Abstract
Epigenetic mechanisms mediated by histone deacetylases (HDACs) have been implicated in a wide-range of CNS disorders and may offer new therapeutic opportunities. In vivo evaluation of HDAC density and drug occupancy has become possible with [11C]Martinostat, which exhibits selectivity for a subset of class I/IIb HDAC enzymes. In this study, we characterize the kinetic properties of [11C]Martinostat in the nonhuman primate (NHP) brain in preparation for human neuroimaging studies. The goal of this work was to determine whether classic compartmental analysis techniques were appropriate and to further determine if arterial plasma is required for future NHP studies. Using an arterial plasma input function, several analysis approaches were evaluated for robust outcome measurements. [11C]Martinostat showed high baseline distribution volume (VT) ranging from 29.9–54.4 mL/cm3 in the brain and large changes in occupancy (up to 99%) with a blocking dose approaches full enzyme saturation. An averaged nondisplaceable tissue uptake (VND) of 8.6 ± 3.7 mL/cm3 suggests high specific binding of [11C]Martinostat. From a two-tissue compartment model, [11C]Martinostat exhibits a high K1 (averaged K1 of 0.65 mL/cm3/min) and a small k4 (average of 0.0085 min−1). Our study supports that [11C]Martinostat can be used to detect changes in HDAC density and occupancy in vivo and that simplified analysis not using arterial blood could be appropriate.
Keywords: Epigenetic regulation, histone deacetylases, PET, kinetic model, occupancy
Introduction
Epigenetics are biomedical processes that alter gene expression as a result of environmental interactions with an individual’s genome. Epigenetic regulations, such as histone methylation and acetylation, impact many aspects of brain function. Histone deacetylases (HDACs) – a family of chromatin-modifying enzymes – are most frequently implicated in epigenetic mechanisms. Inhibition of HDAC enzymes has led to new therapeutics in cancer research. Epigenetic mechanisms have also been linked to the pathophysiology of a wide range of CNS disorders that include schizophrenia, depression, mood disorders, alcohol and drug addiction, and neurodegenerative disorders 1–5. HDAC inhibitors were shown to be effective in preclinical models of mood disorders and memory deficits 2, 6–12, suggesting treatments targeting epigenetic processes could be potentially useful to rectify CNS dysfunction. However, our knowledge of normal HDAC density and distribution in vivo remains extremely limited. How HDAC enzymes change across the life span, and how HDACs are modified in human CNS diseases have yet to be demonstrated in vivo. A better understanding of the roles of HDACs in CNS function may facilitate the development of novel treatments targeting gene regulatory mechanisms. The capability to determine drug-occupancy relationships of promising HDAC medications in vivo could facilitate drug development activities in this area.
The effort of developing positron emission tomography (PET) radiotracers for in vivo visualization of epigenetic processes began with radiolabeling HDAC inhibitors (or their derivatives) that are in use in clinical cancer treatment trials. However, the development of appropriate imaging agents for neuroepigenetic investigations has only resulted in limited success. For example, published radiolabeled HDAC inhibitors, such as [11C]MS-275, [11C]BA, [11C]PBA, [11C]VPA, and [18F]SAHA have exhibited poor brain penetration 13–16, while [18F]FAHA has been shown to be useful for delineating class-II HDAC activity in nonhuman primates 17, 18. We have recently developed a novel HDAC radioligand, [11C]Martinostat, with selectivity for a subset of class I/IIb HDAC enzymes (class I: HDAC 1, 2, 3 and class IIb: HDAC 6)19. [11C]Martinostat, Figure 1, is a hydroxamic acid with uniquely high brain penetration (with a measured in vitro partition coefficient Log D = 2.03) relative to other hydroxamic acids in this class 19, 20. Our initial evaluation in rodents and nonhuman primates demonstrated high binding and appropriate retention of [11C]Martinostat in the brain 11, 19. In addition, [11C]Martinostat binds selectively and reversibly to its target enzymes, implying that [11C]Martinostat could be a useful imaging tool to quantify the drug-occupancy relationship 11. This radiotracer shows a good safety profile in animals and is currently being evaluated in human imaging experiments (eIND #123154, approved July 2014).
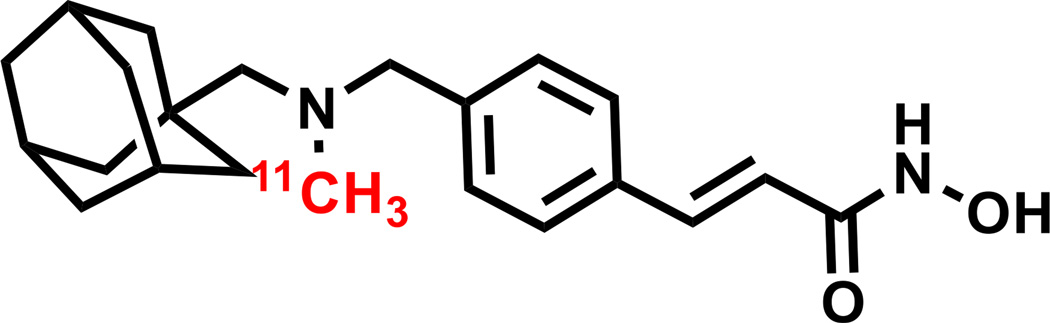
Figure 1.
Chemical structure of radiolabeled [11C]Martinostat.
In light of these promising results and in anticipation of the need to quantify human HDACs using [11C]Martinostat, herein we applied compartmental modeling methods to evaluate the in vivo kinetics of [11C]Martinostat in nonhuman primates. Arterial blood data were obtained for the kinetic analyses. We applied five kinetic models to quantify in vivo HDAC expression using [11C]Martinostat, and the best approach was determined using standard model selection criteria. We further estimated enzyme occupancy for unlabeled Martinostat in nonhuman primates because such a pharmacological study is likely to be more difficult to perform in humans and provides additional validation of the radiotracer and model. This study demonstrates that [11C]Martinostat is a promising PET imaging agent for the in vivo quantification of class I/IIb HDAC binding in the brain.
