a, b, c, d). NK1.1+NKp46+ cells were lost from Tox−/− animals in the colon (Fig. 3e) and small intestine (Fig. 3g, f, h). Intracellular staining for lineage-determining transcription factors in the colon (Supplementary Fig. 3a) and small intestine (Supplementary Fig. 3b) indicated that these cells consisted of cNK, ILC1s, and a minor population of ILC3s.
Figure 3.
TOX is required for the development of mature ILCs. (a) Comparison of lung NK1.1+NKp46+ cells within the Lin−CD45+ population from WT and Tox−/− mice (n=3). (b) Intracellular staining of WT, lung Lin−CD45+NK1.1+NKp46+ population as shown in (a) including cNK and ILC1s (n=3). (c, d) Compiled frequency (c) and absolute numbers (d) of lung NK1.1+NKp46+ population. (e, g) Colon (e) and small intestine (g) lamina propria NK1.1+NKp46+ cells from WT and Tox−/− mice (n=3). (f, h) Frequency of colon (f) and small intestine (h) NK1.1+NKp46+ population. (i) Comparison of lung ILC2s from WT and Tox−/− mice (n=10). (j, k) Compiled frequency (j) and absolute numbers (k) of lung ILC2s. (l) Staining for lung ILC2 in WT and Tox−/− animals treated with IL-33 or PBS (n=3–5). (m, n) Frequency (m) and absolute numbers (n) of ILC2s from WT and Tox−/− lung, treated as indicated. (o) Comparison of ST2 expression on IL-33-expanded lung Lin−CD45+Thy-1+ cells isolated from WT or Tox−/− mice (n=3–5). (p) Comparison of splenic Lin−CD45+CD127+c-Kit+ cells from WT and Tox−/− mice gated as shown (n=4). (q) Absolute numbers of total spleen Lin−CD45+CD127+c-Kit+. (r) Absolute numbers of RORγt+CD4− and CD4+ cells as gated in (p). *P <0.05, **P <0.01, ***P <0.001, NS= non-significant, P >0.05 (Student’s t-test). Symbols (c, d, f, h, j, k, m, n, q, r) represent individual mice analyzed independently; horizontal lines denote the mean.
ILC2s in the lung were significantly reduced in Tox−/− mice (Fig. 3i–k). As is also true for T cell development33, heterozygous loss of Tox had no effect on the presence of ILC2s (Fig. 3j, k). WT ILC2s expanded greatly in lung of IL-33-treated mice (Fig. 3l–n). However, only minimal expansion of ILC2s was observed in lungs of Tox−/− mice, and cell recovery was poor, even compared to WT untreated animals (Fig. 3l–n). Expanded ILC2s from Tox−/− animals also expressed less of the IL-33R ST2 subunit, possibly indicative of abnormal development (Fig. 3o).
Splenic ILC3s contained both CD4− and CD4+ populations, and both were significantly diminished in Tox−/− animals (Fig. 3q, r). Similarly, splenic NKp46+ and NKp46− ILC3s were significantly decreased in the absence of TOX (Supplementary Fig. 4a, b). LP ILC3s are heterogeneous, containing fetal derived LTi-like cells and postnatal NKp46− (both T-bet+ and T-bet−) and NKp46+T-bet+ cells36. Even in the same animals where splenic ILC3s were lost, there was no reduction in the frequency of total ILC3s in Tox−/− LP, or subpopulations defined by NKp46 or T-bet, while ILC2s were absent (Supplementary Fig. 4a, c, d). Moreover, the yield of NKp46+ ILC3s was significantly increased in Tox−/− LP preparations (Supplementary Fig. 4e). NKp46− ILC3s were also isolated in greater numbers from mutant compared to WT animals, although this difference did not reach statistical significance (Supplementary Fig. 4e). In contrast to WT, the Tox−/− NKp46−T-bet− subset did not contain CCR6hi cells (Supplementary Fig. 4e), consistent with loss of the fetal LTi-like subpopulation36.
Loss of TOX results in a near complete block in development of CD4+ T cells in the thymus31 (Supplementary Fig. 4f). Nevertheless, Lin+CD45+Thy-1+CD4+ cells were observed in the LP of these animals and the majority was RORγt+ (Supplementary Fig. 4f). Moreover, >80% of the total Lin+CD45+Thy-1+ cell population was RORγt+ (Supplementary Fig. 4f). This is consistent with an inflammatory environment in the Tox−/− small intestine, potentially caused by loss of T regulatory cells31. Whether TOX-independent lineages of ILC3s exist or, alternatively, if the perturbed immune system in Tox−/− animals leads to expansion of minor populations of both CD4+ T cells and ILC3s remains to be determined.
