Abstract
Activated Leukocyte Cell Adhesion Molecule (ALCAM/CD166), is expressed on osteoblasts (OB) and hematopoietic stem cells (HSC) residing in the hematopoietic niche, and may have important regulatory roles in bone formation. Because HSC numbers are reduced 77% in CD166-/- mice, we hypothesized that changes in bone phenotype and consequently the endosteal niche may partially be responsible for this alteration. Therefore, we investigated bone phenotype and OB function in CD166-/- mice. Although osteoclastic measures were not affected by loss of CD166, CD166-/- mice exhibited a modest increase in trabecular bone fraction (42%), and increases in osteoid deposition (72%), OB number (60%), and bone formation rate (152%). Cortical bone geometry was altered in CD166-/- mice resulting in up to 81% and 49% increases in stiffness and ultimate force, respectively. CD166-/- OB displayed elevated alkaline phosphatase (ALP) activity and mineralization, and increased mRNA expression of Fra 1, ALP, and osteocalcin. Overall, CD166-/- mice displayed modestly elevated trabecular bone volume fraction with increased OB numbers and deposition of osteoid, and increased OB differentiation in vitro, possibly suggesting more mature OB are secreting more osteoid. This may explain the decline in HSC number in vivo because immature OB are mainly responsible for hematopoiesis enhancing activity.
Keywords: ALCAM, CD166, Osteoblasts, Bone Mass, Bone Structure, Hematopoiesis, Bone Microenvironment
Introduction
Activated Leukocyte Cell Adhesion Molecule (ALCAM or CD166) is a transmembrane glycoprotein belonging to the immunoglobulin superfamily. CD166 mediates both strong heterophilic (CD166-CD6) and relatively weaker homophilic (CD166-CD166) cell-cell interactions[1-7]. CD166 was originally implicated more than a decade ago in T cell activation and in early stages of murine[8,9] and human[1] hematopoiesis. Uchida and colleagues[10], as well as others[2,11], described the involvement of hematopoietic cell antigen (HCA later recognized as ALCAM or CD166) in the identification of a subset of human adult bone marrow (BM) and mobilized peripheral blood (PB) CD34+ cells enriched for hematopoietic cell activity. The amino acid sequence of human and murine CD166 display an overall similarity of 93%[12]; and is currently the only known molecule expressed on osteoblasts (OB) and hematopoietic stem cells (HSC) in both humans and mice[11]. It is also important to point out that CD6, the only other ligand of CD166, is not expressed on HSC or OB.
HSC reside in the BM microenvironment that consists of OB, mesenchymal progenitor cells, stromal cells, endothelial cells, adipocytes, osteoclasts, as well as multiple soluble components of the extracellular matrix. Collectively these cellular and extra cellular components constitute the hematopoietic niche, and the complex interplay of signals between HSC and niche elements determine the fate of HSC[13]. Amongst the other cellular components, OB play a significant role in HSC function. Primitive HSC reside in close association with OB in endosteal regions of the niche thereby maintaining critical HSC properties such as quiescence and self-renewal[5,14]. Although OB-HSC interactions in the endosteal niche are crucial for HSC survival and function, the molecular relationships remain poorly understood.
It has been established that OB lineage cells express CD166[1-5,15]. Recently, we began to dissect the hierarchical organization of OB lineage cells to determine which cells are responsible for supporting hematopoiesis. Collectively, our studies identified Lin-Sca1-OPN+ CD166+CD44+CD90+ OB as immature osteoblastic lineage cells promoting hematopoietic progenitor and stem cell [Lineage- Sca-1+ c-kit+ (LSK)] proliferation and function in vitro and in vivo[6,7,16]. Furthermore, our studies illustrated that support of hematopoietic function by OB lineage cells declines as OB mature and the level of CD166 expression declines progressively[16]. Recently, we also reported the significant role of CD166 in the identification of both murine and human primitive hematopoietic progenitor cells, and its role in HSC-niche interactions and HSC engraftment. CD166-/- HSC engrafted poorly in wild-type (WT) recipients and CD166-/- hosts supported only short-term, but not long-term WT HSC engraftment, suggesting that loss of CD166 is detrimental to the competence of the hematopoietic niche[11]. However, it remains unclear whether this hematopoietic defect was caused by disrupting the homophilic interactions between cells expressing CD166, or due to changes in the bone microenvironment. In order to further investigate the role of CD166 in the overall bone microenvironment, we examined its effects on the bone phenotype - as changes to the bone phenotype based on CD166 expression could then be related to altering hematopoiesis. Although there have been many investigations into CD166’s various roles in hematopoiesis[1,5,10], T cell development[8,9,17], and neural migration[18], to our knowledge, this is the first study to date to examine its effects on the bone phenotype.
Methods
Animals
Two day and 6-7 week-old (hereafter referred to as 6 week-old), female and male CD166-/- (B6.129[FVB]-Alcamtm1Jawe/J mice; The Jackson Laboratories stock No. 010635) and C57BL/6J mice (The Jackson Laboratories stock No. 000664, WT controls) were utilized in these studies. Breeding pairs of C57BL/6J and CD166-/- mice were obtained from the Jackson Laboratories (Maine), and breeding colonies of both genotypes were maintained at our facilities. We examined 6 week-old mice to match the age of mice used for our recently reported hematopoietic studies[11]. Adult mice were euthanized by CO2 inhalation followed by cervical dislocation to ensure death, whereas 2-day old pups were euthanized by cervical decapitation. Seven and 14 days prior to sacrifice, 6 week-old mice were injected intraperitoneally with 30 mg/kg of calcein (Sigma Chemical Co., St. Louis, MO) and 50 mg/kg Alizarin Red S (Sigma Chemical Co., St. Louis, MO) respectively, for dynamic histomorphometric measures. Mice were maintained at a designated animal facility at Indiana University School of Medicine, IN and all procedures were approved by the Institutional Animal Care and Use committee of Indiana University and followed NIH guidelines. Briefly, a 12 hr light/12 hr dark cycle was used for housing; maximum of 5 mice per cage; food and water were available ad libitum; and facilities were maintained with minimal noise and vibrations with sterile animal handling.
Bone marrow collection, cell staining, and flow cytometry
BM collection, LSK staining, and sorting were done as described before[19]. Briefly, BM was made from either male or female 6 week-old C57BL/6 WT mice. Long bones from all four limbs of each mouse were flushed thoroughly using 27G1/2 needle with 8-10 mL of heparin medium consisting of Hank’s Balanced Salt Solution (HBSS) supplemented with 1 % Penicillin/Streptomycin, and 20 U/mL heparin. Flushed BM cells were layered on top of Ficoll (hydrophilic polysaccharide) and centrifuged at 1500 rpm for 30 min at room temperature. Low-density BM cells were collected at the interface of medium and Ficoll. These low-density BM cells were washed once with stain wash (PBS, 1% bovine calf serum, and 1% penicillin-streptomycin) followed by antibody staining for 15 min on ice. The following cocktail of antibodies was used: phycoerythrin (PE)-conjugated CD3, CD4, CD45R, Ter119, and Gr1; AF700-conjugated c-Kit (CD117, eBioscience); PE-Cy7-conjugated Sca-1; fluorescein isothiocyanate (FITC)-conjugated CD34; allophycocyanin (APC)-conjugated Flk2 (eBioscience). Unless noted otherwise, all monoclonal antibodies were obtained from BD Biosciences. Cells were washed once more with stain wash buffer, and data was acquired using a BD LSRII (BD Biosciences).
