Abstract
Advances in reading, writing and editing genetic materials have greatly expanded our ability to reprogram biological systems at the resolution of a single nucleotide and on the scale of a whole genome. Such capacity has greatly accelerated the cycles of design, build and test to engineer microbes for efficient synthesis of fuels, chemicals and drugs. In this review, we summarize the emerging technologies that have been applied, or are potentially useful for genome-scale engineering in microbial systems. We will focus on the development of high-throughput methodologies, which may accelerate the prototyping of microbial cell factories.
Keywords: Genome-scale engineering, microbial cell factory, transcriptome engineering, genome synthesis, homologous recombination, high-throughput technology
1. Introduction
Microbial cell factories (MCFs), which convert biomass resources to value-added compounds such as fuels, chemicals, materials and pharmaceuticals, have been proposed as a sustainable and renewable alternative to the traditional petrochemical-based processes (Keasling, 2010, Lee et al., 2012, Rabinovitch-Deere et al., 2013). However, intensive reprogramming of cellular metabolism is required to achieve economically feasible fermentation processes with MCFs. Conventional strain engineering approaches rely on random mutagenesis, which is achieved through chemical mutagens/UV irradiation (Crook and Alper, 2012), prolonged cultivation under selective pressure (Portnoy et al., 2011), transposon insertions (Eckert et al., 2011, Hamer et al., 2001, Hutchison et al., 1999) and genome shuffling (Biot-Pelletier and Martin, 2014, Zhang et al., 2002). Effective in generating improved phenotypes using simple techniques, these methods are widely adopted in industry, especially for those host organisms with poorly defined genetics and limited engineering tools (Crook and Alper, 2012). However, traditional approaches are often labor-intensive, time-consuming, and difficult to analyze and transfer the genetic basis of a selected trait. Recently, the scale, efficiency and precision of genetic analysis and manipulation have been remarkably improved by several enabling technologies, including but not limited to microarray DNA synthesis, next-generation DNA sequencing (NGS), programmable DNA-binding proteins, and in vivo biosensors. Nowadays, billions of genome variants can be created in a directed and/or combinatorial manner, and the mutant strains with the optimal performance can be rapidly isolated. Collectively, these new technologies and their applications exemplify an emerging discipline called ‘genome engineering’ or ‘genome-scale engineering’ (Carr and Church, 2009, Esvelt and Wang, 2013, Jeong et al., 2013, Segal and Meckler, 2013).
The practice of genome-scale engineering can be broadly classified into three categories: genome editing, transcriptome engineering, and genome synthesis. Genome editing precisely or combinatorially modifies the target genome at multiple loci. Modifications are located either in the open-reading frames (ORFs) or in the cis-acting regulatory elements such as promoters and ribosome-binding sites (RBSs). Transcriptome engineering essentially targets trans-acting regulatory elements, such as transcription factors (TFs) or non-coding RNAs (ncRNAs), by mutating endogenous regulators or introducing artificial ones. Genome synthesis involves hierarchical assembly of short chemically synthesized DNA fragments into viral/microbial genomes and yeast chromosomes. Although current synthetic genomes are constructed mainly based on their wild type templates, the ultimate goal is to write genome sequences de novo.
In this review, we first introduce the recent development in genome editing (section 2), transcriptome engineering (section 3), and genome synthesis (section 4). We then highlight how these techniques can facilitate high-throughput genotyping and phenotyping (section 5), which greatly accelerates our understanding and engineering of microbial genomes. In addition, we will discuss several examples on the application of genome-scale engineering to improve MCF performance and provide perspectives on how computational approaches and laboratory automation can be further integrated.
2 Genome editing
Unlike random mutagenesis, targeted genome editing results in elaborative and massive genome modifications with a traceable manner. Homologous recombination (HR) is the core mechanism of most targeted genome editing techniques, and various enzymes have therefore been investigated to either mediate or promote HR in microorganisms.
2.1 Recombinases
Recombinases catalyze exchange of short homologous regions (30~40 bp) of DNA. Site-specific recombinases are grouped into two families, the tyrosine recombinase family and the serine recombinase family (Turan et al., 2013). An early characterized member of the tyrosine recombinase family was λ integrase, which enables incorporation of phage DNA into the bacterial chromosomes. The λ integrase mediates irreversible recombination between the attP and attB sites in the phage and host chromosomes respectively, generating recombinant attL and attR sites (Mizuuchi and Mizuuchi, 1980). Later, Cre (from phage P1) and flippase (FLP, from the 2μ plasmid of yeast Saccharomyces cerevisiae), recognizing the loxP site and the flippase recognition target (FRT) site, respectively, were widely used for efficient recombination in a variety of species (Dymecki, 1996, Nagy, 2000, Sternberg et al., 1981, Turan et al., 2011). With identical recognition sites, Cre and FLP can reversibly invert, integrate or excise DNA sequences between recognition regions. Alternatively, such processes can be made irreversible using a partially mutated recognition site to yield a poorly recognized region after recombination (Albert et al., 1995, Schlake and Bode, 1994). For the serine recombinase family, ϕC31 integrase (from Streptomyces phage ϕC31) was the most well-studied example. Behaving like the λ integrase, ϕC31 was proven to have great potential in eukaryotic genome engineering (Karow and Calos, 2011).
In addition to inversion, integration and excision facilitated by the above-mentioned recombinases, recombinase-mediated cassette exchange (RMCE) is another useful approach in genome engineering. By flanking the target genomic locus with two different spacer mutant (“heterospecific”) sites recognized by the same recombinase or orthogonal sites of different recombinases, the endogenous region will be replaced by a donor cassette with compatible recognition sites (Turan, Zehe, 2013). Many Cre and FLP variants were engineered to recognize different sites with little cross reactivity with the wild type system, allowing efficient directional cassette exchange (Fig. 1A) (Buchholz and Stewart, 2001, Schlake and Bode, 1994, Turan et al., 2010). By exploiting the specific attP×attB recombination event, ϕC31 was also applied to cassette exchange without the requirement of heterospecific att-sites. However, ϕC31 mediated-cassette change was in a unidirectional manner (Turan and Bode, 2011).
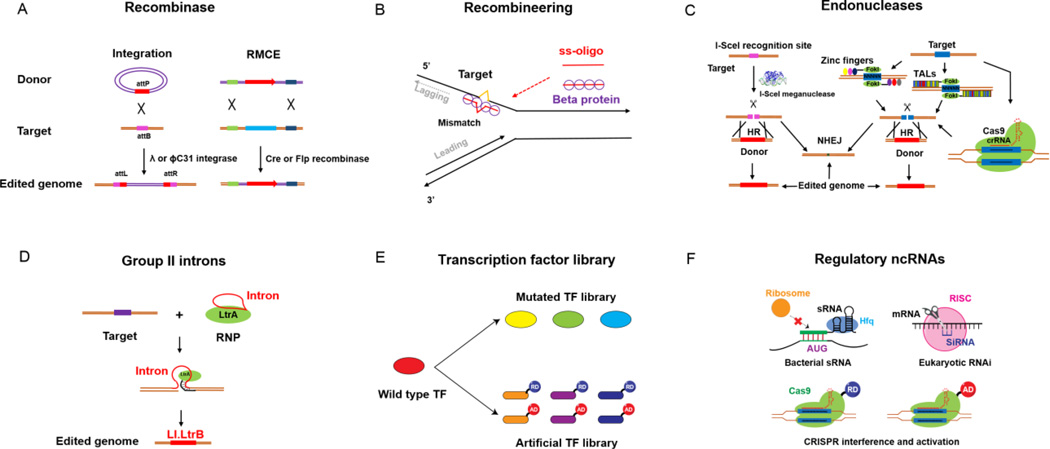
Fig. 1. Overview of various genome-scale engineering tools.
(A) A circular donor cassette can be integrated into the recognition site by an integrase. By flanking a target sequence with heterospecific sites, RMCE enables replacement of a target sequence using a donor cassette flanked by compatible sites. (B) The ss-oligos containing designed mutations are incorporated into the lagging strand of replicating DNA through recombineering. (C) HR or NHEJ is greatly promoted via DSBs using various endonucleases. (D) Site specific insertion can be achieved via group II introns. (E) Transcription factor (TF) libraries can be constructed by mutating the endogenous TFs (gTME) or introducing artificial TFs for large-scale perturbation on transcriptome. (F) Different regulatory non-coding RNAs (ncRNAs), including sRNAs, siRNAs and gRNAs, are used to modulate targeted gene expression in bacteria and yeast.
Notably, pre-existing recognition sites are required for all events mediated by recombinases. Therefore, introduction of recognition sites into the target locus is unavoidable, which limits the application of recombinase-based methods for genome editing. Although much effort has been invested in the directed evolution of recombinases with new target recognition sequences, the engineered enzymes were inefficient in most cases (Gordley et al., 2009).
2.2 Recombination-mediated genetic engineering (Recombineering)
Taking advantage of bacteriophage-based recombination proteins (e.g., Red-Exo, Beta and Gam from λ phage (Murphy, 1998) and Rec E/T from Rac prophage (Zhang et al., 1998)), recombineering enables efficient and large-scale recombination between the transformed DNA fragments and the bacterial genome (Sharan et al., 2009). Recombination through the λ Red system relies on three proteins: Exo (also known as α), Beta and Gam. Exo is a 5’to 3’ exonuclease that digests double-stranded DNA (dsDNA) with 3’ overhangs generated. Beta is a single-stranded DNA (ssDNA) binding protein, which facilitates recombination between target locus and donor DNA (Fig. 1B). Gam inhibits endogenous RecBCD and SbcCD activity, preventing degradation of exogenous DNA by the host (Datta et al., 2008). In the RecE/T system, RecE is a 5’to 3’ exonuclease (similar to Exo) and RecT is a ssDNA-binding protein that stabilizes ssDNA and promotes recombination (similar to Beta). Either dsDNA or ssDNA can be employed for recombineering (Ellis et al., 2001, Sawitzke et al., 2007). With a linear donor DNA flanked by homology sequences as short as 40 bp, efficient recombination can be achieved by the λ red system (Sharan, Thomason, 2009). In addition, the RecET proteins are more efficient than the λ red proteins, especially in case of HR between two linear molecules (Fu et al., 2012). When these bacteriophage proteins were expressed in E. coli, the recombination efficiencies dramatically increased from 1 per 106 cells to 1 per 103–4 cells without optimization (Boyle et al., 2013, Murphy, 1998, Swingle et al., 2010, Zhang, Buchholz, 1998).
With further optimization (e.g., modifying the cellular machinery involved in DNA replication (Lajoie et al., 2012) or optimizing the concentration and length of donor oligos (Sawitzke et al., 2011)), recombineering technology has been exploited for multiplexed genome engineering. Notably, multiplex automated genome engineering (MAGE) and trackable multiplex recombineering (TRMR) accelerated E. coli genome evolution and analysis by creating genome-wide combinatorial genetic modifications in a short time (Wang et al., 2009, Warner et al., 2010). These studies are discussed in greater details in later sections.