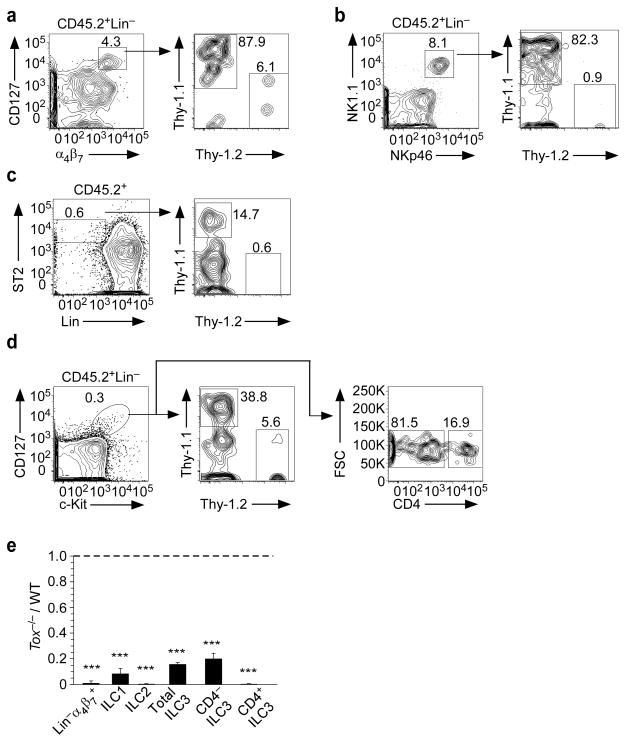
To verify that loss of TOX inhibited ILC development by a cell-intrinsic mechanism, BM chimeras were generated by injecting WT and Tox−/−CLP at equal ratios into NSG mice, which lack all T cells, B cells and ILCs. Of the donor-derived Lin−α4β7+CD127+ progenitors that developed, greater than 90% were derived from WT CLP (Fig. 4a). Similarly, the vast majority of donor-derived ILC1s (Fig. 4b) and ILC2s (Fig. 4c) in the lung, and splenic ILC3s (Fig. 4d) were derived from WT CLP. Figure 4e presents compiled data. In contrast, we did not observe a WT bias during thymocyte development (data not shown). Thus, all subtypes of ILCs, including the common progenitor, are intrinsically TOX dependent.
Figure 4.
TOX regulates the development of ILCs by a cell-intrinsic mechanism. (a–d) Donor-derived (CD45.2+) cells in (a) BM (n=4), (b, c) lung (n=3), and (d) spleen (n=3) were analyzed for allelic markers for WT (Thy-1.1+) or Tox−/− (Thy-1.2+) genotypes. (e) Ratio of Tox−/− to WT cells in the indicated populations. ***P <0.001 (Student’s t-test). Data (± s.d.) are compiled from four (a) or three (b, c, d) experiments, pooling two animals per experiment.
Molecular dissection of TOX-mediated ILC progenitor differentiation
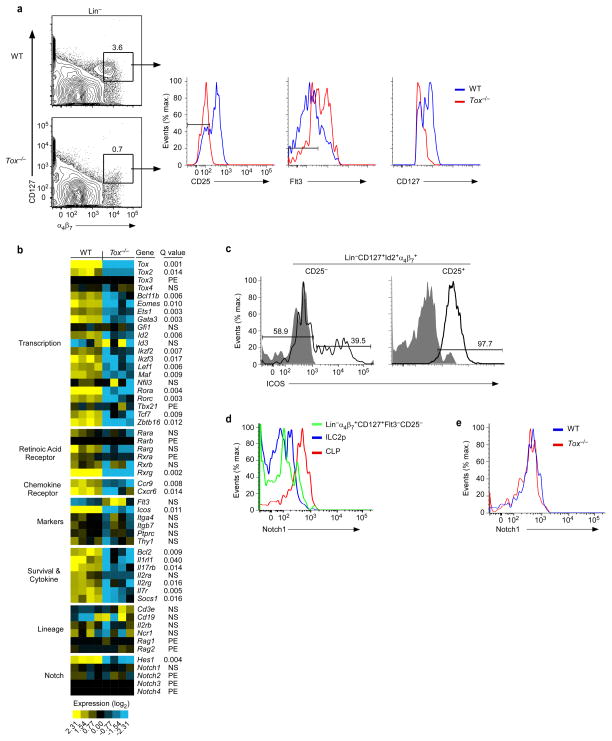
Lin−α4β7+CD127+CD25−Flt3− BM cells, which include the earliest known ILC progenitors3,24, are severely reduced in Tox−/− mice, although not entirely absent (Fig. 2f and Supplementary Fig. 2a–c). We took advantage of this to define the TOX-dependent stage of precursor development by global transcriptome analysis of remaining progenitor cells (Fig. 5a). Notably, while WT progenitor cells had a range of CD127 expression, Tox−/− cells more uniformly expressed lower amounts of this cytokine receptor (Fig. 5a).
Figure 5.
Whole transcriptome analysis of Tox−/− α4β7+ progenitors reveals a block in induction of the ILC gene program. (a) Gating strategy used for isolation of Lin−CD127+α4β7+CD25−Flt3− cells from WT and Tox−/− mice (n=4). (b) Heat map of select gene expression. PE, poorly expressed (average FPKM of both genotypes ≤ 2), NS, not significant (Q value ≥ 0.05) (Student’s t-test, Benjamini and Hochberg correction for multiple tests). (c) Expression of ICOS or isotype control (filled histograms) in CHILP from Id2GFP reporter mice (n=2). (d) WT progenitor cells as gated in (a) were compared to ILC2p and CLP for Notch1 surface expression (n=2). (e) CLP from WT and Tox−/−mice were analyzed for Notch1 (n=2). Data are compiled from four (b) independent experiments, pooling 2–3 animals per experiment per genotype.
Unsupervised hierarchical clustering revealed that coordinated down-regulation of many genes was associated with the Tox−/− phenotype (Supplementary Fig. 5). This is consistent with a primary role for TOX as a transcriptional activator37. 713 well-annotated genes were significantly differentially expressed (>2-fold, Q value= <0.05) in WT and Tox−/− cells (Supplementary Table 1).