Results
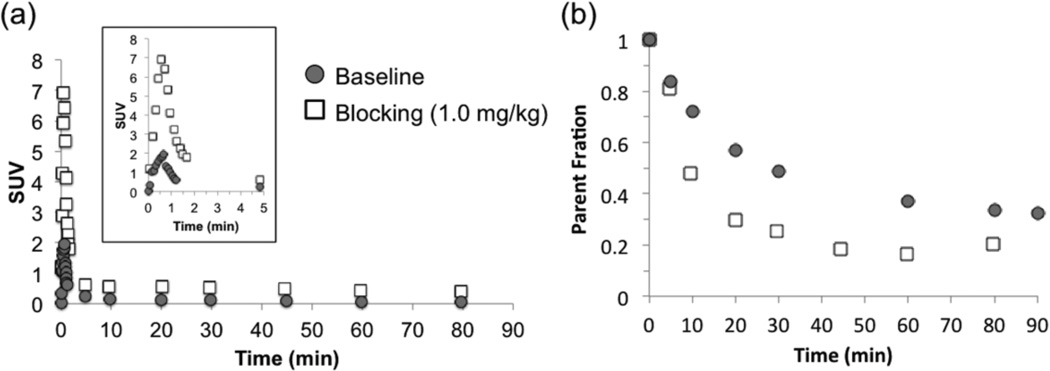
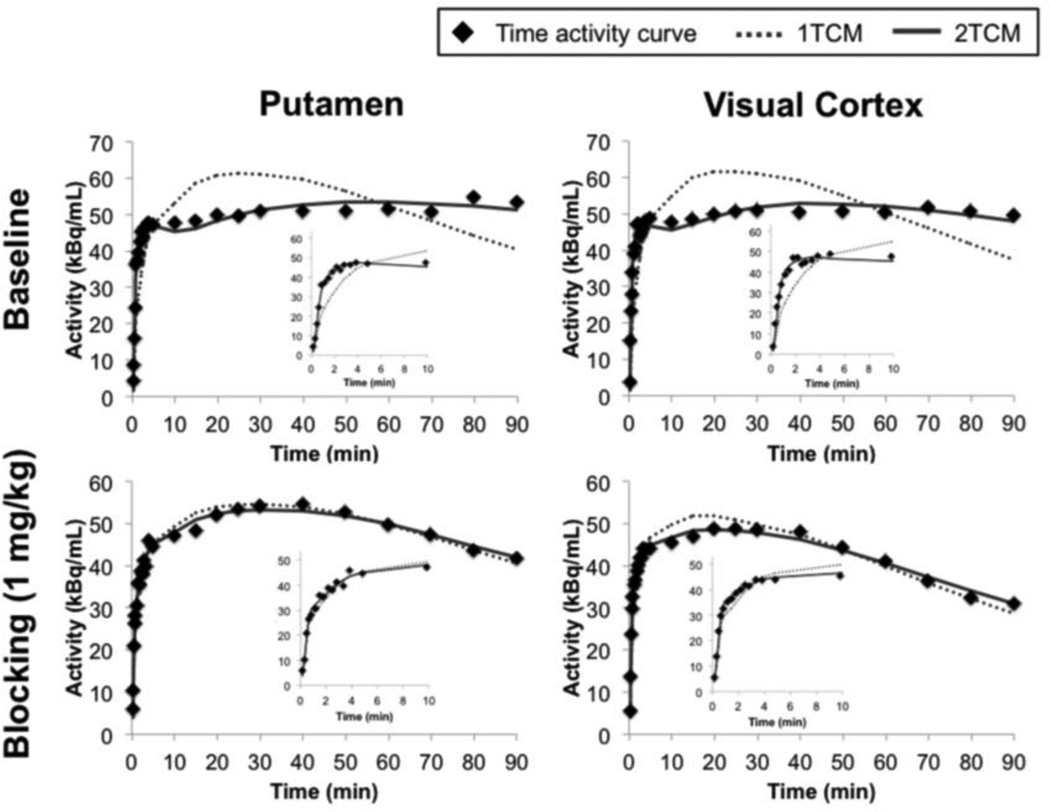
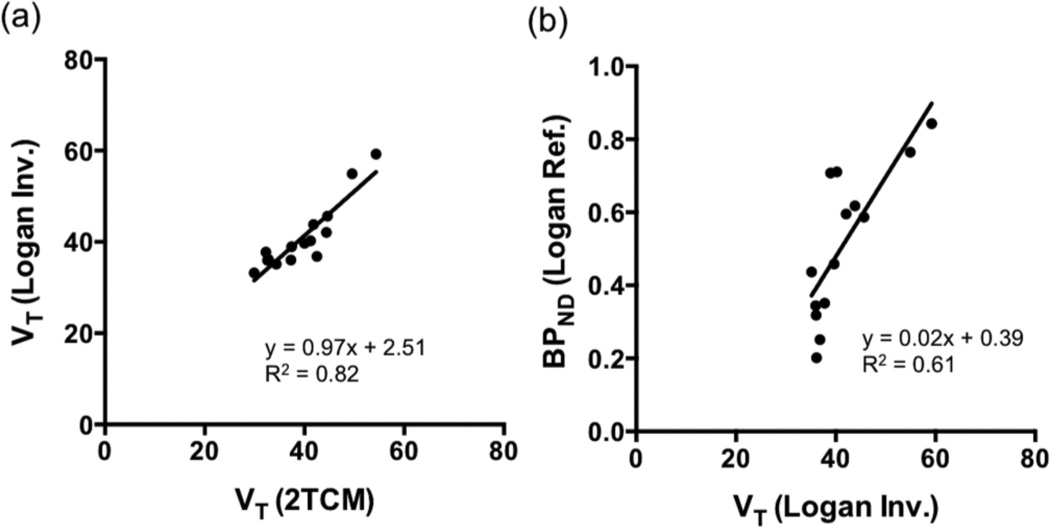
[11C]Martinostat administered at different mass doses did not cause significant changes in blood pressure, heart rate, ETCO2, or oxygen saturation in any animal at any dose. Radioactivity in the arterial plasma peaked within 1 min after radiotracer injection and decreased rapidly thereafter (Figure 2a). When pretreated with unlabeled Martinostat, plasma radioactivity increased ~2–4 fold (dependent on the dose of unlabeled Martinostat) when compared to baseline scans (Figure 2a), suggesting blockade of HDAC binding sites in the brain and the periphery leads to increased plasma exposure. Specifically, the plasma peak activity ratio of blocking:baseline is 2.2 for 0.1 mg/kg, 2.5 for 0.25 mg/kg, 2.3 for 0.5 mg/kg, and 3.8 for 1.0 mg/kg. The averaged parent fraction activity curves reached 0.3 and 0.2 for baseline and blocking scans, respectively (Figure 2b). [11C]Martinostat penetrated the blood-brain barrier and showed high uptake in the cortex, cerebellum, and subcortical regions. The concentration of [11C]Martinostat reached maximum at ~20 min post-injection for the whole brain and decreased slowly over time (Figure 3). The VOI-based regional VT values, AIC, and MSC were tabulated in Table 1. A 2TCM best described dynamic [11C]Martinostat PET data both at baseline and with blocking doses. Figure 3 shows representative TACs and compartmental model fitting results for the putamen and visual cortex at baseline and 1 mg/kg blocking dose. One-tissue compartment model could not sufficiently explain the PET TACs and underestimated VT values in all VOIs examined (Table 1). AIC are the highest in all VOIs evaluated using 1TCM. In addition, MSC for 1TCM (0.3–1.4) are poor and are statistically lower than those of 2TCM (2.2–3.6) (Table 1. Two-way ANOVA with Tukey correction for multiple comparisons, p<0.0001). Therefore, we selected the 2TCM as a parsimonious model that is best suited for [11C]Martinostat analysis, with estimated VT values ranging from 30–54 mL/cm3, which are relative high compared to values typically estimated for neuroreceptor-binding ligand studies. Graphical analysis using the Logan invasive plot resulted in regional VT values that were positively correlated with those derived from a 2TCM (Pearson r = 0.91, two-tailed p<0.0001) (Figure 4a) suggesting that VT values are reliably estimated and graphical methods may also be used to analyze [11C]Martinostat PET data. The mean VT ratio of Logan:2TCM is 1.04 ± 0.08 (mean ± SD across VOIs). This result indicates that VT values derived from Logan plot correspond to those estimated with a 2TCM (~4% overestimation). We further evaluate the Logan non-invasive model by using the white matter as a pseudo-reference tissue to derive binding potential to non-displaceable tissue (BPND)21, 22. Outcome measurements derived from Logan invasive vs. non-invasive plots were positively correlated (Pearson r = 0.78, two-tailed p<0.0001) (Figure 4b). To assess the stability of VT estimates, 2TCM was applied to one baseline dynamic PET dataset (120 min) with increasingly truncated duration. Most VOIs required long scan length (90 min or 100 min) for stable VT estimation.
