Abstract
A20 is an anti-inflammatory protein linked to multiple human diseases, however the mechanisms by which A20 prevents inflammatory disease are incompletely defined. We now find that A20 deficient T cells and fibroblasts are susceptible to caspase independent and RIPK3 dependent necroptosis. Global RIPK3 deficiency significantly rescues the survival of A20 deficient mice. A20 deficient cells exhibit exaggerated formation of RIPK1-RIPK3 complexes. RIPK3 undergoes physiological ubiquitination at lysine 5 (K5), and this ubiquitination event supports the formation of RIPK1-RIPK3 complexes. The catalytic cysteine of A20’s deubiquitinating motif is required for inhibiting RIPK3 ubiquitination and RIPK1-RIPK3 complex formation. These studies link A20 and RIPK3 ubiquitination to necroptotic cell death, and suggest new mechanisms by which A20 may prevent inflammatory disease.
A20 is a deubiquitinating enzyme that inhibits NF-kB activation and restricts TNF-induced apoptosis1–4. A20 is a potent anti-inflammatory protein linked to multiple human autoimmune diseases and to human malignancies5, 6. Polymorphisms in the human TNFAIP3 gene (which encodes the A20 protein) are associated with reduced A20 function or expression that confer susceptibility to autoimmune diseases7, 8. Deletion of A20 in mice leads to widespread tissue inflammation and perinatal lethality2. A20 regulates multiple signaling cascades and as such, plays distinct physiological functions in different cell types5, 6. In myeloid cells, A20 prevents inflammation by restricting the activation of the transcription factor NF-κB downstream signals from TLRs, NOD2 and other innate immune receptors4, 9–14. These signals lead to the production of pro-inflammatory cytokines such as interleukin 6 (IL-6) and TNF and co-stimulatory molecules that activate other innate immune cells and lymphocytes and lead to autoimmune and inflammatory diseases. In A20-deficient B cells, exaggerated B cell receptor- and CD40-triggered NF-κB activation leads to increased B cell survival and autoimmunity15–17. Hence, A20 inhibits NF-κB actvation in various cell types to prevent inflammatory and autoimmune diseases.
The biochemical mechanisms by which A20 restricts signals leading to NF-κB activation are complex and incompletely understood. Ubiquitination of signaling proteins can facilitate their recruitment to non-degradative signaling complexes, often mediated by K63-linked or linear polyubiquitin chains18. A20 is an unusual protein that utilizes two distinct motifs to remove activating K63-linked polyubiquitin chains from substrates and build degradative K48-linked ubiquitin chains3, 4, 19, 20. A20 may also disrupt E2-E3 ubiquitin ligase interactions by destabilizing E2 enzymes21. A20 also possesses ubiquitin binding motifs that support its interaction with RIPK1, E2 and IKKγ19, 22–25. In addition, A20 binds E3 ubiquitin ligases such as TRAF2 and TRAF6, ubiquitin sensors, such as ABIN-1 and ABIN-2, and other proteins (e.g., TAX1BP1) that may collaborate with A20 to perform its critical biochemical functions26. A20’s motifs and protein interactions suggest that A20 regulates multiple signaling cascades by modifying the ubiquitination of key signaling proteins.
Here we have investigated the physiological function of A20 in mouse T cells. We observed decreased expansion of A20-deficient T cells due to exaggerated cell death, and describe a previously unknown function for A20 in protecting T cells against necroptosis, a caspase-independent form of programmed cell death. T cell-specific RIPK3 deficiency restored cell survival in A20-deficient T cells, and global RIPK3 deficiency partially rescued the perinatal lethal phenotype of Tnfaip3−/− (called A20−/− here) mice. RIPK3 ubiquitination at lysine 5 (K5) of RIPK3 supports RIPK1-RIPK3 complex formation and necroptosis. A20 utilizes its deubiquitinating motif to restrict RIPK3 ubiquitination and the formation of necroptotic RIPK1-RIPK3 complexes.
Results
A20 supports the survival of activated T cells
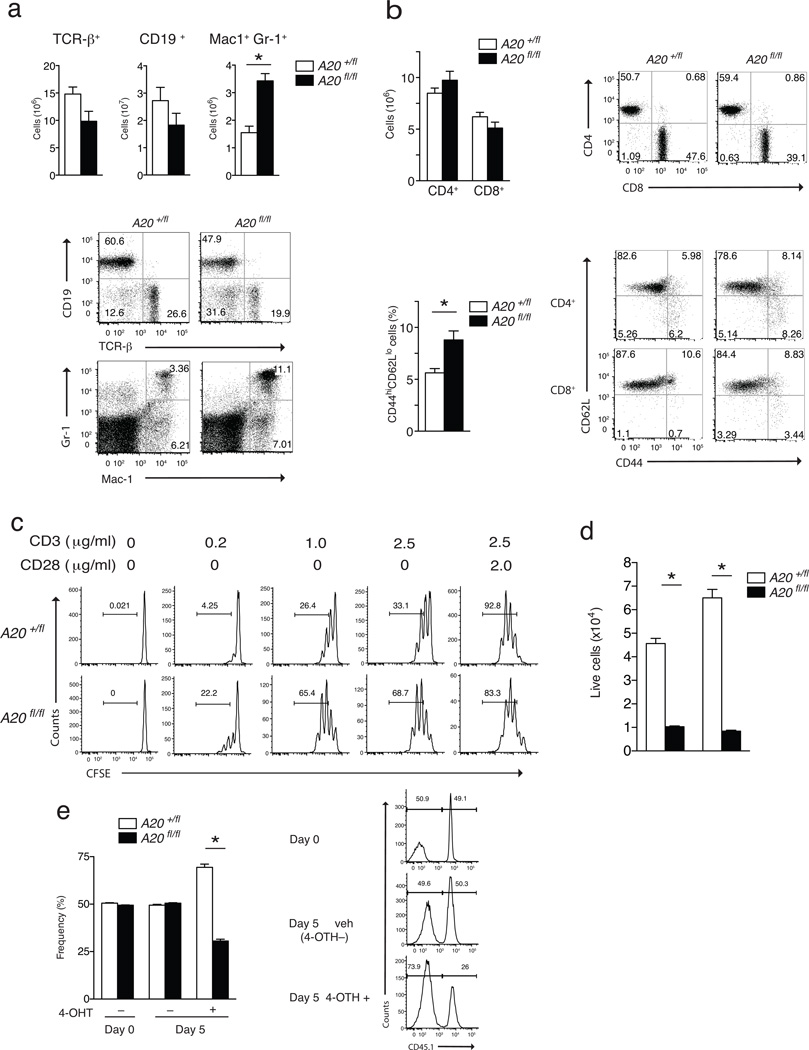
Tnfaip3fl/fl CD4-Cre mice (called A20fl/fl CD4-Cre here) developed normally and survived for at least 6 months without gross disease, suggesting that conditional T cell-specific deletion of A20 does not profoundly perturb cellular immune homeostasis (data not shown). The numbers and frequencies of CD4+CD8+ double positive and CD8+ and CD4+ single positive thymocytes were normal (data not shown). The total numbers of splenic T cells in A20fl/fl CD4-Cre mice were similar to those in control A20+/fl CD4-Cre mice and A20+/+ CD4-Cre mice, suggesting that T cell development occurs normally in the absence of A20 (Fig. 1a and data not shown). Analysis of T cell activation markers revealed a greater frequency of T cells with a memory phenotype (CD44hiCD62lo in A20fl/fl CD4-Cre mice compared to A20+/fl CD4-Cre mice suggesting a role for A20 in regulating T cell responses following antigen exposure (Fig. 1b). Spontaneous T cell activation in A20fl/fl CD4-Cre mice was accompanied by modestly increased myeloid cell (Mac-1+, Gr1+), but not B cell (CD19+), numbers compared to A20+/fl CD4-Cre mice (Fig. 1a). We observed no difference in all these parameters between A20+/fl CD4-Cre mice and A20+/+ CD4-Cre mice, so A20+/fl CD4-Cre mice were used for controls in subsequent experiments.
Figure 1. A20 supports T cell expansion.