Comparative expression of select genes is shown in Figure 5b. Consistent with reporter expression (Fig. 1d), Tox was expressed in WT precursors and acted as an internal control for Tox−/− cells. Family member Tox2 was more moderately expressed in WT precursors compared to Tox, but also exhibited decreased expression in Tox−/− cells. Tox3 expression could not be detected, while expression of Tox4, in general a ubiquitously expressed family member37, did not differ between WT and Tox−/− cells.
Genes encoding lineage markers were poorly, if at all expressed, and did not differ between WT and Tox−/− cells. Genes Itga4 and Itgb7, encoding the α4β7 marker used for cell isolation, were expressed and did not differ between WT and Tox−/− cells. Both cell populations also expressed Ptprc and Thy1 equivalently. Expression of Il7r was significantly lower in Tox−/− progenitors, consistent with cell surface expression of the encoded protein (Fig. 5b), as was Il2rg. Tox−/− cells also failed to express Bcl2 in the amounts seen in WT cells. Il1rl1 and IL17rb mRNA, both expressed by mature ILC2s, were also markedly decreased in Tox−/− progenitors, as was Eomes. How expression of these genes in WT progenitor cells compares to that of mature ILC, however, is unknown. Thus, low expression could represent the onset of locus accessibility.
Id2 was highly expressed in the WT progenitor population, as expected. While Id2 was also detected in Tox−/− cells, there was an 83% reduction in average expression. Expression of genes encoding key transcriptional regulators of ILC development was also significantly decreased in Tox−/− progenitors, including Gata3, Rorα, Rorc, Tcf7, and Zbtb16. In contrast, Gfi1 and Nfil3 were not differentially expressed between WT and Tox−/− cells. Inducible T-cell co-stimulator (ICOS) is expressed early in ILC development and is a downstream target of PLZF38. ICOS was expressed on a subset of CHILP and increased upon lineage specification to ILC2p (Fig. 5c). These data are consistent with CHILP containing a subpopulation of more restricted PLZFhiICOS+ progenitors3. In the absence of TOX, average Icos expression was decreased 98% when compared to WT cells, likely indicating loss of this progenitor subpopulation.
Genes encoding other transcriptional regulators not yet reported to have a role in ILC development were also differentially expressed in WT and Tox−/− cells, including Maf, Lef1 Ets1 and members of the Ikaros family, Ikzf2 and Ikzf3.
A role for retinoic acid (RA) in ILC2 and ILC3 homeostasis as well as LTi maturation has recently been reported39,40. Rxrg was expressed by WT cells, but was undetectable in Tox−/− progenitors. Genes encoding other RXR and RA receptor (RAR) isoforms were either not differentially expressed, or poorly expressed by both cell types. In addition, expression of the chemokine receptor gene Ccr9, a downstream target of RA signaling41, had significantly lower expression in Tox−/− progenitors. The chemokine receptor CXCR6 identifies a non-CLP, early pan-ILC progenitor population27,28 and TOX-deficient progenitors had a significant decrease in Cxcr6 expression.
Adult ILCs are dependent on Notch signaling for their development6,18–20. Consistent with this, the Notch target gene Hes1 was expressed by WT progenitors. Surprisingly, however, Notch genes were poorly or undetectably expressed in these cells. As ILC development may require a transient Notch signal6,18 we considered the possibility that this might be caused by Notch downregulation. To address this, we stained CLP, α4β7+ progenitors, and more restricted ILC2p for Notch1 (Fig. 5d). Indeed, Notch1 was more highly expressed in CLP than downstream progenitors. Tox−/− CLP expressed Notch1 normally (Fig. 5d). Despite this, Tox−/− α4β7+ progenitors failed to upregulate Hes1, as well as the Notch target gene Tcf77 (Fig. 5b). Taken together, TOX-deficient progenitors lack key ILC transcriptional regulators necessary for their subsequent development.
In vitro ILC differentiation requires TOX
RNA-seq analysis suggested there could be poor survival of ILC progenitors in the absence of TOX, as well as failure to fully activate genes downstream of Notch signaling. ILC development can be modeled in vitro by culturing BM-derived CLP with OP9 stromal cells that express the DL1 Notch ligand (OP9-DL1)6. We took advantage of this system, which allows analysis of cell numbers as well as cell differentiation, to determine the expression and role of TOX during ILC specification. CLP were cultured with OP9-DL1 cells or, as a control, OP9 cells; the latter promoting B cell development. CLP from WT and Tox−/− animals showed similar CD127 expression (Supplementary Fig. 6a) and equivalent B cell potential (Supplementary Fig. 6b).
To assess TOX induction, CLP derived from WT or ToxTOM animals were cultured with OP9-DL1 cells. As early as six days in culture, differentiation of CLP resulted in significant cell expansion and appearance of Lin−Thy-1+CD25−, Lin−Thy-1+CD25hi, and Lin−Thy-1hi cell populations, with increasing expression of TOM in these cells, respectively (Fig. 6a). A small subpopulation of Lin−Thy-1−CD25− cells remained in culture and did not express TOM, likely representing undifferentiated CLP (Fig. 6a).
Figure 6.