Figure 2.
Arterial plasma radioactivity and the measured parent fraction of [11C]Martinostat in a baseline and a blocking scans. (a) Arterial plasma activity (corrected for injected dose and animal weight) following i.v. administration with (1 mg/kg blocking, square label) and without (baseline, circle label) unlabeled Martinostat (Image inset zoomed into the first 5 min of the plots). (b) Parent fraction curves of radioactivity from a baseline and a blocking (1 mg/kg) scan.
Figure 3.
Brain regional time-activity curves (TACs) of the putamen and visual cortex and compartmental model fits from a representative subject. 1TCM and 2TCM represent one-tissue and two-tissue compartmental models, respectively. Image insets zoomed into the first 10 min of the plots.
Table 1.
Regional distribution volume (VT) for [11C]Martinostat baseline binding (n=4, 90 min scan) were estimated with one- and two-tissue compartmental models (1TCM and 2TCM) and graphical analysis (Logan Plot and MA1). Goodness of fit for one- and two-tissue compartmental models (1TCM and 2TCM) is evaluated with Akaike information criteria (AIC) and Model selection criteria (MSC).
| VT (mL/cm3) | AIC | MSC | ||||||
|---|---|---|---|---|---|---|---|---|
| Brain Region | 1TCM | 2TCM | Logan | MA1 | 1TCM | 2TCM | 1TCM | 2TCM |
| ACC | 20 ± 4 | 32 ± 9 | 38 ± 13 | 43 ± 10 | 75 ± 9 | 23 ± 6 | 0.4 ± 0.5 | 2.6 ± 0.5 |
| Amygdala | 22 ± 4 | 42 ± 6 | 37 ± 12 | 49 ± 10 | 69 ± 12 | 23 ± 16 | 1 ± 0.7 | 2.9 ± 0.8 |
| Cerebellum | 30 ± 10 | 54 ± 16 | 59 ± 17 | 75 ± 25 | 73 ± 11 | 20 ± 26 | 0.8 ± 0.6 | 3 ± 1.2 |
| DLPFC | 26 ± 5 | 42 ± 10 | 44 ± 12 | 50 ± 6 | 62 ± 9 | 10 ± 15 | 1.4 ± 0.5 | 3.5 ± 0.5 |
| Hippocampus | 21 ± 4 | 37 ± 8 | 36 ± 16 | 51 ± 11 | 72 ± 7 | 37 ± 17 | 0.9 ± 0.4 | 2.3 ± 0.6 |
| M1 | 25 ± 5 | 45 ± 10 | 46 ± 13 | 60 ± 6 | 65 ± 10 | 7 ± 14 | 1.2 ± 0.5 | 3.6 ± 0.6 |
| NAc | 19 ± 5 | 33 ± 13 | 36 ± 13 | 42 ± 13 | 76 ± 9 | 26 ± 24 | 0.5 ± 0.6 | 2.6 ± 1.2 |
| OFC | 19 ± 6 | 34 ± 17 | 35 ± 23 | 41 ± 24 | 72 ± 7 | 32 ± 16 | 0.8 ± 0.3 | 2.5 ± 0.7 |
| PCC | 19 ± 4 | 33 ± 5 | 36 ± 7 | 41 ± 8 | 72 ± 8 | 21 ± 20 | 0.7 ± 0.6 | 2.8 ± 1 |
| Putamen | 29 ± 7 | 50 ± 14 | 55 ± 15 | 63 ± 20 | 68 ± 8 | 15 ± 14 | 1.1 ± 0.3 | 3.3 ± 0.6 |
| SMA | 25 ± 5 | 41 ± 9 | 40 ± 14 | 46 ± 10 | 73 ± 10 | 19 ± 19 | 0.7 ± 0.7 | 2.9 ± 0.9 |
| Thalamus | 24 ± 6 | 40 ± 8 | 40 ± 12 | 45 ± 12 | 68 ± 13 | 15 ± 14 | 1.1 ± 0.7 | 3.3 ± 0.7 |
| V1 | 22 ± 6 | 37 ± 10 | 39 ± 15 | 45 ± 12 | 81 ± 9 | 36 ± 21 | 0.3 ± 0.6 | 2.2 ± 0.9 |
| Caudate | 26 ± 6 | 44 ± 10 | 42 ± 17 | 54 ± 10 | 67 ± 6 | 8 ± 14 | 1.1 ± 0.4 | 3.6 ± 0.7 |
| WM | 17 ± 4 | 30 ± 8 | 33 ± 7 | 36 ± 8 | 63 ± 8 | 5 ± 20 | 1.2 ± 0.5 | 3.6 ± 0.8 |
ACC: anterior cingulate cortex, DLPFC: dorsal lateral prefrontal cortex, M1: primary motor cortex, NAc: nucleus accumbens, OFC: orbitofrontal cortex, PCC: posterior cingulate cortex, SMA: supplementary motor area, V1: primary visual cortex, WM: white matter.
Figure 4.
(a) Correlation of the VOI-based, regional distribution volume values (VT) of [11C]Martinostat derived from a two-tissue compartmental model (2TCM) and the Logan graphical model with arterial plasma as input function (Logan Invasive method). (b) Correlation of the regional VT values derived from a Logan invasive (Logan Inv.) plot and the regional binding potential (BPND) values derived from a Logan non-invasive plot (Logan Ref.) using the white matter as a pseudo-reference tissue. The VT values derived using Logan invasive plot were significantly correlated with those derived using a 2TCM with a correlation coefficient R2=0.82 (Pearson r = 0.91, two-tailed p<0.0001). Outcome measurements estimated with Logan invasive vs. non-invasive plots were also positively correlated (Pearson r = 0.78, two-tailed p<0.0001).
Individual rate constants (K1, k2, k3, k4) estimated from baseline scans using 2TCM are summarized in Table 2. The estimated K1 ranged from 0.39–0.89 mL/cm3/min, with an averaged K1 of 0.65 mL/cm3/min. The average k2 of [11C]Martinostat is 0.85 min−1 (ranging from 0.52–1.50 min−1) and the averaged k3 is 0.34 min−1 (ranging from 0.29–0.40 min−1). The disassociation rate constant, k4, from the specific compartment are estimated to be 0.0071–0.0099 min−1, with an average of 0.0085 min−1 for all brain regions. Although [11C]Martinostat is reversible as demonstrated in our earlier rodent studies 19, its slow washout kinetics warrant evaluation of flow dependency. The fact that k2 is larger than k3 by a factor ~2 (Table 2) indicates that the tracer accumulation is likely insensitive to flow changes. In addition, correlation coefficients between regional K1 vs. k2 and k3 vs. k4 are low (R2 = 0.41 and R2 = 0.02, respectively) suggesting that the rate constants were sufficiently independent as identified.