(a) FACS analyses of splenocytes from unperturbed A20fl/fl CD4-Cre (fl/fl) and control A20+/fl CD4-Cre (+/fl) mice. Quantitation of T (TCR-β+), B (CD19+), and myeloid (Mac-1+, Gr-1+) cell numbers are shown above; representative FACS plots are shown below. (b) FACS analyses of lymph node T cells from unperturbed A20fl/fl CD4-Cre (fl/fl) and control A20+/fl CD4-Cre (+/fl) mice. Quantitation of CD4+ and CD8+ T cell numbers and percentages of CD4 T cells expressing CD44highCD62low are shown at left; representative FACS plots shown at right. * indicates p < 0.05 by Student’s T test; error bars indicate standard deviations; n = 4 and 6 pairs of mice from 2 independent experiments for (a) and (b). (c) Representative FACS data showing CFSE dilution of naïve CD4+ T cells of the indicated genotypes after stimulation with the indicated doses of agonist anti-CD3 (2.5 µg/ml and anti-CD28 (2.0 µg/ml) antibodies in vitro for three days. Numbers in each plot indicate percentages of cells that have diluted CFSE (i.e., divided). n = 2 pairs of mice, 2 independent experiments. (d) Live cell numbers of TCR activated naïve CD4+ T cells from mixed cultures of A20fl/fl CD4-Cre (solid columns) and A20+/fl CD4-Cre (open columns) CD4+ T cells. Numbers of live (DAPI negative) CD4+ T cells of the indicated genotypes 48 and 72 hrs after in vitro stimulation with anti-CD3 and anti-CD28 antibodies as in (c) above. * indicates p < 0.05 by ANOVA; error bars indicate standard deviations; n = 3 pairs of mice, 5 independent experiments. (e) Reduced survival of CD4+ T cells acutely rendered A20 deficient. Naïve CD4+ T cells were purified from CD45 congenic A20fl/fl ROSA/ER-Cre and A20+/fl ROSA/ER-Cre mice, mixed, and treated with 4-OH-tamoxifen in the indicated samples during stimulation with anti-CD3 and anti-CD28 antibodies. Deletion of A20 protein from T cells was confirmed by immunoblotting (data not shown). The percentages of live CD4+ T cell numbers from each of the two genotypes in each sample are measured (cell %) on the indicated days. Means and standard deviations are indicated. * indicates p < 0.05 between genotypes of mice. Representative histograms of CD45.1 expression on DAPI negative (live) cells are shown at right. Numbers within each histogram show the percentage of CD45.1+ (A2flLfl) and CD45.1 (A20+/fl) cells within each sample. n = 2 pairs of mice, 2 independent experiments
To investigate the role of A20 in T cell responses, we purified naïve CD4+ T cells from A20fl/fl CD4-Cre and A20+/fl CD4-Cre mice, and stimulated these cells with anti-CD3 and anti-CD28. A20fl/fl CD4-Cre T cells were activated as readily as control cells, as indicated by the expression of CD69 and CD44 activation markers and by expression of IL-2 (data not shown). TCR-stimulated A20fl/fl CD4-Cre T cells diluted CFSE more than control cells (Fig. 1c), indicating that activated A20-deficient CD4+ T cells cycle more than control CD4-Cre T cells. However, the percentage of A20fl/fl CD4-Cre T cells of the total numbers of live T cells on day 2 and 3 after stimulation was markedly lower compared to control cells (Fig. 1d), indicating that A20 deficiency impairs the survival of activated T cells in a cell-autonomous fashion. To normalize potential differences in cytokine production and investigate the cell-autonomous role of A20 in regulating T cell function, we interbred A20fl CD4-Cre mice with CD45.1 congenic mice and stimulated equal (1:1) mixes of sorted naïve congenic A20fl/fl CD4-Cre and control A20+/fl CD4-Cre CD4+ T cells in vitro. In these mixed cultures, both A20-deficient and A20-sufficient T cells were exposed to the same amounts of IL-2 and other T cell-derived cytokines. These experiments confirmed that A20fl/fl CD4-Cre T cells expanded poorly compared to A20+/fl CD4-Cre CD4+ T cells (data not shown). To eliminate potential development defects contributed by A20 deficiency and to confirm that A20 directly regulates the survival of mature peripheral T cells, we interbred A20fl/fl mice with ROSA26-ER/Cre mice, which allows the 4-OH-tamixifen inducible deletion of A20. A20fl/fl ROSA26-ER/Cre mice had normal lymphoid populations (data not shown). Sorted naïve CD4+ T cells from A20fl/fl ROSA26-ER/Cre and control A20+/fl ROSA26-ER/Cre mice were mixed and stimulated in vitro with anti-CD3 and anti-CD28 antibodies in the presence of 4-OH-tamixifen for three days to induce the efficient deletion of A20 protein (Supplementary Fig. 1). Acute deletion of A20 in A20fl/fl ROSA26-ER/Cre T cells resulted in increased cell death compared to A20+/fl ROSA26-ER/Cre T cells (Fig. 1e), suggesting that A20 directly supports the survival of activated T cells.
Increased RIPK1-RIPK3 complexes in activated A20-deficient T cells
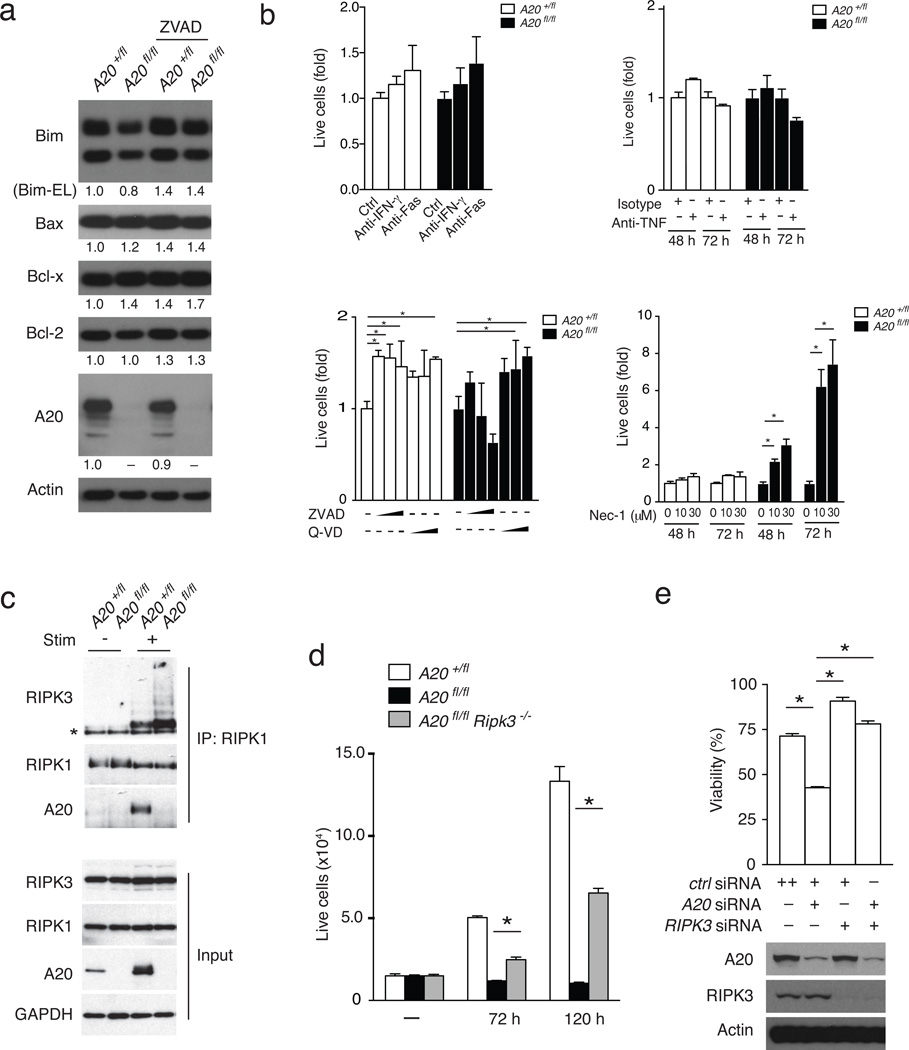
Activated A20-deficient B cells express increased amounts of Bcl-x, which renders them resistant to Fas-mediated death15. To investigate how A20 protects survival of activated T cells, we assessed the expression of Bcl-2 family proteins in A20-deficient T cells. Immunoblotting revealed that the expression of Bim, Bax, Bcl-x and Bcl-2 proteins was similar in activated A20fl/fl CD4-Cre and A20+/fl CD4-Cre T cells (Fig. 2a). Increased A20fl/fl CD4-Cre T cell death was also seen after blockade of TNF, Fas or IFN-γ, suggesting cell death in A20-deficient cells is not triggered by these stimuli (Fig. 2b). Importantly, the pan-caspase inhibitors ZVAD or Q-AD did not abrogate the increased cell death in stimulated A20fl/fl CD4-Cre CD4+ T cells, indicating a caspase-independent death pathway (Fig. 2b). Activated T cells die by necroptosis in the absence of the pro-apoptotic proteins FADD or caspase 827, 28. Necroptosis requires RIPK1 kinase activity and the formation of RIPK1-RIPK3 complexes29–31. The RIPK1 kinase inhibitor necrostatin 1 (Nec-1) selectively rescued the survival and expansion of TCR-activated A20fl/fl CD4-Cre T cells compared to control cells (Fig. 2b), while an inactive form of Nec-1 was less effective in rescuing A20fl/fl CD4-Cre T cell survival (Supplementary Fig. 2). Thus, RIPK1 kinase activity contributes to the increased death of A20-deficient T cells following in vitro activation.
Figure 2. A20 inhibits T cell necroptosis.
(a) Immunoblotting analyses of indicated survival proteins in TCR stimulated CD4+ T cells 13 hours after stimulation. Quantitation of immunoblots normalized to actin are shown below each blot. Actin and A20 protein levels are shown below as controls. Data are representative of 3 experiments using cells from 2 pairs of mice each. (b) Nec-1 sensitive death of A20fl/fl CD4-Cre T cells. Congenically marked A20fl/fl and A20+/fl T cells were co-cultured for 3 days with TCR stimulation in the presence of the indicated inhibitory antibodies or small molecules. In each graph, the numbers of live cells at each condition were normalized to the numbers of cells of the same genotype obtained without blocking antibody or inhibitor. In lower left graph, ZVAD was used at 0, 10, 50 and 100 µM; and QVAD was used at 0, 20, and 40 µM for each genotype. Bars indicate standard deviations; * indicates p < 0.05 by 2-way ANOVA; n = 2 experiments using cells from 2 pairs of mice each. (c) Enhanced RIPK1-RIPK3 complexes in A20fl/fl CD4-Cre T cells. Immunoblotting analyses of RIPK1 associated proteins in A20fl/fl and A2flL/+ CD4-Cre T cells before (−) and eight hours after (+) TCR stimulation. Note induction of A20 by TCR stimulation in A20+/fl CD4-Cre T cells. Note induction of RIPK1 associated RIPK3 in both A20fl/fl and A20+/fl CD4-Cre T cells, with greater RIPK3 recruitment in A20fl/fl CD4-Cre T cells. Data are representative of 3 experiments. (d) Rescue of survival of A20fl/fl CD4-Cre T cells by RIPK3 deficiency. Live cell numbers of CD4+ T cells from indicated genotypes of mice at indicated time points after stimulation with anti-CD3, anti-CD28, plus ZVAD. Bars represent standard deviations; * indicates p < 0.05; n = 3 sets of mice from 2 experiments. (e) Rescue of A20 deficient Jurkat I9.2 cells by RIPK3 deficiency. Caspase 8 deficient Jurkat I9.2 cells were transfected with the indicated combinations of siRNAs, and stimulated 72 hours later with TNF. The percentage of surviving cells compared to non-stimulated Jurkat I9.2 cells is shown. Immunoblots for A20, RIPK3 and actin on treated cells are shown below as controls. Bars indicate standard deviations; * indicates p < 0.05 by one way ANOVA; n = 2 experiments.
