Abstract
The viral V2 protein is one of the key factors that Tomato yellow leaf curl geminivirus (TYLCV), a major tomato pathogen worldwide, utilizes to combat the host defense. Besides suppressing the plant RNA silencing defense by targeting the host SGS3 component of the silencing machinery, V2 also interacts with the host CYP1 protein, a papain-like cysteine protease likely involved in hypersensitive response reactions. The biological effects of the V2-CYP1 interaction, however, remain unknown. We addressed this question by demonstrating that V2 inhibits the enzymatic activity of CYP1, but does not interfere with post-translational maturation of this protein.
Keywords: TYLCV, V2 protein, Papain-like cysteine protease, CYP1, Programmed cell death
1. Introduction
Tomato yellow leaf curl virus (TYLCV), a whitefly-transmitted geminivirus, is a major tomato pathogen worldwide [1–3]. TYLCV has a circular ssDNA genome [4,5] that encodes six ORFs, one of which, termed V2 [5], is essential for infection, but is not directly involved in viral replication or spread [6]. Recently, we have shown that the V2 protein of the TYLCV-Israel isolate (Acc. X15656) is a suppressor of the RNA silencing defense [7] and interacts with the tomato SGS3 [8], a component of the RNA silencing machinery. This suppressor function was also reported for the V2 proteins of TYLCV-China [9] and Cotton leaf curl Multan virus [10].
Intriguingly, the counter-defense function of V2 is not limited to silencing suppression. Our recent data [11] suggest that V2 interacts with a tomato protein, CYP1, that belongs to the family of papain-like cysteine proteases (PLCPs). Cysteine proteases are found in a variety of organisms, where they are involved in protein degradation and turnover, programmed cell death (PCD), and immunity [12–16]. Specifically, CYP1 (AJ003137.1) is a tomato homolog of the Arabidopsis RD21, a PLCP that participates in stress responses and senescence [17–19], as well as in plant immune response [16]. CYP1 is encoded by a single gene and has several conserved domains, i.e., a cysteine protease domain, a proline-rich domain, and a granulin domain [11], that are characteristic of the RD21-type PLCPs. By analogy to RD21 [19], CYP1 is predicted to accumulate in two isoforms: the immature isoform, iCYP1, that contains the granulin domain, and the mature isoform, mCYP1, that lacks the granulin domain due to post translational proteolytic cleavage.
Is there a biological consequence of the V2-CYP1 interaction? As a defense-related factor, CYP1 could simply target V2 to inhibit its suppression of RNA silencing activity. In an alternative scenario, V2 could act to suppress the CYP1 activity; generally, this suppression might occur either by inhibiting CYP1 maturation or by affecting its enzymatic activity. Here, we examined these possibilities and demonstrated that, whereas CYP1 does not interfere with the V2 activity as silencing suppressor, V2 blocks the CYP1 protease activity, but does not affect its maturation.
2. Materials and methods
2.1. Plasmid construction
For all agroinfiltration assays, the CYP1 gene was excised from its pET-based vector [11] and cloned into the BamHI and NotI sites of the binary vector pBin19 (GenBank accession number U09365). GFP-expressing binary plasmid pBin19-GFP was obtained from Dr. B. Epel (Tel Aviv University). TYLCV-Is V2-expressing binary vector was made by cloning a PCR-amplified V2 gene into the KpnI and SalI sites of pCAMBIA 2300 (GenBank accession number AF234315).
2.2. Plant material
Tomato (Solanum lycopersicum) plants used in this study were of the M82 variety obtained from HaZera Genetics, Israel. Nicotiana benthamiana and tomato plants were germinated from seeds for each experiment. Plants were cultivated in growth chambers at 16 h under light at 23 °C.