2.3 Endonucleases
Apart from recombineering, introduction of a double-strand break (DSB) to a chosen site is another important strategy for targeted genome editing. Normally, a DSB is repaired by either HR or non-homologous end joining (NHEJ). Given a donor sequence flanked by homologous regions to target locus, HR can be facilitated by a DSB (Iglehart and Silver, 2009). In some hosts with low HR efficiency (e.g., mammalian cells), error-prone NHEJ is boosted by DSBs, which results in gene disruption (Sun et al., 2012b). Taken together, DSB-promoted HR or NHEJ eases the arduous targeted genome editing in most organisms. As a result, different natural and artificial endonucleases have been developed and engineered to introduce DSBs at desired loci (Fig. 1C).
2.3.1 I-SceI meganucleases
I-SceI is a homing endonuclease from the mitochondria of Saccharomyces cerevisiae and was the first endonuclease used in genome engineering (Silva et al., 2011). Compared to traditional restriction enzymes, I-SceI recognizes a longer sequence (18 bp), rendering it more rare-cutting (once in every 6.9×1010 bp). Indeed, HR promoted by I-SceI has been demonstrated in several prokaryotes and eukaryotes (Cox et al., 2007, Maggert et al., 2008, Yu et al., 2008). However, like recombinases, the requirement to introduce a recognition site into a target locus limits the application of I-SceI-based methods. Directed evolution of I-SceI towards a pre determined site was an alternative to make I-SceI more programmable. However, a substantial challenge arose since the DNA binding domain could not be decoupled from cleavage domain in naturally occurring I-SceI (Moure et al., 2008).
2.3.2 Zinc-finger nucleases (ZFNs)
ZFNs are artificial proteins that combine the DNA binding domain of a zinc finger protein with the non-specific cleavage domain of a FokI endonuclease, providing a general strategy to deliver site-specific DSBs to the chromosome (Li et al., 1992, Pavletich and Pabo, 1991). A zinc finger (ZF) is a ~30 amino acid motif that recognizes 3 nucleotides of DNA. Three ZF repeats are typically contained in an individual ZFN (Pavletich and Pabo, 1991). To strengthen the interaction of two FokI cleavage domains, a pair of zinc fingers is designed to bind neighboring sequences with a 5 to 7 bp spacer to form a dimeric cleavage domain. Such optimal configuration allows dimerization and subsequent cleavage (Bibikova et al., 2001). Successful ZFN-induced genome modifications were reported in many organisms such as plants (Townsend et al., 2009), zebrafish (Meng et al., 2008), frogs (Young et al., 2011), mice (Carbery et al., 2010), and sea urchin (Ochiai et al., 2010). Potential off-target effects were alleviated by utilizing a less toxic nickase (Kim et al., 2012) or additional ZF repeats (four to six) to bind longer sites (Wood et al., 2011). However, it has been shown that each triplet recognized by a ZF cannot be simply assembled to recognize a longer sequence (Ramirez et al., 2008). As a result, synthesizing customized ZFs remains difficult and expensive (Carroll, 2011).
2.3.3 Transcription activator-like effector nucleases (TALENs)
Similar to ZFNs, TALENs fuse the DNA-binding domain of a transcription activator-like effector (TALE) with a catalytic nuclease domain. TALE is a bacterial effector protein for synergistic regulation of gene expression in Xanthamonas sp. and Ralstonia sp. (Fu et al., 2013). Highly conserved 33–35 amino acid TALE repeat domains each bind one nucleotide of DNA with specificity dictated by two hypervariable residues (Fu, Foden, 2013). Unlike ZFNs, this one-to-one code allows the design of proteins with desired DNA-binding specificities by simply concatenating TALE repeats. Due to the more flexible recognition rule, TALENs can in principle be designed to readily target any sequence across the genome. In fact, TALENs have been applied in various organisms such as human cells (Sun et al., 2012c), yeast (Li et al., 2011) and zebrafish (Li, Huang, 2011). According to some preliminary studies, TALENs also seem to have fewer off-target effects compared to the corresponding ZFNs (Mussolino et al., 2011). Nonetheless, the primary drawback of TALENs is the trickiness of assembling a large number of repeats into an array. Several strategies have been reported to address this limitation (Briggs et al., 2012, Liang et al., 2013).
2.3.4 CRISPR nucleases
The type II bacterial Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated proteins (CRISPR-Cas) system has recently been exploited as an efficient genetargeting technology (Cong et al., 2013, Hwang et al., 2013, Jiang et al., 2013, Wang et al., 2013a). Directed by a trans-activating crRNA (tracrRNA):crRNA duplex (or a chimeric guide RNA (gRNA)), the CRISPR nuclease (e.g., Cas9 protein) is able to cleave a target DNA sequence with the protospacer adjacent motifs (PAM) (Mali et al., 2013). The 12 bp of rigorous homology at 5’ of the PAM sequence ensured the activity of the CRISPR nuclease (Cong, Ran, 2013). Diversified short PAM sequences (e.g., NGG (Deltcheva et al., 2011), NAAR (van der Ploeg, 2009), NGGNG (Horvath et al., 2008)) recognized by different CRISPR nucleases permit almost all sequences to be targeted. Rather than protein based recognition by ZFNs or TALENs, the nucleic acid-based recognition by CRISPR nucleases significantly eases the assembly process. CRISPR-Cas assisted genome modifications have been demonstrated in many species such as mammalian cells (Cong, Ran, 2013, Mali, Yang, 2013), yeast (Dicarlo et al., 2013b), E. coli (Jiang et al., 2013) and plants (Feng et al., 2013). In some cases, the HR and NHEJ efficiencies mediated by the CRISPR nuclease were much higher than those obtained by TALENs (Mali, Yang, 2013). Morever, efficient multiple deletions were also achieved in mammalian cells and S. cerevisiae using CRISPR-Cas (Bao et al., 2014, Wang, Yang, 2013a). However, because of the short recognition sequence (12 bp), many off-target cleavages may be generated by CRISPR nucleases, which may be a serious issue in hosts with large genome sizes. In fact, the off-target problem has been mitigated by several approaches in recent studies. With a pair of closely spaced Cas9 variants (Cas9-D10A) nicking adjacent regions on opposite DNA strands rather than introducing DSBs, the mutation frequencies of off-target sites were reduced in mammalian cells (Cho et al., 2014, Shen et al., 2014). To circumvent the difficulties of adopting the paired nicking strategy for multiplex or genome-scale targeting by CRISPR-Cas, Fu et al. successfully improved the targeting specificity using truncated gRNAs with 17 or 18 nucleotides. Most importantly, those truncated gRNAs can function as efficiently as (or even more efficiently than) their matched 20 nucleotides counterparts (Fu et al., 2014).
2.4 Group II introns
A group II intron (also known as a targetron) consists of a self-catalytic RNA and a ribonucleoprotein (RNP), which catalyzes the insertion of an intron into specific DNA sites via retrohoming (Michel and Ferat, 1995) (Fig. 1D). Through base-pairing with the intron RNA, site-specific insertions or disruptions can be accomplished by group II introns. Group II intron based gene disruptions are prevalent in prokaryotes, albeit there are a few applications in eukaryotes (Jeong, Cho, 2013). Group II introns have been used in bacterial hosts with fairly low HR efficiency (e.g., Clostridia) (Shao et al., 2007). In addition, group II introns can promote targeted insertion, deletion, inversion and cassette exchange by delivery of recombinase recognition sites (e.g., loxP sites) to a given locus (Enyeart et al., 2013). Group II introns were also reported to introduce site-specific DSBs (Karberg et al., 2001), which may facilitate HR or NHEJ-based DNA repair. As observed with other programmable nucleases (e.g., TALENs (Aouida et al., 2014) and CRISPR (Bao, Xiao, 2014)), the targeting efficiency mediated by group II introns was also site-dependent (Perutka et al., 2004).
3 Transcriptome engineering
In addition to genome editing, transcriptome engineering provides a complementary strategy for genome-scale engineering. Targeting at trans-acting regulatory elements, genetic modulation is achieved without modifying the target chromosomal loci. This feature eliminates the need for prior knowledge of host genomes, which is required by most genome editing methods that depend on homologous recombination. Transcription factors (TFs) and regulatory non-coding RNAs (ncRNAs) are the most common targets for transcriptome engineering.
3.1 Transcription factor engineering
Thanks to transcriptional regulatory networks, cells can rapidly coordinate the expression of thousands of genes when facing both internal and environmental stimuli (Lopez-Maury et al., 2008). Such networks exhibit pyramid-shaped hierarchical structures, with most transcription factors (TFs) at the bottom and middle levels, and only a few master TFs on the top for global regulation (Yu and Gerstein, 2006). Whereas specific TFs at the bottom levels modulate dozens of genes in the same functional group, the master TFs have global influence over the gene expression profile (Yu and Gerstein, 2006). These features make TFs the ideal targets for transcriptome reprogramming by modulating many genes simultaneously (Lin et al., 2013, Santos and Stephanopoulos, 2008). Two main strategies have been applied to engineer TFs: modulation of native transcriptional machinery and introduction of artificial TFs (Fig. 1E).
As a demonstration of native TF engineering, global transcriptional machinery engineering (gTME) introduces mutations to the master TFs that mainly mediate DNA recognition, based on the assumption that variations in these TFs may exert substantial changes to the promoter preference of the RNA polymerase. As proof of concept, the principal sigma factor in E. coli (σ70) was subjected to error-prone PCR. From the resultant strain libraries, mutants with improved tolerance to sodium dodecyl sulfate (SDS) and ethanol were identified through serial subculturing (Alper and Stephanopoulos, 2007). In S. cerevisiae, the TATA-binding protein Spt15p and a TATA-binding protein-associated factor Taf25p were mutated. The best variant, which harbored three amino acid mutations in Spt15p, conferred a 70% improvement in ethanol productivity (Alper et al., 2006). It has also been demonstrated that gTME is more effective in diversity creation than chemical mutagenesis methods, and therefore increases the possibility to isolate phenotypes that were unattainable through traditional methods (Klein-Marcuschamer and Stephanopoulos, 2008).
On the other hand, artificial transcription factor (ATF) libraries have also been created to generate transcriptional diversities. A minimal ATF may only contain a DNA-binding domain, whose interaction with its target sequence most likely down-regulates the expression of a nearby gene by interfering with transcriptional initiation or elongation (Park et al., 2005). The DNA-binding domain can also be attached to effector (activator/repressor) domains or ligand-binding domains, which permits more sophisticated regulation. Most ATFs reported so far have employed zinc finger proteins (ZFPs) as the DNA-binding domains, and the library of ATFs is constructed through combinatorial assembly of individual zinc fingers with diverse DNA-binding specificities. The first example of such effort was the introduction of over 105 ZFPs fused with effector domains into S. cerevisiae (Park et al., 2003). The library consisted of threeor four-finger ZFPs, which recognize 9 bp or 12 bp DNA sequences with limited randomness constrained by choice of individual zinc fingers (40 and 25 individual zinc fingers for three-and four-finger proteins, respectively). Several ATFs were identified to confer a number of tolerance phenotypes towards heat, osmotic pressure and an antifungal drug ketoconazole. The relatively short recognition sequence (9 bp or 12 bp) permits the ATFs to modulate many genes. For example, one selected artificial transcriptional factor (K7), which conferred ketoconazole resistance, had 14 perfectly matched binding sites in the yeast genome (Park, Lee, 2003). The perturbation scope by these AFTs may be even larger considering off-target effects. A similar strategy has been applied to E. coli to isolate tolerant strains towards heat shock (Park, Jang, 2005) and butanol (Lee et al., 2011). For future development, we envision that a new generation of ATFs can be developed when the DNA-binding domains are changed from ZFPs to TALEs and CRISPR proteins. As the DNA-binding specificity of TALEs and CRISPR proteins is much more predictable than that of ZFPs, transcriptome perturbation by TALE- and CRISPR-derived ATF libraries should be more effective and programmable.