In vivo dual-energy x-ray absorptiometry
Whole body areal bone mineral density (aBMD; g/cm2), and bone mineral content (BMC, g) were measured in vivo at 6 weeks on the day of sacrifice using a small animal DXA (PIXImus, GE Lunar, Madison, WI). Body weight (BW; g) was obtained prior to scanning using a digital scale. For in vivo scanning mice were anesthetized via inhalation of 2.5% isoflurane (OsFlo: Abbott Laboratories) mixed with O2 (1.5L/min). Mice were placed in a prone position on a specimen tray for scanning. Regional analyses of the total right femur and lumbar spine (L1-L5) were obtained from the whole body scan.
Ex vivo micro-computed tomography (μCT)
A desktop microcomputerized tomography machine (Skyscan 1172 high-resolution μCT; SkyScan, Kontich, Belgium) was used to obtain images of the mid-shaft (cortical region) and distal femur (trabecular region) of the right femur at 50kVp, with a 0.5 mm aluminum filter, and 11 mm isotropic voxel size[20-22]. Trabecular bone properties from the distal femur were obtained from a 1 mm region of interest, 0.5 mm proximal to the distal growth plate. Trabecular bone properties acquired included: trabecular bone volume fraction (bone volume [BV]/tissue volume [TV], %), trabecular number (Tb.N, mm), trabecular thickness (Tb.Th, mm), and trabecular separation (Tb.Sp, mm). Cortical bone properties were obtained from a 1 slice region of interest at the mid-shaft which was calculated as 50% of the total length from the head of the femur to the base of the distal condyles. Cortical bone properties included total area (Tt.Ar, mm2), cortical area (Ct.Ar, mm2), medullary area (Me.Ar, mm2), cortical thickness (Ct.Th, mm), and polar moment of inertia (Ip, mm4).
Histomorphometry
Static and dynamic histomorphometric analyses of cortical and trabecular bone were performed on the right femur (mid-shaft and distal regions) as previously described[20,21]. Briefly, bones were embedded, undecalcified in 99% methyl-methacrylate with 3% dibutyl phthalate (Sigma-Aldrich, St. Louis, Mo, USA). Transverse thick sections (~40 microns) were taken from the mid-shaft using a diamond bladed wire saw, and frontal plane thin sections (6 microns) were obtained from the distal femur using a microtome. Histological measurements were made with a semiautomatic analysis system (Bioquant OSTEO 7.20.10, Bioquant Image Analysis Co.) attached to a microscope with an ultraviolet light source (Nikon Optiphot 2 microscope, Nikon). For dynamic measures of bone formation the following data was derived: mineralizing surface (MS/BS, %), mineral apposition rate (MAR, μm/day), and bone formation rate (BFR/BS; μm3/mm2 year). For static measures of the distal femur osteoid surface/bone surface (OS/BS, %), number of osteoblast (N.Ob/T.Ar), and number of osteoclasts (N.Oc/T.Ar) were obtained from von Kossa/McNeal’s stained tissue. Measurements were completed on one von Kossa/McNeal’s stained (static) and one unstained (dynamic) section for each animal. The reader was blinded to the animal genotype.
Biomechanics
Relative bone strength was determined by performing three-point bending of the left femur with a materials testing device (MTS Systems Corporation; Eden Prairie, MN) as previously detailed[20-22]. Briefly, femurs were thawed to room temperature in a saline bath (at 37oC)for two hours. Prior to testing femur length (mm), mid-shaft width (medial to lateral; mm), and mid-shaft height (ventral to dorsal; mm) were obtained using hand-held digital calipers measured to the nearest 0.1 mm. Thereafter, each femur was tested in the anterior-posterior direction and stabilized with a static preload of 1 N before being loaded to failure with a crosshead speed of 10 mm/min. Force versus displacement data were gathered at 100 Hz, and the ultimate force (N), stiffness (N/mm), polar moment of inertia (mm4), ultimate stress (MPa), modulus (MPa), and toughness (MJ/m3) were derived.
Preparation of two-day neonatal calvarial cells (OB) and functional OB assays
Neonatal murine calvarial OB from 2-day old WT and CD166-/- pups were prepared according to a modification of the procedure originally described by Wong et al.[23,24]. Briefly, calvariae from an entire litter of C57BL/6 or CD166-/- mice of less than 48 hr old were dissected out, pretreated with EDTA in PBS for 30 minutes, and then subjected to sequential collagenase digestions (200 U/mL). Fractions 3 to 5 (digestions 20–35 min, 35-50 min, and 50–65 min) were collected and used as OB. These cells are >95% OBs or OB precursors as previously shown[25-27]. OB were seeded at 2x104 cells/ml and were cultured for 4 or 14 days. Cells were maintained in αMEM supplemented with 10% fetal bovine serum and were further supplemented with ascorbic acid (50 μg/ml added on day 0 and at all feedings) and β-glycerophosphate (5 mM added starting on day 7 and all subsequent feedings). Medium was changed twice each week.
With respect to functional OB assays, alkaline phosphatase (ALP) activity was determined by the colorimetric conversion of p-nitrophenol phosphate to p-nitrophenol (Sigma) and normalized to total protein (BCA, Pierce) as previously described[28,29]. Calcium deposition was assessed by eluting Alizarin Red S from cell monolayers as previously described[29,30].
Quantitative real-time PCR
For gene expression analysis, quantitative real-time PCR was conducted as previously detailed[24]. Briefly, total RNA was isolated from OBs using TRIzol reagent (Invitrogen). Total RNA from OBs was treated with DNase I (Qiagen) and used to generate cDNAs by reverse transcription according to the manufacturer’s instructions (first strand cDNA synthesis kit; Roche Applied Science). qPCRs were performed in an MX3000 detection system using SYBR Green PCR reagents as described by the manufacturer (Stratagene). For each primer pair, a calibration curve was performed, and all oligonucleotides were tested to ensure specificity and sensitivity. For each OB sample, arbitrary units were obtained using the standard curve and the expression of GAPDH was used to normalize the amount of the mRNA transcript. The relative expression level was set to 1 for WT OB and the CD166-/- OB expression level was normalized accordingly. [Table 1] details the genes analyzed and their primer sequences. QRT-PCR was used for the analysis of Fra1, a cyclin regulator; Runx2, a transcription factor important for OB differentiation; ALP, the major enzyme mediating OB enzymatic activity; type I collagen or ColIa1, the main protein secreted by OB; osteocalcin or Ocn, a primary marker of OB maturation.