2.3. Transient expression by agroinfiltration
Binary plasmids described above were transferred to Agrobacterium tumefaciens (strain EHA105) [30] by the freeze-thaw transformation method, and the resulting bacteria were cultured overnight at 28 °C in LB medium containing 50 μg/ml kanamycin. Then, bacterial cultures were diluted 1:20 in fresh LB medium without antibiotics, grown to an optical density A600 = 0.5, and resuspended in 20 ml of an infiltration solution containing 10 mM MgCl2, 10 mM MES pH 5.6, and 150 μM acetosyringone. For coexpression of CYP1 with V2, the corresponding bacterial cultures were mixed in 1:1 v/v ratio before agroinfiltration which was achieved by pressure-infiltration of the bacteria into young leaves of three-weeks-old greenhouse-grown N. benthamiana plants. For co-expression of GFP with different V2 or with V2 and CYP1, the corresponding bacterial cultures were mixed in equal v/v ratios before agroinfiltration.
2.4. Extraction of proteins from Agroinfitrated leaves
Agroinfitrated N. benthamiana leaves (40 g) on the third day after agroinfiltration, were sliced into strips and ground in 10 mM Tris pH = 8.0, containing 5 mM DTT, in an ice-cold mortar. After slurry was filtered through Miracloth, insoluble material was further removed by centrifugation at 12,000 rpm for 30 min at 4 °C. Protein extracts were adjusted to a concentration of 1 mg/ml. In E64 treatments, a thin layer of 100 μM E64 was applied on the surface-cleaned leaf before protein extraction was performed. The proteins were separated on 12% polyacrylamide gels (10 μg protein per lane) and stained with Coomassie blue or analyzed by western blotting or used for enzyme activity assay.
2.5. Western blot analysis
Leaf extracted protein homogenate was mixed with sample buffer [31], boiled for 5 min, and its protein content (30 μg) was resolved on a 12% SDS polyacrylamide gel [31] followed by electro-transfer to a nitrocellulose membrane. The membrane was blocked with 5% dry milk in phosphate-buffered saline (PBS) and incubated for overnight at room temperature with the anti-CYP1 primary antibody (obtained as anti-RD21 antibody from Dr. Ikuko Hara-Nishimura, Kyoto University, Japan, diluted 1:1000) in PBS. Immunoreactive bands were visualized using goat anti-rabbit IgG (diluted 1:3000) conjugated to horseradish peroxidase (Bio-Rad) and the ECL western blotting detection kit (Amersham Biosciences).
2.6. Analysis of CYP1 activity
For qualitative analysis of the CYP1 enzymatic activity, we used an assay of Rubisco degradation described previously [20]. When incubated for 1 h (or longer) at room temperature in the presence of SDS, Rubisco is markedly degraded in leaf extracts of wild-type plants, and, in Arabidopsis, this degradation has been demonstrated to occur primarily due to the enzymatic activity of the CYP1 homolog, RD21 [20]. Protein extracts were then resolved on 12% SDS polyacrylamide gels (10 mg protein per lane), and their Rubisco content was detected by staining with Coomassie blue.
The enzymatic activity of CYP1 was quantified as described [32]. Briefly, the activity was monitored using the known PLCP fluorescent substrate carboxybezoxy-l-phenylalanyl-L-arginine-4-methyl-coumaryl-7-amide (Z-Phe-Arg-MCA) (Sigmae-Aldrich), which, when cleaved by protease activity, releases the product that fluoresces at 460 nm [33]. All assays were done in 96-well plates. Each well contained 75 μg total protein in 40 μl reaction buffer (0.2 mM sodium acetate pH 5.2, 4 mM EDTA, 8 mM DTT), 100 μl reaction buffer and 100 μl substrate (Z-Phe-Arg-MCA, 0.2 mM) dissolved in the reaction buffer, and incubated at 37 °C. The released product was quantified using a FL600 Fluorescence Microplate Reader (Bio-Tek, Winooski, VT, USA).