3.2 Regulatory non-coding RNAs
Non-coding RNA molecules (ncRNAs) are increasingly recognized as key regulators across the biological kingdoms (Kang et al., 2014, Qi and Arkin, 2014). Here we will mainly focus on regulatory ncRNAs that have been used in genome-scale engineering (Fig. 1F). To be suitable for genome-wide applications, synthetic ncRNAs should be preferably trans-acting, permitting simple introduction of a genome-wide library with minimal considerations on local genetic context. Also, the interaction between an ncRNA and its DNA or mRNA target should be mainly determined by Watson-Crick base pairing, so that the binding specificity and efficiency can be predictable and programmable.
In bacteria, trans-acting small RNAs (sRNAs) and antisense RNAs (asRNAs) are two main regulatory ncRNAs (Qi and Arkin, 2014). Lee and coworkers recently developed a general framework to design synthetic sRNAs in E. coli for metabolic engineering (Na et al., 2013). The synthetic sRNAs were composed of a scaffold sequence and a target-binding sequence. The scaffold was derived from a naturally occurring sRNA, MicC, and the scaffold can recruit the Hfq protein to facilitate sRNA-mRNA interaction and mRNA degradation. The native target binding sequence of MicC can be replaced by the antisense sequence to the translation initiation region (TIR) of any given gene. Correlation was found between the repression capability and the binding energy of the antisense sequence, which allowed for fine-tuning of the knockdown efficiency. Although the sRNA library constructed in that study only targeted the cadaverine production related genes (Na, Yoo, 2013), it is possible to expand the strategy to a genome-scale.
On the other hand, asRNAs have been used for functional genomics study in a series of bacteria, such as Streptococcus mutans (Wang and Kuramitsu, 2005) and Staphylococcus aureus (Forsyth et al., 2002). However, it has been long recognized that asRNAs are inefficient for gene repression in E. coli (Wagner and Flardh, 2002). Recently, it was found that asRNA molecules with paired-termini have enhanced stability and improved repression capacity (Nakashima et al., 2006). E. coli genomic DNA fragments have been cloned into a paired-termini expression vector to generate a genome-wide asRNA library, which was used to successfully isolate asRNAs that target essential genes and led to conditional growth inhibition (Meng et al., 2012).
As for eukaryotes, the most common ncRNA machinery for gene expression regulation is RNA interference (RNAi), a cellular gene silencing mechanism whereby mRNAs are targeted for degradation by homologous double-stranded RNAs (dsRNAs) (Fire et al., 1998, Hannon, 2002). RNAi proves to be a powerful tool for genome-wide reduction-of-function screen in many higher eukaryotes (Boutros and Ahringer, 2008, Echeverri and Perrimon, 2006), yet its applications in microbes are rare. This is probably due to the lack of a native RNAi pathway in S. cerevisiae, which is the most-widely used microbial eukaryote. Recently, a heterologous RNAi machinery has been reconstituted in S. cerevisiae (Drinnenberg et al., 2009), which opens up the possibility of genome-wide RNAi screen (see Section 6.3).
In addition to naturally-occurring ncRNAs, synthetic RNAs were also used to modulate gene expression in the CRISPR-mediated interference (CRISPRi) and CRISPR-mediated activation technologies (Fig. 1F). By co-expressing of a Cas9 mutant with abolished endonuclease activity (dCas9) and a guide RNA targeting at the non-template DNA strand of a target gene, up to 1,000-fold reduction in gene expression was achieved in E. coli (Qi et al., 2013). In S. cerevisiae, the silencing efficiency may be further improved by fusing the dCas9 protein to transcription repressors or chromatin silencers (Gilbert et al., 2013). For gene activation, transcriptional activators can be delivered by the dCas9-guide RNA complex to the upstream region of a promoter, resulting in up-regulation in E. coli (Bikard et al., 2013) and yeast (Farzadfard et al., 2013, Gilbert, Larson, 2013).
4. Genome synthesis
Genome synthesis is one of the most impressive achievements of synthetic biology, ranging from viral genomes (Cello et al., 2002, Chan et al., 2005) and bacterial genomes (Gibson et al., 2008, Gibson et al., 2010, Karas et al., 2012) to yeast chromosomes (Annaluru et al., 2014, Dymond et al., 2011). Early efforts mainly focused on increasing the scale of the final DNA constructs, from the 7.5 kb cDNA copy of a poliovirus genome (Cello, Paul, 2002) to a 1.08 Mb bacterial genome (Gibson, Glass, 2010). In terms of DNA sequences, the synthetic genomes are almost exact copies of the native ones, except for a few inserted “watermarks” such as the names of the team members (Gibson, Benders, 2008, Gibson, Glass, 2010). A recent report took a step further to build a designer yeast chromosome that was substantially different from its wild-type template (Annaluru, Muller, 2014). Compared to the native chromosome III of S. cerevisiae, the designer chromosome synIII was ~ 14% smaller due to the deletion of some non-essential regions such as transfer RNAs, transposons and introns. However, the design principles are still very simplistic, and it requires substantial technological development before we can write a fully synthetic genome. Therefore, rather than discussing the practice of genome synthesis, we will instead focus on its enabling technologies, such as DNA synthesis and parts engineering, as these technologies are also highly useful in engineering better microbial cell factories.
4.1. DNA synthesis
One fundamental enabling factor for synthetic genomes is the decreasing cost of chemical DNA synthesis in a manner akin to Moore’s law (Mueller et al., 2009). While further reduction of the synthesis cost using the traditional column-based methods is unlikely, microarray-based technologies provide great potential for high-throughput and cost-effective DNA synthesis, as discussed in details elsewhere (Mueller, Coleman, 2009, Tang et al., 2013). Here we will focus on two recent advances in microarray-based DNA synthesis, highlighting the importance of technology integration. One major limitation of chip-synthesized sequences is that they are prone to error. In addition to the optimization of the microarray technology itself, the integration of next-generation sequencing (NGS) has greatly improved the fidelity of synthetic DNA (Matzas et al., 2010). A pool of chip-derived oligonucleotides were attached to individual streptavidin-coated beads and amplified via emulsion PCR. Pyrosequencing was performed, and the beads with correct sequences were sorted in a high-throughput manner by a camera-guided micropipette. The fidelity was estimated to be improved by 500-fold, and the throughput may potentially enable DNA construction at a megabase scale (Matzas, Stahler, 2010). Another challenge lies at the heterogeneity of the microarray-generated oligo pools, as well as the limited amount of DNA synthesized at an individual spot, both of which complicate the subsequent gene assembly. Quan et al. devised an integrated solution to overcome this challenge, by combining the synthesis, amplification and assembly steps (Quan et al., 2011). The microchip was divided into microfluidics aided subarray reactors, each containing oligonucleotides for the assembly of the same gene. After synthesis, the oligos were amplified, released and assembled into gene constructs up to 1 kb each by enzymatic reactions. Together, both methods discussed above demonstrate that novel solutions can be achieved through technology integration, which will continue to be a driving force to provide synthetic DNA with lower price and higher quality.
4.2. Parts engineering
With the DNA synthesis capacity becoming less restrictive, the limited repertoire of genetic parts and a lack of design principles are becoming the two major obstacles in synthetic genome construction (Wang et al., 2013b). Rather than providing comprehensive summary on how to overcome these obstacles, we will only focus on the expansion of part collections, which also provides sources for diversity generation to improve MCF performance. The readers are directed to other excellent reviews for the design framework for functional assembly of biological parts (Wang, Wei, 2013b, Way et al., 2014), as well as experimental protocols for physical assembly of DNA fragments (Chao et al., 2014).
Based on their functional roles, genetic parts can be classified as sensors (take in environmental stimuli, e.g. riboswitches), regulators (perform calculation, e.g. promoters), actuators (generate outputs, e.g. structural proteins) and adapters (connect components, e.g. endoplasmic reticulum (ER) tags) (Wang, Wei, 2013b). Here we will mainly discuss transcriptional regulators, such as promoters, RBSs and terminators, as they are so far still the most available, well-studied and widely-used genetic parts. Given the wide application of microbial biotechnology, the most important task on these building blocks is to discover, engineer and characterize as many biological parts as possible (Wang, Wei, 2013b). There are mainly three methods to enlarge the parts collection: to harvest from nature, to create mutant libraries, and to build by modeling.
Advances in sequencing and bioinformatics permit rapid identification and prototyping of biological parts from genome and megagenome sequences. For example, strong constitutive promoter candidates can be isolated from the upstream sequences of housekeeping genes (such as global transcription/translation factors, glycolytic enzymes, etc) (Shao et al., 2013, Sun et al., 2012a). High-throughput techniques may also allow characterization of all putative promoters in a microbe (Zaslaver et al., 2006). To create a mutant library of biological parts, there are essentially three strategies. First, mutations can be introduced via error-prone PCR, and the resultant variants with desirable performance (e.g. various promoter strengths) are isolated through high-throughput methods (Alper et al., 2005, Nevoigt et al., 2006). Second, the heterogeneous DNA oligonucleotide pools synthesized by microarray can serve as a source for diversity. For example, synthetic intergenic regions were combinatorially assembled from three oligo libraries with overlapping ends, modulating relative expression patterns of multiple enzymes in a synthetic operon (Pfleger et al., 2006). Also, recombineering enabled the replacement of the native ribosome binding sites (RBSs) with degenerate libraries in E. coli (Wang, Isaacs, 2009). Finally, chimeric parts can be constructed by inserting modulating cassettes into native parts. For example, arrays of upstream activation sequences (UASs) were placed in front of a core promoter element to create strong synthetic promoters in S. cerevisiae (Blazeck et al., 2012) and Yarrowia lipolytica (Blazeck et al., 2011). If the genetic basis that controls the part performance is relatively clear, modeling can help guide the construction of synthetic parts predicted parameters (Crook and Alper, 2013). For example, a computational model has been established to predict the translational initiation rate of bacterial RBSs, by quantifying the interactions between the 30S ribosome complex and the target mRNA molecule (Salis et al., 2009). Based on a nucleosome architecture model, purely synthetic yeast promoters were obtained with decent strengths (Curran et al., 2014).