Immunoprecipitation of RIPK1 from TCR-activated CD4+ T cells revealed that A20fl/fl CD4-Cre T cells contain significantly greater amounts of RIPK1-associated RIPK3 than A20+/fl CD4-Cre T cells (Fig. 2c), indicating that RIPK1-RIPK3 complexes accumulate in A20-deficient T cells and suggesting that A20 protects activated T cells from necroptotic death. To further determine the role of RIPK3 in mediating increased cell death in A20fl/fl CD4-Cre T cells, we interbred A20fl/fl CD4-Cre mice with Ripk3−/− mice. Following in vitro TCR stimulation for 72 or 120 hours in the presence of ZVAD, the numbers of A20fl/fl CD4-Cre Ripk3−/− T cells were greater than A20fl/fl T cells, though less than control A20+/fl T cells (Fig. 2d), indicating that RIPK3 deficiency significantly rescued the survival of activated A20-deficient T cells. The viability of TCR-stimulated caspase 8 deficient Jurkat I9.2 T cells was reduced in cells treated with A20 siRNA relative to control siRNA, and RIPK3 siRNA abrogated the increased cell death observed in A20-deficient Jurkat I9.2 T cells (Fig. 2e), suggesting the epistatic relationship between A20 and RIPK3 is preserved in human cells. Thus, exaggerated death of activated A20-deficient T cells is caspase-independent, is associated with increased RIPK1-RIPK3 complex formation and requires the kinase activity of RIPK1 and the expression of RIPK3.
A20 and RIPK3 control T cell survival during in vivo activation
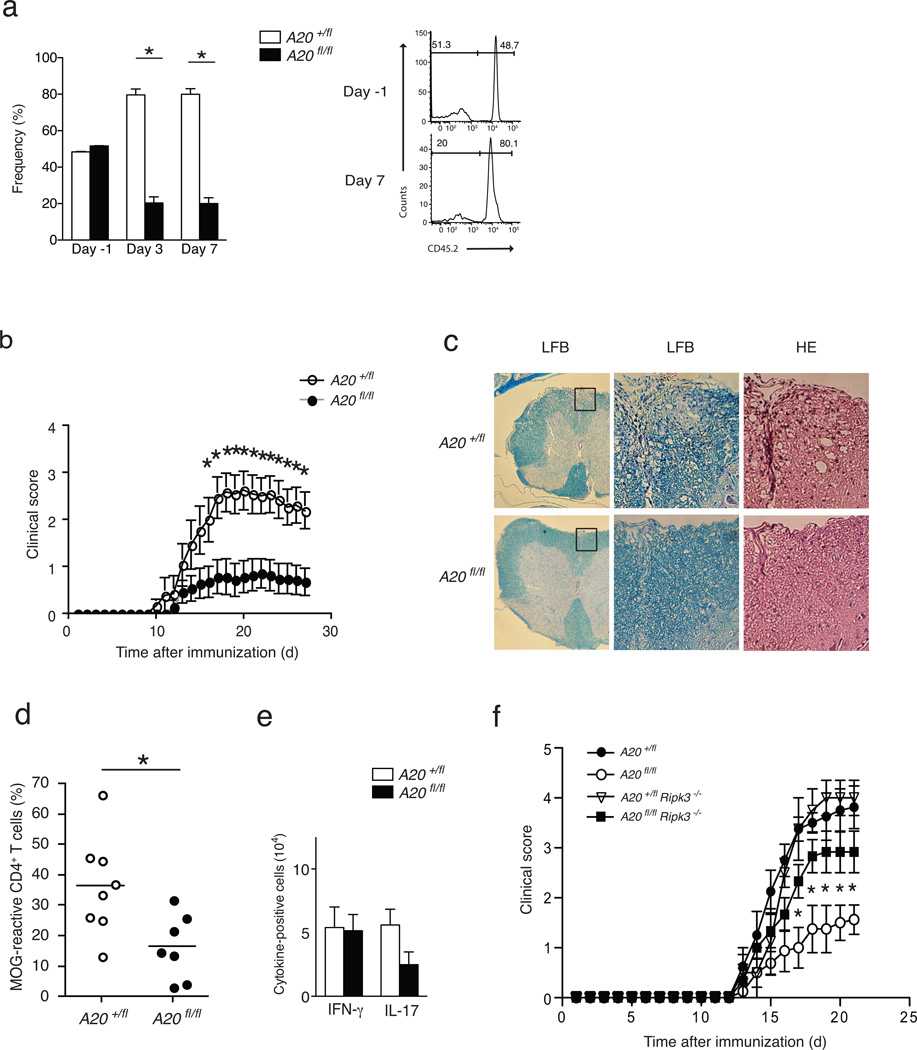
To determine whether A20 regulates T cell survival and responses in vivo, we interbred A20fl mice with OT-II TCR transgenic mice. We adoptively co-transferred naive A20+/fl CD4-Cre OT-II CD4+ (CD45.1/2) T cells and A20fl/fl CD4-Cre OT-II CD4+ (CD45.1) T cells into C57BL/6 (CD45.2) recipients, immunized these mice with LPS plus ovalbumin peptide (OVA) 24 h after transfer (day 0) and quantitated the numbers of splenic OT-II T cells from both donor genotypes at various time points after immunization. This experimental design allowed us to assess the response of A20-deficient and A20-sufficient T cells in the same cytokine and environmental factors milieu. Consistent with the in vitro data, A20fl/fl CD4-Cre OT-II CD4+ T cells expanded poorly relative to control CD4-Cre OT-II CD4+ T cells 3 and 7 days activated within the same miceafter OVA challenge (Fig. 3a). This result indicates that A20 supports expansion of activated T cells in vivo.
Figure 3. A20 supports antigen specific and autoimmune T cell responses in vivo.
(a) Reduced in vivo expansion of A20fl/fl CD4-Cre OT-II CD4+ T cells. FACS purified naïve (CD4+ CD62LHi CD25) CD4+ OT-II cells from congenic A20+/fl CD4-cre CD45.1/45.2 and A20FL/FL CD4-cre CD45.1 mice were mixed 1:1, and co-transferred into congenic CD45.2 C57BL/6J recipients (Day −1). Mice were immunized with LPS plus OVA peptide 24 hours later (Day 0). The percentage of live OT-II CD4+ T cells from each genotype on the indicated days is indicated. Each time point represents data from 3 mice. Representative histograms at right show CD45.2 expression after gating on CD45.1+ (donor) cells. Numbers within each histogram reflect the percentage of CD45.1+ (donor) cells that are either CD45.2 (A20fl/fl) or CD45.2+ (A20+/fl). Data analyzed by two-way ANOVA, error bars represent mean +/− SEM; n = 3 mice of each genotype from 2 independent expts. (b) Reduced EAE in A20fl/fl CD4-Cre mice. Serial EAE clinical scores of A20fl/fl CD4-Cre (fl/fl) and control A20+/fl CD4-Cre (+/fl) mice after immunization with MOG plus CFA. Means and standard errors are indicated. * indicates p < 0.05 between genotypes of mice on the indicated days. Data were obtained from 14 A20fl/fl CD4-Cre and 13 control mice and represent 2 experiments. (c) Representative histopathological analyses of spinal cord sections from the indicated mice 30 days after immunization. Luxol fast blue stains (LFB) highlighting myelin content of spinal cord sections from indicated mice (low magnification, left panels). Higher magnification images of boxed areas showing LFB and hematoxylin and eosin (HE) stains of tissue with infiltrating lymphocytes (right panels). Data are representative of 6 sets of mice and 2 experiments. (d) FACS quantitation of MOG reactive LN CD4+ T cells. Nine days after immunization with MOG, LN T cells from the indicated mice were harvested, labeled with CFSE, and re-stimulated with MOG for 3 days in vitro. The percentage of CFSE labeled T cells in diluted versus non-diluted peaks after 3 days is indicated. Horizontal lines indicate means. * indicates p < 0.05 by Students T test; n = 7 and 8 mice from 2 experiments. (e) Reduced TH17 cells from A20fl/fl CD4-Cre mice during EAE. Lymph node CD4+ T cells from MOG immunized mice of the indicated genotypes were harvested, restimulated in vitro, and quantitated by flow cytometry for intracellular expression of the indicated cytokines. The numbers of CD4+ T cells expressing IFNγ and IL17 are displayed. Data analyzed by Student’s T test; horizontal lines indicate means +/− SEMs; n = 3 sets of mice in 2 experiments. (f) RIPK3 deficiency restores EAE susceptibility of A20fl/fl CD4-Cre mice. Serial EAE clinical scores of RAG-1−/− mice bearing T cells of the indicated genotypes after immunization with MOG. Means and standard errors are indicated. Note that A20fl/fl CD4-Cre RIPK3−/− T cells (filled squares) cause significantly more severe EAE symptoms than A20fl/fl CD4-Cre T cells (open circles). Data analyzed by two way ANOVA; p < 0.05 between A20FL/FL CD4-Cre and A20fl/fl CD4-Cre RIPK3−/− T cells for all days after day 17; n = 5 mice of each genotype in 2 experiments.
We next tested the susceptibility of A20fl/fl CD4-Cre mice to experimental autoimmune encephalomyelitis (EAE), a model of T cell-mediated autoimmunity32. Following immunization with a myelin oligodendrocyte glycoprotein (MOG) peptide in Freund’s complete adjuvant, A20fl/fl CD4-Cre mice developed markedly less severe motor neuron symptoms than control A20+/fl CD4-Cre mice (Fig. 3b). Histological studies revealed fewer lymphoid infiltrates and greater myelin preservation in spinal cord sections from A20fl/fl CD4-Cre mice than control mice (Fig. 3c), while ex vivo MOG stimulation of lymph node CD4+ T cells showed decreased proliferation of A20fl/fl CD4-Cre T cells in vitro compared to A20+/fl CD4-Cre T cells (Fig. 3d). TH17 cells are important in the pathogenesis of EAE33. We thus quantitated the number of TH17 cells from the lymph nodes of MOG-immunized mice and found fewer TH17 cells in the A20fl/fl CD4-Cre lymph nodes compared to A20+/fl CD4-Cre controls (Fig. 3e), which paralleled the reduction of MOG-specific A20-deficient CD4+ T cells (Fig. 3d). Following the in vitro stimulation of naïve CD4+ T cells under TH1 or TH17 polarizing conditions, fewer IFN-γ-producing and IL-17-producing T cells were obtained from A20fl/fl CD4-Cre T cell cultures compared to control A20+/fl CD4-Cre cultures (Supplementary Fig. 3), while TH17 and TH1 differentiation from A20fl/fl CD4-Cre Ripk3−/− T cells was equal to that from A20+/fl CD4-Cre T cells and A20+/fl CD4-Cre Ripk3−/− T cells (Supplementary Fig. 3). Hence, T cell-specific A20 deficiency impairs T cell activation and CD4+ T cell differentiation into TH17 and TH1 cells and renders mice less susceptible to EAE.