Table 2.
Kinetic rate constants estimated from four baseline [11C]Martinostat scans (n=4) with a two-tissue compartmental model.
| Brain Region | K1 (mL/cm3/min) | %COV | k2 (1/min) | %COV | k3 (1/min) | %COV | k4 (1/min) | %COV |
|---|---|---|---|---|---|---|---|---|
| ACC | 0.78 ± 0.25 | 11 ± 5 | 1.5 ± 1.25 | 23 ± 5 | 0.4 ± 0.15 | 14 ± 3 | 0.0089 ± 0.0037 | 18 ± 4 |
| Amygdala | 0.53 ± 0.2 | 9 ± 4 | 0.6 ± 0.41 | 25 ± 9 | 0.3 ± 0.1 | 17 ± 4 | 0.0071 ± 0.003 | 27 ± 9 |
| Cerebellum | 0.83 ± 0.22 | 10 ± 6 | 0.87 ± 0.77 | 26 ± 11 | 0.34 ± 0.23 | 19 ± 12 | 0.008 ± 0.003 | 25 ± 14 |
| DLPFC | 0.6 ± 0.17 | 8 ± 5 | 0.63 ± 0.48 | 23 ± 9 | 0.34 ± 0.21 | 16 ± 7 | 0.0099 ± 0.0037 | 17 ± 4 |
| Hippocampus | 0.56 ± 0.16 | 17 ± 14 | 1.03 ± 1.07 | 38 ± 19 | 0.39 ± 0.23 | 22 ± 7 | 0.0078 ± 0.0035 | 31 ± 14 |
| M1 | 0.59 ± 0.21 | 7 ± 3 | 0.6 ± 0.41 | 19 ± 4 | 0.3 ± 0.19 | 14 ± 5 | 0.0081 ± 0.0026 | 18 ± 4 |
| NAc | 0.67 ± 0.19 | 11 ± 6 | 1.08 ± 0.81 | 24 ± 8 | 0.35 ± 0.17 | 15 ± 5 | 0.0087 ± 0.003 | 20 ± 5 |
| OFC | 0.52 ± 0.15 | 11 ± 6 | 0.76 ± 0.64 | 30 ± 10 | 0.3 ± 0.18 | 22 ± 11 | 0.0085 ± 0.0032 | 27 ± 7 |
| PCC | 0.57 ± 0.22 | 9 ± 3 | 0.75 ± 0.46 | 22 ± 5 | 0.32 ± 0.16 | 15 ± 4 | 0.0084 ± 0.003 | 21 ± 7 |
| Putamen | 0.74 ± 0.12 | 9 ± 5 | 0.88 ± 0.84 | 23 ± 8 | 0.35 ± 0.2 | 16 ± 8 | 0.0084 ± 0.0033 | 18 ± 4 |
| SMA | 0.84 ± 0.33 | 12 ± 8 | 1.07 ± 0.73 | 24 ± 11 | 0.37 ± 0.19 | 15 ± 6 | 0.0086 ± 0.003 | 19 ± 6 |
| Thalamus | 0.63 ± 0.24 | 8 ± 3 | 0.68 ± 0.44 | 23 ± 10 | 0.33 ± 0.13 | 16 ± 7 | 0.0089 ± 0.0036 | 18 ± 4 |
| V1 | 0.89 ± 0.2 | 12 ± 7 | 1.01 ± 0.79 | 26 ± 10 | 0.32 ± 0.17 | 17 ± 5 | 0.0092 ± 0.0031 | 24 ± 11 |
| Caudate | 0.66 ± 0.18 | 7 ± 3 | 0.73 ± 0.47 | 19 ± 6 | 0.35 ± 0.15 | 13 ± 4 | 0.0084 ± 0.0028 | 16 ± 4 |
| WM | 0.39 ± 0.13 | 7 ± 4 | 0.52 ± 0.35 | 19 ± 8 | 0.29 ± 0.18 | 14 ± 4 | 0.0087 ± 0.003 | 17 ± 5 |
| Average | 0.65 | 0.85 | 0.34 | 0.0085 | ||||
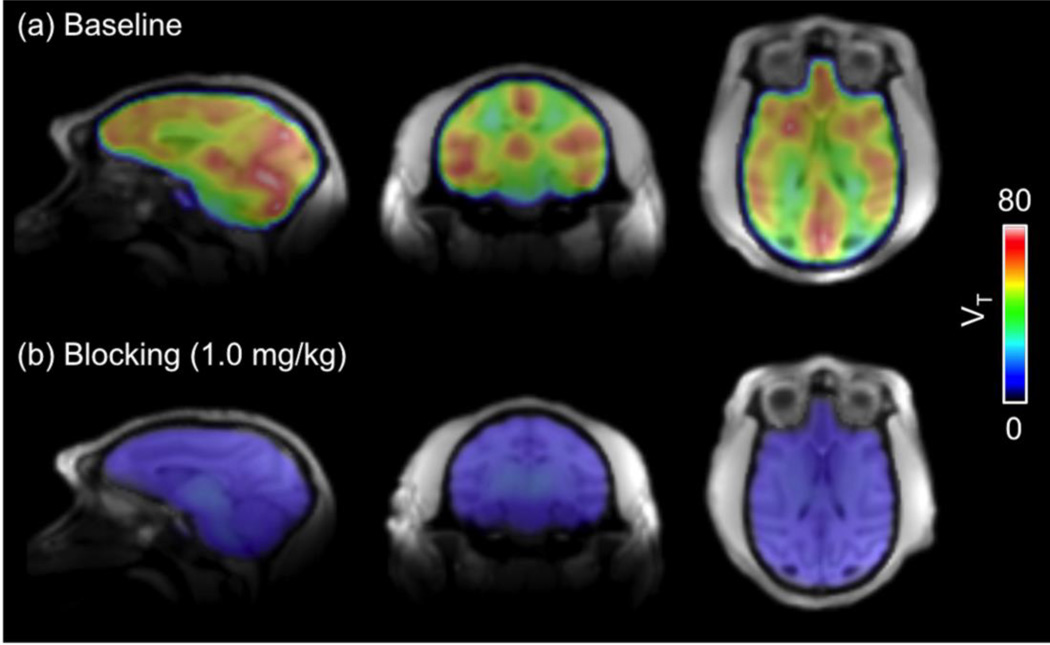
Voxel-wise VT images from a representative subject are shown in Figure 5. The highest radioactivity concentrations were observed in the cortical regions, cerebellum, caudate, putamen, and thalamus under baseline condition. The radioactivity was lower in the white matter than in the grey matter. Regional baseline VT values follow the rank order of cerebellum > putamen > motor cortex > caudate > amygdala > DLPFC > SMA > thalamus > visual cortex > hypothalamus > OFC > PCC > NAc > ACC > WM. To confirm the binding specificity of [11C]Martinostat, we performed a second set of PET imaging studies on each animal following pretreatment with unlabeled Martinostat at different mass doses (0.1, 0.25, 0.5 and 1.0 mg/kg). With all blocking doses, VT values decreased in all brain regions when compared to its baseline values of the same brain region. The mean VT reduction of all VOIs analyzed was 46.3 ± 19.0 % for 0.1 mg/kg, was 67.9 ± 7.2 % for 0.25 mg/kg, was 57.9 ± 12.6 % for 0.5 mg/kg, and was 82.3 ± 5.5 % for 1 mg/kg. Although a 0.5 mg/kg blocking dose shows lower VT reduction than the 0.25 mg/kg blocking dose, these two studies were performed on two separate animals (one male and one female) and may reflect high individual variability of [11C]Martinostat binding and/or sex differences and thus, HDAC level in the brain.