5 High-throughput genotype-phenotype mapping
With limited understanding of complex biological systems, “rational” genetic engineering often encounters challenges. Instead, “inverse metabolic engineering” (IME), which isolates mutant strains with a desirable trait first and then proceeds to determine underlying genetic changes, has proved to be a more effective strategy (Bailey et al., 2002, Santos and Stephanopoulos, 2008). IME not only rapidly improves a target phenotype, but also provides insights to guide future engineering efforts. To fully realize the potential of IME, advanced genotyping and phenotyping techniques are needed to expedite the cycles of creating diversity, selecting best mutants, and mapping relevant genetic changes (Garst et al., 2013). Genotyping helps to create and track comprehensive and/or combinatorial diversity across the genome, whereas phenotyping helps to identify mutants with desirable traits in a high-throughput manner.
5.1. Genotyping
5.1.1. Genome-wide libraries
Genome-wide overexpression/knockout libraries are powerful tools to comprehensively investigate the impact of individual genetic modification on a given phenotype (Fig. 2). For overexpression libraries, either genomic fragments (Lynch et al., 2007) or all the open reading frames (ORFs) under the control of a promoter (Ho et al., 2009) can be cloned into an extrachromosomal vector. After screen/enrichment, the inserts can be identified through microarray analysis (Lynch et al., 2004) or DNA sequencing. Genome-wide knockout libraries can be generated by many strategies. Transposon mutagenesis has been optimized for unbiased integration of an antibiotic marker cassette into the entire genome, hence creating a random knockout library (Alexeyev and Shokolenko, 1995, Badarinarayana et al., 2001). Moreover, all the nonessential genes can be disrupted through homologous recombination, examplified in construction of the yeast deletion collection (Giaever et al., 2002, Winzeler et al., 1999) and the Keio E. coli knockout collection (Baba et al., 2006). In addition to knockout, reduction-of-function screen has also been applied for genome-wide analysis. Examples include the above-mentioned screening with as-RNAs and RNAi. Knockdown libraries are especially important to study essential genes whose deletion mutations are lethal. For example, insertion of an antibiotic marker into the terminator region to destabilize the mRNA enabled knockdown modification on the essential genes in S. cerevisiae (Breslow et al., 2008). Notably, there are also technologies that can create comprehensive genetic libraries including both overexpression and knockdown modifications. For the trackable multiplex recombineering (TRMR), two kinds of synthetic cassettes were designed for promoter replacement: the ‘up’ cassette containing a strong promoter, and the ‘down’ cassette containing an inert sequence to replace the native RBS. Through recombineering, these synthetic cassettes were incorporated in front of every gene in E. coli, which led to either increased or decreased expression of a target gene (Warner, Reeder, 2010). Though not demonstrated to create microbial genome-wide libraries yet, CRISPR-mediated knockout, interference and activation can be readily applied for genome-scale analysis as discussed previously.
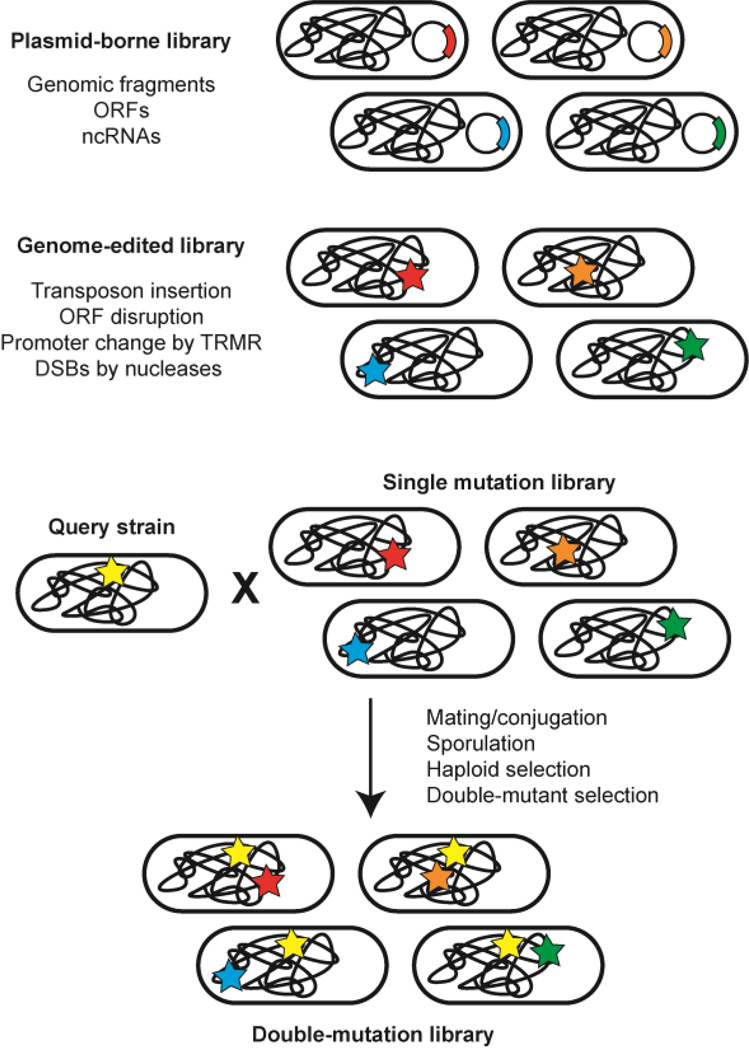
Fig. 2. Genome-wide strain libraries for high-throughput genotyping.
Single mutations can be introduced through plasmid-borne libraries or directed genome editing. A double-mutation strain library can be created using the synthetic genetic array (SGA) method, whereby a query strain harboring the first mutation can be mated with a strain library to incorporate a genome-wide second mutation.
Adding more dimensions to such approaches, i.e. modification at two or even more loci, is necessary because of the non-linear interactions between single genetic variations (Fig. 2). For combinatorial overexpression libraries, the coexpressing genomic libraries (CoGEL) approach was used to construct genomic libraries in a series of vectors (plasmid or fosmid) with compatible replication origins and different resistance markers, which enabled coexistence of two or more genomic inserts in one cell. This approach successfully identified known and novel combinations of genetic changes that conferred improved acid tolerance in E. coli (Nicolaou et al., 2011). On the other hand, construction of a double-mutant library from single loss-of-function collections by mating or conjugation has been demonstrated in model organisms such as E. coli (Butland et al., 2008, Typas et al., 2008) and S. cerevisiae (Pan et al., 2004, Tong et al., 2001). An impressive application was the depiction of a genome-scale digenic interaction network in S. cerevisiae, by examining 5.4 million gene-gene pairs in a double-mutant library (Costanzo et al., 2010). However, current protocols to generate genome-wide double-mutant libraries are quite resource-intensive and time-consuming, as complicated replica-pinning procedures are needed to perform mating, recombination and selection. Therefore, alternative approaches that simplify the introduction of a second mutation on a genome-scale are desirable to speed up the discovery of synergistic modifications. For example, the inherent multiplex capacity of the CRISPR system can be used to create combinatorial genome-wide libraries. As a first step towards this objective, we recently developed a homology integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruptions in S. cerevisiae. The mutagenizing homologous recombination donor is integrated at the 5’ of the guide sequence. To increase the gene disruption efficiency, all the HI-CRISPR elements are embedded on a plasmid with ultrahigh copy number. As proof of concept, the simultaneous disruption efficiency of three genes (CAN1, ADE2 and LYP1) ranged from 27% to 87%, which enabled the identification of desired mutants by random genotyping (Bao, Xiao, 2014).
To facilitate subsequent analysis with these genetic libraries, molecular barcodes have been used to monitor the abundance of every mutant strain in a mixed population. Microarray analysis with complementary probes (Pierce et al., 2006), as well as the “Bar-Seq” method with NGS (Smith et al., 2009), can be used to quantify the dynamics of barcodes and their linked mutants in various screening experiments, enabling high-throughput mapping of relevant genes to a given phenotype. Such high-throughput capacity explains the wide application of molecular barcodes in analyzing overexpression (Ho, Magtanong, 2009), knockout (Giaever, Chu, 2002) and TRMR libraries (Warner, Reeder, 2010).
5.1.2. Genome and transcriptome analysis by next-generation sequencing
Next-generation sequencing (NGS) offers a rapid and inexpensive way to perform genome-scale analysis, whose recent technological advances are reviewed elsewhere (Mardis, 2008, 2013). Here we emphasize on two techniques, comparative genomics (Borneman et al., 2013) and comparative transcriptomic profiling by RNA-seq (Mutz et al., 2013, Ozsolak et al., 2009), which are highly informative in revealing genotype-phenotype relationship, genetic interactions and regulatory networks in microbes.
Comparative studies of genome sequences among laboratory strains, industrial strains and natural isolates of the same species can provide insights on the genetic basis of phenotypic differences (Borneman, Pretorius, 2013). For example, the genomes of four wine and two brewing strains of S. cerevisiae have been sequenced and analyzed against existing S. cerevisiae genome sequences. The variations between different S. cerevisiae strains include single-nucleotide polymorphisms (SNPs), insertions and deletions, and as novel, strain and allele-specific ORFs, and genomic rearrangements (Borneman et al., 2011). Transcriptomic profiling is useful to reveal the impact of different environmental factors on gene expression. For example, transcriptomes of Bacillus subtilis under 104 different environmental and nutritional conditions has been investigated to reveal the structure of transcription network, by grouping 2935 promoters into regulons and then linking these regulons with various transcription factors (Nicolas et al., 2012). For a parental strain and its derivatives obtained from evolutionary engineering or genetic engineering, both genomic and transcriptomic analysis are helpful to understand the genetic basis of acquired traits. Whole genome sequencing (WGS) has been used to track the evolutionary trajectory for aerobic citrate utilization in E. coli (Blount et al., 2012) and efficient xylose fermentation in Scheffersomyces stipitis (Smith et al., 2008). Specifically, mutations that do not affect gene expression can only be identified by genome sequencing. For example, an S. cerevisiae mutant strain with the only lactate transporter gene JEN1 deleted regained the ability to grow on lactate as the sole carbon source during adaptive evolution. Transcriptome analysis provided no clues on the evolved strain; the single-nucleotide changes that conferred an acetate transporter Ady2p with lactate transport activity were only discovered by genome sequencing (de Kok et al., 2012). On the other hand, transcriptional profiling is important to understand the mechanisms on how genetic changes affect a given trait, especially for those mutations resulting in large-scale perturbation in gene expression. For example, differential expression of hundreds of genes caused by mutations in TFs can only be revealed by transcriptome analysis (Alper, Moxley, 2006).
5.2 Reporter-based phenotyping
In addition to generating and detecting modifications on a genome-scale, rapid identification of mutants with desired properties from genome-wide libraries is required in IME. In fact, many high-throughput phenotyping methods, including microplate screening (Behrendorff et al., 2013), surface display (Boder and Wittrup, 1997), compartmentalization (Wang et al., 2014) and fluorescence-activated cell sorting (FACS) (Santoro and Schultz, 2002), have been exploited to detect phenotypes that can be directly screened or selected for (e.g., fluorescence and cell survival, respectively). These methods have been reviewed elsewhere (Leemhuis et al., 2009) and will not be discussed here. However, phenotyping properties that are not amenable to high-throughput screening and/or selection (e.g., bulk and fine chemicals production), remains challenging. To address this limitation, a variety of reporter-based phenotyping methods have been developed to link easily detectable phenotypes to the desired traits.
