Abstract
Introduction
Nuclear RNA foci are molecular hallmarks of myotonic dystrophy type 1 (DM1). However, no designated study has investigated their formation and changes in proliferating cells. Proliferating cells, as stem cells, consist of an important cellular pool in the human body. The revelation of foci changes in these cells might shed light on the effects of the mutation on these specific cells and tissues. In this study, we used human DM1 iPS-cell derived neural stem cells (NSCs) as cellular models to investigate the formation and dynamic changes of RNA foci in proliferating cells.
Materials and Methods
Human DM1 NSCs derived from human DM1 iPS cells were cultured under proliferation conditions and non-proliferation conditions following Mitomycin C treatment. The dynamic changes of foci during the cell cycle were investigated by fluorescence in situ hybridization (FISH).
Results
We found RNA foci formed and dissociated during the cell cycle. Nuclear RNA foci were most prominent in number and size just prior to entering mitosis (early prophase). During mitosis, most foci disappeared. After entering interphase, RNA foci accumulated again in the nuclei. After stopping cell dividing by treatment of Mitomycin C, the number of nuclear RNA foci increased significantly.
Conclusion
DM1 NSC nuclear RNA foci undergo dynamic changes during cell cycle and mitosis is a mechanism to decrease foci load in the nuclei, which may explain why dividing cells are less affected by the mutation. The dynamic changes need to be considered when using foci as a marker to monitor the effects of therapeutic drugs.
Keywords: Muscular Dystrophy, Foci, Molecular Marker, Neural Stem Cell
Introduction
Myotonic dystrophy type 1 (DM1) is a multisystemic disorder and is characterized by progressive weakness and muscle wasting, myotonia, cardiac arrhythmias, central nervous system (CNS) involvement, cataracts and endocrinopathies (Bassez et al., 2004; Groh et al., 2008; Harper, 2001a, b). DM1 is caused by an unstable CTG repeat expansion (>50 repeats) in the 3’-untranslated region of the DMPK gene (Fu et al., 1992). As a result of the expansion, RNA transcripts containing CUG expanded (CUGexp) repeats are retained in the nucleus as a hairpin structure (Jasinska et al., 2003; Kino et al., 2004; Tian et al., 2000) that accumulates as nuclear RNA foci (Jiang et al., 2004; Lin et al., 2006; Mahadevan et al., 2006; Mankodi et al., 2000; Orengo et al., 2008; Seznec et al., 2001; Taneja et al., 1995; Wang et al., 2007). In these foci, CUGexp RNA sequesters splicing factors, including MBNL 1 and 2, which results in splicing dysregulation of many target gene transcripts. Dysregulated splicing has been found to be responsible for specific phenotypes of DM1, including myotonia and insulin insufficiency (Ashizawa and Sarkar, 2011; de Leon and Cisneros, 2008; Lee and Cooper, 2009; Llamusi and Artero, 2008; Ranum and Cooper, 2006; Schara and Schoser, 2006). Thus, nuclear RNA foci containing expanded CUG repeats are the molecular hallmark of the RNA gain-of-function pathomechanism of DM1. Nuclear RNA foci have also been used as a marker for therapeutic approaches targeting the neutralization or degradation of nuclear RNA foci using ribozymes, antisense oligonucleotides (ASOs), peptides (Garcia-Lopez et al., 2011) and small molecule compounds (Childs-Disney et al., 2013; Ketley et al., 2014; Langlois et al., 2003; Lee et al., 2012; Mulders et al., 2009; Parkesh et al., 2012; Warf et al., 2009; Wheeler et al., 2012; Wheeler et al., 2009; Wong et al., 2014). However, no designated study has investigated their formation and changes in proliferating cells. Proliferating cells, as stem cells, consist of an important cellular pool in the human body. The revelation of foci changes in these cells might shed light on the effects of the mutation on these specific cells. We have recently established induced pluripotent stem (iPS) cells from DM1 patients and showed that these iPS cells can be differentiated into neural stem cells (NSCs). Nuclear RNA foci are readily detectable in DM1 pluripotent iPS cells and multipotent NSCs (Xia et al., 2013). In this study, we used human DM1 iPS-cell derived NSCs as cellular models to investigate the formation and dynamic changes of RNA foci in proliferating cells. We found that RNA foci in DM1 NSCs undergo dynamic changes during the cell cycles.
