Abstract
The chemokine receptor CCR5 serves as a co-receptor for the Human Immunodefficiency Virus type-1, HIV-1. The CCR5 N-terminal segment, the second extracellular loop (ECL2) and the transmembrane helices have been implicated in binding the envelope glycoprotein gp120. Peptides corresponding to the sequence of the putative ECL2 as well as peptides containing the ECL1 and ECL3 were found to inhibit HIV-1 infection. The aromatic residues in the C-terminal half of an ECL2 peptide were shown to interact with gp120. In the present study we determined that in aqueous buffer the segment Q188-Q194 in an elongated ECL2 peptide (R168 to K197) forms an amphiphilic helix, which corresponds to the beginning of the fifth transmembrane helix in the crystal structure of CCR5. Two dimensional Saturation Transfer Difference NMR spectroscopy and dynamic filtering studies revealed the involvement of Y187, F189, W190 and F193 of the helical segment, in the interaction with gp120. The crystal structure of CCR5 shows that the aromatic side chains of F189, W190 and F193 point away from the binding pocket and interact with the membrane or with an adjacent CCR5 molecule and therefore, could not interact with gp120 in the intact CCR5 receptor. We conclude that these three aromatic residues of ECL2 peptides interact with gp120 through hydrophobic interactions not representative of the interactions of the intact CCR5 receptor. The HIV-1 inhibition by ECL2 peptides as well as by ECL1 and ECL3 peptides and peptides corresponding to ECL2 of CXCR4, which serves as an alternative HIV-1 co-receptor, suggests that there is a hydrophobic surface in the envelope spike that could be a target for HIV-1 entry inhibitors.
Keywords: gp120, CCR5, ECL2, HIV-1 inhibitors, NMR
INTRODUCTION
Human C-C chemokine receptor 5, CCR5, is a co-receptor for HIV-1 and is a heptahelical membrane protein that belongs to the GPCR superfamily [1]. It interacts with HIV-1 gp120 after this envelope glycoprotein binds to CD4, the primary receptor for the virus on T-cells and macrophages. The N-terminal segment of CCR5, Nt-CCR5, and its second extra cellular loop, ECL2, were implicated in gp120 binding [2-4]. The structure of CCR5 in complex with a small-molecule HIV-1 entry inhibitor reveals interactions between the inhibitor and a binding pocket formed by CCR5 transmembrane helices and by ECL2 [5]. CXCR4 is an alternative coreceptor for X4 strains of HIV-1. Its structure, in complex with an inhibitory peptide, suggests that the CXCR4 binding pocket is formed by the transmembrane helices and ECL2 and that this pocket interacts with the V3 region of gp120 [6, 7].
The structure of the N-terminal segment (Nt-CCR5) encompassing the first 19 residues of CCR5 was not revealed in the X-ray analysis of this receptor. However, the conformation of this segment was elucidated by NMR transferred-NOE studies on peptide surrogates [8, 9] revealing an α-helix formed by residues P8-S17. Saturation-Transfer Difference (STD) NMR experiments identified the Nt-CCR5 residues interacting with gp120 [9]; this data together with biochemical data regarding the gp120 residues interacting with CCR5 were used to dock the Nt-CCR5 peptide into gp120, revealing an Nt-CCR5 orientation that differs by 180° from that found in earlier studies [8]. According to this docking model [9] Nt-CCR5 interacts with the base of the V3 and with the fourth constant region of gp120, C4, which has been identified previously as being involved in forming the co-receptor binding site [10].
Alanine-scanning of charged residues showed that residues in Nt-CCR5 and the three ECLs are important for gp120 binding [11]. Very extensive studies of non-clade B HIV-1 strains revealed that in addition to a large number of residues in Nt-CCR5, residues in ECL1 and in ECL2 contributed to viral entry [12]. In other studies of clade-B viruses, some additional residues in ECL2 and the polar face of TM5 were found to be important [11, 13]. Lagane and co-workers recently carried out a comprehensive site-directed mutagenesis study of CCR5 interactions with gp120 [7] and found residues in all three ECLs and in TM3 and TM7 involved in gp120 binding. All these studies showed that in addition to the established role of Nt-CCR5, residues in the three ECLs, especially in ECL2, and in the trans-membrane helices are important for gp120 binding.
The crystal structures of CXCR4 and CCR5 [5, 6] facilitated modeling of V3 in complex with CCR5 [5, 7, 14]. In these models the tip of the V3 interacts with the CCR5 binding pocket formed by the transmembrane helices and by ECL2. A detailed analysis of V3 interactions with CCR5 revealed that R315 at the tip of the V3 forms a salt bridge with E283 at the bottom of the CCR5 binding pocket and that the N-terminal strand of the V3 interacts with ECL2 [14].
Peptides corresponding to ECL1, ECL2 and ECL3 of CCR5 were found to inhibit infection by different strains [15]. In that study, fusion inhibition by peptides corresponding to sequences of CCR5-ECLs was found to be HIV-1 phenotype specific, with R5 but not X4 clades affected [15]. Specifically, peptides corresponding to residues 90-101 in CCR5 ECL1, residues 168-191 of ECL2 and residues 261-276 of ECL3, inhibited the fusion of R5 viruses with target cells, but did not inhibit fusion of X4 viruses. Among the three peptides, the ECL2 peptide exhibited a slightly stronger fusion inhibition of the R5 virus Ba-L with 50% inhibition at 20 μg/ml concentration [15].
Recently, Bewley and co-workers[16] showed that the same ECL2 peptide (R168-K191) used by Berger and co-workers [15], neutralized both R5 and X4 viruses in the 100 μM range and not only R5 viruses, as previously reported by Berger and co-workers. Bewley and co-workers found a shorter ECL2 peptide (C178-F193) that contained two more ECL2 residues at its C-terminus and was 6-fold more potent in HIV-1 neutralization which was CD4-independent. STD NMR measurements revealed that the aromatic residues H181, F182, Y184, Y187, F189 and W190 interact with a gp120 protein lacking the V1 and V2 loops. The free ECL2 peptide encompassing residues 168-191 was unstructured in solution [16]. Similarly, peptides corresponding to CXCR4 ECL2 were found to inhibit HIV-1 infection by X4 viruses [17, 18].
The goals of the present study were to understand the mechanism by which HIV-1 neutralizing ECL2 peptides interact with gp120, to map the segment of elongated ECL2 involved in binding, especially at its C-terminus, and to examine whether the interactions between ECL2 peptides and gp120 could mimic similar interactions of the intact CCR5 receptor. An ECL2 peptide that is longer by six residues from that characterized by Bewley and co-worker [16] and by Berger and co-workers [15] was chosen for the present study. This elongated ECL2 peptide corresponds to the entire putative ECL2 loop of CCR5 as defined earlier by Agrawal et al. [15], Moreover, residues at the C-terminus of ECL2 and the N-terminus of the fifth transmembrane domain of CCR5 were previously shown to be critical for inhibition of HIV-1 fusion [7, 16]. The added six-residue segment could potentially stabilize secondary structure as indeed was shown to be the case. Moreover, two of the added residues, N192-F193, improved the potency of the ECL2 peptide in HIV-1 neutralization [16]. Specifically, we studied the interactions of gp120 and a peptide containing the 168RSQKEGLHYTASSHFPYSQYQFWKNFQTLK197 sequence. C178 of ECL2 was mutated to alanine or to serine (underlined in the above sequence), to avoid potential complications due to disulfide bond formation. 15N-1H-HSQC titration analysis and dynamic filtering studies mapped the binding site for gp120 to ECL2 segment H181-L196. The ECL2 peptide was found to interact with gp120 lacking the V3 region. The structure of the free ECL2 peptide in aqueous buffer was determined using 3D heteronuclear NMR experiments, revealing that part of the segment implicated in gp120 binding, Q188-Q194, formed a two-turn helix. Examination of the CCR5 structure[5] and results of control experiments, suggests that elongated ECL2 peptides may interact with gp120 through non-specific hydrophobic contacts involving aromatic residues, in a manner that likely does not reflect the interaction of ECL2 in the intact receptor with the HIV envelope spike. Nevertheless, the identification and targeting of an apparently exposed hydrophobic site on gp120 by the ECL2 peptide could be useful in developing strategies for the design of HIV-1 entry inhibitors and/or vaccine development.