In vivo production of a target molecule can be monitored via a TF-promoter based reporter system. Under the regulation of the molecule-responsive TF and its cognate promoter, expression of the reporter gene results in colorimetric, fluorescent or growth-coupled phenotypes, which are correlated to the concentration of the target molecule and are rapidly identifiable (Fig. 3). Using TF-promoter pairs from different microbes, specific activation of transcription factors by dicarboxylic acids and alcohols was coupled to expression of the tetracycline reporter gene. As proof of concept, a biosensor was constructed to identify E. coli mutants with 35% higher specific productivity of 1-butanol. This biosensor was also incorporated in a synthetic selection method that couples 1-butanol biosynthesis with cell fitness, leading to a 120-fold increase in 1-butanol production (Dietrich et al., 2013). On the other hand, a feedback-regulated evolution of phenotype (FREP) system was developed to control mutation rates, which are thought to be associated with the probability of finding improved mutants and therefore speed up the adaptive evolution process (Metzgar and Wills, 2000). In the absence of the target molecule, the sensor assembled from the TF-promoter pair activates expression of the actuator and reporter. Expression of the actuator results in an increase in the mutation rate, which may lead to increased production of the target molecule. In response to the increased concentration of the target molecule, the sensor cannot activate the expression of the actuator and reporter, giving rise to a decrease in mutation rate and thus an increase in hereditary stability. Several synthetic E. coli TF-promoter pairs were accordingly engineered for better response to the metabolic intermediate isopentenyl pyrophosphate (IPP). Adopting such sensors in FREP was able to increase tyrosine and isoprenoid production in E. coli (Chou and Keasling, 2013). With a wide dynamic range, high sensitivity and specificity towards a given ligand, the well-characterized ligand-responsive TF and promoter pair always tends to be adopted in an efficient reporter based screening. Yet, promoter characterization is scant in prokaryotes and almost vacant in eukaryotes to date. Also, the most commonly used TF and promoter pairs are unlikely to be responsive to majority of industrially relevant molecules (Dietrich et al., 2010).
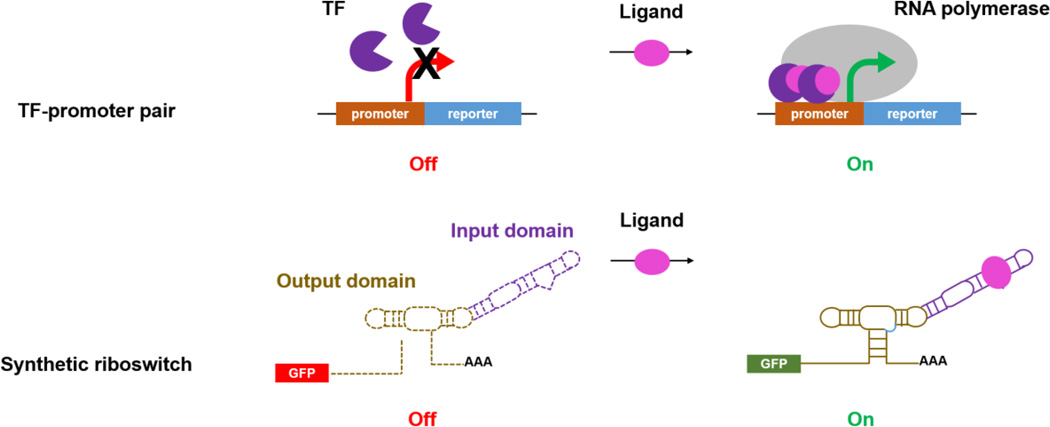
Fig. 3. Reporter based phenotyping.
In the presence of a ligand, the specific activation of transcription factor (TF) is controlled by the expression of the reporter gene. A synthetic riboswitch acts as a biosensor for desired metabolites.
Due to the lack of effective TF-promoter pairs, synthetic riboswitches responsive to various industrially relevant compounds have become an alternative choice for reporter based screening in eukaryotic organisms. Synthetic riboswitch contains an input domain (encoded in an RNA aptamer) and an output domain (encoded in a ribozyme), which is placed in the 3’ untranslated region of a reporter gene. In the absence of a ligand, self-cleavage is catalyzed by the output domain, leading to low gene expression. Binding to a desired molecule by the input domain results in misfolding of the output domain, leading to low cleavage activity and thus high gene expression (Fig. 3). For example, coupled with FACS, an engineered riboswitch was used to identify caffeine demethylase mutants with significantly improved activity and product selectivity (Michener and Smolke, 2012).
6 Notable examples
Recent advances in genome-scale engineering have significantly enhanced the ability to generate and map multiple functional changes across the entire genome. As such, impressive progress in rewiring genomes to elicit robust, complex traits has been achieved with minimal prior knowledge of the genetic determinants. A few notable examples of genome-scale engineering will be highlighted in this section.
6.1 MAGE
Based on the λ Red recombination system, MAGE uses multiple oligos to create combinatorial libraries and optimize gene expression (Fig. 4). Most importantly, the recursive cycles of oligo introduction and allelic replacement are carried out by an automated system, enabling generation of over 4.3 billion genetic variants per day. In one study, the RBSs of 24 genes related to the 1-deoxy-D-xylulose-5-phosphate (DXP) pathway were concurrently modified to increase lycopene production. Mutants with over 5-fold increase in lycopene production were isolated within 3 days (after 35 MAGE cycles), representing a significant improvement over previously reported efforts (Wang, Isaacs, 2009). In another study, MAGE was exploited to insert short DNA sequences into the E. Coli choromosome. After 110 MAGE cycles, 18-nt hexa-histidine tag sequences were successfully inserted into 38 essential genes encoding the entire translation machinery, allowing modification and co-purification of large protein complexes and pathways (Wang et al., 2012).
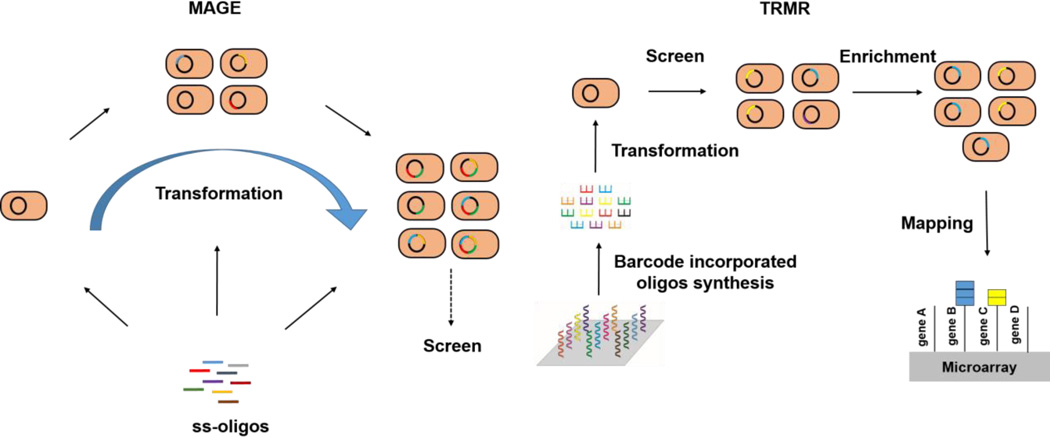
Fig. 4. Multiplex automated genome engineering (MAGE) and trackable multiplex recombineering (TRMR) accelerated E. coli genome evolution.
MAGE enables rapid generation of sequence diversity via continuous delivery of ss-oligos into cells. With barcode incorporated oligos, TRMR enables simultaneous creation and tracking of multiple genetic modifications.
Meanwhile, various MAGE-derived methods were further developed for efficient genome editing. For example, the use of “coselection” MAGE (CoS-MAGE) greatly improves the recombineering efficiency. In the MAGE-generated combinatorial variants, a small portion of cells were observed to harbor multiple mutations (Isaacs, Carr, 2011), which could be selected out in the presence of selective markers. By leveraging co-selection markers around the target site, oligo-mediated allelic replacement efficiency of over 70% per viable progeny was achieved (Carr et al., 2012). Consequently, this approach was used to insert T7 promoter sequences to 12 genomic operons related to aromatic amino acid biosynthesis, permitting rapid generation of promoter libraries (Carr, Wang, 2012). A recently-reported microarray oligonucleotide-MAGE (MO-MAGE) method can cost-effectively amplify thousands of oligos from microarray chips. By adopting such technology, T7 promoters were inserted to the upstream of 2585 operons with an average frequency of 0.02% per locus and 0.4 average insertions per cell (Bonde et al., 2014). Additionally, the hierarchical conjugative assembly genome engineering (CAGE) method was investigated to reprogram the genetic code. The 32 regions of codon modifications (replacing TAG stop codons with TAA) constructed by MAGE were merged into a single genome through conjugation. Because the TAG stop codon was absent in the resultant strain, this liberated codon can be reassigned to a novel amino acid (Isaacs, Carr, 2011).
6.2 TRMR
To reduce the unwanted secondary mutations produced by excessive cycles, MAGE is often limited to create modifications to a subset of relevant genes, which requires a prior knowledge of which genes are to be targeted. A complementary method, TRMR, provides a clue to address this limitation. After the introduction of oligos with unique barcodes for recombineering, cells with desired mutations were enriched in a favorable environment and the corresponding genetic modifications could be quantitatively tracked using the barcoded sequences and microarray analysis (Warner, Reeder, 2010) (Fig. 4). As such, thousands of genes that affected E. coli growth in rich, minimal or cellulosic hydrolysate media in the presence of β-glucoside, D-fucose, valine and methylglyoxal were mapped within one week, permitting identification of large sets of targets for genome engineering endeavors (Warner, Reeder, 2010). TRMR can also assist MAGE to achieve directed genome engineering via identifying the most relevant genetic modifications. By coupling of TRMR with MAGE, barcoded promoter mutant libraries were first introduced to modify gene expression under different challenging environments. Based on the mapping results, RBS mutant libraries were then designed to retarget genes that dramatically affected the cell growth. Consequently, extensive growth-enhancing mutations were identified from different conditions (Sandoval et al., 2012).