Materials and Methods
Reagent
Mitomycin C (M4287) and an anti-alpha-tubulin monoclonal antibody (T9025, clone DM1A) were purchased from Sigma-Aldrich (St. Louis, MO). The NeuroCultTM NS-A proliferation kit (#05751) is from STEMCELL Technologies (Vancouver, Canada).
Cells and culture
DM-03 NSCs were generated from the neural induction of human DM-03 iPS cells (Xia et al., 2013) and expanded in the NeuroCult™ NS-A proliferation medium. Normal control NSCs were derived from normal iPS cells which were generated from a 51-year-old male health donor. DM-03 FBs are from the primary culture of the same subject that was used for generation of iPS cells. FBs were cultured in DMEM with 20% FBS in 5% CO2.
Mitomycin C Treatment
NSCs were cultured under proliferating growth conditions in a 24 well plate in triplicate to densities of ~80%, and ~30% confluence. For the first three wells (~80% confluence), cells were treated with Mitomycin C (10 μg/ml) for 3 hours and then changed to normal growth medium and cultured for 8 days. For the other three wells (~30% confluence), cells were not treated with Mitomycin C and cultured for 8 days when they became 80% confluence. 3 × 103 cells were then transferred to each well of ibidiTreat u-Slides (ibidi GmbH, Am Klopferspitz 19, 85152 Martinsried, Germany) for fluorescent in situ hybridization (FISH) and immunofluorescence (IF) staining.
FISH, foci quantification and immunofluorescence staining of alpha-tubulin
FISH was performed as previously described (Xia et al., 2013). Cells were plated in ibidiTreat μ-slides, fixed in 10% buffered formalin phosphate and then dehydrated in pre-chilled 70% ethanol. The cells were then hybridized with Cy3-labeled (CAG)10 DNA probes (500 pg/μL) after pretreatment. After hybridization, cells were washed three times in pre-warmed 40% formamide/2X SCC buffer for 30 min at 37°C and once in PBS (pH 7.4) followed by IF staining as described (Xia et al., 2013). Cells were incubated overnight at 4°C with antibodies against alpha-tubulin (1:1000). On the following day, slides were washed and incubated with the secondary antibody conjugated with AlexaFluor 488 (Invitrogen) (1:500) for 30 min at room temperature. Finally, the slides were washed with PBS and mounted with Vectashield Mounting Medium containing DAPI. Photomicrographs were taken using Olympus IX81-DSU Spinning Disk confocal microscope. Confocal images were captured in a z-stack interval of 0.5μm increments using a 60X objective to quantify the foci within the cellular volume. Foci number and area in each cell were analyzed by ImageJ. Mitotic phases were identified by mitotic spindle and chromosome morphologies. Even when cells were growing at exponential condition, we still saw some cells that had very large nuclei, which could be terminally differentiated cells or entering senescence and we classified them as G0 cells. Or we saw fragmented nuclei, which we classified as dying cells.
Statistical analysis
Foci number or area during different phases of the cell cycle and in non-proliferating cells was compared by independent-samples t test. P values less than 0.05 are considered statistically significant.