Table 1.
Primers used in Real Time-PCR studies.
| Gene Primer | Primer Sequence |
|---|---|
| Runx2 For | 5’CGACAGTCCCAACTTCCTGT |
| Runx2 Rev | 5’CGGTAACCACAGTCCCATCT |
| ALP For | 5’ GCTGATCATTCCCACGTTTT |
| ALP Rev | 5’ CTGGGCCTGGTAGTTGTTGT |
| Ocn For | 5’ AAGCAGGAGGGCAATAAGGT |
| Ocn Rev | 5’ TTTGTAGGCGGTCTTCAAGC |
| ColIa1 For | 5’ CAGGGAAGCCTCTTTCTCCT |
| ColIa1 Rev | 5’ ACGTCCTGGTGAAGTTGGTC |
| GAPDH For | 5’ CGTGGGGCTGCCCAGAACAT |
| GAPDH Rev | 5’ TCTCCAGGCGGCACGTCAGA |
Statistical analysis
Analyses were performed with the IBM SPSS Statistics software (version 21; SPSS Inc., Chicago, IL). Analyses were two-tailed with a level of significance set at p<0.05, with all data presented as mean ± SE (unless otherwise specified). All in vitro experiments were performed 3 times with triplicate samples in each experiment. For all in vitro studies one-way analysis of variance (ANOVA) was used to identify genotype differences. For the in vivo studies, the sample size is either presented in [Table 2] or the Figure legends. Two-way ANOVA was used to examine sex (male vs. female) and genotype (CD166-/- vs. C57BL/6) differences, with sex being the within-animal, and genotype being the between-animal independent variables. In the advent of a non-significant sex by genotype ANOVA interaction, main effects for genotype were explored. Significant ANOVA interactions were explored using 2 simple effects test to assess for the effect of genotype for each sex on each outcome measured using a paired t-test, with a Bonferroni correction to the level of significance used to maintain the family wise error rate (alpha= 0.05/2= 0.025).
Table 2.
Anthropometric, gross femur geometry, and DXA data of female and male, WT and CD166-/- mice at 6 weeks of age. Results are presented as the mean±SE. *Indicates significant interaction between genotype and sex, with post-hoc analyses indicating sex-specific genotype differences (p<0.05). #Indicates significant main effect for genotype, with no significant interaction found between genotype and sex (p<0.05).
| Strain | Sex | N | Weight (g) | Femur Length (mm) | Femur Width (mm) | Femur Height (mm) | Total Femur aBMD (g/cm2) | Vertebrae aBMD (g/cm2) | Whole Body aBMD (g/cm2) | Total Femur BMC (g) | Vertebrae BMC (g) | Whole Body BMC (g) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | F | 9 | 14.7±0.2 | 12.1±0.2 | 1.63±0.07 | 1.21±0.02 | 0.047±0.001 | 0.039±0.001 | 0.0369±0.0001 | 0.014±0.001 | 0.036±0.001 | 0.22±0.01 |
| CD166-/- | F | 8 | 17.1±0.4# | 12.5±0.2* | 1.65±0.03 | 1.16±0.01 | 0.052±0.003* | 0.044±0.002# | 0.0418±0.0001# | 0.017±0.001* | 0.045±0.002* | 0.31±0.01* |
| WT | M | 10 | 18.3±0.2 | 12.0±0.2 | 1.61±0.05 | 1.26±0.04 | 0.050±0.002 | 0.041±0.001 | 0.0387±0.0006 | 0.016±0.001 | 0.039±0.001 | 0.25±0.01 |
| CD166-/- | M | 7 | 20.7±0.4# | 12.8±0.6* | 1.79±0.04 | 1.27±0.01 | 0.064±0.001* | 0.049±0.001# | 0.0464±0.0001# | 0.021±0.001* | 0.052±0.001* | 0.39±0.01* |
Results
Phenotype of bone marrow hematopoietic progenitor and stem cells
At 6 weeks of age, CD166-/- mice had significantly fewer phenotypically defined LSK and LSKCD34-Flk2- (long-term HSC or LT-HSC) in their BM than did WT controls (Figure 1). We calculated the absolute number of HSC by multiplying the frequency of LSK or LT-HSC with the total number of low-density cells per femur. We found 20,012±885 LSK cells per femur in WT compared to 9,430±892 LSK cells per femur in CD166-/- mice (53% reduction in LSK number). Similarly, we found 2,055±195 LSKCD34-Flk2- cells per femur in WT compared to 467±95 LSKCD34-Flk2- cells per femur in CD166-/- mice (77% reduction in LT-HSC).
Figure 1.
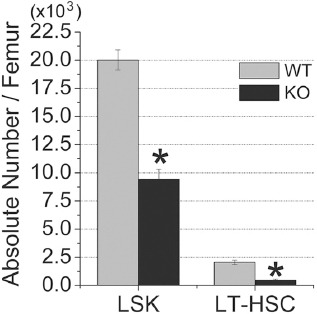
Numbers of phenotypically defined HSC (LSK and LSK CD34-Flk2-) in the BM of 6 week-old, WT and CD166-/- mice under steady state conditions (n=6 mice/group). Results are presented as the mean ± SE. *p<0.05 compared to WT mice.
Anthropometrics, femur geometry, and DXA
[Table 2] summarizes the anthropometric, gross femur geometry, and DXA data of female and male, WT and CD166-/- mice at 6 weeks of age. There was no significant interaction for body weight, or main effects for sex. However, there was a significant main effect for genotype, with CD166-/- mice weighing significantly more (18% greater) than WT mice. With respect to femoral geometry, no significant interactions were detected for femoral length, mid-shaft width (medial to lateral), or mid-shaft height (ventral to dorsal). The main effect for genotype identified CD166-/- mice to have a 4.7% increase in femoral length compared to WT controls, with no significant genotype differences for mid-shaft width or mid-shaft height.
We report a significant sex by genotype interaction for aBMD and BMC of the femur. Post-hoc analyses demonstrated that female and male CD166-/- mice had significantly greater femur aBMD (10.6% and 28.0%, respectively) and BMC (21.4% and 31.3%, respectively) compared to WT mice. There was no significant interaction for aBMD of the lumbar spine or whole body. The main effects for genotype identified CD166-/- mice to have significantly greater lumbar spine aBMD (15.8%) and whole body aBMD (16.1%) compared to WT mice. We report a significant interaction for vertebral and whole body BMC, with female and male CD166-/- mice having significantly greater BMC than WT mice. Specifically, female CD166-/- mice exhibited a significant 25.0% increase in vertebral BMC and a significant 40.9% increase in whole body BMC compared to WT female mice. Similarly, male CD166-/- mice exhibited a significant 33.3% increase in vertebral BMC and a significant 35.9% increase in whole body BMC compared to WT male mice. It should be noted that due to significant differences for body weight between genotypes, we used body weight was used as a covariate.