3. Results
3.1. CYP1 does not affect the RNA silencing suppression activity of V2
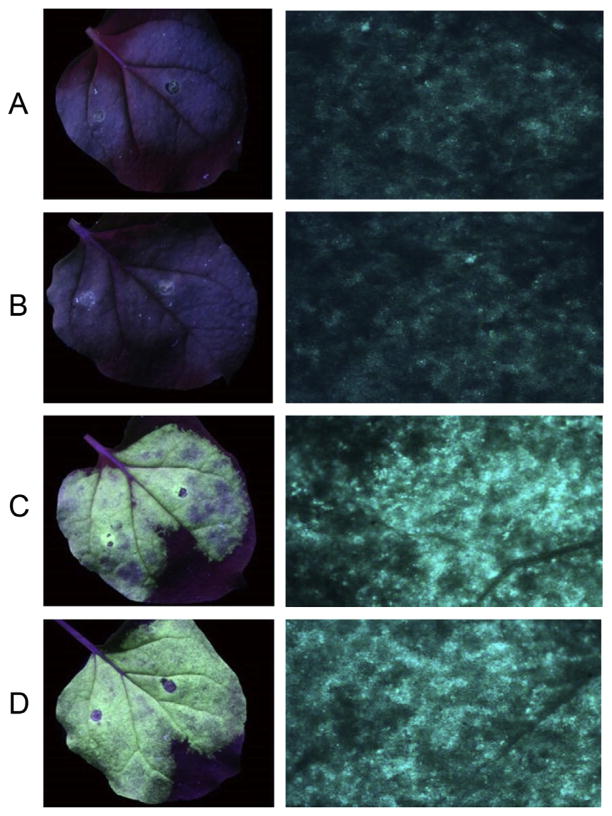
We examined the possibility that CYP1 may defend the plant against the invading virus by targeting V2, one of the viral proteins essential for efficient infection. The hallmark of the V2 function is its ability to suppress RNA silencing; therefore, we assayed this V2-induced suppression in the absence and presence of coexpressed CYP1. In this assay [7], a silencing initiator and reporter GFP is transiently expressed by agroinfiltration in N. benthamiana leaves alone or together with V2, which prolongs the duration of GFP expression in the infiltrated leaf area. Fig. 1 shows that GFP expression was largely silenced at 7 days post infiltration (panel A), and this silencing was not affected by coexpression of CYP1 (panel B). In contrast, coexpression of V2 suppressed silencing, elevating the levels of GFP expression (Fig. 1C). This suppression of GFP silencing by V2 was not affected by coexpression of CYP1 (Fig. 1D). These results indicate that CYP1 does not detectably affect the ability of V2 to suppress RNA silencing.
Fig. 1.
RNA silencing suppression activity of V2. (A) GFP. (B) GFP + CYP1. (C) GFP + V2. (D) GFP + CYP1 + V2. Left panels, view of a whole Agroinfitrated leaf. Right panels, higher magnification of the Agroinfitrated areas. GFP fluorescence was visualized seven days after infiltration.
3.2. V2 does not affect maturation of CYP1
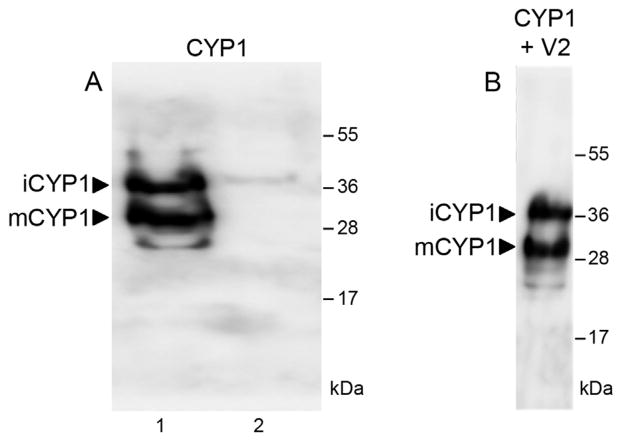
Our western blot analysis of the CYP1 population transiently expressed in tomato leaves (Fig. 2A) revealed that this protein is well expressed and it exists in two isoforms with molecular masses of 38 kDa and 33 kDa (lane 1), which were absent in the absence of transient expression (lane 2). Similar isoforms have been reported for the Arabidopsis CYP1 homolog RD21 [19], where the 38-kDa isoform represented the full-length immature protein and the 33-kDa isoform represented the mature protein with the cleaved-off granulin domain. Thus, the 38-kDa and 33-kDa bands most likely represent the immature and mature forms of CYP1, termed iCYP1 and mCYP1, respectively.
Fig. 2.