We next examined if RIPK3-dependent necroptosis affected the survival of A20-deficient T cells in vivo. RIPK3 is expressed in multiple tissues, and Ripk3fl/fl mice have not yet been described. Hence, we adoptively transferred CD4+ T cells from A20fl/fl CD4-Cre Ripk3−/− or A20fl/fl CD4-Cre Ripk3+/+ control mice into Rag1−/− hosts, which were subsequently immunized with MOG peptide. This experimental design allowed interrogation of the T cell-specific function of A20 and RIPK3 in vivo. Rag1−/− mice that received A20fl/fl CD4-Cre Ripk3−/− T cells developed more severe EAE than RAG-1−/− mice that received A20fl/fl CD4-Cre Ripk3+/+ T cells (Fig. 3f). Taken together, these data indicate that A20 protects activated CD4+ T cells from RIPK3-dependent necroptosis in vivo.
A20 protects mice from RIPK3-dependent necroptosis
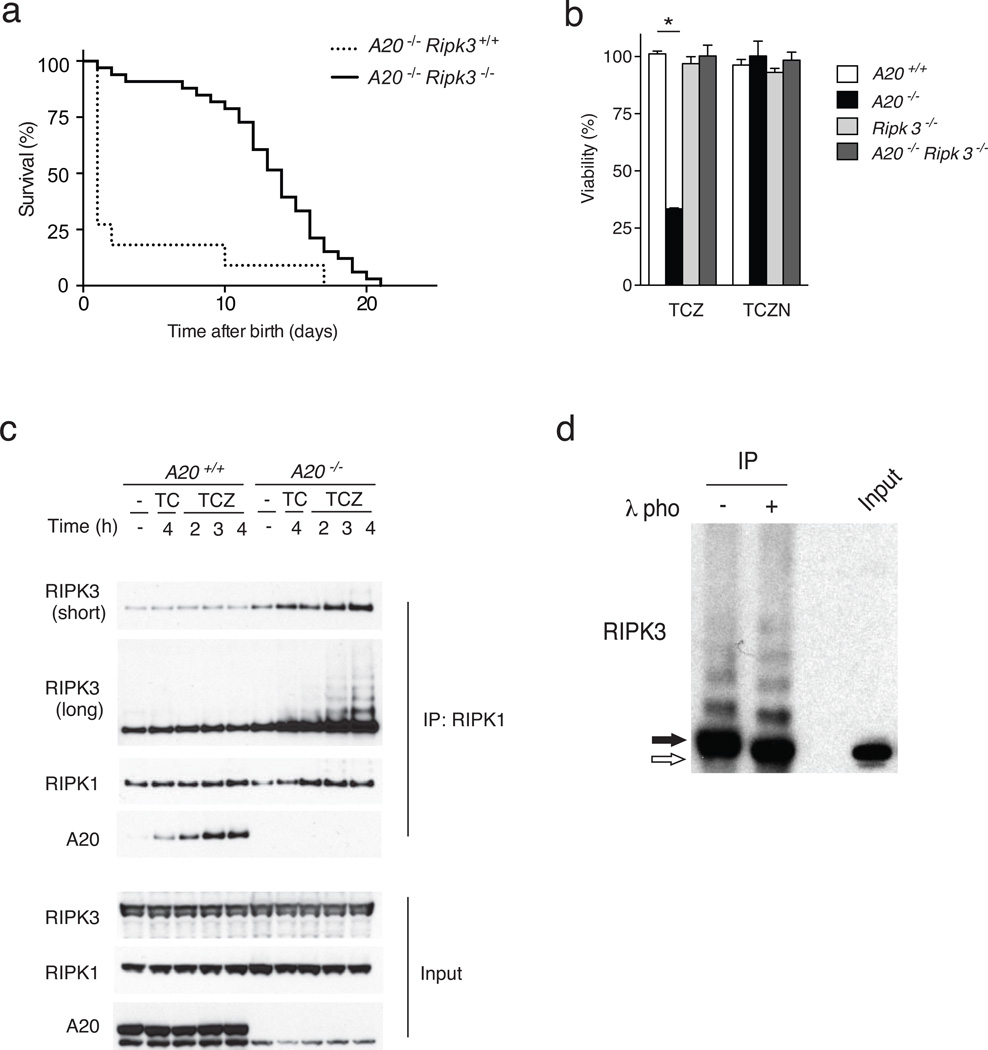
Necroptotic death typically triggers inflammation in vivo due to the release of intracellular molecules from dying cells34. Tissue death and inflammation are hallmarks of A20−/− mice2. To test if A20 might prevent necroptosis in cells other than T cells, we derived mice bearing germline deletion of the floxed A20 exon 2 by interbreeding A20fl/fl mice with B6.EIIA-Cre mice (which deletes floxed alleles in the early mouse embryo)15. Mice bearing germline deletion of A20 exon 2, defined as A20ko2 mice, were bred to C57BL/6 mice to generate A20ko2 mice that lacked the EIIA-Cre transgene. A20ko2 mice were subsequently crossed with Ripk3−/− mice to generate A20ko2/ko2 Ripk3−/− double mutant mice (in the C57BL/6 background). While most A20ko2/ko2 Ripk3+/+ mice died within 1 day of birth, A20ko2/ko2 Ripk3−/− mice survived 2–3 weeks before succumbing to inflammatory death (Fig. 4a). The partial rescue of A20ko2/ko2 mice lethality by RIPK3 deficiency suggests that increased systemic necroptosis contributes to the perinatal lethality observed in A20ko2/ko2 mice.
Figure 4. A20 inhibits RIPK1-RIPK3 complex formation.
(a) Spontaneous survival of A20ko2/ko2 Ripk3−/− (A20−/− Ripk3−/−, solid line) and A20ko2/ko2 Ripk3+/+ (A20−/− Ripk3+/+, dotted line) littermate mice in days after birth. Data represent 33 A20−/− Ripk3−/− mice and 16 A20−/− Ripk3+/+ mice, with each step representing death of one mouse. The mean survival A20−/− Ripk3−/− mice was 14 days, while the mean survival of A20−/− Ripk3+/+ mice was less than 1 day, with enhanced survival of A20−/− Ripk3−/− mice over A20−/− Ripk3+/+ mice statistically significant at p < 0.0001 by Logrank test. (b) Survival of MEFs of the indicated genotypes 4 hours after stimulation with either TNF, CHX, and ZVAD (TCZ), or TNF, CHX, ZVAD, and Nec1 (TCZN). Note that Nec1 rescues cell death of A20ko2/ko2 Ripk3 cells, as does RIPK3 deficiency. Bars indicate the proportion of live cells relative to unstimulated cells of the same genotype. * indicates p < 0.05; n = . (c) RIPK1-RIPK3 complexes in stimulated MEFs. RIPK3 and A20 expression in anti-RIPK1 IPs from A20−/− or wild- type (WT, or A20+/+) MEFs stimulated with the indicated agents for the indicated time periods. Total cell lysate protein levels (input) are shown below as controls. Note the increased RIPK1 associated RIPK3 in A20−/− cells, with laddered, higher molecular weight forms of RIPK3. Note also the A20 recruitment to RIPK1 in WT cells. (d) Phosphorylation of RIPK1 associated RIPK3. A20−/− MEFs were stimulated as in (c) above, after which lysates were immunoprecipitated with anti-RIPK1, treated with lambda phosphatase (+) or control buffer (−), and analyzed by immunoblotting for RIPK3. Whole cell lysate at right (input) shows size of native RIPK3 protein as control. Note that RIPK1-associated RIPK3 is almost entirely phosphatase sensitive in stimulated A20−/− cells undergoing necroptosis.
Because increased T cells necroptosis is unlikely to be the main cause of the perinatal lethality in A20ko2/ko2 mice, we next tested whether A20 directly restricts necroptosis in non-T cells. We isolated mouse embryonic fibroblasts (MEFs) from A20ko2/ko2 and A20+/+ mice, and tested their susceptibility to cell death in response to TNF, cycloheximide (CHX) and ZVAD, a cocktail known to induce caspase-independent necroptosis. In these conditions, as assessed using CellTiter-Glo luminescence, more A20ko2/ko2 MEFs died compared to wild-type MEFs (Fig. 4b). Treatment of TNF, CHX, ZVAD-stimulated MEFs with Nec-1 rescued the viability of A20ko2/ko2 MEFs to that of wild-type MEFs (Fig. 4b), suggesting that the caspase-independent death induced by the TNF-CHX-ZVAD mix was RIPK1 kinase activity-dependent. A20ko2/ko2 Ripk3−/− MEFs were protected from TNF-CHX-ZVAD-induced necroptosis when compared to A20ko2/ko2 MEFs, indicating that A20 inhibits RIPK3-dependent necroptosis in these cells (Fig. 4b). Taken together, these results indicate that A20 protects multiple cell types from necroptosis, and suggests that uncontrolled necroptosis contributes to the perinatally lethal phenotype of A20ko2/ko2 mice.