Results
DM1 ribonucleic foci are dynamic during cell cycles
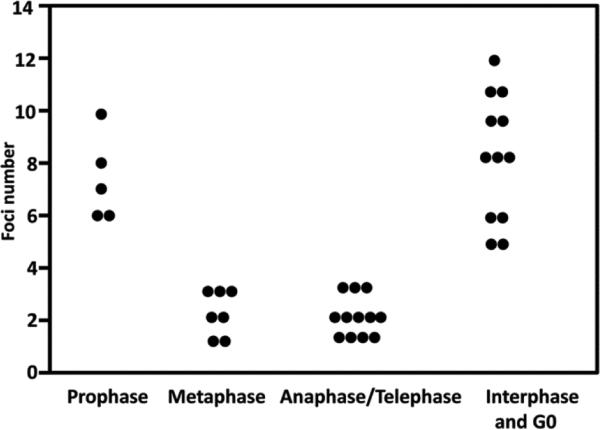
In proliferating DM1 iPS-cell derived NSCs, the number and size of RNA foci varied, consistent with those reported in the literature (Mankodi et al., 2003). Some cells contained more foci than others and cytoplasmic foci were occasionally seen (Figure 1). This variability could be attributed to differences in cell cycle stage with some cells committed to terminal neural differentiation while other cells were in different stages of mitosis. To test this hypothesis, cells were grouped according to cell cycle stage and examined for the number of nuclear RNA foci. As shown in Figure 2, we found RNA foci underwent dynamic changes during the cell cycle with foci most abundant just prior to entering mitosis (early prophase). During mitosis and with the breakdown of the nuclear envelope, the majority of RNA foci disappeared. Following cytokinesis, the remaining foci were randomly distributed to daughter cells and when the nuclear membrane reformed, foci were either enveloped by nuclear membrane or stayed in cytoplasm. Nuclear RNA foci further accumulated in interphase while cytoplasmic RNA foci eventually disappeared prior to entering prophase. In the G0 cells or the aged cells (identified by morphology), nuclear RNA foci accumulated and grew larger and cytoplasmic foci were not detectable in these cells. Significantly more foci were observed in prophase and G0 in contrast to metaphase-telophase (Figure 3). Occasionally, dying cells were seen with abundant nuclear RNA foci. No foci were seen in normal iPS-cell derived NSCs (Figure 4). These findings suggest mitosis is a mechanism to decrease foci load in the nuclei of proliferating cells.
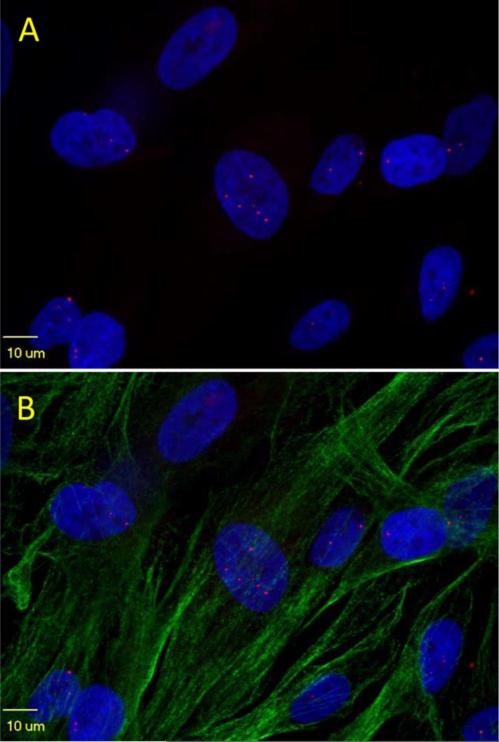
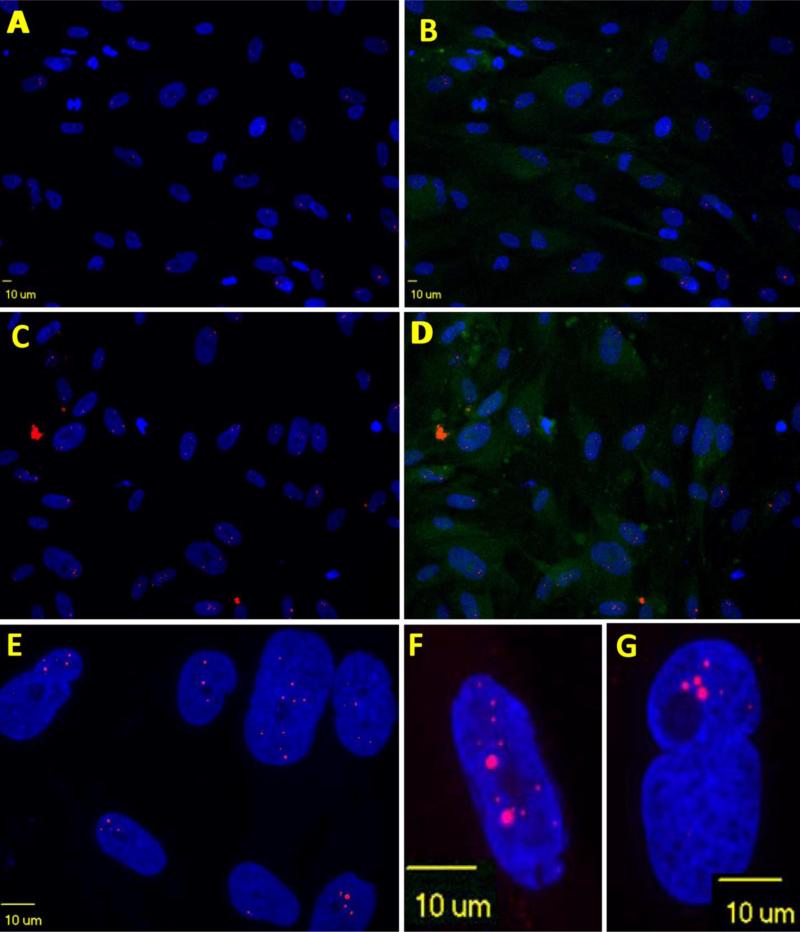
Fig. 1.