Maturation of CYP1 in tomato leaves. (A) Western blot analysis of transiently expressed CYP1. Lane 1, Leaves Agroinfitrated with a CYP1-expressing construct; lane 2, control leaves. (B) Western blot analysis of CYP1 transiently coexpressed with V2. Arrows indicate the positions of iCYP1 (38 kDa) and mCYP1 (33 kDa). The numbers on the right indicate molecular mass standards in thousands of Daltons.
Next, we examine whether this CYP1 maturation is affected by the presence of V2. To this end, we transiently coexpressed CYP1 and V2 in tomato leaves, and detected the presence of iCYP1 and mCYP1 by immunoblotting. Fig. 2B shows that both CYP1 isoforms were detected, and their relative abundance was comparable to that observed in the absence of V2 (Fig. 2A). Thus, the V2 does not interfere with CYP1 maturation.
3.3. V2 inhibits the proteolytic activity of CYP1
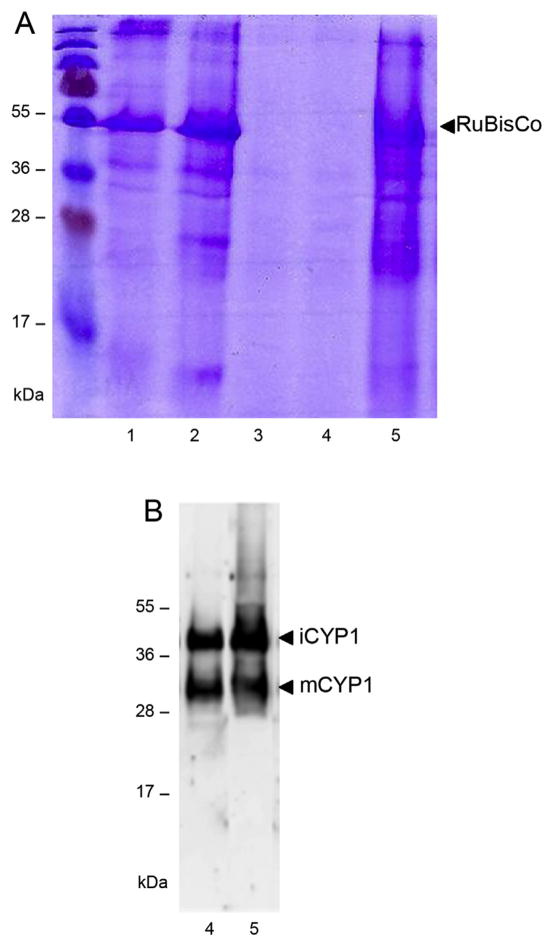
One of the hallmarks of the enzymatic activity of RD21 is its ability to degrade the large subunit of Rubisco within plant extracts [20]. Here, we employed the same assay to monitor the proteolytic activity of CYP1 transiently expressed in N. benthamiana, to rule out any effects of V2 silencing repressor activity on the Rubisco content, the P19 RNA silencing suppressor [21] was expressed in all assay systems. Fig. 3A shows that a large amount of Rubisco was observed in cell extracts in the absence of CYP1 or V2 (lane 1) and that expression of V2 did not decrease this Rucisco content (lane 2). As expected, addition of a commercially available papain protease to the plant extract resulted in an almost complete disappearance of Rubisco (Fig. 3A, lane 3).
Fig. 3.
Effect of V2 on proteolytic activity of CYP1. (A) CYP1-mediated proteolysis of Rubisco. Lane 1, extract from control leaves; lane 2, extract from V2-expressing leaves; lane 3, extract from control leaves with exogenously added papain; lane 4, extract from CYP1-expressing leaves; lane 5, extract from leaves coexpressing CYP1 and V2. Proteins were resolved SDS polyacrylamide gel electrophoresis and detected by Coomassie blue staining. Arrow indicates the position of Rubisco. (B) Western blot analysis of iCYP1 and mCYP1 content. Lane 4, extract from CYP1-expressing leaves; lane 5, extract from leaves coexpressing CYP1 and V2. Arrows indicate the positions of iCYP1 (38 kDa) and mCYP1 (33 kDa). The numbers on the left indicate molecular mass standards in thousands of Daltons.