Results
Characterization of the R168-K197 ECL2 peptide
Initially a 30-residue peptide encompassing the entire putative second extracellular region of CCR5 [15] was investigated. C178 of the peptide was mutated to alanine (168RSQKEGLHYTASSHFPYSQYQFWKNFQTLK197; ECL2) to avoid potential dimerization of the peptide by disulfide bond formation. The use of reducing agent was avoided in order to prevent possible reduction of the multiple disulfide bonds in the gp120 molecule used with the ECL2 peptide for binding studies. The ECL2 peptide was poorly soluble, especially at the neutral pH at which the gp120 molecule was found to be most stable. To increase the peptide solubility, GSGS linkers were added preceding the peptide N-terminus and following the peptide's C-terminus and C178 was mutated to serine rather than to alanine (GSGS-168RSQKEGLHYTSSSHFPYSQYQFWKNFQTLK197-GSGS; ECL2S). A neutral rather than charged solubility tag was chosen in order not to influence possible electrostatic interactions between the peptide and gp120. This peptide was found to be soluble at neutral pH even at 3 mM concentration. However, comparison of the 15N-1H-HSQC spectra of ECL2S at 1.25 mM and 130 μM (data not shown) suggested that the peptide oligomerizes at the higher concentration and therefore, further experiments at pH 7 were carried at concentrations only up to 130 μM. For structure determination of the free ECL2S peptide in solution, NOESY measurements were conducted at pH 4.8. This allowed us to use a 1.5 mM peptide concentration without any significant sign of aggregation as assessed by comparison of HSQC spectra at 130 μM and 1.5 mM concentrations (data not shown). To increase NOE intensities these experiments were carried out at low temperature, 4 °C (277 K).
The ECL2S peptide is helical
Sequential backbone and side-chain assignment of the 38-residue ECL2S peptide was conducted using a standard NOE-based approach utilizing 2D and 3D 15N-separated NOESY and TOCSY experiments recorded at pH 4.8. Backbone amide protons of all residues (excluding the first Gly) were assigned and complete side-chain proton assignments were obtained for the entire peptide.
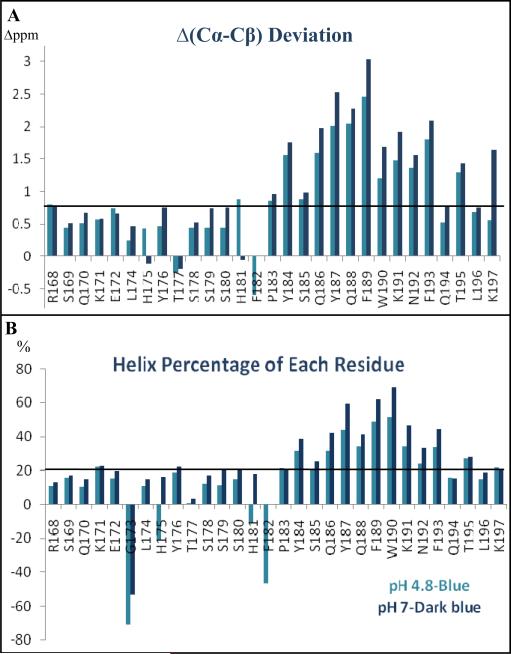
Three-dimensional HNCA, HNCACB, HNCO and CBCA(CO)NH [19] spectra that were acquired at both pH 4.8 and pH 7 were used to verify the sequential backbone assignments and to obtain 13C chemical shifts at both pH conditions. The deviations of Cα and Cβ chemical shifts from random coil values, which provide very reliable information on the secondary structure of peptides and proteins [20, 21] were calculated. Fig. 1A shows that at both pH conditions, positive deviations of Δ(Cα-Cβ) from their random coil values were observed across the peptide sequence for most residues. In particular, the large deviations for residues Y184-T195 suggest the existence of a helical conformation at this C-terminal region of ECL2S. The consistently larger chemical shift deviation values found for this segment at pH 7 suggest a more stable helical conformation in neutral pH solution. The helical content of each ECL2S residue represented in Fig. 1 was calculated based on the observed deviations of Cα chemical shifts from random coil values and average values of these deviations for each amino acid found in a helical conformation [21]. The largest population in a helical conformation was observed for residues F189 and W190 reaching a maximum of 70% helical conformation at pH 7 (Fig. 1B). The region between R168 to P183 appears to be mostly unstructured.
Fig. 1.
Chemical shift deviations from random coil values (CSD) and helical conformation percentage of each residue. (A) ECL2S Δ(13Cα-13Cβ) deviations from random coil values at pH 4.8 (blue) and 7 (dark-blue) are presented. The solid line represents a cut-off value for which higher values are indicative of a helical conformation. (B) Percentage of helical conformation of each residue: the percentage of the molecules found in a helical conformation for each residue was calculated using the equation: (δMeasured - δRandom)/(δAve.-Helix - δRandom). The averaged chemical shift value of each residue in a helical conformation, δAve.-Helix, is according to Wishart et al. (1994, Chemical shifts as a tool for structure determination, Methods Enzymol. 239, 363-392).
While at pH 7 and 150 μM ECL2S concentration only intra-residue and sequential NOE cross peaks were obtained, at pH 4.8 where we were able to use 1.5 mM peptide concentration, numerous medium-range NOEs were detected. A total of 307 NOE connectivities were assigned from the 2D and 3D 15N-separated NOESY spectra of ECL2S recorded at pH 4.8 (Table 1). Of these, 131 are intra-residue NOEs, 108 sequential and 68 are medium-range NOEs. Five dαN(i, i+2) and six dNN(i, i+2) connectivities were observed in the region between S185 to L196. Two medium-range connectivities, one dαN(i, i+3) and one dαN(i, i+4) were observed between residues K191/Q194 and K191/T195, respectively.
Table 1.
NMR constrains and structural statistic of the 10 lowest-energy calculated structures of ECL2S, residues Q188-Q194.
| NMR constrains and statistics | |
|---|---|
| Total distance constraints | 307 |
| Intra-residue NOE constrains | 131 |
| Sequential NOE constrains | 108 |
| Medium NOE constrains | 68 |
| Long NOE constrains | - |
| Hydrogen bond constrains | 6 |
| Φ dihedral angle constrain | 12 |
| Ψ dihedral angle constrain | 10 |
| Maximum individual NOE violation | 0.50 Å |
| rmsd of NOE violation | 0.02 ± 0.001 Å |
| Mean rmsd value of the 10 lowest- energy structures residues 0188-0194 | |
| back bone | 0.27 ± 0.16 Å |
| Heavy | 1.78 ± 0.53 Å |
19 φ and ψ dihedral angles constraints were obtained based on the chemical shift values of Hα, NH, HN and Cα, Cβ, CO using TALOS-N [22]. Three additional φ dihedral angles were constrained to −65° ± 25° for residues with 3JHNHα coupling constants smaller than 6 Hz, which are characteristic of helical conformation.
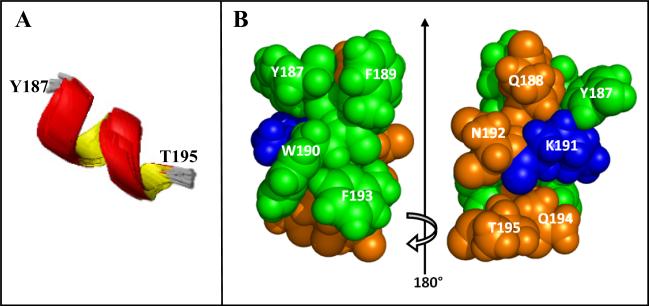
A total of 74 acceptable structures with no violations of the NMR distance constraints larger than 0.5 Å were obtained from 120 runs. The ensemble of the 10 lowest-energy structures is presented (Fig. 2A). An α-helix was obtained from residue Q188 to Q194 with backbone RMSDs of 0.27 ± 0.16 Å. As seen in the space filling model representation, this helix is highly amphiphilic with the side chains forming distinct polar/charged and nonpolar/aromatic faces (Fig. 2B). The helical segment found experimentally is in good agreement with the predicted secondary structure that we calculated for the same ECL2 peptide using different programs, identifying a helix in the segment Q186-Q194 [23-25].
Fig. 2.
The structure of the helical segment of ECL2S. A) Ensemble of the 10 lowest-energy structures for residues Y187-T195. B) Space filling model of the energy-minimized average structure for residues Y187-T195. The amino acids are colored according to their polarity: Green: hydrophobic amino acids Y187, F189, W190 and F193. Dark orange: polar uncharged amino acids Q188, N192, Q194 and T195. Blue: charged amino acid K191.
Binding of ECL2S peptide to mutgp120core
Binding of ECL2S to mutgp120core and to mutgp120core(+V3) [26] in the presence or absence of a 27-residue CD4-mimic peptide, CD4M33 [27], was measured using surface plasmon resonance (SPR) at 25 °C. Biotin was added to the peptide's N-terminus to enable the peptide's immobilization on a streptavidin-coated SPR chip. CD4M33 was used instead of a soluble CD4 molecule to reduce the size of the gp120 complex for NMR studies [9, 26]. Two gp120 molecules were used in the present study, mutgp120core and mutgp120core(+V3). Both molecules were expressed in GnTI- HEK293S cells [9, 28] and therefore were homogenously glycosylated with Man5GlcNAc2 glycans at sites normally occupied by complex or hybrid glycans. The carbohydrates were not cleaved and the glycosylated form of the molecules were used. At 150 mM NaCl, 15 mM MgCl2 and pH 7.0, a KD of 18 ± 1.5 ×10−6 M was measured for mutgp120core in the absence of CD4M33, (Table 2). In the presence of CD4M33 a KD of 6 ± 4 ×10−6 M was determined, offering only a rough estimate of the KD. A reliable KD for mutgp120core(+V3) could not be determined due to partial dimerization of the mutgp120core(+V3)/CD4M33 complex [26]. Similar KD values for ECL2S binding to the mutgp120core/CD4M33 were measured at pH 6 and pH 6.5 (KD of 7 ± 3 and 7.5 ± 3 ×10−6 M, respectively), suggesting that the binding of ECL2S to mutgp120core/CD4M33 is not influenced by pH between pH 6-7.