6.3. RAGE
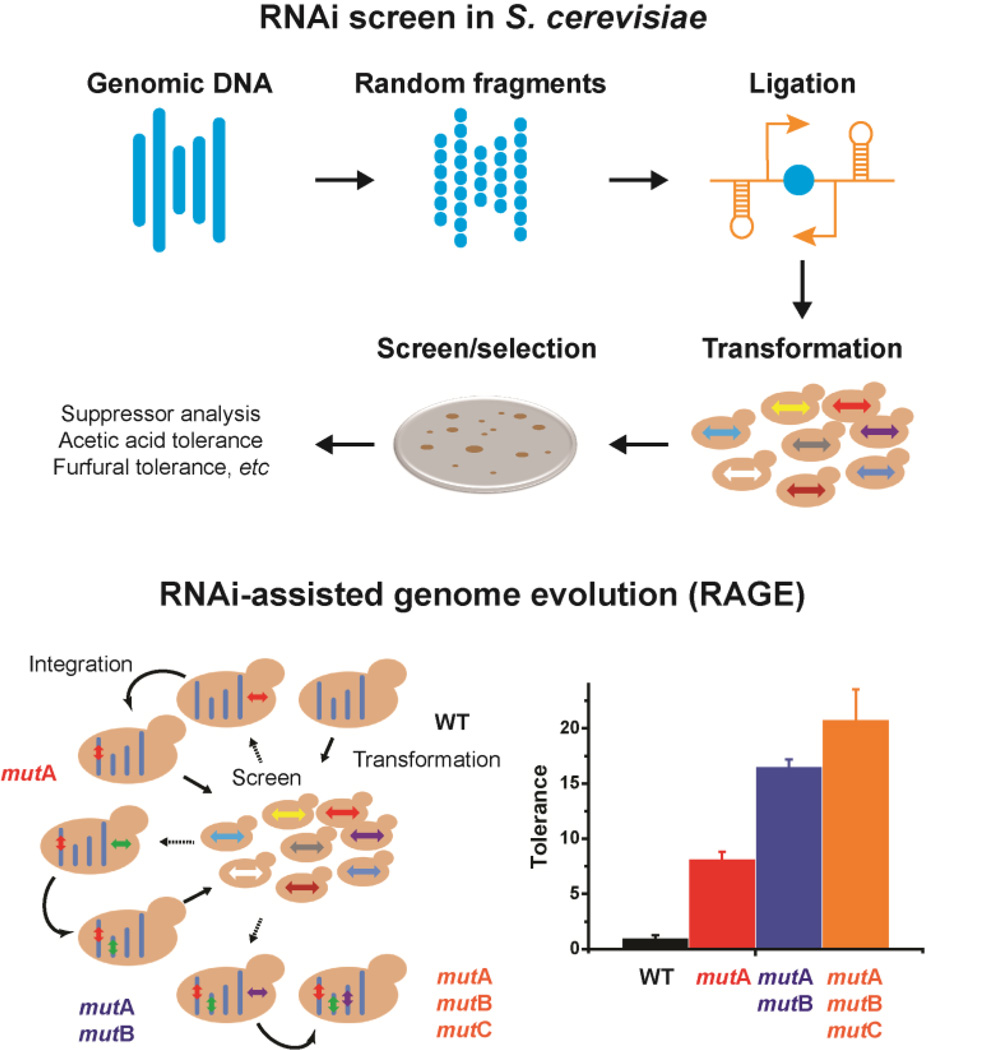
Due to the lack of an efficient recombineering mechanism, it is difficult to apply MAGE or TRMR for genome-scale engineering in eukaryotic microorganisms such as S. cerevisiae (DiCarlo et al., 2013a). On the other hand, RNAi screening has been widely used for functional genomics research in various eukaryotes (Boutros and Ahringer, 2008, Echeverri and Perrimon, 2006), yet its applications in MCF engineering are still rare. Recently, a heterologous RNAi pathway was reconstituted in S. cerevisiae (Drinnenberg, Weinberg, 2009), which enables the use of RNAi screening to rapidly understand and engineer complex phenotypes in this yeast (Fig. 5). By inserting random genomic DNA fragments into a pair of convergent constitutive promoters, double-stranded RNAs were transcribed in vivo to elicit genome-wide knockdown in the presence of the RNAi pathway (Si et al., 2014). The resultant library was used to successfully identify known suppressors of a telomere-defect mutation yku70Δ and genetic determinants for improved resistance towards acetic acid and furfural (Si, Luo, 2014, Xiao and Zhao, 2014). Moreover, compared with the traditional conjugation-based method (Tong, Evangelista, 2001), plasmid-borne RNAi screening is much more convenient in creating genome-wide perturbations in a modified strain background. Therefore, it is possible to apply a directed evolution strategy on a genome-scale to engineer complex phenotypes in S. cerevisiae (Fig. 5). For example, RNAi-assisted genome evolution (RAGE) was developed to identify three knockdown mutations that acted synergistically to improve acetic acid tolerance substantially in S. cerevisiae (Si, Luo, 2014).
Fig. 5. RNAi-assisted genome evolution (RAGE) in S. cerevisiae.
In the presence of a heterologous RNAi pathway, genome-wide knockdown screening can be performed with a double-stranded RNA library derived from genomic DNA. Iterative RNAi screen may help to accumulate beneficial genetic modifications in an evolving yeast genome for continuous improvement of a complex phenotype.
7 Conclusions and perspectives
Recent advances in genome-scale engineering have overcome many barriers that constrain strain engineering and therefore greatly expanded our ability to reprogram biological systems. Owing to their high-throughput, complexity, fidelity, and low cost, these new genome-scale engineering technologies have been able to quickly generate vast libraries of combinatorial variations that may have a greater effect on a given phenotype as well as rapidly map the desired trait. Currently these approaches have heretofore been implemented largely in certain model organisms, but adapting them to other industrial microbes is highly desirable. However, this is not a simple task, as the newly developed methods require effective transformation protocols and certain sets of genetic tools, both of which are often absent for industrial fermentation hosts. Whereas establishing genetic manipulation toolbox in less studied organisms will expand the application of genome-scale engineering in industrial settings, high throughput genotyping and phenotyping technologies, such as whole-genome sequencing, transcriptional profiling, and microfluidic and robotic screening, can be combined with classical strain engineering efforts to decrease the length of time for isolation of improved variants and analysis of underlying mechanisms (Crook and Alper, 2012).
As an alternative strategy, rather than dealing with a large number of variants, genome-scale metabolic models can be used to narrow the search space and create functionally rich libraries for optimizing complex traits. For example, employing designs predicted by in silico models has led to not only enhanced production of desired compounds (Choi et al., 2010, Park et al., 2007), but also discovery of novel drug targets (Kim et al., 2011, Lewis et al., 2010). For a metabolic model, the ability to correctly predict the physiological characteristics of the organism is very important. Hereby, extensive experimental data including high-throughput omics data have been incorporated to validate and improve the quality and the accuracy of a genome-scale metabolic model (Plata et al., 2010). In such a scenario, the massive data obtained from genome-scale engineering could be significant for construction of more refined and complex metabolic models, which may further benefit the genome-scale engineering in return.
Furthermore, laboratory automation may greatly accelerate microbial genome-scale engineering. By eliminating human intervention, laboratory automation promises to improve productivity and reliability, increase throughput, and reduce experimental error rates due to human factors (Linshiz et al., 2013). Pharmaceutical industry has heavily relied on automation technologies to identify new drug lead compounds by screening small-molecule libraries (Nettekoven and Thomas, 2002). Automation platforms have also been developed for bottom-up construction of genetic circuits and metabolic pathways from modular parts (Densmore and Hassoun, 2012, Dharmadi et al., 2014), integrating software for assembly algorithm and data management, as well as hardware for liquid handling and DNA construct analysis. For biological system engineering, the promise of automation has been demonstrated by several recent examples (Esvelt et al., 2011, Wang, Isaacs, 2009). Phage-assisted continuous evolution (PACE) executed 200 rounds of protein evolution in 8 days, during which targeted activities effectively emerged from undetectable levels (Esvelt, Carlson, 2011). Moreover, MAGE created over 4.3 billion combinatorial variants per day, enabling 5-fold increase in lycopene production in 3 days (Wang, Isaacs, 2009). Future development of automation-friendly protocols, including both strain variant generation and phenotypic screening methods, may help to increase the use of laboratory automation in genome-scale engineering of microbial cell factories. Finally, we envision that, with endeavors on but not limited to the above mentioned aspects, genome-scale engineering can accomplish intensive reprogramming of microbial metabolism for multiple engineering purposes.