A20 inhibits pro-necroptotic RIPK1-RIPK3 complexes
RIPK3-dependent necroptosis involves RIPK1 kinase and RIPK3 dependent formation of RIPK1-RIPK3 complexes29, 30. Accordingly, we tested whether A20 restricted the formation of RIPK1-RIPK3 complexes in a cell autonomous fashion. We immunoprecipitated RIPK1 from WT and A20ko2/ko2 MEFS treated with either TNF-CHX or TNF-CHX-ZVAD and immunoblotted for RIPK3. Treatment with TNF-CHX- ZVAD induced higher amounts of RIPK1-associated RIPK3 in A20ko2/ko2 MEFs compared to A20+/+ MEFs (Fig. 4c). Because phosphorylation of RIPK3 is required for the formation of necroptotic RIPK1-RIPK3 complexes, we tested the phosphorylation status of RIPK3 in A20ko2/ko2 MEFs29, 30. We stimulated A20ko2/ko2 MEFs with TNF-CHX-ZVAD, immunoprecipitated RIPK1, and then treated these RIPK1 immunoprecipitates with lambda phosphatase (or buffer control). Immunoblot analyses of RIPK3 expression in these samples revealed that RIPK1-associated RIPK3 was phosphorylated (Fig. 4d), suggesting that A20 inhibits the formation of pro-necroptotic RIPK1-RIPK3 complexes that contain phosphorylated RIPK3.
RIPK3 ubiquitination supports RIPK1-RIPK3 complexes and necroptosis
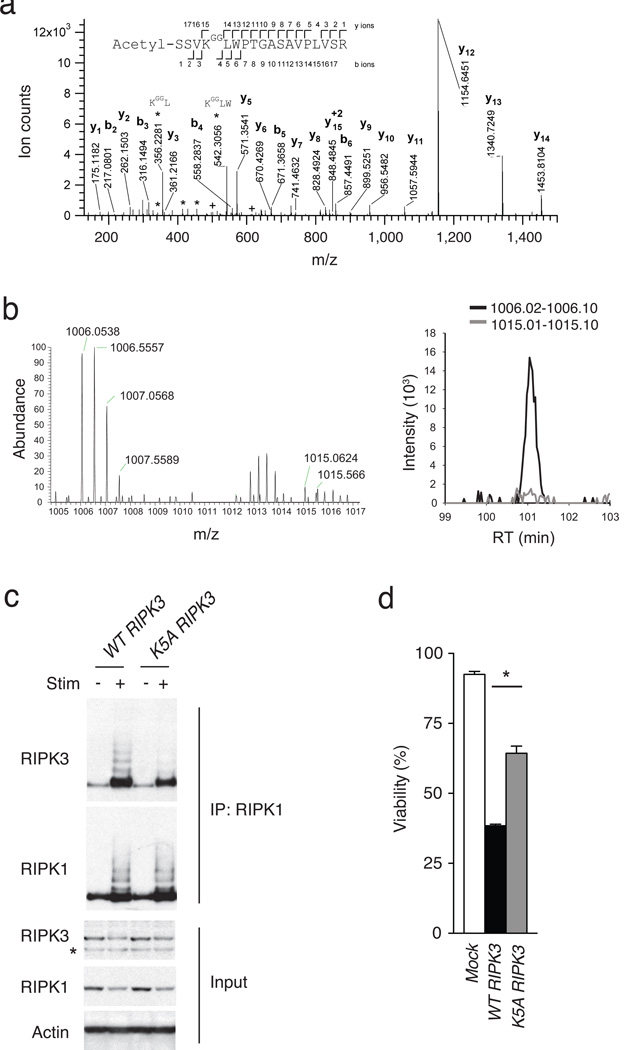
The laddered species at ~8kd intervals detected with an anti-RIPK3 antibody in the RIPK1 immunoprecipitates suggests that the RIPK3 proteins, rather than non-covalently associated co-precipitated proteins, are modified (Fig. 4c, d). To more precisely define RIPK3 ubiquitination, we used anti-gly-gly antibody assisted mass spectrometry (termed “ubiscan”) to search for ubiquitinated RIPK3 peptides in necroptotic MEFs35. We stimulated A20ko2/ko2 MEFs with TNF-CHX-ZVAD, trypsinized whole cell protein lysates, immunoprecipitated the tryptic lysates with an antibody directed against gly-gly remnants of ubiquitinated lysines and performed mass spectrometric analyses of the affinity-purified ubiquitinated peptides. Tandem mass spectra from A20ko2/ko2 MEF lysates repeatedly revealed a precursor ion with an m/z value of 1006.0525+2 (Fig. 5a). A data base search identified this species as a peptide spanning the amino acids 2 to 19 of RIPK3, which was acetylated at the N-terminus and carried a di-glycine remnant at the lysine 5 (K5) residue (theoretical monoisotopic m/z= 1006.0522+2). Because this modification was observed in endogenous proteins from TNF-CHX-ZVAD-stimulated A20ko2/ko2 MEFs, these observations indicate that RIPK3 ubiquitination at position K5 is a physiological modification.
Figure 5. RIPK3 ubiquitination supports RIPK1-RIPK3 complexes and necroptosis.
(a) Site specific ubiquitination of RIPK3. A20−/− MEFs were stimulated with TNF, CHX and ZVAD, and lysed after 4 hours. Lysates were immunoprecipitated with anti-gly-gly antibody, after which immunoprecipitates were analyzed by nanoflow-UPLC-HCD-MSMS. A HCD tandem mass spectrum obtained from a precursor ion with m/z value 1006.0525+2 from A20−/− cells undergoing necroptosis is shown. A data base search showed the spectra to identify the peptide spanning amino acids 2 to 19 of RIPK3, acetylated in the N terminus and carrying a di-glycine remnant on K5 (theoretical monoisotopic m/z= 1006.0522+2). Sequence ions are labeled in the figure, and indicated with marks over (C-terminal ions) and below (N-terminal ions) the sequence of the peptide. Prominent ions corresponding to internal fragments are labeled (*). Ions labeled (+) show fragments with masses 501.2667 and 614.3508, corresponding to ions b4 and b5 undergoing a second fragmentation, with a loss of 57.02146 mass units, corresponding to the terminal glycine on the ɛ-amino of RIPK3 K5. (b) SILAC based mass spectrometric quantitation of RIPK3 ubiquitination. Heavy (A20+/+) and light (A20−/−) isotope labeled MEFs were stimulated to undergo necroptosis, after which cell lysates were mixed and then analysed by ubiscan assisted mass spectrometry as in (a). The di-glycine remnant of RIPK3 K5 from light isotope labeled cells (A20−/− cells) was again identified at m/z = 1006, while the heavy isotope labeled cells (A20+/+ cells) contained the RIPK3 K5 peptide at m/z = 1015 (spectra on left). The relative abundance of these two forms of the ubiquitinated K5 peptide is displayed in the histogram at right. (c) K5 ubiquitination supports RIPK1-RIPK3 complexes. WT or mutant K5A RIPK3 expression constructs were transduced into A20−/− RIPK3−/− MEFs using a GFP-expressing lentivirus, after which cells were FACS-sorted to obtain pure populations of transduced (GFP+) cells. Cells were then stimulated for four hours, lysed and analysed for RIPK1-associated RIPK3 expression by immunoblotting. “+” indicates treatment with TNF, CHX, and ZVAD; “-” indicates media control. RIPK1 immunoblot of RIPK1 IP is shown below RIPK3 immunoblot as control for IP; immunoblots for RIPK3, RIPK1, and actin on whole cell lysates are shown below as controls. * indicates non-specific band below specific RIPK3 band. (d) K5 ubiquitination supports necroptosis. A20−/− Ripk3−/− MEFs complemented with RIPK3 constructs in (c) above analyzed for sensitivity to necroptosis. Survival of MEFs expressing the indicated forms of RIPK3 is shown relative to unstimulated cells. Bars indicate standard deviation. * indicates p < 0.05 when compared with cells expressing WT RIPK3.
To quantify if RIPK3 ubiquitination at position K5 is dependent upon A20, we performed stable isotope labeling (SILAC) of A20ko2/ko2 and wild-type MEFs35. A20ko2/ko2 MEFs were grown in “light” (normal) isotope labeled media and wild-type MEFs were grown in “heavy” isotope labeled media for two weeks. Complete replacement of endogenous amino acids by isotope labeled residues was confirmed by mass spectrometry (data not shown). Labeled MEFs were stimulated with TNF-CHX-ZVAD for three hours, lysates from A20ko2/ko2 and wild-type MEFs were mixed, trypsinized and immunoprecipitated for ubiquitinated peptides prior to LC-MS mass spectrometric analysis. Tandem mass spectra revealed species with m/z ~ 1006, indicating the presence of RIPK3 peptide ubiquitinated at position K5 in normal (“light”)-labeled A20ko2/ko2 MEFs (Fig. 5b). These spectra also revealed species at m/z ~1015, indicative of RIPK3 peptide modified at position K5 with “heavy” isotope labeled residues in wild-type MEF cells (Fig. 5b, left). Direct comparison of the relative intensity of these peaks showed that m/z~1006 species were ~15-fold more abundant than m/z~1015 species, thereby indicating that A20ko2/ko2 MEF cells contained 15-fold greater amounts of K5-ubiquitinated RIPK3 peptide than wild-type MEF cells (Fig. 5b, right). As A20ko2/ko2 MEFs express similar amounts of total RIPK3 protein as wild-type MEF cells (Fig. 4c), these findings indicate increased RIPK3 ubiquitination at position K5 in A20-deficient cells. K5-ubiquitinated RIPK3 peptides were not detected in lysates from unstimulated A20ko2ko2 or control MEFs (data not shown), supporting the notion that ubiquitination at position K5 of RIPK3 is induced during necroptosis.
To determine the functional importance of RIPK3 ubiquitination at position K5 in necroptosis, we mutated K5 to alanine (K5A) or arginine (K5R). Because the K5R RIPK3 mutant was not stable (data not shown), we tested the ability of K5A RIPK3 to form RIPK1-RIPK3 complexes in necroptotic cells. We virally transduced wild-type or K5A RIPK3 into A20ko2/ko2 Ripk3−/− MEFs, sorted transduced cells expressing similar amounts of RIPK3 and analyzed the amount of RIPK1-associated RIPK3 expression as well as the amount of cellular necroptosis following TNF-CHX-ZVAD stimulation. MEFs expressing the K5A RIPK3 exhibited decreased RIPK1-RIPK3 complexes when compared to cells expressing wild-type RIPK3 (Fig. 5c). Furthermore, cells expressing K5A RIPK3 survived significantly better than cells expressing wild-type RIPK3 (Fig. 5d), indicating that ubiquitination of RIPK3 at position K5 supports the formation of RIPK1-RIPK3 complexes and necroptosis.