Table 2.
Dissociation constants of ECL2S bound to mutgp120core and mutgp120core/CD4M33 at different pH. The KD values were determined by Surface Plasmon Resonance (SPR) at 20 mM Tris-HCl, 0.05% NaN3, 0.005% Tween, 15 mM MgCl2 with 150 mM NaCl at 25 °C and at the indicated pH.
| Ligand | pH | Binding affinity (KD) |
|---|---|---|
| mutgp120core | 7.0 | 18 ± 1.5 μM |
| mutgp120core/CD4M33 | 7.0 | 6 ± 4 μM |
| mutgp120core/CD4M33 | 6.5 | 7.5 ± 3 μM |
| mutgp120core/CD4M33 | 6.0 | 7 ± 3 μM |
Mapping the ECL2S region interacting with gp120
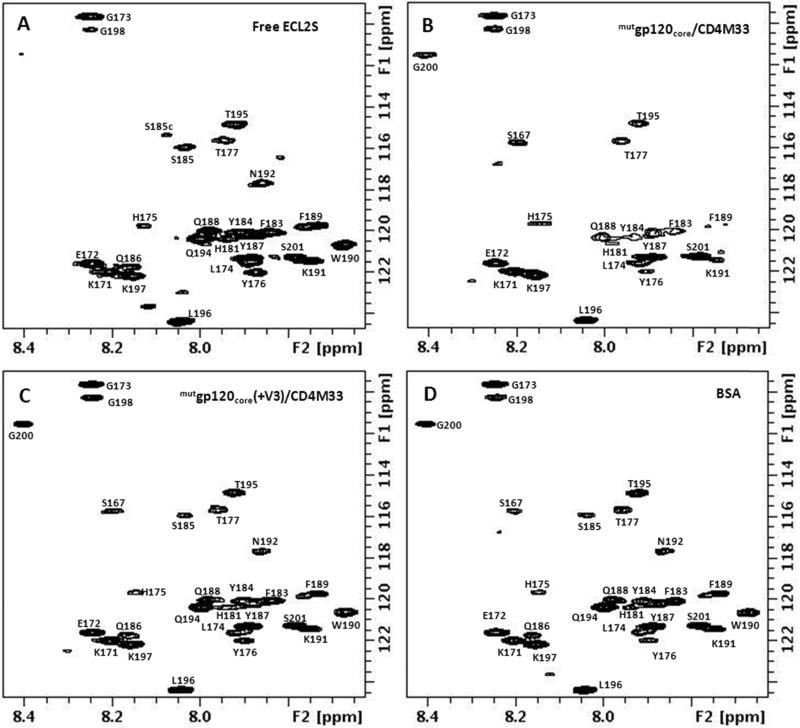
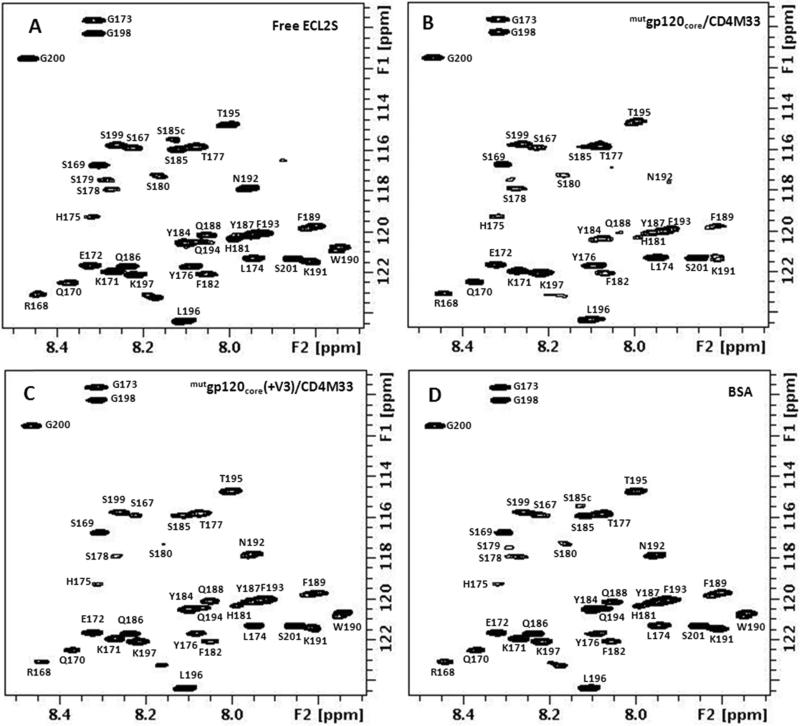
The 1H-15N HSQC spectrum of U-15N labeled ECL2S in the presence of equimolar concentration of mutgp120core/CD4M33 or mutgp120core(+V3)/CD4M33 complex (Fig. 3 A-D) can be used to distinguish between residues located in the ECL2S segment interacting with gp120 and residues outside this segment, similar to the dynamic-filtering approach that we used in the past [29]. The mobility of peptide residues in the region interacting with a large protein is very much restricted upon binding and their T2 relaxation times are similar to those of the protein residues. As a result, the cross peaks of the residues in the peptide segment interacting with the protein undergo a large reduction in their cross peak intensity upon protein binding and practically are not observable under the measurement conditions. Indeed, as shown in Fig. 3B, a large number of ECL2S cross peaks exhibited pronounced reduction in their intensities upon mutgp120core/CD4M33 binding. No noticeable changes in ECL2S chemical shifts upon binding were observed, suggesting slow-exchange and no averaging of chemical shift between the bound and free peptide.
Fig. 3.
1H-15N-HSQC spectra of 15N-labeled ECL2S: free (A) and in the presence of a 1:1 molar ratio of mutgp120core/CD4M33 (B), mutgp120core/CD4M33(+V3) (C) and BSA (D), at pH 7, 50 mM NaCl, 15 °C.
The small remaining intensity observed after protein binding for residues exhibiting the largest decrease in cross peak intensities is probably due to free peptide molecules found in equilibrium with the bound peptide. For example, for a KD of 6 μM and gp120 and ECL2S concentration of 50 μM, the percentage of the free peptide is expected to be 29%. Reliable quantification of the bound and free peptide concentrations is not possible.
Residues that are outside the region of a peptide interacting with a large protein exhibit minor or no changes in chemical shift upon binding to the protein and their T2 relaxation times decreases gradually closer to the core epitope. These residues retain strong cross peak intensities even after binding to the protein.
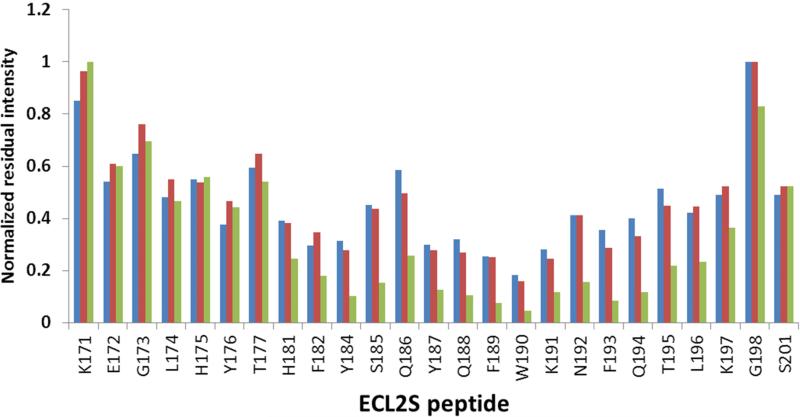
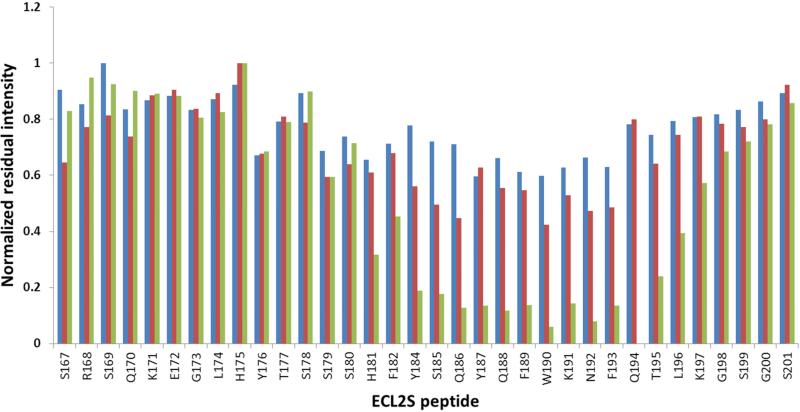
The diagram presented in Fig. 4 shows the residual cross peak intensity, normalized vs. the cross peaks with the smallest reduction in intensity upon gp120/CD4M33 binding. The HSQC measurements were carried out at 15 °C, because higher temperatures cause the disappearance of a large number of HSQC cross peaks due to increased solvent exchange of the amide protons (Fig. 3A). The cross peaks of S178-S180 could not be observed for the free peptide at pH 7 even at 15 °C and therefore the effect of mutgp120core/CD4M33 binding on these residues could not be assessed. Inspection of the residual intensities indicates that for mutgp120core/CD4M33 the largest reduction in intensity occurs for the H181-L196 segment (Fig. 4). Therefore, we conclude that this region of the peptide is interacting with the gp120/CD4M33 complex.