In vitro ILC specification is blocked early in the absence of TOX. (a) Subpopulations at day six of Lin− cells subgated on Thy-1 and CD25 were analyzed for TOM expression (n=3). (b) Calculated fold expansion over number of input WT or Tox−/− CLP at day six. (c) Differentiation of WT and Tox−/− CLP at day six was assessed by expression of Thy-1 and CD25 (n=6). (d, e) Frequency (d) and number (e) of Lin−Thy-1hi cells (% of Lin−) gated as in (c). (f) Lin−Thy-1hiCD25−ICOS+ WT and Tox−/− cells at day six in culture (n=6). (g) Lin−Thy-1lo and Lin−Thy-1hi cell populations were isolated by cell sorting and expression of indicated genes was determined by qRT-PCR. Data was normalized to Hprt expression and presented relative to in vivo, IL-33-expanded ILC2s. *P <0.05, **P <0.01, ***P <0.001, NS= non-significant, P >0.05 (Student’s t-test). Symbols (b, d, e) represent individual mice analyzed independently; horizontal lines denote the mean. Data (± s.d.) are compiled from four mice (g), using CLP pooled from two animals per experiment.
To determine the effect of loss of TOX in this system, differentiation of WT and Tox−/− CLP was compared. Tox−/− CLP showed significant deficits in cell expansion (Fig. 6b) and differentiation (Fig. 6c–e). We also noted that Lin−Thy-1hi cells derived from Tox−/− CLP failed to downregulate CD25 (Fig. 6c, f) and upregulate ICOS (Fig. 6f).
We compared the Lin−Thy-1loCD25+ to Lin−Thy-1hi transition for expression of transcriptional regulators involved in ILC development4,6,7,21,24. Significant upregulation of Id2, Gata3, Rorα, Tcf7, and Zbtb16 was detected in WT Lin−Thy-1hi cells, consistent with developmental progression of these cells to an ILC fate (Fig. 6g). However, expression of Id2, Gata3, and Rorα, was well below that of mature WT ILC2s (used as an internal comparison for gene expression in Fig. 6g). Upregulation of Tox in Lin−Thy-1hi cells was consistently observed in individual experiments, although it did not reach overall statistical significance (Fig. 6g). However, in conjunction with measurement of TOM expression (Fig. 6a), Tox is induced early in these cultures with likely increased expression as cells transit to the Lin−Thy-1hi stage.
To determine if TOX was required for establishing aspects of the ILC gene program, irrespective of poor production of Thy-1hi cells, we analyzed cell populations six days after initiation of culture. Rorα, Tcf7, and Zbtb16 were significantly decreased in Tox−/− Lin−Thy-1hi cells relative to their WT counterparts (Fig. 6g). Upregulation of Id2 and Gata3, in contrast, was not significantly different in WT and Tox−/− cells at this early stage of differentiation (Fig. 6g).
To further delineate the developmental potential of TOX-deficient CLP into ILC lineages, in vitro-generated WT and Tox−/− Lin−Thy-1hi cells were cultured at an equal ratio in ILC-promoting conditions. These cells differentiated into ILC1 and ILC2 lineages (Fig. 7a, b) and retained Thy-1hi expression. However, greater than 90% of ILC1s and ILC2s were WT derived (Fig 7a, b), despite eliminating the growth and differentiation (i.e. transition to Thy-1hi) advantage of WT cells. Interestingly, NK1.1−ICOS− cells that remained in these cultures showed no bias towards a WT phenotype (Fig 7a, b), pointing towards a specific defect in differentiation in the absence of TOX, and not solely diminished cell growth or survival.
Figure 7.
In the absence of TOX, there is a cell-intrinsic differentiation defect in the generation of ILC1s and functional ILC2s. (a) Frequency of WT (CD45.2+) and Tox−/− (CD45.2−) in vitro differentiated non-ILCs (ICOS−NK1.1−), ILC2s (ICOS+NK1.1−T-bet−GATA-3+) and ILC1s (ICOS+NK1.1+T-bet+GATA-3−) (n=4). (b) Transcription factor staining for WT ILC1s and ILC2s (n=4). (c) WT to Tox−/− ratio of populations in (a). (d) CLPs were isolated from WT and Tox−/− BM, cultured for six, 12, or 18 days, and numbers of Lin−Thy-1hi cells determined. Data are calculated based on input number of CLP. (e) Analysis of day 18 cultures as shown (n=7). (f) Gene expression in day 18 Lin−Thy-1hiICOS− or ICOS+ cells, determined as in Fig. 6. (g, h) Frequency (% of Lin−Thy-1hi) (g) and cell numbers (h) of WT culture-generated ICOS+ cells gated as in (e). Cell numbers are expressed as total cells recovered at day 18 per 5,000 cells plated at day 12. (i, j) Quantification of secreted IL-5 (i) or IL-13 (j) from in vitro differentiated CLP. (k, l) Expression of α4β7 (k) and PLZF (l) on Lin−Thy-1hi cells. Histograms display results from isotype control, ICOS− and ICOS+ cells (n=3). *P <0.05, **P <0.01, ***P <0.001, NS= non-significant, P >0.05 (Student’s t-test). Symbols (c, g, h, I, j) represent individual mice analyzed independently; horizontal lines denote the mean. Data are compiled from seven (d) or three (f) independent experiments using two animals pooled per experiment.