A20 utilizes its deubiquitinating motif to restrict necroptosis
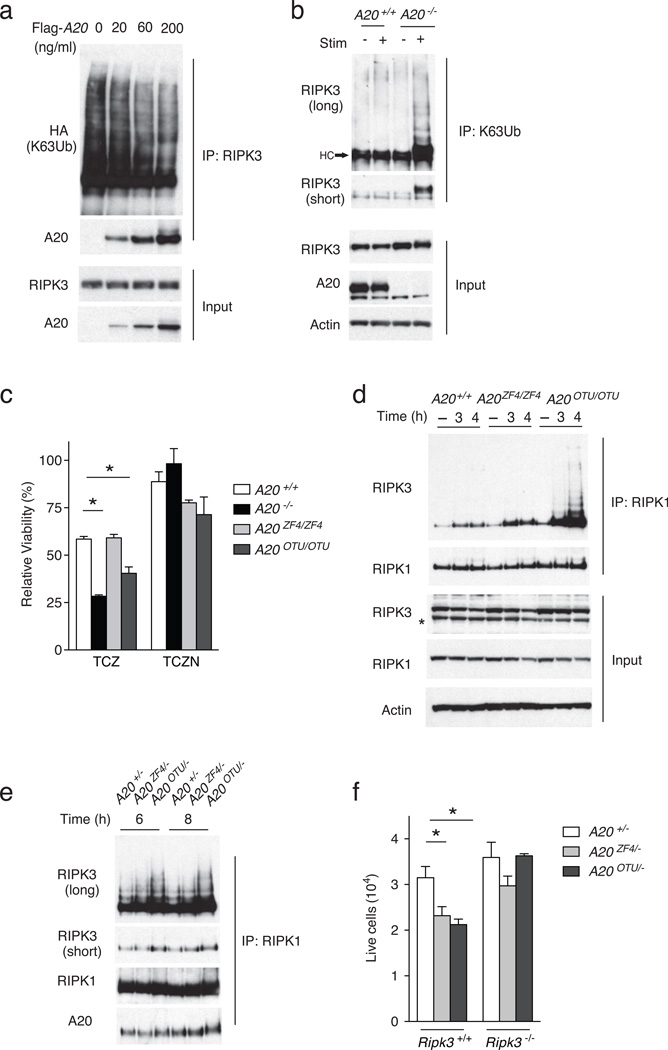
Polyubiquitin chains of various conformations are added to target proteins during cell signaling18. Because A20 restricts RIPK1 ubiquitination with K63-linked polyubiquitin chains, we investigated whether A20 also inhibits RIPK3 ubiquitination with K63-linked polyubiquitin chains. 293T cells were transfected with plasmids expressing RIPK3, and HA-K63-Ub (ubiquitin in which all ubiquitin lysines except K63 are mutated). Lysates were immunoprecipitated with anti-RIPK3 and immunoblotted with anti-HA to reveal a ladder of K63-ubiqitinated RIPK3 (Fig. 6a). Addition of increasing amounts of Flag-A20 plasmid to the transfection led to a progressive decrease in the amount of HA-K63-Ub signal (Fig. 6a), indicating that A20 inhibited the K63-linked ubiquitination of RIPK3 in a dose-dependent fashion. In addition, increased K63-linked ubiquitination of RIPK3 was found in A20ko2/ko2 MEFs compared to wild-type MEFs following stimulation with TNF-CHX-ZVAD (Fig. 6b). Thus, A20 restricts K63-linked ubiquitination of RIPK3.
Figure 6. A20 utilizes its C103 motif to inhibit RIPK1-RIPK3 complexes.
(a) A20 mediated deubiquitination of K63-ubiquitinated RIPK3. 293T cells were transfected with plasmids expressing RIPK3, HA-K63-Ub (HA-tagged ubiquitin in which all ubiquitin lysines except K63 were mutated to arginine), and FLAG-A20, immunoprecipitated with anti-RIPK3, and immunoblotted for HA (K63-ubiquitin) and A20. Immunoblots of whole cell lysates for RIPK3 and A20 are shown below as controls. (b) K63-ubiquitination of RIPK3 in A20−/− cells. A20+/+ and A20−/− MEFs were stimulated with TNF, CHX and ZVAD (TCZ) (“stim +”), and lysed under denaturing conditions (6M urea). Lysates were diluted, imunoprecipitated with anti-K63 ubiquitin antibody, and analyzed for RIPK3 expression. Short exposure of RIPK3 immunoblot shown to demonstrate increased and phosphorylated RIPK3; long exposure shown to demonstrate increased K63-ubiquitination of RIPK3. Immunoblots of whole cell lysates for RIPK3, A20 and actin are shown on whole cell lysates below as controls. Note the increased K63-ubiquitinated RIPK3 in stimulated A20−/− cells. Data are representative of 3 independent experiments. (c) Cell survival of the indicated genotypes of MEFs after stimulation with TNF, CHX and ZVAD (TCZ) or TNF, CHX, ZVAD and Nec1 (TCZN) for five hours. Bars indicate standard deviation. * indicates p < 0.05 by ANOVA when compared with WT cells. (d) RIPK1-RIPK3 complexes in indicated genotypres of MEFs treated with TCZ as in (c) above. RIPK1 expression in IPs is shown as control. RIPK1, RIPK3 and actin expression in whole cell lysates shown below as controls. (e) RIPK1-RIPK3 complexes in indicated genotypes of T cells stimulated with anti-CD3, anti-CD28, and ZVAD for the indicated time periods. Short and long exposures of RIPK1 associated RIPK3 shown; RIPK1 and A20 expression in RIPK1 IPs are shown as controls. (f) Live cell numbers of indicated genotypes of T cells after TCR stimulation as in (e) for 72 hours. Bars indicate standard deviation. * indicates p < 0.05 when compared with WT cells. Data are representative of 3 independent experiments.
To determine the mechanism by which A20 restricts RIPK3 ubiquitination, we used MEFs from two lines of knockin mice bearing point mutations that abrogate the enzymatic activity of each domain. A20OTU mice bear a mutation in the catalytic cysteine 103 (C103) residue of A20 that mediates A20’s deubiquitinating activity19. A20ZF4 mice bear point mutations (C609, C612) in the ZF4 domain that mediates A20’s binding to ubiquitinated RIP119. A20ZF4/ZF4 or wild-type MEFs showed comparable viability in response to TNF-CHX- ZVAD, whereas A20OTU/OTU MEFs had reduced viability (Fig. 6c), indicating that A20 uses its catalytic deubiquitinating Cys103 residue to inhibit necroptosis.
To understand how A20’s C103 and ZF4 motifs regulate necroptotic signaling complexes, we analyzed the formation of RIPK1-RIPK3 complexes in A20ZF4ZF4 and A20OTUOTU MEFs. After stimulation of wild-type, A20ZF4ZF4 and A20OTUOTU MEFs with TNF-CHX-ZVAD for 3 or 4 hours, protein lysates were immunoprecipitated with anti-RIPK1 and immunoblotted with anti-RIPK3. A20OTU/OTU MEFs contained markedly increased RIPK1-RIPK3 complexes when compared to wild-type or A20ZF4/ZF4 MEFs (Fig. 6d), while A20ZF4/ZF4 MEF cells contained modestly elevated RIPK1-RIPK3 complexes when compared to wild-type MEFs (Fig. 6d). RIPK1-associated RIPK3 showed a laddered pattern indicative of ubiquitination in A20OTU/OTU MEFs (Fig. 6d). A20OTU/− T cells also exhibited greater amounts of RIPK1-RIPK3 association and greater RIPK3 ubiquitination than either A20ZF4/− or A20+/− T cells following TCR stimulation (Fig. 6e), indicating that Cys103 regulates RIPK3 ubiquitination and RIPK1-RIPK3 complex formation in T cells as well as in MEFs. In addition, A20OTU/− T cells and A20ZF4/− survival was decreased compared to A20+/− T cells following 72 hours of stimulation with anti-CD3, anti-CD28, and ZVAD for 72 hours (Fig. 6f). TCR-ZVAD stimulated A20OTU/OTU and A20ZF4/ZF4 T cells exhibited similar relative phenotypes as A20OTU/− T cells and A20ZF4/− T cells (data not shown). Finally, A20OTU and A20ZF4 mice were crossed with Ripk3−/− mice and the responses to TCR-ZVAD stimulation in naïve CD4+ T cells isolated from these mice were tested. Following TCR-ZVAD stimulation for 72 hours, expansion of A20OTU/− Ripk3−/− CD4+ T cells was the same as A20+/− T cells, and expansion of A20ZF4/− Ripk3−/− CD4+ T cells was partially restored to that of A20+/− T cells (Fig. 6f), indicating that RIPK3-deficiency restored expansion of TCR-ZVAD stimulated A20OTU/−, and to a lesser extent, A20ZF4/− CD4+ T cells. This indicates that while the ZF4 domain of A20 is also required for normal A20 activity during T cell activation, the Cys103-dependent deubiquitination function of A20 is required to inhibit the formation of RIPK1-RIPK3 complexes and RIPK3-dependent necroptosis.
Discussion
Our study reveals that A20 restricts necroptosis in multiple cell types. These findings suggest A20 regulates cellular and tissue homeostasis as well as immune homeostasis. We have discovered that RIPK3 undergoes a specific ubiquitination event that supports the formation of RIPK1-RIPK3 complexes in cells undergoing necroptosis, and that A20 potently inhibits this event. Our studies have broad implications for the molecular regulation of cell death signaling as well as the physiological mechanisms by which A20 prevents inflammatory diseases.