Fig. 4.
Dynamic-filtering mapping of the ECL2S determinant interacting with gp120 at neutral pH. Plot of the intensities of the 1H-15N HSQC cross peaks in the presence of a 1:1 molar ratio of the different proteins divided by the intensity of the corresponding cross peak of the free ECL2S peptide obtained at the same peptide concentration. Intensity ratios are shown for ECL2S cross peaks after titration with a 1:1 molar ratio of BSA (blue), mutgp120core(+V3)/CD4M33 (red) or mutgp120core/CD4M33 (green). The intensities ratios were normalized against the residue exhibiting the lowest reduction in intensity (G198 or K191) upon protein binding. The HSQC spectra were recorded at pH 7 and 15° C. The estimated error in these measurements is ± 15%.
Interestingly the amide cross peaks of residues S167 and G200 do not appear in the HSQC spectrum of the free peptide measured at pH 7 and 15 °C. However, these cross peaks do appear in the presence of mutgp120core, mutgp120core(+V3) and BSA. These two residues are part of the N- and C-terminal solubility tags. Their appearance in the presence of the proteins but not in the spectrum of the free peptide indicates some partial solvent protection in the presence of the proteins. S185 contributes two cross peaks, probably due to cis-trans isomerization of P183.
Addition of mutgp120core(+V3)/CD4M33 complex to the labeled ECL2S peptide resulted in smaller changes in cross peak intensities (Fig. 3C and red bars Fig. 4). The segment Y187-K191 underwent the most pronounced decrease in cross peak intensities, indicating that it interacts with mutgp120core(+V3)/CD4M33. The involvement of residues H181-Q186 and N192-L196 could not be determined with confidence, due to smaller changes in cross peak intensities in comparison with those observed for residues Y187-K191. The higher residual intensity of the peptide resonances in the segment interacting with mutgp120core(+V3)/CD4M33 compared to what was found in case of mutgp120core/CD4M33 binding suggests somewhat lower affinity of mutgp120core/CD4M33.
The measurements were repeated at pH 6, where even at 27 °C the vast majority of the free peptide residues could be observed (Fig. 5 A-D). These measurements enabled to define more precisely the ECL2S segment interacting with mutgp120core/CD4M33 (Fig. 6). While similar reduction in ECL2S cross peak intensities upon binding to mutgp120core/CD4M33 was observed at pH 6, considerably smaller decrease in cross peak intensities were observed upon mutgp120core(+V3)/CD4M33 to ECL2S at this in comparison with pH 7, suggesting weaker binding at the lower pH (compare Fig. 4 and Fig. 6).
Fig. 5.
1H-15N-HSQC spectra of 15N-labeled ECL2S: free (A) and in the presence of a 1:1 molar ratio of mutgp120core/CD4M33 (B), mutgp120core(+V3)/CD4M33 (C) and BSA (D), at pH 6, 50 mM NaCl, 27 °C.
Fig. 6.
Dynamic-filtering mapping of the ECL2S determinant interacting with gp120 at pH 6. Plot of the intensities of the 1H-15N HSQC cross peaks in the presence of a 1:1 molar ratio of the different proteins, divided by the intensity of the corresponding cross peak of the free ECL2S peptide measured at the same peptide concentration. Intensity ratios are shown for ECL2S cross peaks after titration with a 1:1 molar ratio of BSA (blue), mutgp120core(+V3)/CD4M33 (red) or mutgp120core/CD4M33 (green). The residual intensities were normalized against the residue exhibiting the lowest reduction in intensity (H175 or S169) upon protein binding. The HSQC spectra were recorded at pH 6 and 27°C. The estimated error in these measurements is ± 15%.
From the comparison of spectra of ECL2S in the presence of a 1:1 molar ratio of mutgp120core/CD4M33 or mutgp120core(+V3)/CD4M33 we conclude that the V3 loop does not contribute to gp120 binding to the ECL2S peptide and in fact actually decreases binding affinity especially below physiological pH. This is seemingly in contrast to biochemical data which suggested that ECL2 in the intact receptor is involved in V3 binding [7, 11-13].
ECL2S Aromatic residues directly interact with gp120
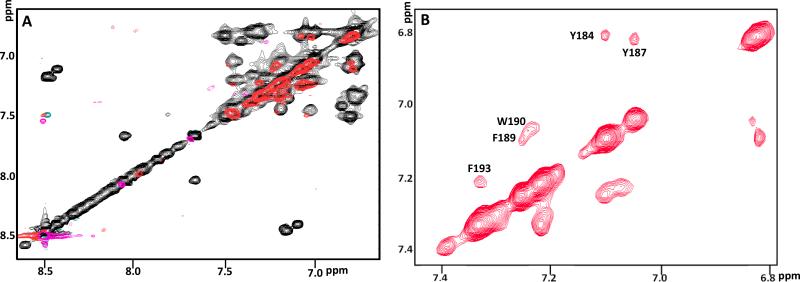
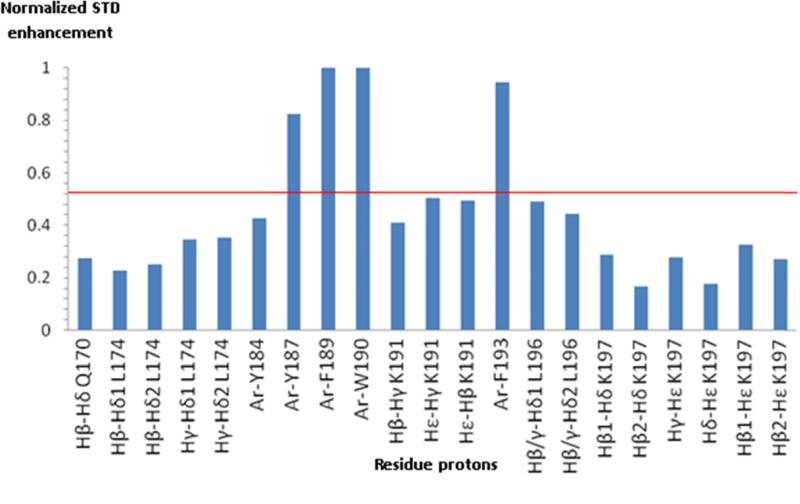
Saturation transfer difference (STD) technique [30] enables identification of ligand-residues that are interacting directly with a macromolecule. 2D-TOCSY-STD (Fig. 7 A and B and Fig. 8) of ECL2S in the presence of mutgp120core/CD4M33 recorded at pH 5.3 reveals that the aromatic amino acids Y187, F189, W190 and F193 in the H181-L196 segment in the ECL2S peptide exhibit significant effects. Protons of non-aromatic amino acids exhibit saturation transfer below the threshold level set in (Fig. 8). These results are in agreement with those obtained by Bewley and co-workers for a shorter ECL2 peptide (C178-K191) except for H181 and F182, which exhibited very weak saturation transfer in the previous studies by Bewley and co-workers [16]. This minor difference could be due to the fact that at the lower pH at which our STD measurements were carried out, histidine is positively charged, resulting in weaker interactions of residues surrounding H181.
Fig. 7.
2D-Saturation Transfer Difference (STD) of ECL2S in the presence of mutgp120core/CD4M33. Overlay of 2D TOCSY (black) and 2D-STD TOCSY (red) of the aromatic region (A). Expansion of 2D-STD TOCSY of this aromatic region (B). The experiments were recorded for a sample containing 30 μM mutgp120core/CD4M33 with 600 μM ECL2S (1:20 molar ratio), 50 mM deuterated acetate pH 5.3, 50 mM NaCl, 1 mM d-EDTA, 1:1000 protease inhibitors cocktail and 0.05% NaN3 at 27 °C.
Fig. 8.