Expression of CYP1 also completely depleted Rubisco from the cell extract (Fig. 3A, lane 4). Importantly, extracts from plants coexpressing of V2 and CYP1 largely retained their Rubisco content (Fig. 3A, lane 5), indicating that the presence of V2 substantially compromised the CYP1 proteolytic activity. In control experiments, the presence of CYP1 in the corresponding plant extracts was confirmed by western blotting. Fig. 3B shows that the extracts from plants expressing CYP1 or both CYP1 and V2 (lanes 4 and 5, respectively) contained comparable amounts of iCYP1 and mCYP1 isoforms, consistent with the noninterference of V2 in CYP1 maturation. Collectively, these data suggest that the V2-CYP1 interaction inhibits the enzymatic activity of CYP1.
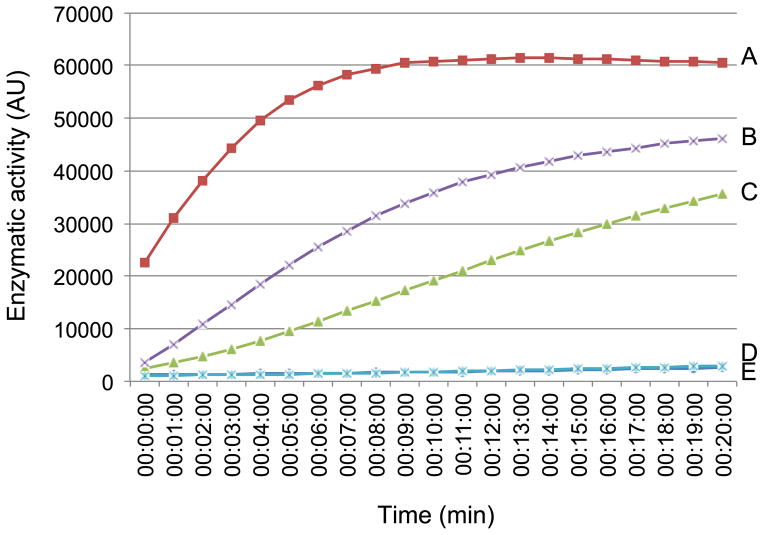
To investigate the inhibitory effect of V2 on CYP1 enzymatic activity in more detail, we performed enzyme kinetic studies. Cell extracts from N. benthamiana leaves transiently expressing CYP1 were assayed for protease activity using the fluorogenic substrate Z-Phe-Arg-MCA [22]. Fig. 4 shows a representative experiment in which the substrate conversion was monitored continuously as function of time of reaction and expressed in arbitrary units (AU) of enzymatic activity when one AU was defined as change in absorption at 460 nm by 1.0 OD unit per minute at 37 °C and pH 5.2. In these experiments, CYP1 expressed alone exhibited enzymatic activity of 70 AU/μg protein (Fig. 4A). This activity was substantially reduced by E64, a specific inhibitor of RD21 [20], to 28 AU/μg protein (Fig. 4B). A comparable inhibition of the CYP1 enzymatic activity to 21 AU/μg protein was observed in cell extracts that coexpressed V2 (Fig. 4C). Interestingly, addition of the E64 inhibitor to this latter assay system completely blocked the reaction (Fig. 4D), suggesting that V2 and E64 inhibit CYP1 in a synergistic fashion. As expected, no reaction occurred without CYP1 expression, i.e., in extracts from tissues Agroinfitrated with an empty expression vector (Fig. 4E).
Fig. 4.
Enzyme kinetics of CYP1 proteolytic activity. (A) Extract from CYP1-expressing leaves. (B) Extract from CYP1-expressing leaves with exogenously added E64. (C) Extract from leaves coexpressing CYP1 and V2. (D) Extract from leaves coexpressing CYP1 and V2 with exogenously added E64. (E) Extract from control leaves. All experiments were repeated at least three times.