Acknowledgments
We gratefully acknowledge financial support from National Institutes of Health (GM077596), Defense Advanced Research Projects Agency (DARPA), Department of Energy (DE-SC0008743), and National Academies Keck Futures Initiative on genome engineering.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7:649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- Alexeyev MF, Shokolenko IN. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram-negative bacteria. Gene. 1995;160:59–62. doi: 10.1016/0378-1119(95)00141-r. [DOI] [PubMed] [Google Scholar]
- Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci U S A. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- Alper H, Stephanopoulos G. Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Annaluru N, Muller H, Mitchell LA, Ramalingam S, Stracquadanio G, Richardson SM, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouida M, Piatek MJ, Bangarusamy DK, Mahfouz MM. Activities and specificities of homodimeric TALENs in Saccharomyces cerevisiae. Curr Genet. 2014;60:61–74. doi: 10.1007/s00294-013-0412-z. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badarinarayana V, Estep PW, 3rd, Shendure J, Edwards J, Tavazoie S, Lam F, et al. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat Biotechnol. 2001;19:1060–1065. doi: 10.1038/nbt1101-1060. [DOI] [PubMed] [Google Scholar]
- Bailey JE, Sburlati A, Hatzimanikatis V, Lee K, Renner WA, Tsai PS. Inverse metabolic engineering: a strategy for directed genetic engineering of useful phenotypes. Biotechnol Bioeng. 2002;79:568–579. doi: 10.1002/bit.10441. [DOI] [PubMed] [Google Scholar]
- Bao Z, Xiao H, Liang J, Zhang L, Xiong X, Sun N, et al. Homology-Integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth Biol. 2014 doi: 10.1021/sb500255k. [DOI] [PubMed] [Google Scholar]
- Behrendorff JB, Vickers CE, Chrysanthopoulos P, Nielsen LK. 2,2-Diphenyl-1-picrylhydrazyl as a screening tool for recombinant monoterpene biosynthesis. Microb Cell Fact. 2013;12:76. doi: 10.1186/1475-2859-12-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biot-Pelletier D, Martin VJJ. Evolutionary engineering by genome shuffling. Appl Microbiol Biotechnol. 2014;98:3877–3887. doi: 10.1007/s00253-014-5616-8. [DOI] [PubMed] [Google Scholar]
- Blazeck J, Garg R, Reed B, Alper HS. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol Bioeng. 2012;109:2884–2895. doi: 10.1002/bit.24552. [DOI] [PubMed] [Google Scholar]
- Blazeck J, Liu L, Redden H, Alper H. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl Environ Microbiol. 2011;77:7905–7914. doi: 10.1128/AEM.05763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Bonde MT, Kosuri S, Genee HJ, Sarup-Lytzen K, Church GM, Sommer MO, et al. Direct mutagenesis of thousands of genomic targets using microarray-derived oligonucleotides. ACS Synth Biol. 2014 doi: 10.1021/sb5001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, et al. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Pretorius IS, Chambers PJ. Comparative genomics: a revolutionary tool for wine yeast strain development. Curr Opin Biotechnol. 2013;24:192–199. doi: 10.1016/j.copbio.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- Boyle NR, Reynolds TS, Evans R, Lynch M, Gill RT. Recombineering to homogeneity: extension of multiplex recombineering to large-scale genome editing. Biotechnol J. 2013;8:515–522. doi: 10.1002/biot.201200237. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, et al. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- Butland G, Babu M, Diaz-Mejia JJ, Bohdana F, Phanse S, Gold B, et al. eSGA: E. coli synthetic genetic array analysis. Nat Methods. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, et al. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- Carr PA, Wang HH, Sterling B, Isaacs FJ, Lajoie MJ, Xu G, et al. Enhanced multiplex genome engineering through co-operative oligonucleotide co-selection. Nucleic Acids Res. 2012;40:e132. doi: 10.1093/nar/gks455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol Syst Biol. 2005;1:2005 0018. doi: 10.1038/msb4100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao R, Yuan Y, Zhao H. Recent advances in DNA assembly technologies. FEMS Yeast Res. 2014 doi: 10.1111/1567-1364.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Lee SY, Kim TY, Woo HM. In silico identification of gene amplification targets for improvement of lycopene production. Appl Environ Microbiol. 2010;76:3097–3105. doi: 10.1128/AEM.00115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Keasling JD. Programming adaptive control to evolve increased metabolite production. Nat Commun. 2013;4:2595. doi: 10.1038/ncomms3595. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Layton SL, Jiang T, Cole K, Hargis BM, Berghman LR, et al. Scarless and site-directed mutagenesis in Salmonella enteritidis chromosome. BMC Biotechnol. 2007;7:59. doi: 10.1186/1472-6750-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook N, Alper HS. Engineering complex phenotypes in industrial strains. John Wiley & Sons, Inc.; 2012. Classical Strain Improvement; pp. 1–33. [Google Scholar]
- Crook N, Alper HS. Model-based design of synthetic, biological systems. Chem Eng Sci. 2013;103:2–11. [Google Scholar]
- Curran KA, Crook NC, Karim AS, Gupta A, Wagman AM, Alper HS. Design of synthetic yeast promoters via tuning of nucleosome architecture. Nat Commun. 2014;5 doi: 10.1038/ncomms5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci U S A. 2008;105:1626–1631. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok S, Nijkamp JF, Oud B, Roque FC, de Ridder D, Daran JM, et al. Laboratory evolution of new lactate transporter genes in a jen1Delta mutant of Saccharomyces cerevisiae and their identification as ADY2 alleles by whole-genome resequencing and transcriptome analysis. FEMS Yeast Res. 2012 doi: 10.1111/j.1567-1364.2012.00787.x. [DOI] [PubMed] [Google Scholar]
- Deiters A, Cropp TA, Mukherji M, Chin JW, Anderson JC, Schultz PG. Adding amino acids with novel reactivity to the genetic code of Saccharomyces cerevisiae. J Am Chem Soc. 2003;125:11782–11783. doi: 10.1021/ja0370037. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densmore D, Hassoun S. Design automation for synthetic biological systems. Design & Test of Computers, IEEE. 2012;29:7–20. [Google Scholar]
- Dharmadi Y, Patel K, Shapland E, Hollis D, Slaby T, Klinkner N, et al. High-throughput, cost-effective verification of structural DNA assembly. Nucleic Acids Res. 2014;42:e22. doi: 10.1093/nar/gkt1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Conley AJ, Penttila M, Jantti J, Wang HH, Church GM. Yeast oligo-mediated genome engineering (YOGE) ACS Synth Biol. 2013a;2:741–749. doi: 10.1021/sb400117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013b;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich JA, McKee AE, Keasling JD. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu Rev Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- Dietrich JA, Shis DL, Alikhani A, Keasling JD. Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis. ACS Synth Biol. 2013;2:47–58. doi: 10.1021/sb300091d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, et al. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci U S A. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymond JS, Richardson SM, Coombes CE, Babatz T, Muller H, Annaluru N, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user's guide. Nat Rev Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- Eckert SE, Dziva F, Chaudhuri RR, Langridge GC, Turner DJ, Pickard DJ, et al. Retrospective application of transposon-directed insertion site sequencing to a library of signature-tagged mini-Tn5Km2 mutants of Escherichia coli O157:H7 screened in cattle. J Bacteriol. 2011;193:1771–1776. doi: 10.1128/JB.01292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci U S A. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyeart PJ, Chirieleison SM, Dao MN, Perutka J, Quandt EM, Yao J, et al. Generalized bacterial genome editing using mobile group II introns and Cre-lox. Mol Syst Biol. 2013;9:685. doi: 10.1038/msb.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Wang HH. Genome-scale engineering for systems and synthetic biology. Mol Syst Biol. 2013;9:17. doi: 10.1038/msb.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol. 2012;30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- Fu YF, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YF, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst A, Lynch M, Evans R, Gill RT. Strategies for the multiplex mapping of genes to traits. Fact. 2013;12:99. doi: 10.1186/1475-2859-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordley RM, Gersbach CA, Barbas CF., 3rd Synthesis of programmable integrases. Proc Natl Acad Sci U S A. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer L, DeZwaan TM, Montenegro-Chamorro MV, Frank SA, Hamer JE. Recent advances in large-scale transposon mutagenesis. Curr Opin Chem Biol. 2001;5:67–73. doi: 10.1016/s1367-5931(00)00162-9. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, Natarajan P, et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat Biotechnol. 2009;27:369–377. doi: 10.1038/nbt.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, et al. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu YF, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in Zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglehart JD, Silver DP. Synthetic lethality--a new direction in cancer-drug development. N Engl J Med. 2009;361:189–191. doi: 10.1056/NEJMe0903044. [DOI] [PubMed] [Google Scholar]
- Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Cho N, Jung D, Bang D. Genome-scale genetic engineering in Escherichia coli. Biotachnol Adv. 2013;31:804–810. doi: 10.1016/j.biotechadv.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Z, Zhang CZ, Zhang JL, Jin P, Zhang J, Du GC, et al. Small RNA regulators in bacteria: powerful tools for metabolic engineering and synthetic biology. Appl Microbiol Biotechnol. 2014;98:3413–3424. doi: 10.1007/s00253-014-5569-y. [DOI] [PubMed] [Google Scholar]
- Karas BJ, Tagwerker C, Yonemoto IT, Hutchison CA, 3rd, Smith HO. Cloning the Acholeplasma laidlawii PG-8A genome in Saccharomyces cerevisiae as a yeast centromeric plasmid. ACS Synth Biol. 2012;1:22–28. doi: 10.1021/sb200013j. [DOI] [PubMed] [Google Scholar]
- Karberg M, Guo H, Zhong J, Coon R, Perutka J, Lambowitz AM. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol. 2001;19:1162–1167. doi: 10.1038/nbt1201-1162. [DOI] [PubMed] [Google Scholar]
- Karow M, Calos MP. The therapeutic potential of PhiC31 integrase as a gene therapy system. Expert Opin Biol Ther. 2011;11:1287–1296. doi: 10.1517/14712598.2011.601293. [DOI] [PubMed] [Google Scholar]
- Keasling JD. Manufacturing molecules through metabolic engineering. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res. 2012;22:1327–1333. doi: 10.1101/gr.138792.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Kim SY, Jeong H, Kim TY, Kim JJ, Choy HE, et al. Integrative genome-scale metabolic analysis of Vibrio vulnificus for drug targeting and discovery. Mol Syst Biol. 2011;7:460. doi: 10.1038/msb.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Marcuschamer D, Stephanopoulos G. Assessing the potential of mutational strategies to elicit new phenotypes in industrial strains. Proc Natl Acad Sci U S A. 2008;105:2319–2324. doi: 10.1073/pnas.0712177105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie MJ, Gregg CJ, Mosberg JA, Washington GC, Church GM. Manipulating replisome dynamics to enhance lambda Red-mediated multiplex genome engineering. Nucleic Acids Res. 2012;40:e170. doi: 10.1093/nar/gks751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol. 2012;8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yang KS, Jang SA, Sung BH, Kim SC. Engineering butanol-tolerance in Escherichia coli with artificial transcription factor libraries. Biotechnol Bioeng. 2011;108:742–749. doi: 10.1002/bit.22989. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Kelly RM, Dijkhuizen L. Directed evolution of enzymes: library screening strategies. IUBMB Life. 2009;61:222–228. doi: 10.1002/iub.165. [DOI] [PubMed] [Google Scholar]
- Lewis NE, Schramm G, Bordbar A, Schellenberger J, Andersen MP, Cheng JK, et al. Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat Biotechnol. 2010;28:1279–1285. doi: 10.1038/nbt.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wu LP, Chandrasegaran S. Functional domains in Fok I restriction endonuclease. Proc Natl Acad Sci U S A. 1992;89:4275–4279. doi: 10.1073/pnas.89.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Chao R, Abil Z, Bao Z, Zhao H. Fairytale: a high-throughput TAL effector synthesis platform. ACS Synth Biol. 2014;3:67–73. doi: 10.1021/sb400109p. [DOI] [PubMed] [Google Scholar]
- Lin Z, Zhang Y, Wang J. Engineering of transcriptional regulators enhances microbial stress tolerance. Biotechnol Adv. 2013;31:986–991. doi: 10.1016/j.biotechadv.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Linshiz G, Stawski N, Poust S, Bi C, Keasling JD, Hillson NJ. PaR-PaR laboratory automation platform. ACS Synth Biol. 2013;2:216–222. doi: 10.1021/sb300075t. [DOI] [PubMed] [Google Scholar]
- Lopez-Maury L, Marguerat S, Bahler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- Lynch MD, Gill RT, Stephanopoulos G. Mapping phenotypic landscapes using DNA micro-arrays. Metab Eng. 2004;6:177–185. doi: 10.1016/j.ymben.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Lynch MD, Warnecke T, Gill RT. SCALEs: multiscale analysis of library enrichment. Nat Methods. 2007;4:87–93. doi: 10.1038/nmeth946. [DOI] [PubMed] [Google Scholar]