Nuclear RNA foci number varies among DM1-03 NSCs cultured in proliferating medium. Occasionally, cytoplasmic foci are seen. A. DAPI only. B. merged picture with microtubules (Alpha-tubulin) staining of cytoskeleton. Most of the cells in this field are in interphase (lacking architecture of spindle fibers of mitosis)
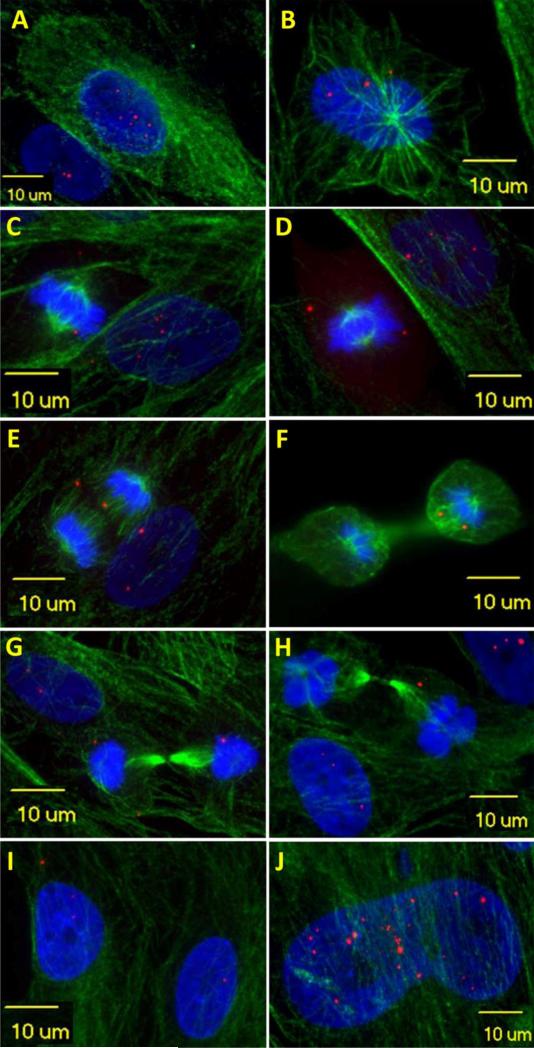
Fig. 2.
Nuclear RNA foci undergo dynamic changes during cell cycles. In this set of figures, cytoskeleton is stained green with alpha-tubulin to show the structure of mitosis. In early prophase (A), cells start rounding up. Nucleolus fades. Chromatin condenses to form chromosomes. Cells contain more nuclear RNA foci and no foci are seen in cytoplasm. In late prophase (or prometaphase) (B), spindle fibers form. Nuclear envelop starts breaking down. Foci are seen released from nucleus (appearing outside of DAPI area). In metaphase (C, D), chromosomes align with their centromeres on the equator across the center of the cell, spindle fibers attach to centromere. The nuclear envelop disappears. Foci vary in size and number but in general are dissipating. Through anaphase (E) and telophase (F), chromatids of each chromosome separate into daughter chromosomes and move to opposite poles. Remaining foci are randomly distributed between the two daughter cells. In cytokinesis (G, H), cell membrane pinches in around the middle of the cells. A contractile ring cleaves the cells into two daughter cells. Foci drop in number and size and remaining foci are randomly distributed to daughter cells at the end of this stage. After cytokinesis, microtubules reorganize into a new cytoskeleton for the return to interphase. A new nuclear envelop forms around each region of chromosomes. Foci can be trapped in the nucleus or left in the cytoplasm (I). If cells get out of cell cycle, foci will further accumulate and grow larger (J)
Fig. 3.