To assess the role of TOX in generation of functional ILC, cultures were maintained under ILC2 conditions and ICOS was used as a marker6. Tox−/− CLP were poor generators of Lin−Thy-1hi cells (Fig 7c). Nevertheless, re-plating equivalent numbers of mutant and WT cells at day six and then every six days onward allowed a comparative analysis at day 18. At this time point, a well-defined population of Lin−Thy-1hiICOS+CD25lo/neg cells was present in WT cultures (Fig. 7d). A comparison of ICOS− and ICOS+ WT cell populations demonstrated significant upregulation of Id2, Gata3, Rorα, and Zbtb16 in the latter, now reaching levels equivalent to or exceeding that found in mature ILC2s (Fig. 7e). Nfil3, a factor required for normal development of cNK42 and other ILCs25–27,43, was also significantly upregulated in Lin−Thy-1hiICOS+ WT cells (Fig. 7e). In contrast, neither Tox nor Tcf7 was differentially expressed between ICOS− and ICOS+ WT cell populations (Fig. 7e), consistent with an earlier induction peak (Fig. 6g).
In addition to the severe differentiation deficit of mutant CLP to the Lin−Thy-1hi stage, few ICOS+CD25lo/neg Tox−/− cells formed in these cultures (Fig. 7d, f, g). Moreover, WT cells secreted IL-5 (Fig. 7h) and IL-13 (Fig. 7i), presumably in response to IL-33 in the culture, confirming the development of functional ILC2s. In contrast, Tox−/− culture supernatants contained no detectable IL-5 and IL-13 (Fig. 7h, i), mirroring TOX-dependence of ILC2 development observed in vivo.
ILC progenitors express α4β73,24, and we observed initial upregulation of this integrin at day six on Thy-1hi cells (data not shown). At day 18, the majority of WT ICOS+ cells further upregulated α4β7 (Fig. 7j). In contrast, Tox−/− cultures contained a low frequency of ICOS+ cells, and these cells failed to fully upregulate α4β7 (Fig. 7j). PLZF is expressed in α4β7 + ILC progenitors that can give rise to ILC1s, ILC2s, and ILC3s and is required for development of some ILC lineages24. Poor upregulation of Zbtb16 (encoding PLZF) at day six, coupled with inhibition of later development of ICOS+ and α4β7hi cells, led us to ask whether PLZF expression would also be compromised in the absence of TOX. In WT cultures, the majority of ICOS+ cells were PLZFhi, with lower expression in ICOS− cells (Fig. 7e). In Tox−/− cultures, PLZFhi cells fail to develop, including in the minor population of Lin−Thy-1hiICOS+ cells (Fig. 7k). Therefore, TOX-deficient CLP fails to develop into ILCs even in the presence of sustained Notch signaling.
DISCUSSION
The identification and characterization of ILCs has led to considerable advances in understanding the cellular mechanisms that underlie immunological responses during homeostasis and disease. Overlaps in the transcriptional networks that regulate T effector subtypes and ILCs have been reported, yet the developmental changes that specifically lead to an ILC fate have not been well established. Here, we identified an obligatory role for the transcription factor TOX in the development of ILC1, ILC2, and ILC3 lineages, due to a block in the transition of CLP to common ILC progenitors. By whole transcriptome sequencing and TOX reporter generation, we identified the post-CLP stage of ILC development in which TOX is required. Furthermore, our data offers a window into other potential regulators of the ILC lineage, including a number of transcriptional regulators not previously implicated in ILC development.
The Id2hiCD25− subset of α4β7+ progenitors includes CHILP, which can give rise to all adult ILC with the exception of cNK3. All CHILP expressed Tox by reporter. A minor subset of CXCR6+ cells within the α4β7 progenitor population was recently reported to have in vivo potential to produce ILC1, ILC2, ILC3, and cNK, and to be dependent on the transcriptional regulator NFIL327. NFIL3 was reported to be required for TOX induction in adult BM27, although other data suggest a primary role for NFIL3 in regulating Id2 in CHILP 43. Surprisingly, TOX-deficient but not NFIL3-deficient mice lack LTi cells required for lymph node organogenesis and thus the exact relationship between these two factors remains to be elucidated. In addition, the relationship between CHILP and CXCR6+ progenitors is unclear. CXCR6+ cells may be a small subset of CHILP, making detection of the cNK cell fate difficult to discern in this mixed population or alternatively, there could be lower expression of Id2 in CXCR6+ cells compared to CHILP and thus represents a distinct cell population.
Isolated Tox−/− cells were lower for cell surface CD127 expression compared to the majority of their WT counterparts and expressed less Il7r mRNA. The transition of fetal liver LTi precursors from RORγt− to RORγt+ is associated with upregulation of CD12744. In addition, a proportion of isolated WT α4β7+ progenitors are CD127lo. Thus, we propose that Tox−/− α4β7+ progenitors represent a post-CLP but pre-CHILP transitional cell state, with intermediate expression of Id2 and CD127, and that these cells cannot undergo lineage specification in the absence of TOX.
Like T cells, ILCs are dependent on Notch signaling, with a transient Notch signal favoring the ILC lineage6. Expression of Notch gene targets Hes1 and Tcf17, and expression of potential downstream gene targets of TCF1, Bcl11b and Gata3,7 were significantly reduced in Tox−/− α4β7+ progenitors. Down regulation of Notch1 mRNA is also associated with upregulation of CXCR6 on α4β7+ progenitors28. It is possible that downregulation of Notch genes during early ILC development ensures a transient Notch signal. Moreover, these data may indicate that one primary deficit in Tox−/− cells may be an inability to activate downstream gene targets of Notch signaling. Interestingly, binding of TOX to the HES1 gene has been detected in HEK-293T cells 45.
Maf, a transcription factor that plays a critical role in the differentiation of various T helper cell subsets46, was differentially expressed in WT and Tox−/− cells and possibly represents a new example of overlap in the transcriptional landscape of ILC and T-helper cell lineages. In the absence of TOX, expression of Rxrg and the RA target gene Ccr9 was also significantly reduced, pointing to the potential for modulation of ILC development by RA signaling in ILC progenitor populations, as has been reported for mature ILC subtypes39,47.