Figure 5.
Voxel-wise volume of distribution (VT) maps overlaid on a high-resolution structural MRI at (a) baseline and (b) with an 1 mg/kg blocking dose.
Using the occupancy plot, it was estimated that the doses of Martinostat given in the current study achieves high target occupancy (>80%). An averaged VND of 8.6 ± 3.7 mL/cm3 suggests a high level of specific binding of [11C]Martinostat. Note that the thalamus has been excluded from the occupancy analysis because it does not show a dose-dependent change in VT, suggesting minimum level of changes in occupancy due to self-blocking. Cerebellum was also excluded from the analysis because of large uncertainty in VT estimation (%COV > 30%) in the one animal lacking measured blood data.
Discussion
Epigenetic modifications mediated by HDACs play critical roles in normal CNS function. Recently, multiple lines of preclinical and clinical evidence suggest that HDAC inhibitors may represent promising new therapeutics in CNS disorders 6. Noninvasive detection and quantification of HDACs expression in living humans using PET imaging will provide the first translational insights into the role of HDAC in brain dysfunction. Moreover, it will provide a tool to assess target engagement and occupancy for understanding treatment efficacy of novel HDAC inhibitors and drugs. Our recently developed [11C]Martinostat possesses great potential for first in man translational imaging due to its ideal properties.
[11C]Martinostat exhibits high brain uptake and high VT values throughout the brain (Figure 5), and has appropriate kinetic binding properties for human imaging. Because VT = VND (1+BPND), the high [11C]Martinostat VT values together with a low estimated VND of 8.6 ± 3.7 mL/cm3 reflect the high density of HDACs in the brain. A high affinity of the radiotracer might also contribute the VT high values. By pretreating with cold Martinostat, the blockade confirms the specific binding of [11C]Martinostat. As anticipated, the four blocking doses of unlabeled Martinostat resulted in high (~79–99%) occupancy and large changes in regional VT values (~40–90%), while metrics of animal physiology were not significantly changed.
Individual rate constants estimated through kinetic modeling from baseline datasets show that K1 is large and k4 is small for [11C]Martinostat. Large K1 means rapid uptake of [11C]Martinostat from the blood stream, which is consistent with the excellent BBB penetration property of [11C]Martinostat 20. Because the majority of early PET radiotracers targeting neuroepigenetic failed to cross the BBB, [11C]Martinostat was specifically designed to overcome this challenge. White matter VOI showed the lowest K1 value among all brain regions analyzed (0.39 vs. 0.52–0.89 mL/cm3/min, Table 2). The slow washout kinetics (k4 = 0.0085 min−1) of [11C]Martinostat could be due to slow peripheral clearance, high affinity of the radiotracer, and/or high density of target binding sites 23. High densities of HDAC expression in the periphery have been reported. Our previous experience with [11C]Martinostat in nonhuman primates indeed demonstrated high saturable binding of [11C]Martinostat in the heart, kidney, spleen, and pancreas 19. In addition, in the current study, arterial blood data from the blocking experiments showed elevated plasma radioactivity (~2–4 folds) compared to the baseline scans (Figure 2a). Taken together, it is likely that slow peripheral clearance contributes to the slow washout of [11C]Martinostat. For radiotracers with a slow kinetics, the uptake of radiotracer might become flow-dependent when k3 >> k2. Based on our estimation of kinetic rate constants of [11C]Martinostat (Table 2), k2 is larger than k3 by a factor ~2 suggesting that [11C]Martinostat is not flow-dependent. In addition, we have demonstrated in a previous study that [11C]Martinostat efflux from the brain is not related to the thexenobiotic pump, P-glycoprotein (P-gp) 19. Despite slow tracer kinetics, [11C]Martinostat has been shown in our previous rodent studies to bind reversibly with a competition binding experiment 19. Radiotracers with slow kinetics are usually more difficult to quantify than fast tracers. Although we have only acquired 90–120 min of dynamic PET data in the present study, we obtained stable VT measurements as determined by the fact that a 2TCM fit resulted in small % coefficient of variance (on the order of 10–20%). Future studies in human may require longer scans in order to assure stable outcome measurements and to assess potential sources of errors in quantification. In addition, carefully designed test-retest studies in human will be valuable to determine within-subject variability and set the stage for future application in monitoring disease progression and treatment efficacy.
Compartmental analysis of regional brain radioactivity concentrations demonstrated that a 2TCM provided statistically better fits than the 1TCM for all brain regions. A 2TCM provided reliable estimates of VT values as determined by small % coefficient of variances. The VT values obtained with Logan invasive plot corresponded with the 2TCM estimates. Reference tissue models have the advantage of reducing technical demands by alleviating the need for arterial blood sampling during PET scans and improving PET study feasibility in clinical settings. However, we did not find a brain region devoid of specific binding, based upon the fact that no brain regional VT value was unchanged in self-blocking studies. This was not surprising given the existing understanding of the widespread, robust expression of HDAC 1, 2, 3 and 6 in brain 24. Future human studies with an estimation of test-retest variability of [11C]Martinostat may support or disapprove the existence of a reference tissue for [11C]Martinostat. Recently, a method using a brain region with specific binding as reference tissue to derive a “pseudo BPND” or estimate drug occupancy has been proposed 21, 22. It would be most reasonable to choose a brain region that shows the lowest baseline VT value and/or is minimally affected by varying blocking doses. We chose the white matter as a potential pseudo-reference tissue in this study, and showed appropriate outcome measurements estimated with Logan non-invasive plot. Further human studies with a carefully evaluated test-retest variability of [11C]Martinostat might be interesting and finding an appropriate surrogate outcome measurements to eliminate the need for arterial blood sampling warrants future investigation.
Conclusion
We performed kinetic analysis of the novel HDAC radiotracer, [11C]Martinostat, and determined that it can be used to quantify in vivo HDAC expression and changes in occupancy in the brain. Based on our kinetic modeling results, we determined that a two-tissue compartmental model best describes in vivo characteristics of [11C]Martinostat. [11C]Martinostat showed high baseline VT values with heterogeneity consistent with known distribution of HDAC in rodents. [11C]Martinostat binds reversibly and dose-dependently in the brain and a dose of 1 mg/kg reaches approximately 99% occupancy. Although the radiotracer demonstrated slow washout kinetics, stable outcome measurements (VT values) can be obtained with compartmental model (2TCM) and graphical methods. We are currently clarifying the contribution of each HDAC isoform to the [11C]Martinostat signal in vivo.