Scatter plot to show nuclear RNA foci number reaches the highest just before entering mitosis (prophase). During mitosis (metaphase, anaphase and telophase), nuclear RNA foci number drops. If cells get out of cell cycle (G0), nuclear RNA foci further accumulate
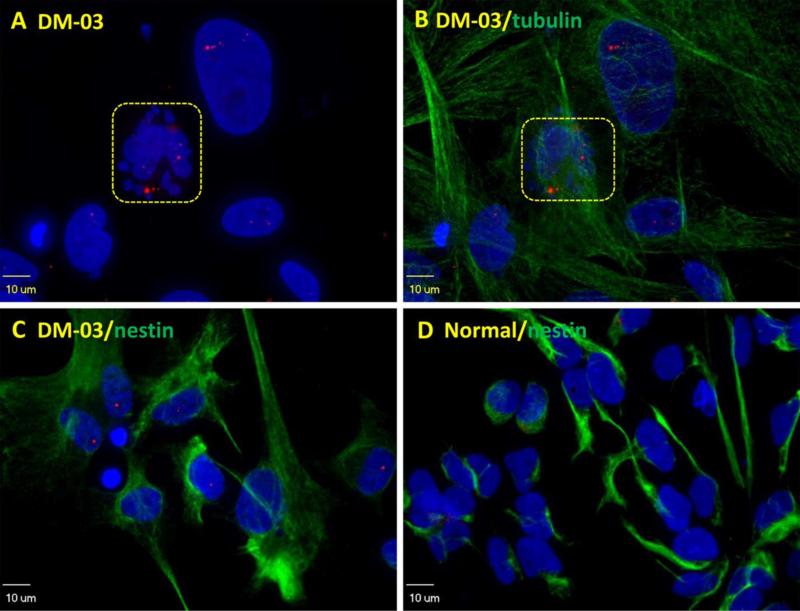
Fig. 4.
Dying cell (squared) is shown with dissembled nuclei and abundant nuclear RNA foci (A, B) in comparison to dividing DM1 NSCs identified by nestin staining (C). No foci are seen in normal iPS-cell derived NSCs (D).
Nuclear RNA foci progressively increase in number and become larger in size when cells get out of cell cycle and start aging
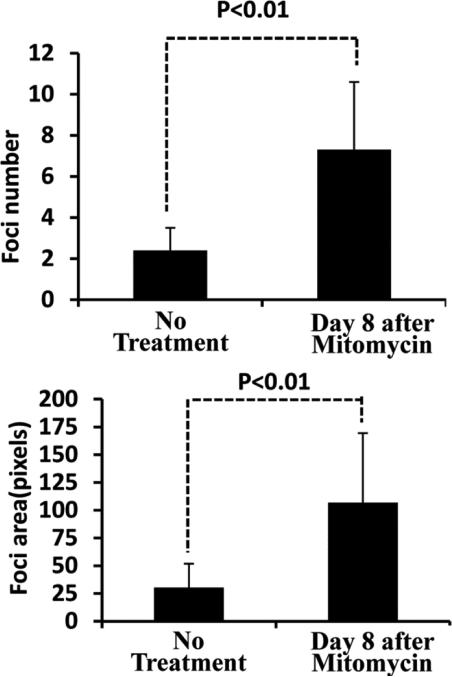
We next examined how foci changed when DM1 NSCs stopped dividing and started aging. After we stopped cell division with treatment of Mitomycin C, we found that nuclear RNA foci started accumulating in number, size and fluorescence intensity as time passed (Figure 5). Foci had significantly increased on day 8 after treatment with Mitomycin C (P<0.01) (Figure 6). The biggest foci could reach to 2 μm in diameter comparable to the largest foci found in autopsy brain neurons (Jiang et al., 2004). We never observed cytoplasmic foci on day 8 after Mitomycin C treatment.