4. Discussion
Here, we demonstrate a novel ability of the TYLCV V2 protein to inhibit the enzymatic activity of a host PLCP CYP1. What is the biological rationale that might underline this V2 function? Viral infection often results in profound, sometimes even catastrophic, changes in the host plant cell physiology. In the case of TYLCV, one such effect would be destruction of the host cell vacuole. Indeed, following plant viral infection, the vacuolar membrane is known to collapse [23], leading to the release of vacuolar hydrolytic enzymes, including PLCPs that participate in immune response against pathogens [16]. These enzymes include CYP1, the homolog of which, RD21, is involved in plant immunity [16] and is negatively regulated by direct interaction [24] with serpins, members of a large superfamily encoded by most plant species, among them tomato [25,26]. Potentially, V2 may represent a viral functional homolog of the cellular serpins, acting to down-regulate the CYP1 activity. This is by analogy to many other plant and animal pathogen-encoded proteins which have acquired functional features of a host protein required for infection [27–29]. These pathogen factors usually do not exhibit sequence similarities with the eukaryotic factor they mimic, making their functional annotation difficult [29], and requiring experimental identification of their activities.
The invading virus, however, requires a living host cell for successful infection and subsequent spread. Thus, it makes biological sense for the virus to evolve a strategy for suppression of the host cell death. It is tempting to speculate that V2 represents such strategy for TYLCV. In this scenario, TYLCV infection promotes vacuole rupture and release of CYP1 into the cytoplasm. Although the infected cell might survive this event utilizing its endogenous serpins to bind and down regulate CYP1, the virus facilitates cell survival by augmenting the serpin action with the CYP1-inhibiting activity of V2. V2, therefore, represents a multifunctional viral anti-host-defense factor: it suppresses the host gene silencing defense by interacting with the SGS3 silencing machinery component [8], and it suppresses the host cell death/hypersensitive response by interacting with the programmed cell death machinery component CYP1.
Acknowledgments
This work was supported by the US–Israel Binational Agricultural Research and Development (BARD) grant IS-4605-13C and by the US–Israel Binational Science Foundation (BSF) grant 2011070, to V.C. and Y.G. V.C. is also supported by NIH, grant R01 GM50224, USDA/NIFA, grant 2013-02918 and NSF, grant MCB1118491. This paper is a contribution from the Agricultural Research Organization, The Volcani Center, Bet Dagan, Israel, No. 216/2014.
Footnotes
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2015.03.063.
References
- 1.Glick E, Levy Y, Gafni Y. The viral etiology of Tomato yellow leaf curl disease—a review. Plant Prot Sci. 2009;45:81–97. [Google Scholar]
- 2.Moriones E, Navas-Castillo J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000;71:123–134. doi: 10.1016/s0168-1702(00)00193-3. [DOI] [PubMed] [Google Scholar]
- 3.Nakhla NK, Maxwell DP. Epidemiology and management of tomato yellow leaf curl disease. In: Hadidi A, Khetarpal RK, Koganezawa H, editors. Plant Virus Disease Control. APS Press; St. Paul, Minnesota: 1998. pp. 565–578. [Google Scholar]
- 4.Kheyr-Pour A, Bendahmane M, Matzeit V, Accotto GP, Crespi S, Gronenborn B. Tomato yellow leaf curl virus from Sardinia is a whitefly-transmitted monopartite geminivirus. Nucleic Acids Res. 1991;19:6763–6769. doi: 10.1093/nar/19.24.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navot N, Pichersky E, Zeidan M, Zamir D, Czosnek H. Tomato yellow leaf curl virus: a whitefly-transmitted geminivirus with a single genomic component. Virology. 1991;185:151–161. doi: 10.1016/0042-6822(91)90763-2. [DOI] [PubMed] [Google Scholar]
- 6.Wartig L, Kheyr-Pour A, Noris E, De Kouchkovsky F, Jouanneau F, Gronenborn B, Jupin I. Genetic analysis of the monopartite tomato yellow leaf curl geminivirus: roles of V1, V2, and C2 ORFs in viral pathogenesis. Virology. 1997;228:132–140. doi: 10.1006/viro.1996.8406. [DOI] [PubMed] [Google Scholar]