STD enhancements of ECL2S in the presence of mutgp120core/CD4M33. 2D-STD TOCSY enhancements calculated by dividing the peak intensities in the 2D-STD TOCSY by the corresponding peak intensities in a TOCSY spectrum. Cross-peaks intensities were normalized relative to the cross peak that exhibited the largest enhancement. Sample of 30 μM mutgp120core/CD4M33 with 600 μM ECL2S (1:20 molar ratio), 50 mM deuterated acetate pH 5.3, 50 mM NaCl, 1 mM deuterated-EDTA, 1:1000 Protease inhibitors cocktail and 0.05% NaN3 measured at 27 °C.
ECL2S binding to Bovine serum albumin
As a control experiment, binding of ECL2s to Bovine Serum Albumin (BSA) was tested. BSA is a multi-transporter serum protein that has multiple binding sites and binds to a large number of hydrophobic molecules including tryptophan, hormones, fatty acids, degradation products such as bilirubin and a large number of drugs such as diazepam, ibuprofen and diclofenac. The affinities to these compounds vary over a wide range with KD values between 1 to 50 μM [31, 32].
The binding of BSA to the immobilized ECL2S peptide was too weak to be detected by SPR. This is reasonable considering that KD values higher than 50-100 μM are beyond the SPR detection limit. However, the HSQC titration experiment carried out at 50 μM concentration revealed that at pH 7 and 15 °C, BSA binds the ECL2s peptide with an affinity comparable to that of the mutgp120core(+V3)/CD4M33 complex (Figs. 3-6). Stronger binding to ECL2S, in comparison with BSA, was observed for mutgp120core/CD4M33 lacking V3. The binding of ECL2S to BSA, albeit weak, is not surprising considering that the peptide has a tryptophan and an adjacent phenylalanine residue that could present a hydrophobic surface for BSA binding. The weak ECL2S binding to BSA demonstrates that this peptide is capable of binding to other proteins due to the large hydrophobic surface presented by its amphiphilic helical structure (Fig. 2).
DISCUSSION
The C-terminal segment of ECL2S adopts a helical conformation that binds to gp120 via nonpolar interactions
In the present investigation we set out to further investigate the conformation of a CCR5 ECL2 peptide and its interactions with gp120. For that purpose we used the peptide ECL2S containing CCR5 residues R168-K197, corresponding to the entire putative ECL2. Similar peptides were found to be HIV-1 entry inhibitors [15]. Originally, residues N192-K197 were predicted to be part of ECL2 [15] but the recent CCR5 crystal structure showed that these residues correspond to the N-terminal residues of the 5th transmembrane helix of CCR5, TM5 [5]. Two of these residues, namely N192 and F193, increased the neutralization potency of ECL2 peptides [16]. Here we found that the segment Q188-Q194 of ECL2S folds into a helical structure, both at pH 4.8 and pH 7 (Fig. 2A). The structure of the peptide segment Q188-Q194 is in excellent agreement with that of the same region in the CCR5 crystal structure [5] with a backbone RMSD of 0.29 Å. The remaining residues of the ECL2S peptide are mostly unstructured. The ECL2S helical region is amphiphilic with a predominantly polar, uncharged surface opposing one that is predominantly apolar and aromatic (Fig. 2B). Thus, it is reasonable to conjecture that the interaction of the ECL2S peptide with gp120 is strengthened by this helical tendency and that the aromatic residues, previously found by STD measurements to interact with YU2gp120 [16], form a large continuous surface that can bind to a hydrophobic region of this envelope protein.
ECL2S binding to gp120 is due to non-specific hydrophobic interactions involving helical residues near the C-terminus of the peptide
The binding of ECL2S to two gp120 (R5) constructs was investigated in the present study. Both constructs lack the V1/V2 loop. Gp120core(+V3) contains the V3 loop while gp120core does not [26]. Using STD and dynamic filtering analyses, the interacting sequence of ECL2S with both gp120core(+V3) and gp120core was mapped to the CCR5 segment H181-L196 that contains the helix (Figs. 3-8). Within this segment, the aromatic residues Y187, F189, W190 and F193 directly interact with gp120, in good agreement with a previous study by Bewley and co-workers [16]. Strikingly, the inhibition results on ECL2 peptides showed that removal of W190-K191 from the peptide (#3 in [16]) resulted in nearly an order of magnitude decrease in HIV-1 inhibition, and removal of Q188FWK191 abolished the inhibitory activity. Addition of N192-F193 improved HIV-1 inhibition by 2-fold [16]. It is reasonable to conclude that the residues found in the present study to be part of the amphiphilic helix in ECL2S are critical for the previously observed antiviral activities.
To probe whether the interactions of ECL2S with gp120 represent those of this region in the intact CCR5 receptor, we examined the recently published crystal structure of CCR5 in complex with Maraviroc [5]. According to this structure, Y184 and Y187 point towards the CCR5 binding pocket while the side chains of F189, W190 and F193 point away from the binding pocket and most probably interact with the hydrophobic alkyl chain of the cell membrane phospholipids (Fig. 9) [5]. In the NMR structure of the ECL2S peptide (Fig. 2) the side chains of F189, W190 and F193 are located in a mostly hydrophobic surface in the amphiphilic helix formed by the C-terminal segment (Fig. 2B), and are at the center of the ECL2S segment that we found to interact with gp120 (Figs. 3 and 4). In view of the orientation of the side chains of F189, W190 and F193 in the CCR5 crystal structure, as shown in Fig. 9 A and B, we conclude that in the intact receptor F189, W190 and F193 cannot interact with gp120. However, in our ECL2S peptide, these three residues clearly interact with gp120 (Figs. 3-8). The fact that similar interactions were found with BSA (Fig. 3 and 4) suggests that the ECL2S peptide, which contains residues from TM5 at its carboxyl terminus, interacts with gp120/CD4M33 through non-specific hydrophobic interactions, which are not representative of the native interactions of ECL2 in the cellular CCR5 receptor with gp120.
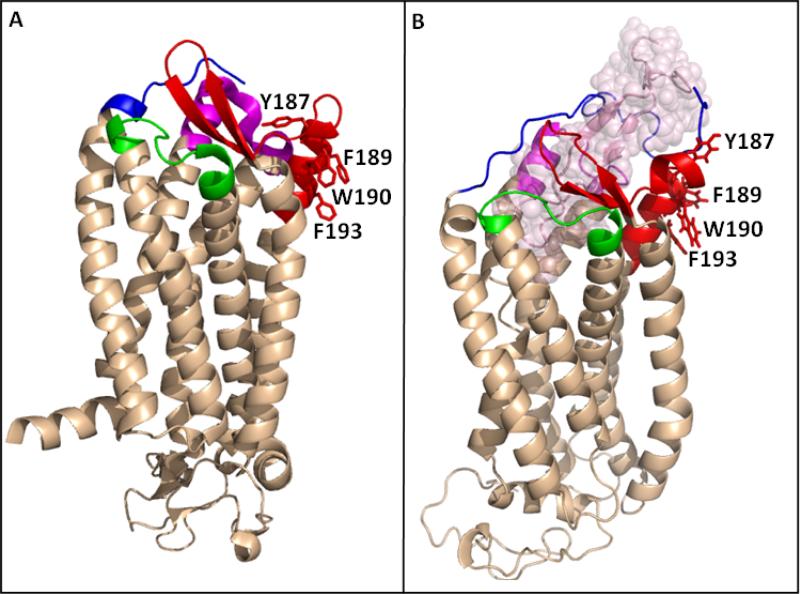
Fig. 9.
The orientation of the side chains of Y187, F189, W190 and F193 in the crystal structure of CCR5. (A) Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex (Tan, Q. et al. (2013) Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex, Science 341, 1387-1390.). The CCR5 N-terminus (residues 20-27) is colored in blue. ECL1 (90AAAQWDFGNTM101C) is presented in green, ECL2 region, residues R168-K197, is colored in red and ECL3 (260FQEFFGLNNCSSSNRL276D) is presented in magenta. (B) A model of CCR5 in complex with gp120 V3 region (Tamamis & Floudas (2014) Molecular Recognition of CCR5 by an HIV-1 gp120 V3 Loop, PlosOne 9, e95767). Color code as in (A), with the V3 region presented in light-pink spheres. F189, W190 and F193 are presented in sticks in (A) and (B) revealing that in the crystal structure these hydrophobic amino acids point away from the receptor binding pocket and towards the putative location of the cell membrane.
The presence of V3 is detrimental to the binding of elongated ECL2 peptides to gp120
Unexpectedly, in the present study we found that the V3 region is not necessary for the interaction of mutgp120core(+V3) and mutgp120core with the ECL2S peptide, and was even detrimental to binding at acidic pH (below pH 7). This conclusion is seemingly in contrast to previous biochemical studies, which suggested that the V3 crown interacts with ECL2 in the intact receptor [4], and to models of V3-CCR5 interactions based on the crystal structure [5, 14]. According to the HSQC-titration measurements, the binding of mutgp120core(+V3) to ECL2S was comparable to BSA binding to ECL2S while the binding of mutgp120core to the peptide was much stronger as judged by the dynamic filtering experiments (Figs. 3 and 4). We suggest that the absence of a V3 contribution to the binding of our elongated ECL2 peptide is consistent with the report that interaction of ECL2 peptide R178-K191 with HIV-1 is CD4 independent [16]. Interaction of gp120 with CD4 is understood to trigger a conformational change that uncovers the V3 loop for further interaction with its co-receptors. Therefore, the CD4 independence reported for R178-K191 in a standard neutralization assay with HIV-1 YU2 suggests this peptide does not interact with V3. This would support our conjecture above that elongated ECL2 peptide surrogates interact primarily through non-polar contacts with the gp120 core.