Materials and Methods
Animal Preparation
A paired baseline/blocking PET/MR study was performed on each of four baboons (two males and two females, Papio Anubis, 13.6 ± 4.7 kg) for a total of 8 scans with approval of the Institutional Animal Care and Use Committee at the Massachusetts General Hospital. All animals were deprived of food for 12 h prior to the study. Anesthesia was induced with intramuscular (i.m.) ketamine (10 mg/kg) and xylazine (0.5 mg/kg). For maintenance of anesthesia throughout the study, the baboon was provided 1%–1.5% isoflurane in oxygen while a dose of yobine (0.11mg/kg, i.m.) was given to reverse the effects of ketamine/xylazine before the start of the scan. A saphenous vein of the baboon was catheterized for radiotracer injection and a radial arterial line was placed for arterial blood sampling to enable determination of the metabolite-corrected plasma input function. Vital signs including end-tidal CO2 (ETCO2), O2 saturation, heart rate, and respiration rate were monitored continuously (recorded every 15 min) and were maintained within normal physiological ranges.
Radiosynthesis of [11C]Martinostat
[11C]CO2 was obtained via the 14N(p, α)11C reaction (Siemens Eclipse cyclotron), and trapped with TRACERlab FX-M synthesizer (General Electric). [11C]CH4 was obtained by the reduction of [11C]CO2 and passed through an oven containing I2 to produce [11C]CH3I via a radical reaction. [11C]CH3I was trapped in the TRACERlab FX-M synthesizer preloaded with a solution of precursor (1.0 mg) in dry DMSO (300 µL). The solution was stirred at 100 °C for 4 min and water (1.2 mL) was added. The solution was then purified by reverse phase semi-preparative HPLC. The final product was reformulated by loading onto a solid-phase exchange (SPE) C-18 cartridge, rinsing with 1M NaOH aq (5 mL), eluting with EtOH (1 mL), and diluting with 50 µL acetic acid in saline (0.9%, 9 mL). The identity of the product was confirmed by analytical HPLC with additional co-injection of unlabeled Martinostat. The average time required for the synthesis was 35 min. The average radiochemical yield was 3–5% (non-decay corrected to trapped [11C]CH3I; n = 3). Chemical and radiochemical purities were ≥ 95 % with a specific activity 34 ± 5 MBq/nmol at the time of injection.
Arterial Plasma and Metabolite Analysis
Blood samples were drawn from the arterial line at 10 sec intervals for 3 min (~1 mL each), followed by additional samples at 5, 10, 20, 30, 45, 60, and 80 min (~3 mL each) post-injection for plasma and metabolite analyses. Additional blood samples at 100 and 120 min post-injection were obtained from the two animals that underwent longer scans (120 min). The collected samples were centrifuged to obtain plasma, which was then removed (200µL for samples collected during the first 3min; 600µL for all later samples) and placed in an automatic gamma counter that was cross-calibrated with the PET scanner. The analysis of radiolabeled metabolites was conducted on a custom automated robot, fitted with Phenomenex Strata-X 500 mg SPE cartridges that were primed with ethanol (2 mL) and deionized water (20 mL). Beginning with the arterial sample acquired at 5 min after radiotracer administration, an aliquot (300 µL) of plasma was added to acetonitrile (300 µL) and centrifuged for 1 min to obtain protein-free plasma (PFP). An aliquot (300 µL) of PFP/acetonitrile solution was diluted into deionized water (3 mL), loaded onto the C18 cartridge, and removed of polar metabolites with 100% water. Next, a series of extractions were performed using water and acetonitrile in quantities: 95:5, 90:10, 85:15, 80:20, 70:30, 60:40, 30:70 and 100% acetonitrile at a volume of 4 mL. A control experiment was performed before metabolite analysis to determine the retention of the parent compound by injection of a small amount of [11C]Martinostat onto a test series of extraction cartridges. Each sample was counted in a WIZARD2 Automatic Gamma Counter to determine the presence of radiolabeled metabolites. Final total plasma radioactivity was interpolated linearly and corrected for the fraction of radiometabolites. The metabolite-corrected plasma activity curve was used as the arterial input function for kinetic modeling. In one animal (120 min scans, baseline and 0.1 mg/kg pretreatment), metabolite analysis was unsuccessful due to technical difficulty. An averaged fractional metabolite curve was generated from the other three animals for the baseline scan and the blocking scan separately. Because radiometabolites measured from the three blocking scans (0.25 mg/kg, 0.5 mg/kg, and 1 mg/kg) did not show a dose-dependent difference, therefore, an averaged fractional metabolite curve was appropriate to correct for radiometabolites in the 0.1 mg/kg pretreatment scan.
PET/MR Image Acquisition
PET and MRI images were acquired on a 3T Siemens TIM-Trio with a BrainPET insert (Siemens, Erlangen, Germany). A custom PET/MRI compatible 8-channel array coil for nonhuman primate brain imaging was used to improve image signal and quality than using a clinical human head coil. Dynamic PET image acquisition was initiated followed by administration of ~185 MBq (180 ± 15 MBq averaged from 8 scans) of [11C]Martinostat as a manual bolus over about 30 sec to the baboon. A baseline [11C]Martinostat PET scan was first carried out on each animal, followed by a second PET scan (i.e. the blocking scan) at 10 min after the injection of unlabeled Martinostat. One dose of unlabeled Martinostat (0.1, 0.25, 0.5, or 1 mg/kg; 10% DMSO/10% Tween 80 /Saline (V/V)) was intravenously administered to each animal in order to determine specific binding and quantify occupancy of [11C]Martinostat. Each of these doses was expected to result in high occupancy due to the high affinity of Martinostat and its high brain-plasma penetraion 19. Both baseline and blocking scans were carried out in the same animal on the same day, with the baseline scan acquired first. The two scans were separated by at least 2.5 hours.
Dynamic PET data were collected and stored in list mode for 90 min in two animals (0.5 and 1 mg/kg) and 120 min in the other two animals (0.1 and 0.25 mg/kg). Baseline and blocking scans were acquired for the same duration on an imaging session. The corresponding images were reconstructed using the 3D ordinary Poisson expectation maximization algorithm (32 iterations) with detector efficiency, decay, dead time, attenuation, and scatter corrections applied. PET data were reconstructed with gradually increasing intervals (6 × 10 sec, 6 × 20 sec, 2 × 30 sec, 1 × 1 min, 5 × 5 min, 6 × 10 min (or 9 × 10 min for those with 120 min scans)). The highest image resolution was on the order of 2–3 mm for BrainPET. The final image volumes were reconstructed into 76 slices with 128 × 128 pixels and a 2.5-mm isotropic voxel size. A high-resolution anatomical scan using multi-echo MPRAGE sequence (TR = 2530ms, TE1/TE2/TE3/TE4 = 1.64/3.5/5.36/7.22 ms, TI = 1200 ms, flip angle =7°, and 1mm isotropic) was obtained at about 30 min into the baseline scan.
Image Analysis
PET data were registered to the Black baboon brain atlas 25 using JIP tools optimized for nonhuman primate data processing (www.nitrc.org/projects/jip). The high-resolution T1-weighted anatomical MRI image was first registered to the baboon brain atlas using a mutual information approach and the transformation parameters were then applied to the simultaneously collected dynamic PET data. Thirteen volumes of interest (VOIs) were defined according to the Black baboon brain atlas 25. Common VOIs were applied to all scans. Time-activity curves (TACs) were extracted from the anterior cingulate cortex (ACC), posterior cingulate cortex (PCC), amygdala, thalamus, caudate, putamen, nucleus accumbens (NAc), hippocampus, primary motor cortex (M1), primary visual cortex (V1), dorsal lateral prefrontal cortex (DLPFC), orbital frontal cortex (OFC), supplementary motor area (SMA), whole cerebellum, and white matter (WM) VOIs for analysis.