We have found that uncontrolled RIPK3-dependent necroptosis in the absence of A20 inhibited T cell expansion in vitro and in vivo. Exaggerated necroptosis in caspase-8 deficient and FADD mutant T cells has been reported to compromise anti-LCMV or anti-Toxoplasma responses27, 28. Hence, our studies reinforce the concept that, in certain pathological contexts, necroptosis can restrict the expansion of activated T cells. Our studies also indicate that A20 has dual roles in T cells, restricting both cellular activation and cell death. Activation and survival signals appear to be integrated differently in activated T cells compared to activated B cells, in which A20 deficiency causes increased NF-κB-dependent Bcl-x expression and resistance to Fas-mediated death13. These differences re-emphasize the importance of studying the cell type-specific functions of pleiotropically expressed molecules to understand their physiological roles in disease pathogenesis.
Our finding that A20KO2/KO2 RIPK3−/− mice live longer than A20KO2/KO2 mice suggested that A20 protects multiple cell types from necroptosis. As such, loss of A20 from other cells (e.g., stromal cells) may render mice more susceptible to necroptotic tissue damage. Our observations are consistent with prior experiments showing that A20 protects L929 fibrosarcoma cells from TNF-induced death36. RIPK3 deficiency might also abrogate inflammasome activity in A20−/− mice37. Because A20 also protects cells against apoptotic death2, 38, the anti-necroptotic function of A20 positions this molecule as a potent pro-survival protein. The notion that necroptosis of target organs may contribute to disease severity has been suggested in inflammatory bowel disease and other conditions39. Considered together with observations that A20 polymorphisms are associated with more severe nephritic complications of SLE patients and increased disease severity in psoriasis vulgaris and cystic fibrosis patients, A20 may partly prevent human disease by protecting against necroptotic death in non-lymphoid tissues40–42.
Necroptosis is known to occur in cells lacking pro-apoptotic molecules such as caspase 8 and FADD43–46. This has been explained by the cleavage of RIPK1 by caspase 8, which prevents RIPK1 kinase-dependent formation of RIPK1-RIPK3 complexes. Virally infected cells may also undergo necroptosis when viral proteins abrogate apoptosis30. Hence, necroptosis has been viewed as a form of cell death that occurs predominantly in cells rendered caspase deficient. By contrast, we have found that A20-deficient MEFs exhibit increased sensitivity to both apoptosis and necroptosis2. Therefore, our findings suggest that A20 restricts necroptosis in a fashion that is distinct from caspase 8 or FADD.
RIPK1 ubiquitination inhibits RIPK1 kinase activity, and the deubiquitinating enzyme CylD supports necroptosis by deubiquitinating RIP146, 47. Like CylD, A20 inhibits RIPK1 ubiquitination and NF-κB signaling3, 19. However, unlike CylD, our current findings show that A20 inhibits necroptosis. This divergence suggests that A20 inhibits necroptosis via a mechanism other than inhibiting RIPK1 ubiquitination. Our finding that A20 restricted RIPK3 ubiquitination provide such a mechanism. K63-linked ubiquitination of RIPK1 and RIP2 occur during TNF- and NOD2-induced NF-κB signaling, respectively, and A20−/− MEFs and macrophages exhibit increased RIPK1 and RIP2 ubiquitination, respectively when stimulated through these pathways3, 10, 19. Hence, A20 appears to restrict RIPK1, RIP2 and RIPK3 ubiquitination in distinct signaling contexts. The cIAP E3 ligases can ubiquitinate RIP family proteins in vitro, and cIAP1 and cIAP2 may be responsible for activating RIPK1 and RIP2 by supporting their ubiquitination with K63-linked chains following TNF and NOD2 pathway activation48, 49. Hence, one might speculate that cIAPs similarly activate RIPK3 through ubiquitination during necroptosis. However, cIAPs appear to limit necroptosis in macrophages48. Thus, if cIAPs activate RIPK3 and support RIPK1-RIPK3 complex formation and necroptosis, they likely also perform a separate function that restricts necroptosis (e.g., cIAPs could ubiquitinate RIPK1). Future studies will be needed to identify the physiological E3 ligases that ubiquitinate RIPK3.
Our mass spectrometric screen identified K5 of RIPK3 as a physiological ubiquitination site, and our SILAC experiments revealed that A20−/− MEFs contain 15-fold more RIPK3 peptide ubiquitinated at K5 than wild-type MEFs. Mutation of this residue abrogated RIPK3 ubiquitination and the formation of RIPK1-RIPK3 complexes and necroptosis. In other signaling complexes, K63-linked ubiquitination facilitates the recruitment of K63-ubiquitin sensor proteins18. Hence, K63-linked ubiquitination of RIPK3 could similarly support the recruitment of other ubiquitin sensors. These ubiquitin binding proteins could in turn enhance RIPK3 kinase activity, support RIPK1-RIPK3 complexes, recruit downstream mediators such as MLKL51, and/or support higher-order amyloid complexes comprised of RIPK1 and RIPK3 proteins31. Structurally, K5 resides in an unstructured portion of RIPK3, and is thus likely to be accessible for ubiquitination52. In addition, the K5 residue is conserved in human RIPK3, and RIPK3 deficiency abrogated increased necroptosis of A20-deficient human Jurkat I9.2 T cells. Thus, RIPK3 ubiquitination may support necroptosis in human cells. Therefore, ubiquitination can support necroptosis, and A20 strongly inhibits this pro-necroptotic ubiquitination event.
We have found that A20’s C103 deubiquitinating motif was required for inhibiting RIPK3 ubiquitination and forming RIPK1-RIPK3 complexes. The C103 motif cleaves unanchored K48 ubiquitin chains as well as RIPK1- or TRAF6- anchored K63 chains in cell-free experiments3, 4, 20. This motif has also been shown to inhibit E2-E3 interactions in cells21. Both these C103-dependent activities could inhibit the K63-linked polyubiquitination of RIPK3. In contrast, A20’s ZF4, which binds ubiquitin and supports E3 ligase activity, appeared less critical than C103 for inhibiting RIPK3 ubiquitination during necroptosis. One possible explanation for this difference could be that A20, which relies upon ZF4 to bind ubiquitinated RIPK1, may rely more on other ubiquitin binding motifs, such as ZF7, to bind ubiquitinated RIPK3. Such distinct roles for A20’s biochemical motifs can provide important insights into how A20 performs its physiological functions.
In conclusion, we have discovered that A20 restricts RIPK3 dependent necroptosis. Our studies show that RIPK3 is physiologically ubiquitinated at K5 during necroptosis in a manner that supports the formation of RIPK1-RIPK3 complexes. A20’s regulation of necroptosis should be integrated with A20’s previously described roles in restricting NFkB signaling and cellular activation. As hypomorphic A20 expression and function are linked to a wide range of human inflammatory and autoimmune diseases, and as necroptosis can trigger inflammation, our studies suggest a new pathway by which A20 prevents human disease.
Online Methods
Mice and in vivo experiments
The generation of mice bearing a “floxed” allele of A20 (A20fl mice) has been described (9, 13). Mice lacking A20 specifically in T cells were generated by breeding A20fl mice with CD4-Cre transgenic mice. Mice bearing gene-targeted point mutations in the A20 gene, A20OTU mice and A20ZF4 mice, were recently described19. Ripk3−/− mice were kindly provided by X. Wang. A20−/− mice were generated by breeding A20fl mice with B6.EIIA-Cre mice, and subsequently breeding to C57BL/6J mice and selecting for mice bearing the predicted A20 exon 2 deletion but not the EIIA-Cre transgene. RAG1−/− mice were purchased from Jackson Labs. All mice were generated and maintained on a C57BL/6 inbred background, and all mouse experiments were performed according to UCSF institutional guidelines.
For antigen specific T cell responses, naïve TCR transgenic OT-II CD4+ T cells were flow cytometrically purified from congenically marked mice, mixed 1:1, CFSE labeled, and adoptively transferred into C57BL/6J mice. Mice were immunized with OVAp (200 mg) and LPS (20ug) 24 hours after cell transfer, and analyzed at various time points. For EAE experiments, mice were immunized with MOG peptide p35–55 (50 µg) in CFA with pertussis toxin (200 ng). Clinical symptoms and histological analyses were performed as previously described53.
In vitro T cell analysis
For in vitro T cell assays, naïve (CD4+ CD25− CD44Lo CD62LHi) T cells were purified by bead enrichment and/or flow cytometric sorting from spleens and lymph nodes from the indicated strains of mice prior to stimulation with plate bound anti-CD3 (2.5 µg/ml; UCSF Monoclonal Antibody Core) and anti-CD28 (2 µg/ml; UCSF Monoclonal Antibody Core). In addition, mIL-12 (20ng/ml, Peprotech) and anti-IL-4 (10µg/ml, 11B11:Tonbo) were used for TH1 polarization, while hTGFβ (5ng/ml, Peprotech), mIL-6 (20ng/ml, Peprotech), anti-IL-4 (10µg/ml) and anti-IFNγ (10µg/ml, XMG1.2, UCSF hybridoma core) were used for TH17 polarization. In selected experiments, T cells were labeled with 3 mM CFSE (Invitrogen). siRNA mediated reduction of RIPK3 in Jurkat I9.2 cells (ATCC) was performed using one pulse of 2200 V and 20 ms, using Neon Transfection System (Invitrogen) according to manufacturer’s instructions.
Flow Cytometry
Cell survival of MEFs was quantitated using the CellTiter-Glo Luminescent Cell Viability Assay per manufacturer’s instructions (Promega). The following antibodies were purchased (BD bioscience) and used for FACS studies: anti-CD4-PEcy7, anti-CD4-APC, anti-CD8-PEcy7, anti-CD8-APC, anti-CD11b–PEcy7, anti-Gr1-APC, anti-CD45.1-APC, anti-CD45.1-FITC, anti-CD45.2-APC, anti-CD45.2-FITC, anti-CD44-FITC, anti-CD62L–PE, anti-CD62L–APC, anti-TCRβ-FITC. Live cells were quantitated by flow cytometry of DAPI-negative cells. For intracellular cytokine expression, cells were incubated with PMA (50 ng/ml, Sigma-Aldrich), ionomycin (250 ng/ml, Sigma-Aldrich) and GolgiPlug (1 µl/ml, BD Bioscience) for 4 hrs before harvesting. Cells were stained using anti-CD4-e450 (Tonbo), anti-IFNγ−PE (BD Bioscience), anti-IL17A–Alexa647 (BD Bioscience), Live/Dead cell stain kit (Invitrogen) and Cytofix/Cytoperm kit (BD Bioscience).