How V3-independent ECL2S-gp120 interactions can be explained
Peptide surrogates have been widely used to understand the interaction of regions of proteins with receptors and other protein partners. We have published extensively on the use of peptides to provide information on the structure of antigenic regions in proteins. In particular, we used V3 peptides and HIV-1 neutralizing antibodies to determine that this loop assumed a β-hairpin structure [33-35]. This was later confirmed by X-ray studies [8, 36, 37]. Despite the successes in the literature, caution should be applied when results with peptides are applied to these same sequences in the intact protein.
In the present investigation we have concluded that the elongated ECL2 peptides examined by us and others [15, 16], likely interact with a different region of gp120 than does the ECL2 loop in the intact receptor. Models of the V3 in complex with CCR5 and CXCR4 (Fig. 9B) [14] show that the primary interactions involve polar or charged residues located in the CCR5 TM5 helix (Y187, Q188 and K191), in the V3 β-hairpin (S306, N308) and to a lesser degree in the β-hairpin region of the ECL2 loop [5, 14]. Almost no interactions involving the hydrophobic residues in the ECL2 peptides (CSSHFPYSQFWKNFQTLK) were found in these models [5], in contrast to the ECL2S interaction with gp120.
The solvent exposure of hydrophobic aromatic TM5 residues in the ECL2S peptide, in contrast to the protection of these same TM5 residues in CCR5 by burial in the cell membrane, provides an entropic driving force leading to non-specific interactions of the ECL2S peptide with hydrophobic surfaces on gp120. These hydrophobic interactions, which are responsible for an affinity in the 10 μM range to gp120, compete with and appear to prevent the more specific interactions found in CCR5-V3 models for the ECL2 loop segment preceding the TM5 residues [5, 14].
It is known that the V3 region is highly positively charged and therefore is expected to repel the positively charged ECL2S peptide. This electrostatic repulsion would be increased by the presence of the positively charged gp120 fourth constant region, C4, an important component of the co-receptor binding site. Such electrostatic repulsion would explain why mutgp120core(+V3) binds much more weakly at pH lower than 6 while mutgp120core lacking the V3 is considerably less pH dependent. By comparison, the native gp120 contains the V1/V2 loops and sialic acid moieties in its glycosyl groups that add negative charges in native gp120. The negative charges contributed by V1/V2 and by the sialic acid may neutralize the positive regions in gp120, such as the V3 and the C4, and thereby weaken the repulsion between the ECL2S peptide and the native envelope spike on virions. This may enable the ECL2S peptide to interact more strongly with the HIV-1 envelope spike and to serve as an HIV-1 entry inhibitor due to the hydrophobic interactions with the native virion.
Implications for R5 and X4 HIV-1 inhibition by ECL peptides
ECL1 (90AAAQWDFGNTMC101) and ECL3 (260FQEFFGLNNCSSSNRLD276) peptides were found to inhibit HIV with a potency comparable to that of the ECL2 peptide [15]. While PL2 (ECL2 peptide) was found to be specific for R5 strains, PL1 (ECL1 peptide) and PL3 (ECL3 peptide) neutralized also the R5/X4 strain 89.6. None of these peptides neutralized the X4 strain LAV[15]. According to the CCR5 crystal structure, ECL1 and ECL3 are outside the receptor surfaces predicted to be involved in co-receptor binding. The ECL1 and ECL3 peptides contain segments of TM2 and TM3 in case of PL1 and TM7 in case of PL3 (residues underlined above) and the loops connecting the TMs contain hydrophobic residues (residues in italic). Thus, the free peptides have a high hydrophobic content that could be involved in non-specific interactions with gp120. These hydrophobic residues are buried or partially buried in the structure of CCR5 due to interactions with other receptor regions. We suggest that similarly to the extended ECL2 peptides, the extended ECL1 and ECL3 peptides may be important for HIV-1 inhibition but would carry out this inhibition through interactions that are different from those of these same regions in intact CCR5 with gp120.
Scrutiny of the literature also shows that peptides corresponding to ECL1 (99VANWYFGNFLSKA111) and ECL2 (175ANVSEADDRYISDRFYPNDLWVVVFQFQH203) of CXCR4 were found to inhibit infection by X4 HIV viruses [18]. The ECL1 peptide used in the Hashimoto study [18] contained segments of TM2 and TM3 while ECL2 peptide contained a long TM5 hydrophobic segment (underlined in the given sequences). Similarly to the CCR5 ECL peptides, these two CXCR4 peptides could inhibit X4 HIV-1 strains due to non-specific hydrophobic interactions of the ECL peptides with gp120 and the envelope spike. Based on this conclusion we suggest that peptides containing sequences from CCR5 and CXCR4 loops overlapping the contiguous transmembrane regions may prevent viral infection by binding to hydrophobic patches on gp120 from both R5 and X4 viruses.
Conclusions
In summary, we have solved the solution structure for a peptide corresponding to the entire ECL2 loop and part of TM5 of CCR5. This peptide interacts with gp120 through a primarily hydrophobic surface that is presented on the amphiphilic helix at the carboxyl end. The binding of the ECL2 peptides with gp120 is different from the native binding of the ECL2 loop of CCR5 with the HIV-1 envelope glycoprotein. Specifically, the ECL2 peptide-gp120 interaction is judged by NMR not to involve the V3 region. Comparisons with previous results and crystal structures help to explain the reason CCR5 ECL2 peptides inhibit HIV-1 entry across X4 and R5 strains and in a CD4 independent manner. The finding of a hydrophobic exposed “hot spot” on HIV-1 may prove useful in drug and vaccine design. The low efficacy and the specificity of the ECL2 peptide could be improved by linkage to a moiety targeting other regions of gp120 such as the binding surface for the N-terminal segment of CCR5 which is involved in the first step of CCR5 binding to gp120 [38, 39].
Experimental procedures
Peptide chemical synthesis
The 30-residue ECL2 peptide R168-K197 with a C178A mutation (R168SQKEGLHYTASSHFPYSQYQFWKNFQTLK197) and a biotinylated ECL2S used in the SPR studies (Biotin-GSGS-R168SQKEGLHYTSSSHFPYSQYQFWKNFQTLK197GSGS) were chemically synthesized by automated solid phase peptide synthesis using Fmoc protection, and HBTU/HOBt activation. Biotin was attached directly using a single coupling cycle. Both peptides were cleaved from the resin using a mixture of 95% trifluoroacetic acid (TFA), 2.5% water and 2.5% triisopropilsilane at room temperature for 2 hours. For the ECL2 peptide the cleavage yield was 89% and the target peptide content in the crude material was estimated to be 30% by HPLC. ECL2 was purified on a Waters DeltaPak C-18 column (19 × 300 mm) using a 25 to 50% gradient of acetonitrile (+0.1% TFA)/water (+0.1% TFA) per 90 min with flow rate 5 mL/min at room temperature. Peptide with 96% purity and MW 3707.83 (Calculated 3708.14) was obtained. Yield of pure ECL2 peptide after preparative HPLC purification was 23% relative to crude peptide weight. The longer biotinylated-ECL2S peptide, with 95% purity, MW 4526.40 (Calculated 4526.97) and 40% yield relative to crude weight, was obtained similarly.
Expression and purification of recombinant ECL2S
The CCR5 ECL2S peptide (R168-K197) with a C178S mutation, including N- and C-terminal 4 amino-acid long solubility tags (Gly-Ser-Gly-Ser), was expressed in E. coli as a TrpΔLE fusion protein [40]. Expression was induced by 1 mM isopropyl β-D-thiogalactopyranoside (IPTG, Fermentas Ontario, Canada), followed by incubation for 18 hours at 37 °C. The plasmid coding for the TrpΔLE-ECL2S fusion protein was transformed into BL21 (DE3) pLysS (Novagen) cells which were subsequently grown in LB medium +100μg/ml Kan. For uniform labeling the cells were grown in M9 minimal media containing 15NH4Cl for 15N labeling or 13C glucose and 15NH4Cl for 15N and 13C labeling. The cells were induced at A595 = 0.7 by adding IPTG to 1 mM. After overnight growth at 37 °C the cells were harvested by centrifugation. The cell pellet was resuspended in 20 ml Lysis Buffer (100 mM Tris 8, 5 mM EDTA, and 5 mM DTT) + 0.05% Triton X-100. The suspended cells were passed through a French Press, two times and the resulting mixture was sonicated six times using a 10 sec pulse followed by a 30 sec pulse off interval and then centrifuged at 20,000g for 40 minutes to isolate inclusion bodies.