Kinetic modeling was performed using PMOD 3.4 (PMOD Technologies Ltd., Zurich, Switzerland). Compartmental models of increasing complexity (one-tissue (1TCM) and two-tissue (2TCM) compartments) and graphical analysis approaches (Logan plot with arterial plasma as input function (Logan Invasive) and Logan reference tissue model) were applied using the metabolite-corrected arterial blood as input function 26, 27. Volume of distribution (VT) measures specific receptor binding, nonspecific binding, and free radiotracer (VT = VS + VND), where the sum of nonspecific binding and free radiotracer can be represented as the distribution volume of nondisplaceable compartment (VND) and can be estimated separately (see below). Standard compartmental models with rate constants as previously described 28 were applied. Where VT = K1/k2 for 1TCM and VT = K1/k2 (1 + k3/k4) for 2TCM. In all compartmental models, the VOI blood volume contribution was fixed to be 5%. Akaike information criteria (AIC) and the model selection criteria (MSC) were chosen as objective quantities to use to compare model performance, with the most appropriate model yielding the smallest AIC and the largest MSC values 29, 30. For graphical methods, the linearity start time (t*) was chosen to be 35 min post-injection, based on visual inspection of the Logan plots, although tissue:plasma TAC ratios did not begin to plateau until after 70–80 min. A t* of 35 min provided a minimum of 6 points for the linear regression. Lastly, voxel-wise VT maps were calculated using Logan plot (t*=35) after spatially smoothed the original images with a 6 mm FWHM 3D Gaussian kernel.
Minimum Scan Duration
To determine the minimum scan duration needed to obtain stable outcome measures (VT), we analyzed baseline PET data using the 2TCM from one animal (animal #3) by removing time portions from the end of each scan. Data were analyzed with 10 min decrement intervals from the full length (0–120 min) to a truncated 0–50 min. Regional VT values determined using varying scan durations were compared with VT derived using the full scan length and percent differences in VT were calculated. An estimated VT of a VOI was considered to be stable (at a given scan duration) if the mean percent VT ratios across all studies was between 90% and 110%.
Occupancy
Occupancy with exogenous drug can be estimated using the Lassen plot with the assumption that nonspecific binding and occupancy are homogeneous in the brain 31, 32. The term Occ is the occupancy of drug at the target-binding site, which is presumed to be class I/IIb (HDAC 1, 2, 3, and to a lesser extent, HDAC 6) for Martinostat based on in vitro recombinant protein binding assays (13). The Lassen Plot is given by
where VT,Baseline and VT,Drug are the regional VT values estimated at baseline and following a drug challenge, respectively, while VND represents nondisplaceable tissue uptake (free plus nonspecifically bound). In the present study, we performed paired baseline and self-blocking experiments by pretreating with vehicle or unlabeled Martinostat in each study. Regional VT was estimated using a 2TCM as the tissue model which best fit the PET data (see Results). In order to minimize potential inter-subject variation, the Occ and VND were estimated for each animal (i.e. each blocking dose) separately.
Statistical Methods
All group results are reported as mean ± SD. Statistical analysis were performed using GraphPad Prism (Prism6, GraphPad Software Inc., La Jolla, CA, USA). Two-way ANOVA (α = 0.05, two-sided with Tukey’s multiple comparisons correction) has been carried out to compare performance (i.e. AIC and MSC values) between compartmental models for each VOI. Pearson correlation analysis was performed to compare regional VT values estimated with Logan plot and MA1 to those derived with a 2TCM (α = 0.05, two-sided). Linear regression analysis without constraining the intercept was performed to estimate Occ and VND from each paired baseline and blocking study.
Acknowledgements
This research was supported by the National Institute of Drug Abuse (NIDA) of the National Institutes of Health under grant number R01DA030321 (J.M.H.). H.Y.W. and C.W. were supported by the Harvard / MGH Nuclear Medicine Training Program from the Department of Energy (DE-SC0008430). H.Y.W. is supported by NIDA K99DA037928. This research was carried out at the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital, using resources provided by the Center for Functional Neuroimaging Technologies (P41EB015896), a P41 Regional Resource supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health. This work also involved the use of instrumentation supported by the NIH Shared Instrumentation Grant Program and/or High-End Instrumentation Grant Program; specifically, grant numbers: S10RR017208, S10RR026666, S10RR022976, S10RR019933, S10RR029495.
Footnotes
Disclosure/Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Akbarian S, Nestler EJ. Epigenetic mechanisms in psychiatry. Neuropsychopharmacology. 2013;38:1–2. doi: 10.1038/npp.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fass DM, Reis SA, Ghosh B, Hennig KM, Joseph NF, Zhao W-N, Nieland TJ, Guan J-S, Kuhnle CEG, Tang W, Barker DD, Mazitschek R, Schreiber SL, Tsai L-H, Haggarty SJ. Crebinostat: a novel cognitive enhancer that inhibits histone deacetylase activity and modulates chromatin-mediated neuroplasticity. Neuropharmacology. 2013;64:81–96. doi: 10.1016/j.neuropharm.2012.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology. 2013;38:138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nature reviews. Neuroscience. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun H, Kennedy PJ, Nestler EJ. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2013;38:124–137. doi: 10.1038/npp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroeder FA, Lewis MC, Fass DM, Wagner FF, Zhang Y-L, Hennig KM, Gale J, Zhao W-N, Reis S, Barker DD, Berry-Scott E, Kim SW, Clore EL, Hooker JM, Holson EB, Haggarty SJ, Petryshen TL. A selective HDAC 1/2 inhibitor modulates chromatin and gene expression in brain and alters mouse behavior in two mood-related tests. PLoS ONE. 2013;8:e71323. doi: 10.1371/journal.pone.0071323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakovcevski M, Bharadwaj R, Straubhaar J, Gao G, Gavin DP, Jakovcevski I, Mitchell AC, Akbarian S. Prefrontal Cortical Dysfunction After Overexpression of Histone Deacetylase 1. Biological psychiatry. 2013 doi: 10.1016/j.biopsych.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahari-Javan S, Maddalena A, Kerimoglu C, Wittnam J, Held T, Bahr M, Burkhardt S, Delalle I, Kugler S, Fischer A, Sananbenesi F. HDAC1 regulates fear extinction in mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:5062–5073. doi: 10.1523/JNEUROSCI.0079-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2647–2652. doi: 10.1073/pnas.1213364110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroeder FA, Wang C, Van de Bittner GC, Neelamegam R, Takakura WR, Karunakaran A, Wey HY, Reis SA, Gale J, Zhang YL, Holson EB, Haggarty SJ, Hooker JM. PET Imaging Demonstrates Histone Deacetylase Target Engagement and Clarifies Brain Penetrance of Known and Novel Small Molecule Inhibitors in Rat. ACS chemical neuroscience. 