Signaling assays
Stimulated T cells were incubated in lysis buffer. For signaling complex analyses, cell lysates were pre-cleared with Protein G beads prior to immunoprecipitation with anti-RIPK1 antibody. Antibodies and reagents used for immunoprecipitation and immunoblotting studies included: anti-RIPK1, anti-Bim, anti-Bclx, (BD Biosciences), nec1, anti-Bcl2, β-actin (Merck chemicals), anti-RIPK3 (ProSci Incorporated), anti-ubiquitin, anti-Bax, anti-RIPK3 (Santa Cruz Biotechnology), anti-RIPK1, anti-A20 (Cell signaling Technology), QVD, ZVAD (Enzo Life Science).
Signaling studies in MEFs were performed as described19. RIPK1 immunoprecipitations were performed using anti-RIPK1 (Cell Signaling) and Dynabeads M270 Epoxy (Invitrogen). K63-ubiquitin immunoprecipitations were performed using anti-K63 (Millipore) and Dynabead Protein A (Invitrogen). Studies of RIPK3 mutants in A20−/− Ripk3−/− MEFs were performed by generating RIPK3 lysine mutants, introducing mutant RIPK3-encoding cDNAs into GFP expressing lentiviral constructs, and infecting A20−/− Ripk3−/− MEFs with these viruses. Productively infected cells were FACS-sorted to obtain pure populations of RIPK3-expressing cells prior to being tested in necroptosis assays. Cell survival of MEFs was quantitated using the CellTiter-Glo Luminescent Cell Viability Assay per manufacturer’s instructions (Promega).
Mass spectrometry
Pellets of stimulated cells were solubilized in urea, ammonium bicarbonate and TCEP, alkylated with iodoacetic acid, and digested overnight with TPCK modified trypsin. Tryptic peptides were enriched for ubiquitinated peptides with the PTMScan Ubiquitin Remnant Motif kit (Cell Signaling Technology) according to the manufacturer’s protocol. Affinity purified ubiquitinated peptides were separated by nano-flow liquid chromatography, the eluate coupled to a ion trap-Orbitrap mass spectrometer, and peptides analyzed using PAVA in-house software. For quantitative (SILAC) mass spectrometry experiments, ko2 and A20+/+ MEFs were passaged in the presence of dialyzed serum supplemented with either heavy (A20+/+) or light (A20ko2/ko2) isotope labeled amino acids for two weeks prior to stimulation. Lysates from A20+/+ and A20ko2/ko2 cells were mixed, trypsinized and immunoprecipitated with anti-gly-gly (ubiscan) prior to mass spectrometry analysis35.
Supplementary Material
Acknowledgements
This work was supported by the NIH (DK071939 and DK095693 [A.M.]; AI073737 and NS063008 [S.S.Z.]), the Kenneth Rainin Foundation (A.M.), the NMSS (RG 4768 and RG 5180 [S.S.Z.]), the Guthy Jackson Charitable Foundation and Maisin Foundation (S.S.Z.). M.O. and S.O. were partially supported by the CCFA; U.S. was supported by the NMSS. We thank Juan Carlos Patarroyo and Nicolas Molnarfi for assistance with EAE experiments. We thank Scott Oakes for helpful discussions. Mass spectrometry analysis was provided by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A.L. Burlingame, Director) supported by funding from the Biomedical Technology Research Centers program of the NIH National Institute of General Medical Sciences, NIH NIGMS 8P41GM103481 and Howard Hughes Medical Institute.
References
- 1.Opipari AW, Jr, Boguski MS, Dixit VM. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J. Biol. Chem. 1990;265:14705–14708. [PubMed] [Google Scholar]
- 2.Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 4.Boone DL, et al. The ubiquitin modifiying enzyme A20 is essential for terminating TLR signaling. Nat. Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 5.Ma A, Malynn BA. A20: Linking ubiquitination with immunity and human diseases. Nat. Rev. Immunol. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catrysse L, Vereecke L, Beyaert R, van Loo G. A20 in inflammation and autoimmunity. Trends in Immunol. 2014;35:22–31. doi: 10.1016/j.it.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Musone S, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat. Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adrianto I, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat. Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turer EE, et al. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J. Exp. Med. 2008;205:451–464. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitotsumatsu O, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kool M, et al. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity. 2011;35:82–96. doi: 10.1016/j.immuni.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Hammer GE, et al. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat. Immunol. 2011;12:1184–1193. doi: 10.1038/ni.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matmati M, et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat. Genet. 2011;43:908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- 14.Heger K, et al. A20 deficient mast cells exacerbate inflammatory responses in vivo. PLoS Biol. 2014;12:e1001762. doi: 10.1371/journal.pbio.1001762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tavares RM, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu Y, et al. B cells lacking the tumor suppressor TNFAIP3/A20 display impaired differentiation and hyperactivation and cause inflammation and autoimmunity in aged mice. Blood. 2011;117:2227–2236. doi: 10.1182/blood-2010-09-306019. [DOI] [PubMed] [Google Scholar]
- 17.Hövelmeyer N, et al. A20 deficiency in B cells enhances B-cell proliferation and results in the development of autoantibodies. Eur. J. Immunol. 2011;41:595–601. doi: 10.1002/eji.201041313. [DOI] [PubMed] [Google Scholar]
- 18.Chen ZJ, Sun L. J Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Lu TT, et al. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity. 2013;38:896–905. doi: 10.1016/j.immuni.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin SC, et al. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J. Mol. Biol. 2008;376:526–540. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosanac I, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol. Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Skaug B, et al. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol. Cell. 2011;44:559–71. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokunaga F, et al. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhelst K, et al. A20 inhibits LUBAC-mediated NF-kB activation by linear polyubiquitin chains via its zinc finger 7. EMBO J. 2012;31:3845–3855. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beyaert R, Heyninck K, Van Huffel S. A20 and A20 binding proteins as cellular inhibitors of NF-kB dependent gene expression and apoptosis. Biochem. Pharm. 2000;60:1143–1151. doi: 10.1016/s0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 27.Ch’en IL, Tsau JS, Molkentin JD, Komatsu M, Hedrick SM. Mechanisms of necroptosis in T cells. J. Exp. Med. 2011;208:633–641. doi: 10.1084/jem.20110251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborn SL, et al. Fas-associated death domain (FADD) is a negative regulator of T-cell receptor mediated necroptosis. Proc. Natl. Acad. Sci. USA. 2010;107:13034–13039. doi: 10.1073/pnas.1005997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He S, et al. Receptor interacting protein kinase 3 determines cellular necroptotic responses to TNFa. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 30.Cho Y, et al. Phosphorylation-driven assembly of the RIPK1-RIPK3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamvil SS, Steinman L. The T lymphocyte in experimental allergic encephalomyelitis. Ann. Rev. Immunol. 1990;8:579–621. doi: 10.1146/annurev.iy.08.040190.003051. [DOI] [PubMed] [Google Scholar]
- 33.Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 34.Linkermann A, Green DR. Necroptosis. N. Engl. J. Med. 2014;370:455–465. doi: 10.1056/NEJMra1310050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Udeshi ND, Mertins P, Svinkina T, Carr SA. Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Prot. 2013;8:1950–1960. doi: 10.1038/nprot.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanlangenakker N, et al. TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell Death Dis. 2011;2:e230. doi: 10.1038/cddis.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duong B, et al. A20 restricts ubiquitination of pro-interleukin-1β protein complexes and suppresses NLRP3 inflammasome activity. Immunity. 2015;42:55–67. doi: 10.1016/j.immuni.2014.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Opipari AW, Hu HM, Yabkowitz R, Dixit VM. The A20 zinc finger protein protects cells from tumor necrosis factor cytotoxicity. J. Biol. Chem. 1992;267:12424–12427. [PubMed] [Google Scholar]
- 39.Welz, et al. FADD prevents RIPK3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–334. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- 40.Bates JS, et al. Meta-analysis and imputation identifies a 109 kb risk haplotype spanning TNFAIP3 associated with lupus nephritis and hematologic manifestations. Genes Immun. 2009;10:470–477. doi: 10.1038/gene.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang X, et al. Expression of tumor necrosis factor alpha-induced protein 3 mRNA in peripheral blood mononuclear cells negatively correlates with disease severity in psoriasis vulgaris. Clin. Vaccine Immunol. 2012;19:1938–1942. doi: 10.1128/CVI.00500-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelly C, et al. Expression of the inflammatory regulator A20 correlates with lung function in patients with cystic fibrosis. J. Cyst. Fibros. 2013;12:411–415. doi: 10.1016/j.jcf.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Kaiser WJ, et al. RIPK3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oberst A, et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, et al. Functional complementation between FADD and RIPK1 in embryos and lymphocytes. Nature. 2011;471:373–376. doi: 10.1038/nature09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Donnell MA, et al. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat. Cell Biol. 2011;13:1437–1442. doi: 10.1038/ncb2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moquin DM, et al. CYLD deubiquitinates RIP1 in the TNFα-induced necrosome to facilitate kinase activation and programmed necrosis. PLoS One. 2013;8:E76841. doi: 10.1371/journal.pone.0076841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertrand MJ, et al. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30:789–801. doi: 10.1016/j.immuni.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 49.Bertrand MJ, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIPK1 ubiquitination. Mol. Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 50.McComb S, et al. cIAP1 and cIAP2 limit macrophage necroptosis by inhibiting Rip1 and Rip3 activation. Cell Death Differ. 2012;19:1791–1801. doi: 10.1038/cdd.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun L, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIPK3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 52.Xie T, et al. Structural insights into RIPK3-mediated necroptotic signaling. Cell Rep. 2013;5:70–78. doi: 10.1016/j.celrep.2013.08.044. [DOI] [PubMed] [Google Scholar]
- 53.Molnarfi N, et al. MHC class II-dependent B cell APC function is required for induction of CNS autoimmunity independent of myelin-specific antibodies. J. Exp. Med. 2013;210:2921–2937. doi: 10.1084/jem.20130699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.