Dissolving buffer (6 M Guanidine HCl, 10 mM Imidazole, 50 mM Na2 HPO4, pH 7.4) was added to the isolated inclusion bodies and the resulting mixture was incubated overnight at room temperature and centrifuged for 30 minutes at 13,000 rpm.
The supernatant was removed and incubated with Ni-NTA beads for 30 minutes. The beads were loaded on a column, washed with at least 200 mL of dissolving buffer and eluted with elution buffer (6 M Guanidine HCl, 0.8 M imidazole, 0.5 M NaCl, 50 mM Buffer acetate pH 5.9). The eluted protein was dialyzed twice against 5 L of H2O +50 mM EDTA with 3500 Da SnakeSkin and the protein was lyophilized. Generally, ~150 mg of lyophilized crude protein were recovered per liter of growth culture.
The crude ECL2S fusion protein was dissolved in 70% TFA to a concentration of 10 mg/ml. 3M CNBr was prepared in 70% TFA and added at time 0 in an amount to give a 400:1 molar ratio to the fusion protein and again after 4 h for a total 8 h cleavage reaction. Rotary evaporation was used to remove most of the TFA, after which the peptide was dissolved in an organic/aqueous mixture (~85% water: 15% acetonitrile : 0.1% TFA) and lyophilized. The peptide was purified by high pressure liquid chromatography (HPLC) (Thermo Separation Products, NJ, USA) using a C4 reverse-phase preparative column (Grace Vydac). With the following eluents (buffer A: 99.9% H20, 0.1% TFA; buffer B: 75% acetonitrile, 24.9% H20, 0.1% TFA) and a linear gradient form 21% B to 29% B in 50 min. 10-15 mg of purified ECL2S was recovered per 1L of culture. Molecular weight was verified by electrospray ionization (ESI) mass spectrometry. The final peptide was >95% homogeneous by analytical HPLC.
gp120 constructs used in the study
A truncated 88-492gp120JR-FL lacking the V1 and V2 variable loops was expressed in human embryonic kidney (HEK) 293 cells. To increase the affinity of the modified gp120JR-FL molecule to CD4, two glycosylation sites were mutated and removed, N301Q and T388A (88-492gp120ΔV1/V2(N301Q;T388A)) [41]. Additionally, 4 residues in the first α- helix (N99, E106, D107 and D113) were mutated to glutamine to create non-aggregating gp120, termed mutgp120core(+V3) [26]. A second construct, mutgp120core, in which residues R298-A330 of the V3 loop of mutgp120core(+V3) were replaced by a GAG sequence, was also used. mutgp120core(+V3) and mutgp120core were expressed in HEK293 cells lacking N-acetyl glucosaminyl transferase I enzymatic activity (GnTI- HEK293S cells [9, 28]). Proteins expressed in cells lacking this enzyme are homogenously glycosylated with Man5GlcNAc2 glycans at sites normally occupied by complex or hybrid glycans. The carbohydrate chains were not removed in the gp120 molecules used in the present study. Expression and purification of the gp120 molecules was done as described previously [26].
Surface Plasmon Resonance (SPR) studies of ECL2S
Dissociation constants (KD) were determined by SPR using a ProteOn XPR36 Protein Interaction Array System (Bio-Rad, Haifa, Israel). All experiments were carried out at 25 °C using SPR buffer: 20 mM Tris-HCl, 0.05% NaN3, 0.005% Tween, 15 mM MgCl2 with 150 mM NaCl. Binding was measured at different pH values of 6.0, 6.5 and 7.0. Biotinylated long-ECL2S was immobilized on modified alginate polymer layer pre-coated with NeutrAvidin® (ProteOn NLC sensor chip). The immobilization level for ligand peptides was within the following ranges of RU (response units; 1 RU = change of surface concentration ~1 pg of peptide/mm2): 240 RU up to 1000 RU. Two blank sensor surfaces, i.e., without an immobilized peptide, served as a ‘control’ and as a reference for the binding interaction. Binding was then measured using serial dilutions of glycosylated gp120 both complexed with CD4M33 or by itself in SPR buffer (concentration ranging from few hundreds of nM to few μM,), which were simultaneously injected (analyte) over the peptide-immobilized (ligand) surface at a flow rate of 70 μl/min. The surface regeneration step consisted of an 18 sec pulse of 10 mM NaOH. Regeneration steps were followed by two running buffer washes. Sensograms were analyzed with the ProteOn Manager software after subtraction of a blank channel (“inter-spots”). Analysis type was either ‘equilibrium’ or ‘kinetic’ Langmuir 1:1 model.
NMR sample preparation and measurements
NMR samples of ECL2S were dissolved in 350 μL of a 90% H2O/10% D2O mixture, containing 150 mM NaCl, 0.05% NaN3, 1 mM D16-EDTA, 10 mM D18-HEPES buffer. NMR spectra were recorded at different pH and temperatures. Free peptide sample contained 130 μM ECL2S. Sample for studying the interaction of ECL2S with the envelope glycoprotein contained 140 μM of ECL2S:gp120:CD4M33 in 1:1:1 concentration ratio. For dynamic filtering experiments, 1H-15N-HSQC spectra of ECL2S were recorded in the presence or absence of gp120/CD4M33 complex at 1:1:1 molar ratio. Experiments were carried out at 50 mM sodium acetate pH 6, 50 mM NaCl, 5% D2O, 0.05% NaN3 with 38-48 μM ECL2S and measured at 27°C or at 50 mM d-Tris HCl pH 7, 50 mM NaCl, 5% D2O, 0.05% NaN3 with 50 μM ECL2S measured at 15 °C.
All spectra were recorded using a Bruker AVIII-800 spectrometer equipped with a 5 mm inverse triple-resonance TCI CryoProbe with z gradients, and a Bruker DMX-500 spectrometer equipped with a 5 mm z gradient inverse triple-resonance TXI CryoProbe. 3D 15N-separated HSQC-TOCSY and 3D 15N-separated HSQC-NOESY spectra were measured on [15N]-ECL2S using mixing times of 90 ms and 150 ms, respectively, for sequential and side chain assignment, at 4 °C, pH 4.8. For completing the assignment, HNCA, HN(CO)CA,HNCACB and CBCA(CO)NH spectra were measured on a sample of [13C,15N]-ECL2S. The data were processed and analyzed using nmrDraw/nmrPipe [42], Bruker’s TopSpin and nmrView [43].
Structure determination
The structure of ECL2S was calculated using XPLOR-NIH [44] in the simulated-annealing protocol. The calculation started from an extended strand conformation. The distance-constraints were obtained from NOESY experiments and the dihedral angles constraints were generated by TALOS-N according to the Hα, and Cα and Cβ chemical shifts input or determined using the 3JHNHα coupling constants During the refinement process, hydrogen bond constraints were gradually introduced into the calculation if a hydrogen bond was present in 50% or more of the calculated structures as analyzed by MOLMOL [45]. The secondary structure elements and RMSD values were calculated with the MOLMOL program. Validation of the structures was carried on the 10 lowest-energy averaged structure, after carrying out a minimization step with Procheck-NMR[46].
2D Saturation transfer difference
2D-TOCSY-STD was acquired on the AVIII800 spectrometer using irradiation power of 1.2 kHz for 2 seconds at −0.65 ppm (or at 30 ppm for the control experiment). The sample contained 30 μM mutgp120core/CD4M33 with a 20-fold excess of the ECL2S peptide. The experiment was carried out at 27 °C, pH 5.3.
Acknowledgement
‡ This study was supported by Minerva and Israel Science Foundation grants, by NIH Grants GM53329 (J.A.) and GM22087 (F.N.) and by the Kimmelman center. J.A. is the Dr. Joseph and Ruth Owades Professor of Chemistry.
Abbreviations
- AIDS
acquired immune deficiency syndrome
- CCR5
human C-C chemokine receptor 5
- CD4
cluster of differentiation 4
- CD4M33
a peptide mimic of CD4 used in this study
- dNN(i,i+x)
distance connectivities between HN of residues at position i and i+X along the protein sequence
- dαN(i,i+x)
distance connectivities between Hα of residue at position i and HN of residue at position i+x along the protein sequence
- ECL2S
peptide constituting residues R168-K197 of the extracellular loop 2 of human CCR5 receptor with a C178S mutation and flanked N- and C-terminally by a GSGS solubility tag
- 3JHNHα
the three-bond coupling-constant between the amide and the α proton of the same amino acid
- EDTA
ethylene diamine tetraacetic acid
- gp120
the extra cellular subunit of the HIV-1 envelope protein
- GSGS
Gly-Ser-Gly-Ser tag
- HIV-1
human immuno-deficiency virus type 1
- HPLC
high pressure liquid chromatography
- HSQC
hetero-nuclear single quantum coherence
- mutgp120core
a gp120 core molecule lacking V1, V2 and V3 containing four mutations that alleviated the aggregation problem
- NMR
nuclear magnetic resonance
- NOE
Nuclear Overhauser effect
- Nt-CCR5
N-terminal part of CCR5 receptor (from M1 to R30)
- STD NMR
Saturation Transfer difference NMR
- SPR
surface plasmon resonance
- V3
variable loop number 3 of HIV-1 gp120
Footnotes
Author Contributions
MA, AM, OT and NK helped to design the experiments, prepared reagents, carried out the experiments and analyzed the results. BA prepared essential reagents. TS helped supervising the experiments, analyze the results and writing the manuscript. FN and JA discussed the results and wrote the manuscript. JA designed the experiments and helped in the analysis.