Trabecular bone phenotype of 6 week CD166-/- mice: μCT and histology
μCT and histomorphometric trabecular bone data from 6 week-old CD166-/- and WT mice are reported in Figures 2 and 3, respectively. As illustrated in Figure 2, which depicts the μCT results, there were no significant interactions for distal femur BV/TV, Tb.N, Tb.Th, or Tb.Sp. However, post-hoc analyses for genotype found CD166-/- mice to have significantly greater distal femur BV/TV (24% increase) and lower Tb.Sp (85% decrease) than WT. No significant genotype differences were identified for Tb.N or Tb.Th.
Figure 2.
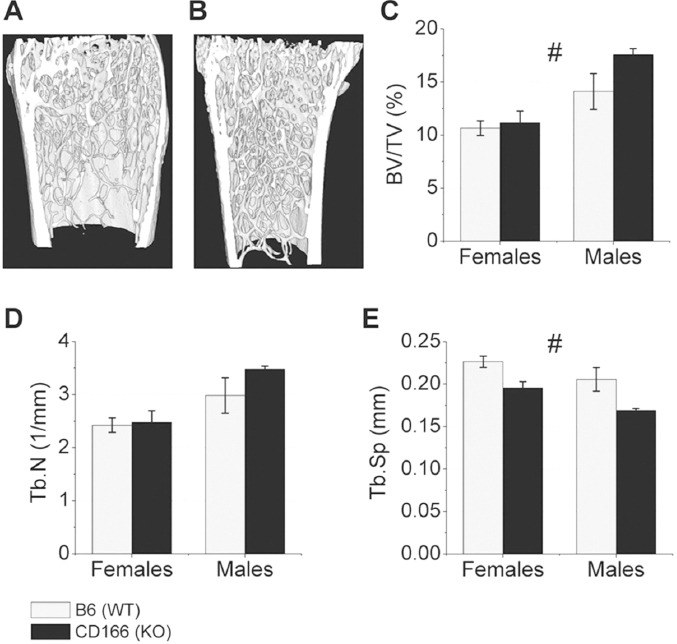
μCT was used to examine the trabecular bone phenotype of the distal femur of 6 week-old (A) WT and (B) CD166-/- mice. (C) BV/TV, (D) Tb.N, and (E) Tb.Sp. n=7-10 mice/group. Results are presented as the mean ± SE. #Indicates significant main effect for genotype, with no significant genotype and sex interaction (p<0.05).
Figure 3.
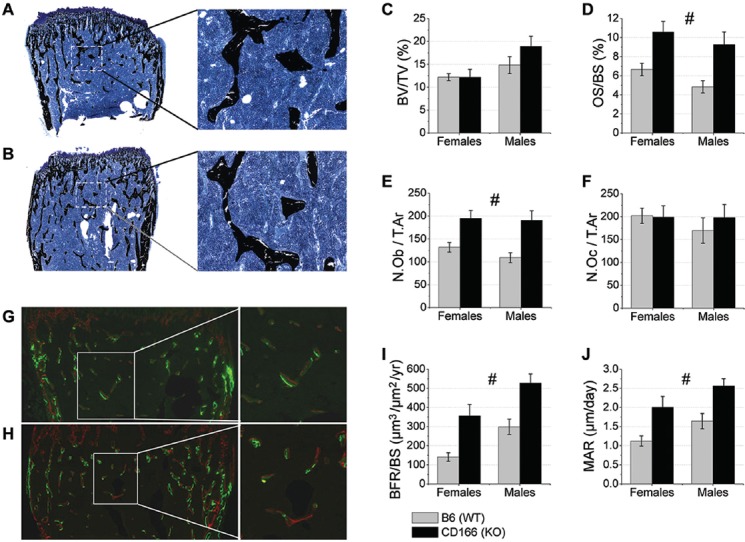
Static and dynamic trabecular bone phenotype histomorphometric analysis of 6 week-old, WT and CD166-/- distal femurs. Trichrome staining of (A) WT and (B) CD166-/- distal femurs, original magnification, 10X (inset 20X). (C) BV/TV, (D) OS/BS (%), (E) N.Ob/T.Ar, and (F) N.Oc/T.Ar. Dual fluorochrome labeling of (G) WT and (H) CD166-/- distal femurs, original magnification 10X (inset 20X). (I) BFR/BS and (J) MAR. n=6-8 mice/group. Results are presented as the mean ± SE. #Indicates significant main effect for genotype, with no significant genotype and sex interaction (p<0.05).
As shown in Figure 3A-F, there was no significant interaction for distal femur BV/TV, and post-hoc genotype differences showed a modest, non-significant, increase in BV/TV. Static histological measures revealed no significant interaction for OS/BS, N.Ob/T.Ar, or N.Oc/T.Ar. Post-hoc analyses of the main effects for genotype revealed CD166-/- mice to have greater OS/BS than WT mice (72% greater). Furthermore, there were significantly more OB [N.Ob/T.Ar], in CD166-/- compared to WT mice (60% increase). However, no statistically significant differences in the numbers of osteoclasts/tissue area (N.Oc/T.Ar) were observed between CD166-/- and WT mice.
For dynamic trabecular bone histomorphometric analyses of the distal femur (Figure 3G-J) there was no significant interaction for BFR/BS or MAR. Post-hoc analyses of the main effect for genotype revealed BFR/BS and MAR were significantly increased in CD166-/- compared to WT mice (104% increase and 64% increase, respectively).
Cortical bone phenotype of 6 week CD166-/- mice: μCT and histology
Figure 4 depicts measures obtained by μCT for cortical bone from the mid-shaft femur. Figures 4 A&B depict representative images of cortical bone from A: WT and B: CD166-/- mice. We report a significant interaction for Ct.Ar, Ct.Th, and polar moment of inertial (Ip). Female CD166-/- had significantly more Ct.Ar (7.8% more) and greater Ct.Th (6.7% greater) than WT controls, with no difference for medullary area (Me.Ar) or polar moment of inertia (Ip) between genotypes. Male CD166-/- had significantly greater cortical Ct.Ar (33% greater), cortical thickness (25% greater), and polar moment of inertia (53% greater). Post-hoc analyses of the main effect for genotype revealed total area (T.Ar) was greater in CD166-/- mice than WT controls (18% greater).
Figure 4.