Fig. 5.
Nuclear RNA foci start accumulating in number and size with time after they stopped dividing. A, B. DM1 NSCs cultured in proliferating condition. C-G. 8 days after Mitomycin C treatment. More and larger foci are seen in the nuclei. The biggest foci reach to 2μm in diameter (F). In some cells with fewer foci, large foci tend to localize to one pole of the cells (E, G). No cytoplasmic foci were seen on day 8 after Mitomycin C treatment. B, D: pictures were taken with some green background to show the cell contour
Fig. 6.
Nuclear RNA foci had significantly increased in number (A) and area (B) on day 8 after being stopped dividing by treatment with Mitomycin C (P<0.01)
Discussion
The current study investigated nuclear RNA foci changes in NSCs differentiated from human DM1 iPS cells. Previous studies of nuclear RNA foci in human DM1 CNS were mainly on postmortem autopsy brains (Jiang et al., 2004). Using the DM1 iPS-cell derived NSCs enabled us to demonstrate the dynamic changes of CUG repeat foci in cultured human neural lineage cells. In NSCs, foci start accumulating in interphase and reach the most abundant level before prophase. When cells enter mitosis, most foci dissipate while some of the remaining foci randomly move to cytoplasm after mitosis. The cytoplasmic foci will eventually dissipate. A previous study has shown that cytoplasmic CUG foci are not sufficient to elicit key DM1 features (Dansithong et al., 2008). It is plausible that a cytoplasm environment is not compatible for expanded CUG RNA repeats to form stable foci. The remaining nuclear RNA foci appear to grow in size and accompany formation of additional new foci as the cells enter another cycle. The same findings were also found in DM1 fibroblast in this study (data not shown). These observations raise a concern in using proliferating cells, such as myoblast (Dansithong et al., 2005; Langlois et al., 2003; Mulders et al., 2009) , fibroblast (Wheeler et al., 2009) and NSCs (here) and engineered cells (HeLa cells and COSM6 cells ) (Lee et al., 2012; Parkesh et al., 2012; Warf et al., 2009), as cellular models for DM1 study, especially when using foci as a marker for drug screening (Ketley et al., 2014). The natural dynamic changes need to be considered in the evaluation. Ideally, cells should be synchronized to G0 stage.
The dynamic changes of foci were also found in DM2 fibroblast cells (Giagnacovo et al., 2012). It will be interesting to see whether similar phenomenon exists in other microsatellite expansion disorders which have also been shown to have a RNA gain-of-function. These include fragile X-associated tremor ataxia syndrome (FXTAS, CGG repeat), Huntington's disease-like 2 (HDL2, CUG repeat), spinocerebellar ataxias type 8 (SCA8, CUG/CAG repeat), type 31 (SCA 31, UGGAA repeat) and type 10 (SCA 10, AUUCU repeat) and familial ALS with GGGGCC repeat expansion in C9ORF72 gene (Daughters et al., 2009; Rudnicki et al., 2007; Sato et al., 2009; Sellier et al., 2010; White et al., 2010) (DeJesus-Hernandez et al., 2011). The lower load of nuclear RNA foci in NSCs suggests these cells are protected by either low expression of DMPK or high a proliferating rate, which will not allow foci to grow.
We found nuclear RNA foci increase in number and size as cells grow. Even though the sequestration of MBNL proteins into the CUG foci is considered to play the major role in the DM1 disease mechanism, the increasing of foci size may imply further gain of function. The larger nuclear RNA foci may have different quaternary structure in non-proliferating cells. ASO or small molecules may not have the same effect on reducing the foci burden compared to newly-formed nuclear RNA foci in highly proliferating cells. The current study offers a model to study the effect of therapeutic drugs on foci in different cell cycle stages.
We have adapted the iPS-cell derived DM1 NSCs to be cultured in routine culture medium (20% FBS in DMEM). These cells are easily maintained and have been passed to passage 23 and are still in healthy proliferating condition. Furthermore, iPS cell can provide unlimited cell resources. They are relatively homogeneous. We found nuclear RNA foci in these cells vary in a wide range in abundance (number, size and immunofluorescence density). With the advancement of imaging techniques and quantification software as newly developed Volocity 3D Image Analysis Software (PerkinElmer), foci can be more precisely and reliably quantified. We think these cells can be used as an ideal cellular model for therapeutic drug development by monitoring nuclear RNA foci as molecular marker.