The small intestine contains a mix of fetal-derived LTi-like cells that express CCR6 and a heterogeneous mix of postnatal ILC36. Here, the frequencies of major populations of small intestine LP ILC3 were comparable between WT and Tox−/− mice. These data may suggest a TOX-independent pathway of gut-resident ILC3 development. LP contain CLP-like cells that can give rise to NK and RORγt+ cells in culture28, and thus the relative contribution of BM progenitors to LP ILC3 development is not clear. Alternatively, it is possible that the gut microenvironment is particularly conducive to expansion of any ILC3 that may form, even in small numbers. A proportion of CCR6−T-bet+ ILC3 in LP are proliferative36, and in general, small intestine LP ILC have a transcriptome associated with activated cells when compared to ILC in other locations47. Post-natal proliferation of LP ILC3 could be particularly pronounced in Tox−/− mice, which have a block in CD4+ T cell, including T regulatory cell, development31. In support of a disturbed gut microenvironment in these animals, we can detect Lin+CD4+RORγt+ cells in Tox−/− mice despite the thymic block, and a high frequency of other Lin+RORγt+ cells. This potential inflammatory environment could influence homeostasis of ILC3 populations. A definitive molecular dissection of small intestine ILC3 development awaits further experimentation.
Based on these data, we propose the following events regarding the role of TOX in ILC development. Notch signaling in CLP leads to upregulation of α4β7 and early factors TOX and Id2. TOX may be required for translation of Notch signals to downstream gene expression, including induction of Tcf7 and Hes1. Relatively rapid downregulation of Notch genes at a TOXloId2loCD127lo transitional stage may extinguish continued Notch signaling, promoting an ILC rather than a T cell fate. Loss of TOX at this early stage results in a cascading failure to initiate programs of cell survival and differentiation required for continued ILC lineage specification through CXCR6+, CHILP, and PLZFhi progenitor stages, and ultimately, failure of ILC lineage commitment.
METHODS
Mice
All mice were bred at the Cedars-Sinai Medical Center and kept under specific pathogen-free conditions. Mice used for experiments were between 6–14 weeks of age, of either sex. ToxTOM reporter mice were generated by insertion of tandem dimer Tomato (TOM) coding sequence lacking a transcriptional stop at the start ATG of Tox exon 1 by standard methodology (inGenious Targeting Laboratory). A neomycin cassette within FRT sites was removed by breeding with FLPase mice. FLPase negative mice were selected in a subsequent generation, and heterozygous reporter mice maintained on a C57BL6/J background for experiments. Tox −/− mice were generated as described previously, and maintained by Tox −/− x Tox+/− breeding31. B6.129S (Cg)-Id2tm2.1Blh/ZhuJ Id2GFP reporter mice were obtained from The Jackson Laboratory and bred to ToxTOM reporter animals to generate a double reporter strain. Mice heterozygous for both reporters were used in experiments. NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were obtained commercially (The Jackson Laboratory). Mice were selected for experimental groups based only on genotype within the age range indicated above. Within experiments, animals of each genotype were age-matched as closely as possible within constraints of availability. No blinding or specific randomization protocol was used. All animal procedures were performed in accordance with protocols approved by the CSMC Institutional Animal Care and Use Committee.
Isolation of lymphocytes from BM, lung and lamina propria
Whole lung was removed following a 5 ml injection of PBS into the right ventricle of the heart. Lungs were minced and digested in HBSS containing 1 mg/ml Liberase with 10 U/ml DNase I (Roche Diagnostics). Cells were filtered using a 70 μm cell strainer and red blood cells lysed. For CD27 labeling, 0.5 mg/ml collagenase type VIII (Sigma) was used. For IL-33 induced ILC2 expansion in vivo, animals were injected intraperitoneally once daily with 500 ng IL-33 (BioLegend) or with PBS for four days.
Colons were removed, opened longitudinally and washed in PBS to remove fecal content. Intestines were cut into two centimeter segments, transferred into HBSS supplemented with 2% FBS and 2 mM EDTA, and shaken for 15 min at 37°C. This step was repeated twice in order to remove epithelial cells and fat tissue. The remaining intestinal tissue was washed, cut in small segments and incubated in digestion media consisting of RPMI 1640, 2% FBS, 0.5 mg/ml collagenase type VIII, 5 U/ml DNase, 100 IU/ml penicillin and 100 μg/ml streptomycin for 40 min at 37°C by gentle shaking. Digested suspensions were filtered, washed in PBS and cells were collected for analysis.