2014;5:1055–1062. doi: 10.1021/cn500162j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy PJ, Feng J, Robison AJ, Maze I, Badimon A, Mouzon E, Chaudhury D, Damez-Werno DM, Haggarty SJ, Han M-H, Bassel-Duby R, Olson EN, Nestler EJ. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nature Neuroscience. 2013;16:434–440. doi: 10.1038/nn.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendricks JA, Keliher EJ, Marinelli B, Reiner T, Weissleder R, Mazitschek R. In vivo PET imaging of histone deacetylases by 18F-suberoylanilide hydroxamic acid (18F-SAHA) Journal of medicinal chemistry. 2011;54:5576–5582. doi: 10.1021/jm200620f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooker JM, Kim SW, Alexoff D, Xu Y, Shea C, Reid A, Volkow N, Fowler JS. Histone deacetylase inhibitor, MS-275, exhibits poor brain penetration: PK studies of [C]MS-275 using Positron Emission Tomography. ACS chemical neuroscience. 2010;1:65–73. doi: 10.1021/cn9000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SW, Hooker JM, Otto N, Win K, Muench L, Shea C, Carter P, King P, Reid AE, Volkow ND, Fowler JS. Whole-body pharmacokinetics of HDAC inhibitor drugs, butyric acid, valproic acid and 4-phenylbutyric acid measured with carbon-11 labeled analogs by PET. Nuclear medicine and biology. 2013;40:912–918. doi: 10.1016/j.nucmedbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo YJ, Muench L, Reid A, Chen J, Kang Y, Hooker JM, Volkow ND, Fowler JS, Kim SW. Radionuclide labeling and evaluation of candidate radioligands for PET imaging of histone deacetylase in the brain. Bioorganic & medicinal chemistry letters. 2013;23:6700–6705. doi: 10.1016/j.bmcl.2013.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid AE, Hooker J, Shumay E, Logan J, Shea C, Kim SW, Collins S, Xu Y, Volkow N, Fowler JS. Evaluation of 6-([(18)F]fluoroacetamido)-1-hexanoicanilide for PET imaging of histone deacetylase in the baboon brain. Nuclear medicine and biology. 2009;36:247–258. doi: 10.1016/j.nucmedbio.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh H-H, Tian M, Hinz R, Young D, Shavrin A, Mukhapadhyay U, Flores LG, Balatoni J, Soghomonyan S, Jeong HJ, Pal A, Uthamanthil R, Jackson JN, Nishii R, Mizuma H, Onoe H, Kagawa S, Higashi T, Fukumitsu N, Alauddin M, Tong W, Herholz K, Gelovani JG. Imaging epigenetic regulation by histone deacetylases in the brain using PET/MRI with 18F-FAHA. NeuroImage. 2013;64:630–639. doi: 10.1016/j.neuroimage.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Schroeder FA, Wey H-Y, Borra R, Wagner FF, Reis SA, Kim SW, Holson EB, Haggarty SJ, Hooker JM. In vivo imaging of histone deacetylases (HDACs) in the central nervous system and major peripheral organs. Journal of Medicinal Chemistry. 2014;57:7999–8009. doi: 10.1021/jm500872p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Eessalu TE, Barth VN, Mitch CH, Wagner FF, Hong Y, Neelamegam R, Schroeder FA, Holson EB, Haggarty SJ, Hooker JM. Design, synthesis, and evaluation of hydroxamic acid-based molecular probes for in vivo imaging of histone deacetylase (HDAC) in brain. American journal of nuclear medicine and molecular imaging. 2014;4:29–38. [PMC free article] [PubMed] [Google Scholar]
- 21.Gunn RN, Murthy V, Catafau AM, Searle G, Bullich S, Slifstein M, Ouellet D, Zamuner S, Herance R, Salinas C, Pardo-Lozano R, Rabiner EA, Farre M, Laruelle M. Translational characterization of [11C]GSK931145, a PET ligand for the glycine transporter type 1. Synapse. 2011;65:1319–1332. doi: 10.1002/syn.20966. [DOI] [PubMed] [Google Scholar]
- 22.Turkheimer FE, Selvaraj S, Hinz R, Murthy V, Bhagwagar Z, Grasby P, Howes O, Rosso L, Bose SK. Quantification of ligand PET studies using a reference region with a displaceable fraction: application to occupancy studies with [(11)C]-DASB as an example. Journal of cerebral blood flow and metabolism. 2012;32:70–80. doi: 10.1038/jcbfm.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirvonen J, Terry GE, Halldin C, Pike VW, Innis RB. Approaches to quantify radioligands that wash out slowly from target organs. European Journal of Nuclear Medicine and Molecular Imaging. 2010;37:917–919. doi: 10.1007/s00259-010-1426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C, Schroeder FA, Hooker JM. Visualizing epigenetics: current advances and advantages in HDAC PET imaging techniques. Neuroscience. 2014;264:186–197. doi: 10.1016/j.neuroscience.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black KJ, Snyder AZ, Koller JM, Gado MH, Perlmutter JS. Template images for nonhuman primate neuroimaging: 1. Baboon. NeuroImage. 2001;14:736–743. doi: 10.1006/nimg.2001.0752. [DOI] [PubMed] [Google Scholar]
- 26.Ichise M, Toyama H, Innis RB, Carson RE. Strategies to improve neuroreceptor parameter estimation by linear regression analysis. Journal of cerebral blood flow and metabolism. 2002;22:1271–1281. doi: 10.1097/01.WCB.0000038000.34930.4E. [DOI] [PubMed] [Google Scholar]
- 27.Logan J. Graphical analysis of PET data applied to reversible and irreversible tracers. Nuclear medicine and biology. 2000;27:661–670. doi: 10.1016/s0969-8051(00)00137-2. [DOI] [PubMed] [Google Scholar]
- 28.Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, Holden J, Houle S, Huang S-C, Ichise M, Iida H, Ito H, Kimura Y, Koeppe RA, Knudsen GM, Knuuti J, Lammertsma AA, Laruelle M, Logan J, Maguire RP, Mintun MA, Morris ED, Parsey R, Price JC, Slifstein M, Sossi V, Suhara T, Votaw JR, Wong DF, Carson RE. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. Journal of cerebral blood flow and metabolism. 2007;27:1533–1539. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- 29.Turkheimer FE, Hinz R, Cunningham VJ. On the undecidability among kinetic models: from model selection to model averaging. Journal of cerebral blood flow and metabolism. 2003;23:490–498. doi: 10.1097/01.WCB.0000050065.57184.BB. [DOI] [PubMed] [Google Scholar]
- 30.Akaike H. A new look at the statistical model identification. Automatic Control, IEEE Transactions on. 1974;19:716–723. [Google Scholar]
- 31.Cunningham VJ, Rabiner EA, Slifstein M, Laruelle M, Gunn RN. Measuring drug occupancy in the absence of a reference region: the Lassen plot re-visited. Journal of cerebral blood flow and metabolism. 2010;30:46–50. doi: 10.1038/jcbfm.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lassen NA, Bartenstein PA, Lammertsma AA, Prevett MC, Turton DR, Luthra SK, Osman S, Bloomfield PM, Jones T, Patsalos PN. Benzodiazepine receptor quantification in vivo in humans using [11C]flumazenil and PET: application of the steady-state principle. Journal of cerebral blood flow and metabolism. 1995;15:152–165. doi: 10.1038/jcbfm.1995.17. [DOI] [PubMed] [Google Scholar]