In summary, the current study suggests nuclear RNA foci of DM1 neural stem cells undergo dynamic changes during cell cycle. Mitosis is a mechanism to decrease foci load. When cells get out of cell cycle, foci abundance progressively increases. The dynamic changes need to be considered when using foci as a marker to follow the effects of therapeutic drugs.
Acknowledgement
The work was supported in part by NIH grant K08AR064836-01(GX).
References
- Ashizawa T, Sarkar PS. Myotonic dystrophy types 1 and 2. Handb Clin Neurol. 2011;101:193–237. doi: 10.1016/B978-0-08-045031-5.00015-3. [DOI] [PubMed] [Google Scholar]
- Bassez G, Lazarus A, Desguerre I, Varin J, Laforet P, Becane HM, Meune C, Arne-Bes MC, Ounnoughene Z, Radvanyi H, Eymard B, Duboc D. Severe cardiac arrhythmias in young patients with myotonic dystrophy type 1. Neurology. 2004;63:1939–1941. doi: 10.1212/01.wnl.0000144343.91136.cf. [DOI] [PubMed] [Google Scholar]
- Childs-Disney JL, Stepniak-Konieczna E, Tran T, Yildirim I, Park H, Chen CZ, Hoskins J, Southall N, Marugan JJ, Patnaik S, Zheng W, Austin CP, Schatz GC, Sobczak K, Thornton CA, Disney MD. Induction and reversal of myotonic dystrophy type 1 pre-mRNA splicing defects by small molecules. Nat Commun. 2013;4:2044. doi: 10.1038/ncomms3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansithong W, Paul S, Comai L, Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J Biol Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- Dansithong W, Wolf CM, Sarkar P, Paul S, Chiang A, Holt I, Morris GE, Branco D, Sherwood MC, Comai L, Berul CI, Reddy S. Cytoplasmic CUG RNA foci are insufficient to elicit key DM1 features. PLoS One. 2008;3:e3968. doi: 10.1371/journal.pone.0003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon MB, Cisneros B. Myotonic dystrophy 1 in the nervous system: from the clinic to molecular mechanisms. J Neurosci Res. 2008;86:18–26. doi: 10.1002/jnr.21377. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YH, Pizzuti A, Fenwick RG, Jr., King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, de Jong P, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- Garcia-Lopez A, Llamusi B, Orzaez M, Perez-Paya E, Artero RD. In vivo discovery of a peptide that prevents CUG-RNA hairpin formation and reverses RNA toxicity in myotonic dystrophy models. Proc Natl Acad Sci U S A. 2011;108:11866–11871. doi: 10.1073/pnas.1018213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giagnacovo M, Malatesta M, Cardani R, Meola G, Pellicciari C. Nuclear ribonucleoprotein-containing foci increase in size in non-dividing cells from patients with myotonic dystrophy type 2. Histochem Cell Biol. 2012;138:699–707. doi: 10.1007/s00418-012-0984-6. [DOI] [PubMed] [Google Scholar]
- Groh WJ, Groh MR, Saha C, Kincaid JC, Simmons Z, Ciafaloni E, Pourmand R, Otten RF, Bhakta D, Nair GV, Marashdeh MM, Zipes DP, Pascuzzi RM. Electrocardiographic abnormalities and sudden death in myotonic dystrophy type 1. N Engl J Med. 2008;358:2688–2697. doi: 10.1056/NEJMoa062800. [DOI] [PubMed] [Google Scholar]
- Harper P. Myotonic Dystrophy(ed3) Saunders; Philadelphia, PA: 2001a. Major problems in neurology. [Google Scholar]
- Harper P. Myotonic Dystrophy. 3rd ed. WB Saunders; London: 2001b. [Google Scholar]
- Jasinska A, Michlewski G, de Mezer M, Sobczak K, Kozlowski P, Napierala M, Krzyzosiak WJ. Structures of trinucleotide repeats in human transcripts and their functional implications. Nucleic Acids Res. 2003;31:5463–5468. doi: 10.1093/nar/gkg767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Ketley A, Chen CZ, Li X, Arya S, Robinson TE, Granados-Riveron J, Udosen I, Morris GE, Holt I, Furling D, Chaouch S, Haworth B, Southall N, Shinn P, Zheng W, Austin CP, Hayes CJ, Brook JD. High-content screening identifies small molecules that remove nuclear foci, affect MBNL distribution and CELF1 protein levels via a PKC-independent pathway in myotonic dystrophy cell lines. Hum Mol Genet. 