RNA-seq and data analysis
We performed a customized low input RNA-seq protocol to allow whole transcriptome measurements with as few as 250 cells. Isolated bone marrow cell populations were sorted into a 96-well plate containing lysis buffer (SMARTer® Ultra™ Low Input RNA v3 kit, Takara) for sequencing which was also used for constructing cDNA libraries per manufacturer’s instructions. The protocol for the Ion Torrent Proton was modified by enzymatically fragmenting the resulting double-stranded cDNA libraries and ligating sequencing adapters from the Ion Xpress™ Plus Fragment Library Kit (Life Technologies). cDNA libraries were amplified onto Ion Sphere Particles using Ion PI™ template OT2 200 Kit v3 (Life Technologies) and then sequenced on the Ion Proton™. Samples were sequenced with the Ion PI™ Sequencing 200 v3 kit (Life Technologies) to a depth of 12 to 25 million reads with less than 2.5% of reads coming from ribosomal RNA and over 90% of reads mapping to the mouse genome. The raw reads were filtered and trimmed by FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) then aligned to mouse reference genome (mm10) using TMAP with mouse reference Gencode version 19 reference genome annotation (http://www.gencodegenes.org). FPKM (fragment per kilobase of gene per million reads sequenced) values were calculated for 23,847 genes using Cufflinks 2.0.8. Poorly measured raw FPKM values less than 1 were increased to a floor threshold of 1. Raw FPKM values were then log2 transformed. Genes with an average of 1 or less in addition to miRNAs and SNORDs where removed from further analysis. Unsupervised analysis was performed by filtering FPKM values for any three of eight samples that demonstrated at least five-fold greater expression than any other sample. For visualization of coordinated gene expression in samples, we performed 2-way hierarchical clustering with samples and genes. Genes were mean centered and dendrograms were calculated using hclust and plotted using heatmap.2 (v2.12) in gplots (v 2.14.2) in R (v 3.0.2). A two-tailed t-test was used to assess the significance of gene expression differences and then corrected for multiple hypotheses by calculating the q value using the Benjamini and Hochberg method. Data from well-annotated genes that had significant gene expression differences with a false discovery rate below 5% (defined as a q value < 0.05) are presented (Supplementary Table 2). Data were deposited in Gene Expression Omnibus at NCBI (http://www.ncbi.nlm.nih.gov/geo/) and can be obtained using accession code GSE65850.
Generation of bone marrow chimeras
CLPs were sorted from Tox−/− (Thy-1.2+, CD45.2+) and B6.PL-Thy1a/CyJ (Thy-1.1+, CD45.2+) animals and mixed at a 1:1 ratio using 3.0 × 104 cells per genotype. NSG (CD45.1+) mice were reconstituted with 6.0 × 104 CLPs by injection into the tail vein. Mice were analyzed two weeks later.
In vitro differentiation assay
Whole BM was extracted as described above. CLPs were isolated by cell sorting directly into complete DMEM containing IL-7 (20 ng/ml) and IL-33 (20 ng/ml) (Biolegend) for ILC2 conditions or IL-7 (10 ng/ml), SCF (50 ng/ml) (R&D Systems) and Flt3L (5 ng/ml) (R&D Systems) for B cell conditions. Following isolation, cells were immediately transferred to mitomycin c treated (Sigma) confluent OP9-DL1 (ILC2 conditions) or OP9 (B-cell conditions) at a density of 3000–7500 cells/well in 48 well plates. Cells were passaged and added to fresh OP9 or OP9-DL1 monolayers every six days. For competitive in vitro ILC differentiation, BM was extracted as described above from Tox−/− and B6.SJL-Ptprca Pepcb/BoyJ (Ly5.2) mice. CLPs were isolated by cell sorting directly into complete DMEM containing IL-7 (20 ng/ml) and immediately transferred to mitomycin c treated, confluent OP9-DL1 at a density of 8000 cells/well in 48 well plates. Six days later, Lin−Thy-1hi cells were sorted directly into complete DMEM containing IL-7 (20 ng/ml) and SCF (50 ng/ml), were mixed at a 1:1, WT (CD45.2+):Tox−/− (CD45.2−) ratio, and immediately transferred to confluent OP9-DL1 at a density of 8000 cells/well in a 48 well plate. Cells were analyzed 6 days later.
IL-13 and IL-5 determination
Whole culture medium from in vitro generated ILC2 was collected at day 18 and analyzed for IL-13 and IL-5 by ELISA per manufacturer’s instructions (eBioscience).
Antibodies and flow cytometry
All antibodies used for flow cytometry were purchased from eBioscience, Biolegend or BD Biosciences and are listed in Supplementary Table 3 with clone designations in parentheses. Lineage markers used for each population analyzed can be found in Supplementary Table 3. RORγt, GATA-3, T-bet, Eomes and PLZF intracellular staining was conducted using the FoxP3 staining kit as per manufacturer’s instructions (eBioscience). All samples were pre-incubated with anti-CD16/32 (93) to block FcR. Cells were analyzed using a LSRII and isolated using a FACS Aria III (BD Biosciences). CD1d (PBS57) was generously provided by the NIH tetramer core facility.
qRT-PCR
All data was normalized to Hprt and presented relative to expression determined from IL-33 expanded lung ILC2 cell mRNA. cDNA was generated using Superscript VILO (Life Technologies) and amplified using QuantiTect SYBR green. All primer sets were purchased from QIAGEN (QuantiTect), except for Tox and Rorα primers, which were as follows: Tox F: 5′-TGCCTGGACCCCTACTATTG and Tox R: 5′-CTGGCTGGCACATAGTCCTG. Rora F: 5′-TCTCCCTGCGCTCTCCGCAC and R: 5′-TCCACAGATCTTGCATGGA48.
Statistics
Means, standard deviations, and the probability (P) associated with a Student’s t-test using a two-tailed distribution of equal variance are shown in some figures. P values of <0.05 were considered to represent means with a statistically significant difference. Statistical analysis was performed on groups with similar variance and limited variance was observed within sample groups. Sample or experiment sizes were determined empirically for sufficient statistical power. No statistical tests were used to predetermine the size of experiments and no samples were excluded specifically from analysis.