References
- 1.Berson JF, Doms RW. Structure-function studies of the HIV-1 coreceptors. Semin Immunol. 1998;10:237–48. doi: 10.1006/smim.1998.0130. [DOI] [PubMed] [Google Scholar]
- 2.Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, Sodroski J. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–4. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cormier EG, Tran DN, Yukhayeva L, Olson WC, Dragic T. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J Virol. 2001;75:5541–9. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cormier EG, Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76:8953–7. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, Li T, Ma L, Fenalti G, Zhang W, Xie X, Yang H, Jiang H, Cherezov V, Liu H, Stevens RC, Zhao Q, Wu B. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–90. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–71. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Perez J, Rueda P, Alcami J, Rognan D, Arenzana-Seisdedos F, Lagane B, Kellenberger E. Allosteric model of maraviroc binding to CC chemokine receptor 5 (CCR5). J Biol Chem. 2011;286:33409–21. doi: 10.1074/jbc.M111.279596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–4. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnur E, Noah E, Ayzenshtat I, Sargsyan H, Inui T, Ding FX, Arshava B, Sagi Y, Kessler N, Levy R, Scherf T, Naider F, Anglister J. The conformation and orientation of a 27-residue CCR5 peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide. J Mol Biol. 2011;410:778–97. doi: 10.1016/j.jmb.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding [see comments]. Science. 1998;280:1949–53. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 11.Doranz BJ, Lu ZH, Rucker J, Zhang TY, Sharron M, Cen YH, Wang ZX, Guo HH, Du JG, Accavitti MA, Doms RW, Peiper SC. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–14. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson DA, Cormier EG, Dragic T. CCR5 and CXCR4 usage by non-clade B human immunodeficiency virus type 1 primary isolates. J Virol. 2002;76:3059–64. doi: 10.1128/JVI.76.6.3059-3064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross TM, Bieniasz PD, Cullen BR. Multiple residues contribute to the inability of murine CCR-5 to function as a coreceptor for macrophage-tropic human immunodeficiency virus type 1 isolates. J Virol. 1998;72:1918–24. doi: 10.1128/jvi.72.3.1918-1924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamamis P, Floudas CA. Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop. PLoS One. 2014;9:e95767. doi: 10.1371/journal.pone.0095767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agrawal L, VanHorn-Ali Z, Berger EA, Alkhatib G. Specific inhibition of HIV-1 coreceptor activity by synthetic peptides corresponding to the predicted extracellular loops of CCR5. Blood. 2004;103:1211–7. doi: 10.1182/blood-2003-08-2669. [DOI] [PubMed] [Google Scholar]
- 16.Dogo-Isonagie C, Lam S, Gustchina E, Acharya P, Yang Y, Shahzad-ul-Hussan S, Clore GM, Kwong PD, Bewley CA. Peptides from second extracellular loop of C-C chemokine receptor type 5 (CCR5) inhibit diverse strains of HIV-1. J Biol Chem. 2012;287:15076–86. doi: 10.1074/jbc.M111.332361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chevigne A, Fievez V, Szpakowska M, Fischer A, Counson M, Plesseria JM, Schmit JC, Deroo S. Neutralising properties of peptides derived from CXCR4 extracellular loops towards CXCL12 binding and HIV-1 infection. Biochim Biophys Acta. 2014;1843:1031–41. doi: 10.1016/j.bbamcr.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto C, Nomura W, Narumi T, Fujino M, Tsutsumi H, Haseyama M, Yamamoto N, Murakami T, Tamamura H. Anti-HIV-1 Peptide Derivatives Based on the HIV-1 Co-receptor CXCR4. ChemMedChem. 2013 doi: 10.1002/cmdc.201300289. [DOI] [PubMed] [Google Scholar]
- 19.Bax A, Grzesiek S. Methodological advances in protein NMR. Acc Chem Res. 1993;26 [Google Scholar]
- 20.Spera S, Bax A. Empirical Correlation Between Protein Backbone Conformation and C-Alpha and C-Beta C-13 Nuclear-Magnetic-Resonance Chemical- Shifts. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- 21.Wishart DS, Sykes BD. Chemical shifts as a tool for structure determination. Methods Enzymol. 1994;239:363–92. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 22.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–23. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollastri G, McLysaght A. Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics. 2005;21:1719–20. doi: 10.1093/bioinformatics/bti203. [DOI] [PubMed] [Google Scholar]
- 26.Moseri A, Schnur E, Noah E, Zherdev Y, Kessler N, Singhal Sinha E, Abayev M, Naider F, Scherf T, Anglister J. NMR observation of HIV-1 gp120 conformational flexibility resulting from V3 truncation. FEBS J. 2014;281:3019–31. doi: 10.1111/febs.12839. [DOI] [PubMed] [Google Scholar]
- 27.Martin L, Stricher F, Misse D, Sironi F, Pugniere M, Barthe P, Prado-Gotor R, Freulon I, Magne X, Roumestand C, Menez A, Lusso P, Veas F, Vita C. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat Biotechnol. 2003;21:71–6. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- 28.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–24. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zvi A, Kustanovich I, Feigelson D, Levy R, Eisenstein M, Matsushita S, Richalet Secordel P, Regenmortel MH, Anglister J. NMR mapping of the antigenic determinant recognized by an anti-gp120. human immunodeficiency virus neutralizing antibody. Eur J Biochem. 1995;229:178–87. doi: 10.1111/j.1432-1033.1995.tb20453.x. [DOI] [PubMed] [Google Scholar]
- 30.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem Int Ed Engl. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 31.Kragh-Hansen U, Chuang VT, Otagiri M. Practical aspects of the ligand-binding and enzymatic properties of human serum albumin. Biol Pharm Bull. 2002;25:695–704. doi: 10.1248/bpb.25.695. [DOI] [PubMed] [Google Scholar]
- 32.Curry S. Lessons from the crystallographic analysis of small molecule binding to human serum albumin. Drug Metab Pharmacokinet. 2009;24:342–57. doi: 10.2133/dmpk.24.342. [DOI] [PubMed] [Google Scholar]
- 33.Tugarinov V, Zvi A, Levy R, Anglister J. A cis proline turn linking two beta-hairpin strands in the solution structure of an antibody-bound HIV-1IIIB V3 peptide. Nat Struct Biol. 1999;6:331–5. doi: 10.1038/7567. [DOI] [PubMed] [Google Scholar]
- 34.Sharon M, Kessler N, Levy R, Zolla-Pazner S, Gorlach M, Anglister J. Alternative Conformations of HIV-1 V3 Loops Mimic beta Hairpins in Chemokines, Suggesting a Mechanism for Coreceptor Selectivity. Structure (Camb) 2003;11:225–36. doi: 10.1016/s0969-2126(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 35.Rosen O, Sharon M, Quadt-Akabayov SR, Anglister J. Molecular switch for alternative conformations of the HIV-1 V3 region: implications for phenotype conversion. Proc Natl Acad Sci U S A. 2006;103:13950–5. doi: 10.1073/pnas.0606312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal Structure of a Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013;342:1477–83. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinlan BD, Joshi VR, Gardner MR, Ebrahimi KH, Farzan M. A double-mimetic peptide efficiently neutralizes HIV-1 by bridging the CD4- and coreceptor-binding sites of gp120. J Virol. 2014;88:3353–8. doi: 10.1128/JVI.03800-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acharya P, Dogo-Isonagie C, Lalonde JM, Lam SN, Leslie GJ, Louder MK, Frye LL, Debnath AK, Greenwood JR, Luongo TS, Martin L, Watts KS, Hoxie JA, Mascola JR, Bewley CA, Kwong PD. Structure-Based Identification and Neutralization Mechanism of Tyrosine Sulfate Mimetics That Inhibit HIV-1 Entry. ACS Chem Biol. 2011;6:1069–77. doi: 10.1021/cb200068b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biron Z, Khare S, Quadt SR, Hayek Y, Naider F, Anglister J. The 2F5 epitope is helical in the HIV-1 entry inhibitor T-20. Biochemistry. 2005;44:13602–11. doi: 10.1021/bi0509245. [DOI] [PubMed] [Google Scholar]
- 41.Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313:387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 42.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based On Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 44.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 45.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]