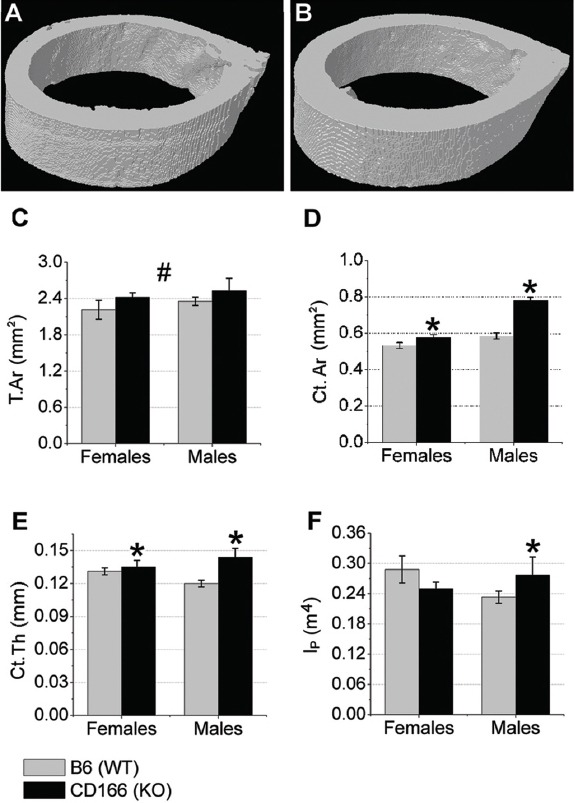
μCT was used to examine the cortical bone phenotype from the mid-shaft femur of 6 week-old, (A) WT and (B) CD166-/- mice. Cortical (C) T.Ar, (D) Ct.Ar, (E) Ct.Th, and (F) IP. n=7-10 mice/group. Results are presented as the mean ± SE. *Indicates significant interaction between genotype and sex, with post-hoc analyses indicating sex-specific genotype differences (p<0.05). #Indicates significant main effect for genotype, with no significant interaction found between genotype and sex (p<0.05).
Static and dynamic cortical bone histomorphometric data collected from the mid-shaft femur are shown in Figure 5. We report a significant interaction for static measures of T.Ar and Ct.Ar, with no significant interaction for the dynamic measures of periosteal and endosteal BFR/BS. As might be expected based on the gross femur measurements, tissue area (T.Ar, μm2), as assessed by histomorphometry, was not different between CD166-/- and WT female femurs; however, T.Ar was significantly increased in CD166-/- male femurs relative to that of WT male femurs (21% greater). Ct.Ar was significantly higher in CD166-/- femurs compared to WT femurs in both genders: 43% greater in females and 86% greater in males. Interestingly, periosteal BFR/BS was significantly lower in CD166-/- femurs compared to WT femurs (38% reduction). Conversely, endosteal BFR/BS revealed no significant differences between genotypes.
Figure 5.
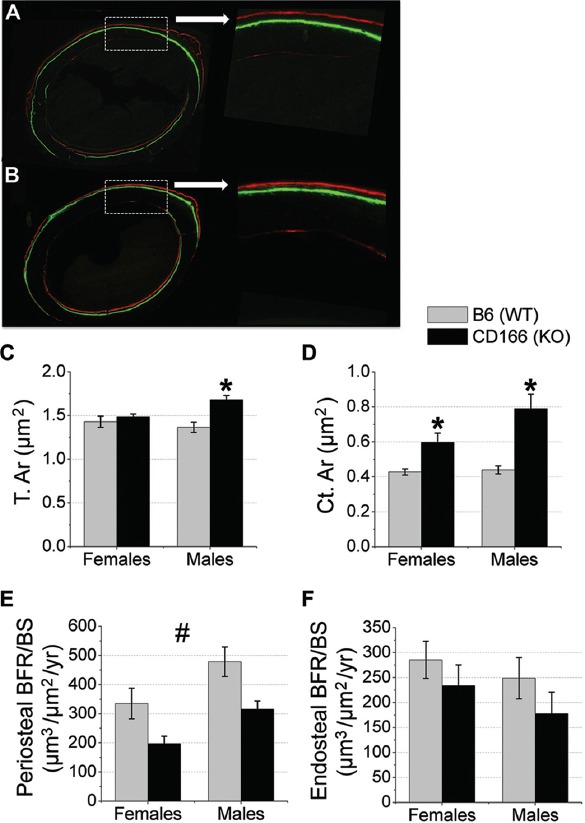
Cortical bone histomorphometric analysis of 6 week-old WT and CD166-/- mid-shaft femurs. Dual fluorochrome labeling of (A) WT and (B) CD166-/- femurs, original magnification, 10X (inset 20X). (C) T.Ar., (D) Ct.Ar, (E) Periosteal BFR/BS, and (F) Endosteal BFR/BS. n=6-8 mice/group. Results are presented as the mean ± SE. *Indicates significant interaction between genotype and sex, with post-hoc analyses indicating sex-specific genotype differences (p<0.05). #Indicates significant main effect for genotype, with no significant interaction found between genotype and sex (p<0.05).
Biomechanical and biomaterial properties of 6 week CD166-/- mice
Figure 6 depicts differences in biomechanical properties between CD166-/- and WT femurs for female and male mice. With a similar or larger outer geometry but a significant increase in Ct.Ar, it would be predicted that the CD166-/- bones would be stronger and stiffer than WT bones. However, we identified a significant sex by genotype interaction for ultimate force and polar moment of inertia for males only. No significant interaction was found for stiffness or toughness. Post hoc analyses found female femurs ultimate force was significantly increased 28% and in male femurs ultimate force was significantly increased 49% in CD166-/- mice as compared to WT controls. Femoral Ip was increased in male CD166-/- mice as compared to WT controls, with no genotype difference for females. The main effects for genotype identified CD166-/- mice to have significantly increased femoral stiffness compared to WT (81% increase). However, there was no significant genotype main effect difference between CD166-/- and WT mice for femoral toughness.
Figure 6.
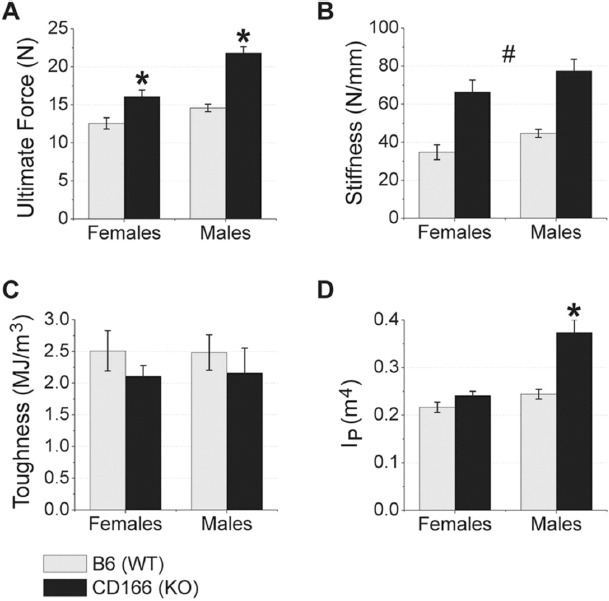
Biomechanical data of 6 week-old WT and CD166-/- femurs from 3-point bending. (A) ultimate force, (B) stiffness, (C) toughness, and (D) IP. n=7-10 mice/group. Results are presented as the mean ± SE. *Indicates significant interaction between genotype and sex, with posthoc analyses indicating sex-specific genotype differences (p<0.05). #Indicates significant main effect for genotype, with no significant interaction found between genotype and sex (p<0.05).