- Maggert KA, Gong WJ, Golic KG. Methods for homologous recombination in Drosophila. Methods Mol Biol. 2008;420:155–174. doi: 10.1007/978-1-59745-583-1_9. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, et al. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509:385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- Mardis ER. Next-generation sequencing platforms. Annu Rev Anal Chem (Palo Alto Calif) 2013;6:287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- Matzas M, Stahler PF, Kefer N, Siebelt N, Boisguerin V, Leonard JT, et al. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing. Nat Biotechnol. 2010;28:1291–1294. doi: 10.1038/nbt.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Kanzaki G, Meas D, Lam CK, Crummer H, Tain J, et al. A genome-wide inducible phenotypic screen identifies antisense RNA constructs silencing Escherichia coli essential genes. FEMS Microbiol Lett. 2012;329:45–53. doi: 10.1111/j.1574-6968.2012.02503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in Zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzgar D, Wills C. Evidence for the adaptive evolution of mutation rates. Cell. 2000;101:581–584. doi: 10.1016/s0092-8674(00)80869-7. [DOI] [PubMed] [Google Scholar]
- Michel F, Ferat JL. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- Michener JK, Smolke CD. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch. Metab Eng. 2012;14:306–316. doi: 10.1016/j.ymben.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M, Mizuuchi K. Integrative recombination of bacteriophage lambda: extent of the DNA sequence involved in attachment site function. Proc Natl Acad Sci U S A. 1980;77:3220–3224. doi: 10.1073/pnas.77.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moure CM, Gimble FS, Quiocho FA. Crystal structures of I-SceI complexed to nicked DNA substrates: snapshots of intermediates along the DNA cleavage reaction pathway. Nucleic Acids Res. 2008;36:3287–3296. doi: 10.1093/nar/gkn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Coleman JR, Wimmer E. Putting synthesis into biology: a viral view of genetic engineering through de novo gene and genome synthesis. Chem Biol. 2009;16:337–347. doi: 10.1016/j.chembiol.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutz KO, Heilkenbrinker A, Lonne M, Walter JG, Stahl F. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol. 2013;24:22–30. doi: 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Na D, Yoo SM, Chung H, Park H, Park JH, Lee SY. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol. 2013;31:170–174. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Nakashima N, Tamura T, Good L. Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli. Nucleic Acids Res. 2006;34:e138. doi: 10.1093/nar/gkl697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettekoven M, Thomas AW. Accelerating drug discovery by integrative implementation of laboratory automation in the work flow. Curr Med Chem. 2002;9:2179–2190. doi: 10.2174/0929867023368764. [DOI] [PubMed] [Google Scholar]
- Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- Nevoigt E, Kohnke J, Fischer CR, Alper H, Stahl U, Stephanopoulos G. Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl Environ Microbiol. 2006;72:5266–5273. doi: 10.1128/AEM.00530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou SA, Gaida SM, Papoutsakis ET. Coexisting/Coexpressing Genomic Libraries (CoGeL) identify interactions among distantly located genetic loci for developing complex microbial phenotypes. Nucleic Acids Res. 2011;39:e152. doi: 10.1093/nar/gkr817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- Ochiai H, Fujita K, Suzuki K, Nishikawa M, Shibata T, Sakamoto N, et al. Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells. 2010;15:875–885. doi: 10.1111/j.1365-2443.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, et al. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, et al. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Park JH, Lee KH, Kim TY, Lee SY. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc Natl Acad Sci U S A. 2007;104:7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K-S, Lee D-k, Lee H, Lee Y, Jang Y-S, Kim YH, et al. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat Biotechnol. 2003;21:1208–1214. doi: 10.1038/nbt868. [DOI] [PubMed] [Google Scholar]
- Park KS, Jang YS, Lee H, Kim JS. Phenotypic alteration and target gene identification using combinatorial libraries of zinc finger proteins in prokaryotic cells. J Bacteriol. 2005;187:5496–5499. doi: 10.1128/JB.187.15.5496-5499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Perutka J, Wang W, Goerlitz D, Lambowitz AM. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol. 2004;336:421–439. doi: 10.1016/j.jmb.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- Pierce SE, Fung EL, Jaramillo DF, Chu AM, Davis RW, Nislow C, et al. A unique and universal molecular barcode array. Nat Methods. 2006;3:601–603. doi: 10.1038/nmeth905. [DOI] [PubMed] [Google Scholar]
- Plata G, Hsiao TL, Olszewski KL, Llinas M, Vitkup D. Reconstruction and flux-balance analysis of the Plasmodium falciparum metabolic network. Mol Syst Biol. 2010;6:408. doi: 10.1038/msb.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy VA, Bezdan D, Zengler K. Adaptive laboratory evolution - harnessing the power of biology for metabolic engineering. Curr Opin Biotechnol. 2011;22:590–594. doi: 10.1016/j.copbio.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Qi LS, Arkin AP. A versatile framework for microbial engineering using synthetic non-coding RNAs. Nat Rev Microbiol. 2014;12:341–354. doi: 10.1038/nrmicro3244. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan JY, Saaem I, Tang N, Ma SM, Negre N, Gong H, et al. Parallel on-chip gene synthesis and application to optimization of protein expression. Nat Biotechnol. 2011;29:449-+. doi: 10.1038/nbt.1847. [DOI] [PubMed] [Google Scholar]
- Rabinovitch-Deere CA, Oliver JWK, Rodriguez GM, Atsumi S. Synthetic biology and metabolic engineering approaches to produce biofuels. Chem Rev. 2013;113:4611–4632. doi: 10.1021/cr300361t. [DOI] [PubMed] [Google Scholar]
- Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval NR, Kim JY, Glebes TY, Reeder PJ, Aucoin HR, Warner JR, et al. Strategy for directing combinatorial genome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2012;109:10540–10545. doi: 10.1073/pnas.1206299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc Natl Acad Sci U S A. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos CN, Stephanopoulos G. Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol. 2008;12:168–176. doi: 10.1016/j.cbpa.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Sawitzke JA, Costantino N, Li XT, Thomason LC, Bubunenko M, Court C, et al. Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J Mol Biol. 2011;407:45–59. doi: 10.1016/j.jmb.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 2007;421:171–199. doi: 10.1016/S0076-6879(06)21015-2. [DOI] [PubMed] [Google Scholar]
- Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- Segal DJ, Meckler JF. Genome engineering at the dawn of the golden age. Annu Rev Genomics Hum Genet. 2013;14:135–158. doi: 10.1146/annurev-genom-091212-153435. [DOI] [PubMed] [Google Scholar]
- Shao L, Hu S, Yang Y, Gu Y, Chen J, Jiang W, et al. Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum. Cell Res. 2007;17:963–965. doi: 10.1038/cr.2007.91. [DOI] [PubMed] [Google Scholar]
- Shao Z, Rao G, Li C, Abil Z, Luo Y, Zhao H. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold. ACS Synth Biol. 2013;2:662–669. doi: 10.1021/sb400058n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- Si T, Luo Y, Bao Z, Zhao H. RNAi-assisted genome evolution in Saccharomyces cerevisiae for complex phenotype engineering. ACS Synth Biol. 2014 doi: 10.1021/sb500074a. [DOI] [PubMed] [Google Scholar]
- Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Heisler LE, Mellor J, Kaper F, Thompson MJ, Chee M, et al. Quantitative phenotyping via deep barcode sequencing. Genome Res. 2009;19:1836–1842. doi: 10.1101/gr.093955.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Quinlan AR, Peckham HE, Makowsky K, Tao W, Woolf B, et al. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 2008;18:1638–1642. doi: 10.1101/gr.077776.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Hamilton D, Hoess R. Bacteriophage-p1 site-specific recombination .2. Recombination between loxp and the bacterial chromosome. J Mol Biol. 1981;150:487–507. doi: 10.1016/0022-2836(81)90376-4. [DOI] [PubMed] [Google Scholar]
- Sun J, Shao Z, Zhao H, Nair N, Wen F, Xu JH, et al. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol Bioeng. 2012a;109:2082–2092. doi: 10.1002/bit.24481. [DOI] [PubMed] [Google Scholar]
- Sun N, Abil Z, Zhao H. Recent advances in targeted genome engineering in mammalian systems. Biotechnol J. 2012b;7:1074–1087. doi: 10.1002/biot.201200038. [DOI] [PubMed] [Google Scholar]
- Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol Biosyst. 2012c;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- Swingle B, Markel E, Costantino N, Bubunenko MG, Cartinhour S, Court DL. Oligonucleotide recombination in Gram-negative bacteria. Mol Microbiol. 2010;75:138–148. doi: 10.1111/j.1365-2958.2009.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Ma SY, Tian JD. New tools for cost-effective DNA synthesis. Synthetic Biology: Tools and Applications. 2013:3–21. [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, et al. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan S, Bode J. Site-specific recombinases: from tag-and-target- to tag-and-exchange-based genomic modifications. FASEB J. 2011;25:4088–4107. doi: 10.1096/fj.11-186940. [DOI] [PubMed] [Google Scholar]
- Turan S, Galla M, Ernst E, Qiao J, Voelkel C, Schiedlmeier B, et al. Recombinase-mediated cassette exchange (RMCE): traditional concepts and current challenges. J Mol Biol. 2011;407:193–221. doi: 10.1016/j.jmb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Turan S, Kuehle J, Schambach A, Baum C, Bode J. Multiplexing RMCE: versatile extensions of the Flprecombinase-mediated cassette-exchange technology. J Mol Biol. 2010;402:52–69. doi: 10.1016/j.jmb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Turan S, Zehe C, Kuehle J, Qiao J, Bode J. Recombinase-mediated cassette exchange (RMCE) - a rapidly-expanding toolbox for targeted genomic modifications. Gene. 2013;515:1–27. doi: 10.1016/j.gene.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Typas A, Nichols RJ, Siegele DA, Shales M, Collins SR, Lim B, et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Methods. 2008;5:781–787. doi: 10.1038/nmeth.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg JR. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology. 2009;155:1966–1976. doi: 10.1099/mic.0.027508-0. [DOI] [PubMed] [Google Scholar]
- Wagner EG, Flardh K. Antisense RNAs everywhere? Trends Genet. 2002;18:223–226. doi: 10.1016/s0168-9525(02)02658-6. [DOI] [PubMed] [Google Scholar]
- Wang B, Kuramitsu HK. Inducible antisense RNA expression in the characterization of gene functions in Streptococcus mutans. Infect Immun. 2005;73:3568–3576. doi: 10.1128/IAI.73.6.3568-3576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BL, Ghaderi A, Zhou H, Agresti J, Weitz DA, Fink GR, et al. Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption. Nat Biotechnol. 2014;32:473–478. doi: 10.1038/nbt.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013a;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Huang PY, Xu G, Haas W, Marblestone A, Li J, et al. Multiplexed in vivo His-tagging of enzyme pathways for in vitro single-pot multienzyme catalysis. ACS Synth Biol. 2012;1:43–52. doi: 10.1021/sb3000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894-U133. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Wei KY, Smolke CD. Synthetic biology: advancing the design of diverse genetic systems. Annu Rev Chem Biomol Eng. 2013b;4:69–102. doi: 10.1146/annurev-chembioeng-061312-103351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LB, Gill RT. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol. 2010;28:856–862. doi: 10.1038/nbt.1653. [DOI] [PubMed] [Google Scholar]
- Way JC, Collins JJ, Keasling JD, Silver PA. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell. 2014;157:151–161. doi: 10.1016/j.cell.2014.02.039. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Zhao H. Genome-wide RNAi screen reveals the E3 SUMO-protein ligase gene SIZ1 as a novel determinant of furfural tolerance in Saccharomyces cerevisiae. Biotechnol Biofuels. 2014;7:78. doi: 10.1186/1754-6834-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, et al. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2011;108:7052–7057. doi: 10.1073/pnas.1102030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BJ, Kang KH, Lee JH, Sung BH, Kim MS, Kim SC. Rapid and efficient construction of markerless deletions in the Escherichia coli genome. Nucleic Acids Res. 2008;36:e84. doi: 10.1093/nar/gkn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. Proc Natl Acad Sci U S A. 2006;103:14724–14731. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WP, del Cardayre SB. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415:644–646. doi: 10.1038/415644a. [DOI] [PubMed] [Google Scholar]