2014;23:1551–1562. doi: 10.1093/hmg/ddt542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum Mol Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- Langlois MA, Lee NS, Rossi JJ, Puymirat J. Hammerhead ribozyme-mediated destruction of nuclear foci in myotonic dystrophy myoblasts. Mol Ther. 2003;7:670–680. doi: 10.1016/s1525-0016(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Lee JE, Bennett CF, Cooper TA. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc Natl Acad Sci U S A. 2012;109:4221–4226. doi: 10.1073/pnas.1117019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Llamusi B, Artero R. Molecular Effects of the CTG Repeats in Mutant Dystrophia Myotonica Protein Kinase Gene. Curr Genomics. 2008;9:509–516. doi: 10.2174/138920208786847944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan MS, Yadava RS, Yu Q, Balijepalli S, Frenzel-McCardell CD, Bourne TD, Phillips LH. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Teng-Umnuay P, Krym M, Henderson D, Swanson M, Thornton CA. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann Neurol. 2003;54:760–768. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, Wieringa B, Wansink DG. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci U S A. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo JP, Chambon P, Metzger D, Mosier DR, Snipes GJ, Cooper TA. Expanded CTG repeats within the DMPK 3' UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, Hoskins J, Tran T, Housman D, Thornton CA, Disney MD. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J Am Chem Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Rudnicki DD, Holmes SE, Lin MW, Thornton CA, Ross CA, Margolis RL. Huntington's disease--like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- Sato N, Amino T, Kobayashi K, Asakawa S, Ishiguro T, Tsunemi T, Takahashi M, Matsuura T, Flanigan KM, Iwasaki S, Ishino F, Saito Y, Murayama S, Yoshida M, Hashizume Y, Takahashi Y, Tsuji S, Shimizu N, Toda T, Ishikawa K, Mizusawa H. Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am J Hum Genet. 2009;85:544–557. doi: 10.1016/j.ajhg.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schara U, Schoser BG. Myotonic dystrophies type 1 and 2: a summary on current aspects. Semin Pediatr Neurol. 2006;13:71–79. doi: 10.1016/j.spen.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, Hagerman PJ, Charlet-Berguerand N. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. Embo J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seznec H, Agbulut O, Sergeant N, Savouret C, Ghestem A, Tabti N, Willer JC, Ourth L, Duros C, Brisson E, Fouquet C, Butler-Browne G, Delacourte A, Junien C, Gourdon G. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum Mol Genet. 2001;10:2717–2726. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, White RJ, Xia T, Welle S, Turner DH, Mathews MB, Thornton CA. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. Rna. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci U S A. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MC, Gao R, Xu W, Mandal SM, Lim JG, Hazra TK, Wakamiya M, Edwards SF, Raskin S, Teive HA, Zoghbi HY, Sarkar PS, Ashizawa T. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6:e1000984. doi: 10.1371/journal.pgen.1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CH, Nguyen L, Peh J, Luu LM, Sanchez JS, Richardson SL, Tuccinardi T, Tsoi H, Chan WY, Chan HY, Baranger AM, Hergenrother PJ, Zimmerman SC. Targeting toxic RNAs that cause myotonic dystrophy type 1 (DM1) with a bisamidinium inhibitor. J Am Chem Soc. 2014;136:6355–6361. doi: 10.1021/ja5012146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, Santostefano KE, Goodwin M, Liu J, Subramony SH, Swanson MS, Terada N, Ashizawa T. Generation of neural cells from DM1 induced pluripotent stem cells as cellular model for the study of central nervous system neuropathogenesis. Cell Reprogram. 2013;15:166–177. doi: 10.1089/cell.2012.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]