CD166-/- positively regulates osteoblast proliferation
To begin to understand the role of CD166 expression in OB function, we sought to determine whether CD166 expression affected N.Ob or cell cycle regulation. We previously reported that in vitro CD166-/- OB exhibit a significant increase in total number of viable OB early in culture with a similar increase in the percentage of cells in active phases of cell cycle[11] compared to WT OB. Figure 7A shows that, consistent with an increase in proliferation, cyclin regulator, Fra1, expression was significantly elevated (40% increase) in the CD166-/- OB after 4 days in culture compared to WT OB.
Figure 7.

OB were isolated from CD166-/- and WT littermate control mice. (A) Real-time PCR was used to examine mRNA expression of Fra1, Runx2, ALP, ColIa1, and Ocn. OB differentiation was also assessed using (B) ALP and (C) mineralization assays (bound calcium). Results are presented as the mean ± SD. *p<0.05 compared to WT OB.
CD166-/- positively regulates osteoblast differentiation in vitro
Next, we examined the effects of CD166 expression on OB differentiation. As shown in Figures 7B&C, we assessed ALP activity and measured bound calcium as a functional measure of mineralization after 14 days. ALP activity and the amount of bound calcium were found to be significantly increased (54% and 32% greater, respectively) in the CD166-/- OB, which suggests a greater degree of OB differentiation.
To further examine the effects of CD166 expression in OB differentiation, we cultured WT and CD166-/- OB under osteogenic conditions as above and examined mRNA expression on day 14 for the following genes: Runx2, ALP, ColIa1, and Ocn. As detailed in Figure 7A, ALP and Ocn expression were significantly elevated in CD166-/- OB compared to WT on day 14 (175% and 130% increase, respectively). However, Runx2 and ColIa1 expression were not significantly different between CD166-/- and WT OB.
Discussion
Our results indicate that the loss of CD166 impacts HSC numbers in CD166-/- mice compared to WT controls. Both 6 week-old male and female CD166-/- mice weighed significantly more, and also displayed an increased whole body aBMD and BMC. Although BV/TV was modestly increased, Tb.N and Tb.Th, N. Oc were not altered. Tb.Sp was decreased along with the increased levels of N.Ob, OS/BS, BFR/BS, and MAR in the trabecular bone of CD166-/- mice. Cortical bone analyses revealed that polar moment of inertia (males only), ultimate force, and stiffness were significantly elevated in CD166-/- femurs, though toughness was not altered. In vitro, OB from CD166-/- mice were more differentiated than their WT counterparts, suggesting that loss of CD166 expression impacts bone biology in general and implying that the CD166-/- bone microenvironment may contain more mature OB which in turn may negatively regulate hematopoiesis.
We were initially interested in examining whether the expression of CD166 altered HSC numbers in the BM. We previously tested and reported that CD166 expression is completely negative in both OB and HSC isolated from CD166-/- mice[11]. Although CD166-/- mice have no apparent major defects in terms of their viability and fertility, we found that CD166-/- mice had reduced numbers of phenotypically defined HSC, suggesting that CD166 expression may play an important role in hematopoiesis (Figure 1 and 11). Since HSC numbers and function are not the focus of this communication, these observations were not further investigated, but several of our findings related to the impact of CD166 on the function and engraftment of hematopoietic stem cells and the competence of the hematopoietic niche are reported in our recent paper[11]. Briefly, we reported that hematopoietic progenitor cells are significantly lower in CD166-/- BM compared to WT mice. CD166 negative HSC derived from either murine or human umbilical cord blood engrafted poorly in appropriate recipients. WT HSC failed to engraft in CD166-/- recipients[11]. Similarly, Jeannet et al.[31] also reported that CD166-/- HSC have an engraftment defect, although young CD166-/- mice displayed normal hematopoietic counts and numbers of phenotypically defined HSC.
Bone phenotype analyses were performed on 6 week-old CD166-/- and WT mice. Body weight, whole body aBMD and BMC were all significantly higher (13-56%) in CD166-/- mice compared to gender matched WT controls (Table 2). As cortical bone is the primary contributor to bone density, our aBMD measurements are suggestive of changes in cortical bone properties. To further understand cortical bone properties in CD166-/- mice we conducted static and dynamic bone histological measures of the femur mid-shaft of CD166-/- and WT mice. Although total cortical area (periosteal area - endocortical area) was significantly elevated in CD166-/- compared to WT mice, periosteal BFR of the femur mid-shaft was significantly lower in CD166-/- mice, with no differences in endosteal BFR. To have an increase in Ct.Ar given this BFR data, it may be that prior to week 5 the periosteal BFR/BS was higher in CD166-/- mice, but in week 5 (when the fluorescent labels were administered) the WT mice were beginning to “catch-up”. An explanation such as this is reasonable with the larger T.Ar observed in the femurs of male mice (although no difference was observed in the femurs of female mice). Consistent with an increase in cortical Ct.Ar, biomechanical data demonstrated that CD166-/- femurs were stronger and stiffer than WT femurs upon three point bending (Figure 6).
Although examination of cortical bone of long bones is important in understanding the overall bone phenotype we would also like to note, that no gross differences were observed in other bones such as the thickness of the calvaria. Turning back to the femur analyses, arguably the trabecular bone in the endosteal region is more relevant to hematopoiesis as it is the endosteal regions where quiescent HSC reside. Therefore, we used μCT and histomorphometric analyses to examine the trabecular bone phenotype of CD166-/- mice. Femoral μCT analysis (Figure 2) revealed a significant increase in BV/TV but not in Tb. N or Tb.Th between CD166-/- and WT mice, and Tb.Sp was reduced in CD166-/-. Dynamic and static histomorphometric analyses of the trabecular bone phenotype (Figure 3) provide a more detailed, specific survey of CD166’s effects on the trabecular bone phenotype. Although trabecular BV/TV was modestly elevated it was not significantly different between CD166-/- and WT mice. That μCT and histomorphometric data show similar data trends, but significance was reached with μCT but not histomorphometric analyses may suggest that a slightly higher sample size was needed for histomorphometric studies. Of note, CD166-/- femurs contained significantly more OB (N.Ob/T.Ar), and the OB of these mice produce significantly higher levels of osteoid (unmineralized bone matrix, OS/BS). Dynamic histomorphometric analysis of the trabecular phenotype demonstrated that BFR/BS and MAR are enhanced in the distal femur of CD166-/- mice, which suggests a greater degree of osteogenesis. That there were more OB that generated more osteoid indicates an alteration in the hematopoietic niche of CD166-/- mice. That said, these data also point to a potential defect in mineralization as the amount of unmineralized bone matrix is higher in the CD166-/- mice. Ongoing studies in our laboratory are exploring this possibility. However, no significant differences in the N. Oc were observed in CD166-/- mice. This finding is somewhat surprising given that OB and osteoclasts are normally coupled, and with an increased N.Ob in CD166-/- mice an increased N. Oc would have been predicted.