Supplementary Material
Acknowledgments
We thank the CSMC Flow Cytometry Core members, especially G. Hultin and L. Dieu, for assistance with cell isolations, Dr. G. Martins for assistance with TH2 cell generation, and Dr. A. Seksenyan and A. Kadavallore for useful discussions. We also thank the CSMC Biomedical Sciences and Translational Medicine Graduate Program. The NIH Tetramer Core Facility (contract HHSN272201300006C) provided CD1d tetramers and J.C. Zuniga-Pflucker (University of Toronto) generously provided OP9-DL1 cells. I.D.I. is supported by NIH grant DK098310. This work was supported by NIH grant 5R01AI054977 to J.K.
Footnotes
Accession codes.
mRNA-seq GSE65850
AUTHOR CONTRIBUTIONS
C.S. and J.K. were responsible for overall design and execution of experiments and data analysis. C.S. performed the bulk of experiments. P.A. designed and performed characterization of Tox knock-in reporter mice. B.D. provided technical assistance for animal experiments. I.I. aided in isolation and analysis of lamina propria cells. L.S. and V.F. performed RNA-seq and data analysis. C.S and J.K. wrote the manuscript, with input from all other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 2.Monticelli LA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klose CS, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 4.Hoyler T, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yagi R, et al. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity. 2014;40:378–388. doi: 10.1016/j.immuni.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong SH, et al. Transcription factor RORalpha is critical for nuocyte development. Nat Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Q, et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nussbaum JC, et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–248. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molofsky AB, et al. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J Exp Med. 2013;210:535–549. doi: 10.1084/jem.20121964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halim TY, et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40:425–435. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Z, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 12.Kim MY, et al. Heterogeneity of lymphoid tissue inducer cell populations present in embryonic and adult mouse lymphoid tissues. Immunology. 2008;124:166–174. doi: 10.1111/j.1365-2567.2007.02750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luci C, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 14.Mielke LA, et al. TCF-1 controls ILC2 and NKp46+RORgammat+ innate lymphocyte differentiation and protection in intestinal inflammation. J Immunol. 2013;191:4383–4391. doi: 10.4049/jimmunol.1301228. [DOI] [PubMed] [Google Scholar]
- 15.Mortha A, et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4+ Lymphoid Tissue-Inducer Cells Promote Innate Immunity in the Gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magri G, et al. Innate lymphoid cells integrate stromal and immunological signals to enhance antibody production by splenic marginal zone B cells. Nat Immunol. 2014;15:354–364. doi: 10.1038/ni.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORgammat sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Q, Saenz SA, Zlotoff DA, Artis D, Bhandoola A. Cutting edge: Natural helper cells derive from lymphoid progenitors. J Immunol. 2011;187:5505–5509. doi: 10.4049/jimmunol.1102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rankin LC, et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokota Y, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 22.Satoh-Takayama N, et al. IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+ cell subsets from Id2-dependent precursors. J Exp Med. 2010;207:273–280. doi: 10.1084/jem.20092029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seillet C, et al. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med. 2014 doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiger TL, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med. 2014;211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X, et al. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife. 2014;3 doi: 10.7554/eLife.04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Possot C, et al. Notch signaling is necessary for adult, but not fetal, development of RORgammat(+) innate lymphoid cells. Nat Immunol. 2011;12:949–958. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson B, et al. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol. 2002;3:272–280. doi: 10.1038/ni767. [DOI] [PubMed] [Google Scholar]
- 30.O’Flaherty E, Kaye J. TOX defines a conserved subfamily of HMG-box proteins. BMC Genomics. 2003;4:13. doi: 10.1186/1471-2164-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205:245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aliahmad P, de la Torre B, Kaye J. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue-inducer cell and NK cell lineages. Nat Immunol. 2010;11:945–952. doi: 10.1038/ni.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aliahmad P, Kadavallore A, de la Torre B, Kappes D, Kaye J. TOX is required for development of the CD4 T cell lineage gene program. J Immunol. 2011;187:5931–5940. doi: 10.4049/jimmunol.1101474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 36.Klose CS, et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 37.Aliahmad P, Seksenyan A, Kaye J. The many roles of TOX in the immune system. Curr Opin Immunol. 2012;24:173–177. doi: 10.1016/j.coi.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gleimer M, von Boehmer H, Kreslavsky T. PLZF Controls the Expression of a Limited Number of Genes Essential for NKT Cell Function. Front Immunol. 2012;3:374. doi: 10.3389/fimmu.2012.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer SP, et al. Adaptation of Innate Lymphoid Cells to a Micronutrient Deficiency Promotes Type 2 Barrier Immunity. Science. 2014;343:432–437. doi: 10.1126/science.1247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Pavert SA, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 2014;508:123–127. doi: 10.1038/nature13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwata M, et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Gascoyne DM, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 43.Xu W, et al. NFIL3 Orchestrates the Emergence of Common Helper Innate Lymphoid Cell Precursors. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.057. [DOI] [PubMed] [Google Scholar]
- 44.Tachibana M, et al. Runx1/Cbfbeta2 complexes are required for lymphoid tissue inducer cell differentiation at two developmental stages. J Immunol. 2011;186:1450–1457. doi: 10.4049/jimmunol.1000162. [DOI] [PubMed] [Google Scholar]
- 45.Artegiani B, et al. Tox: a multifunctional transcription factor and novel regulator of mammalian corticogenesis. EMBO J. 2014 doi: 10.15252/embj.201490061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 47.Robinette ML, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015 doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.