Turning from in vivo to in vitro findings, we previously reported that a lower percentage of CD166-/- OB were in G0/G1 phase compared to WT OB cells during the first 2 days of in vitro cultures confirming the increased proliferation of CD166-/- OB[11]. Consistent with these data, in this study, we found an enhanced expression of Fra1 (Figure 7A) confirming that OB proliferation is enhanced in CD166-/- OB early in culture (4 days). Of importance, Fra1 is known to be critical in regulating cyclin expression and cell-cycle progression, serving to induce cell proliferation. The ALP activity assay and the calcium mineralization assay represent the degree of OB differentiation (Figures 7B&C). Both assays demonstrated that the CD166-/- OB cultures are more mature or are further along in the differentiation process than their WT counterparts. Consistent with this observation, mRNA expression also demonstrates increased levels of both ALP and Ocn. Noteworthy is the 127% increase in Ocn expression observed in CD166-/- OB as compared to WT OB, as Ocn expression is well known to be upregulated in more mature OB. Of note, OB cultures were generated from a mixture of male and female pups. As it appears that CD166-/- males exhibit a more pronounced bone phenotype than females that the cells are derived from a mixed population may result in smaller differences than would possibly be observed had male mice alone been used for the generation of OB cells.
We have previously demonstrated[5-7,16] that immature OB lineage cells expressing high levels of CD166 and Runx2 enhance HSC maintenance and function significantly better than mature OB expressing higher levels of ALP and Ocn. In this study, the increased N.Ob in CD166-/- mice might have resulted from the accumulation of what appears to be more mature cells (low Runx2, with high ALP and Ocn expression) that are most likely less capable of supporting HSC maintenance, thus leading to the observed low numbers of LT-HSC in knock-out mice. Despite of higher N.Ob, the absence of homophilic interactions between OB and HSC through CD166 could be another detrimental factor to the competence of the hematopoietic niche. On the other hand, the increased deposition of unmineralized bone matrix (osteoid) in CD166-/- mice suggests that CD166 may negatively regulate OB differentiation and proliferation such that more mature OB in CD166-/- mice divide more rapidly and produce more osteoid, accounting for the increased N.Ob. However, further study is necessary to investigate these novel theories about the role of CD166 in OB maturation and osteoid production.
Taken together these studies show that global CD166 deficiency results in 3 main alterations within the BM cavity including: 1) a marked reduction in HSC; 2) a marked increase in mature OB; and 3) a subsequent striking increase in osteoid. The latter 2 observations suggest that the bone microenvironment is altered and may contribute to the hematopoiesis defect. That CD166 is expressed not only on HSC and OB, but importantly has functional consequences on bone is a novel finding. Indeed, expression of CD166 and several of its functional roles in the context of hematopoiesis have been identified over the past decade, including: T cell activation, early stages of hematopoiesis, and for identifying a population of human CD34+ cells enriched for hematopoietic progenitors[1,8-10]. More recently[31], loss of CD166 on murine hematopoietic cells was implicated in several hematopoietic defects including reduced engraftment potential of long-term HSC. Several groups[1-4], including ours[11,16] have also identified that CD166 is expressed on OB lineage cells. However, to our knowledge, results in this communication, as well as our previously published results[11], represent the only set of data implicating a single molecule, namely, CD166, in the function of HSC, the competence of the hematopoietic niche, and also significantly alters bone phenotype and biology. In conclusion, our present and published data[11,16,32] suggest that CD166 expression directly and significantly impacts bone phenotype and thus may play a key role in modulating HSC-OB interactions in the niche that are crucial for normal hematopoiesis and maintenance of HSC.
Acknowledgements
The authors thank the operators of the Indiana University Melvin and Bren Simon Cancer Center Flow Cytometry Resource Facility for their outstanding technical help and support. This work was supported by the Medical Student Affairs Summer Research Program in Academic Medicine, Indiana University School of Medicine funded in part by NIH grant HL110854 (PHE) and the Department of Orthopaedic Surgery, Indiana University School of Medicine (MAK). In addition, research reported in this publication was supported in part by the NHLBI R01 HL55716 (EFS), NIAMS R01 AR060332 (MAK), and the Indiana Center for Excellence in Molecular Hematology (NIDDK P30 DK090948). The Flow Cytometry Research Facility is partially funded by NCI P30 CA082709. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Edited by: F. Rauch
Refrences
- 1.Cortes F, Deschaseaux F, Uchida N, et al. HCA, an immunoglobulin-like adhesion molecule present on the earliest human hematopoietic precursor cells, is also expressed by stromal cells in blood-forming tissues. Blood. 1999;93:826–37. [PubMed] [Google Scholar]
- 2.Nelissen JM, Torensma R, Pluyter M, et al. Molecular analysis of the hematopoiesis supporting osteoblastic cell line U2-OS. Exp Hematol. 2000;28:422–32. doi: 10.1016/s0301-472x(00)00127-2. [DOI] [PubMed] [Google Scholar]
- 3.Arai F, Ohneda O, Miyamoto T, Zhang XQ, Suda T. Mesenchymal stem cells in perichondrium express activated leukocyte cell adhesion molecule and participate in bone marrow formation. J Exp Med. 2002;195:1549–63. doi: 10.1084/jem.20011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai F, Nakamura Y, Gomei Y, Suda T. Characterization of the niche complex molecules in bone marrow. Exp Hematol. 2008;36:S25. [Google Scholar]
- 5.Chitteti BR, Cheng YH, Poteat B, et al. Impact of interactions of cellular components of the bone marrow microenvironment on hematopoietic stem and progenitor cell function. Blood. 2010;115:3239–48. doi: 10.1182/blood-2009-09-246173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chitteti BR, Cheng YH, Streicher DA, et al. Osteoblast lineage cells expressing high levels of Runx2 enhance hematopoietic progenitor cell proliferation and function. J Cell Biochem. 2010;111:284–94. doi: 10.1002/jcb.22694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng YH, Chitteti BR, Streicher DA, et al. Impact of maturational status on the ability of osteoblasts to enhance the hematopoietic function of stem and progenitor cells. J Bone Miner Res. 2011;26:1111–21. doi: 10.1002/jbmr.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami Y, Hirata H, Miyamoto Y, et al. Isolation of cardiac cells from E8.5 yolk sac by ALCAM (CD166) expression. Mechanisms of Development. 2007;124:830–9. doi: 10.1016/j.mod.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Ohneda O, Ohneda K, Arai F, et al. ALCAM (CD166): its role in hematopoietic and endothelial development. Blood. 2001;98:2134–42. doi: 10.1182/blood.v98.7.2134. [DOI] [PubMed] [Google Scholar]
- 10.Uchida N, Yang Z, Combs J, et al. The characterization, molecular cloning, and expression of a novel hematopoietic cell antigen from CD34+human bone marrow cells. Blood. 1997;89:2706–16. [PubMed] [Google Scholar]
- 11.Chitteti BR, Kobayashi M, Cheng Y, et al. CD166 regulates human and murine hematopoietic stem cells and the hematopoietic niche. Blood. 2014;124:519–29. doi: 10.1182/blood-2014-03-565721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowen MA, Bajorath J, D’Egidio M, et al. Characterization of mouse ALCAM (CD166): the CD6-binding domain is conserved in different homologs and mediates cross-species binding. Eur J Immunol. 1997;27:1469–78. doi: 10.1002/eji.1830270625. [DOI] [PubMed] [Google Scholar]
- 13.Raaijmakers MH, Scadden DT. Evolving concepts on the microenvironmental niche for hematopoietic stem cells. Curr Opin Hematol. 2008;15:301–6. doi: 10.1097/MOH.0b013e328303e14c. [DOI] [PubMed] [Google Scholar]
- 14.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- 15.Nakamura Y, Arai F, Iwasaki H, et al. Isolation and characterization of endosteal niche cell populations that regulate hematopoietic stem cells. Blood. 2010;116:1422–32. doi: 10.1182/blood-2009-08-239194. [DOI] [PubMed] [Google Scholar]
- 16.Chitteti BR, Cheng YH, Kacena MA, Srour EF. Hierarchical organization of osteoblasts reveals the significant role of CD166 in hematopoietic stem cell maintenance and function. Bone. 2013;54:58–67. doi: 10.1016/j.bone.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel DD, Wee SF, Whichard LP, et al. Identification and characterization of a 100-kD ligand for CD6 on human thymic epithelial cells. J Exp Med. 1995;181:1563–8. doi: 10.1084/jem.181.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekine-Aizawa Y, Omori A, Fujita SC. MuSC, a novel member of the immunoglobulin superfamily, is expressed in neurons of a subset of cranial sensory ganglia in the mouse embryo. Eur J Neurosci. 1998;10:2810–24. doi: 10.1111/j.1460-9568.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- 19.Chitteti BR, Bethel M, Voytik-Harbin SL, Kacena MA, Srour EF. In vitro construction of 2D and 3D simulations of the murine hematopoietic niche. Methods Mol Biol. 2013;1035:43–56. doi: 10.1007/978-1-62703-508-8_5. [DOI] [PubMed] [Google Scholar]
- 20.Feher A, Koivunemi A, Koivunemi M, et al. Bisphosphonates do not inhibit periosteal bone formation in estrogen deficient animals and allow enhanced bone modeling in response to mechanical loading. Bone. 2010;46:203–7. doi: 10.1016/j.bone.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 21.Warden SJ, Nelson IR, Fuchs RK, Bliziotes MM, Turner CH. Serotonin (5-hydroxytryptamine) transporter inhibition causes bone loss in adult mice independently of estrogen deficiency. Menopause. 2008;15:1176–83. doi: 10.1097/gme.0b013e318173566b. [DOI] [PubMed] [Google Scholar]
- 22.Weatherholt AM, Fuchs RK, Warden SJ. Cortical and trabecular bone adaptation to incremental load magnitudes using the mouse tibial axial compression. doi: 10.1016/j.bone.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong GL, Cohn DV. Target cells in bone for parathormone and calcitonin are different: enrichment for each cell type by sequential digestion of mouse calvaria and selective adhesion to polymeric surfaces. Proc Natl Acad Sci U S A. 1975;72:3167–71. doi: 10.1073/pnas.72.8.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kacena MA, Eleniste PP, Cheng YH, et al. Megakaryocytes Regulate Expression of Pyk2 Isoforms and Caspase-mediated Cleavage of Actin in Osteoblasts. J Biol Chem. 2012;287:17257–68. doi: 10.1074/jbc.M111.309880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horowitz MC, Fields A, DeMeo D, Qian HY, Bothwell AL, Trepman E. Expression and regulation of Ly-6 differentiation antigens by murine osteoblasts. Endocrinology. 1994;135:1032–43. doi: 10.1210/endo.135.3.7520861. [DOI] [PubMed] [Google Scholar]
- 26.Jilka RL, Cohn DV. Role of phosphodiesterase in the parathormone-stimulated adenosine 3’,5’-monophosphate response in bone cell populations enriched in osteoclasts and osteoblasts. Endocrinology. 1981;109:743–7. doi: 10.1210/endo-109-3-743. [DOI] [PubMed] [Google Scholar]
- 27.Simmons DJ, Kent GN, Jilka RL, Scott DM, Fallon M, Cohn DV. Formation of bone by isolated, cultured osteoblasts in millipore diffusion chambers. Calcif Tissue Int. 1982;34:291–4. doi: 10.1007/BF02411253. [DOI] [PubMed] [Google Scholar]
- 28.Hughes FJ, Aubin JE. Culture of Cells of the Osteoblast Lineage. In: Arnett TR, Henderson B, editors. Methods in Bone Biology. 1st ed. London, UK: Chapman & Hall; 1998. pp. 1–49. [Google Scholar]
- 29.Ciovacco WA, Goldberg CG, Taylor AF, et al. The role of gap junctions in megakaryocyte-mediated osteoblast proliferation and differentiation. Bone. 2009;44:80–6. doi: 10.1016/j.bone.2008.08.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanford CM, Jacobson PA, Eanes ED, et al. Rapidly forming apatitic mineral in an osteoblastic cell line (UMR 106-01 BSP) J Biol Chem. 1995;270:9420–8. doi: 10.1074/jbc.270.16.9420. [DOI] [PubMed] [Google Scholar]
- 31.Jeannet R, Cai Q, Liu H, Vu H, Kuo YH. Alcam regulates long-term hematopoietic stem cell engraftment and self-renewal. Stem Cells. 2013;31:560–71. doi: 10.1002/stem.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chitteti BR, Bethel M, Kacena MA, Srour EF. CD166 and regulation of hematopoiesis. Curr Opin Hematol. 2013;20:273–80. doi: 10.1097/MOH.0b013e32836060a9. [DOI] [PubMed] [Google Scholar]