- 7.Zrachya A, Glick E, Levy Y, Arazi T, Citovsky V, Gafni Y. Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus-Israel. Virology. 2007;358:159–165. doi: 10.1016/j.virol.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 8.Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc Natl Acad Sci U S A. 2008;105:157–161. doi: 10.1073/pnas.0709036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Dong J, Xu Y, Wu J. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 2012;163:51–58. doi: 10.1016/j.virusres.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Amin I, Hussain K, Akbergenov R, Yadav JS, Qazi J, Mansoor S, Hohn T, Fauquet CM, Briddon RW. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol Plant Microbe Interact. 2011;24:973–983. doi: 10.1094/MPMI-01-11-0001. [DOI] [PubMed] [Google Scholar]
- 11.Bar-Ziv A, Levy Y, Hak H, Mett A, Belausov E, Citovsky V, Gafni Y. The Tomato yellow leaf curl virus (TYLCV) V2 protein interacts with the host papain-like cysteine protease CYP1. Plant Signal Behav. 2012;7:983–989. doi: 10.4161/psb.20935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Hoorn RA. Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- 13.Lam E, Kato N, Lawton M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;411:848–853. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
- 14.Avrova AO, Stewart HE, De Jong WD, Heilbronn J, Lyon GD, Birch PR. A cysteine protease gene is expressed early in resistant potato interactions with Phytophthora infestans. Mol Plant Microbe Interact. 1999;12:1114–1119. doi: 10.1094/MPMI.1999.12.12.1114. [DOI] [PubMed] [Google Scholar]
- 15.Hara-Nishimura I, Hatsugai N. The role of vacuole in plant cell death. Cell Death Differ. 2011;18:1298–1304. doi: 10.1038/cdd.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shindo T, Misas-Villamil JC, Horger AC, Song J, van der Hoorn RA. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS One. 2012;7:e29317. doi: 10.1371/journal.pone.0029317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I. A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 2001;42:894–899. doi: 10.1093/pcp/pce144. [DOI] [PubMed] [Google Scholar]
- 18.Koizumi M, Yamaguchi-Shinozaki K, Tsuji H, Shinozaki K. Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene. 1993;129:175–182. doi: 10.1016/0378-1119(93)90266-6. [DOI] [PubMed] [Google Scholar]
- 19.Yamada K, Matsushima R, Nishimura M, Hara-Nishimura I. A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 2001;127:1626–1634. [PMC free article] [PubMed] [Google Scholar]
- 20.Gu C, Shabab M, Strasser R, Wolters PJ, Shindo T, Niemer M, Kaschani F, Mach L, van der Hoorn RA. Post-translational regulation and trafficking of the granulin-containing protease RD21 of Arabidopsis thaliana. PLoS One. 2012;7:e32422. doi: 10.1371/journal.pone.0032422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu W, Park JW, Scholthof HB. Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol Plant Microbe Interact. 2002;15:269–280. doi: 10.1094/MPMI.2002.15.3.269. [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi Y, Saika H, Yuasa K, Nagahama M, Tsuji A. Isolation and biochemical characterization of two forms of RD21 from cotyledons of daikon radish (Raphanus sativus) J Biochem. 2008;144:789–798. doi: 10.1093/jb/mvn132. [DOI] [PubMed] [Google Scholar]
- 23.Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science. 2004;305:855–858. doi: 10.1126/science.1099859. [DOI] [PubMed] [Google Scholar]
- 24.Lampl N, Budai-Hadrian O, Davydov O, Joss TV, Harrop SJ, Curmi PM, Roberts TH, Fluhr R. Arabidopsis AtSerpin1, crystal structure and in vivo interaction with its target protease RESPONSIVE TO DESICCATION-21 (RD21) J Biol Chem. 2010;285:13550–13560. doi: 10.1074/jbc.M109.095075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts TH, Marttila S, Rasmussen SK, Hejgaard J. Differential gene expression for suicide-substrate serine proteinase inhibitors (serpins) in vegetative and grain tissues of barley. J Exp Bot. 2003;54:2251–2263. doi: 10.1093/jxb/erg248. [DOI] [PubMed] [Google Scholar]
- 26.Roberts TH, Hejgaard J. Serpins in plants and green algae. Funct Integr Genomics. 2008;8:1–27. doi: 10.1007/s10142-007-0059-2. [DOI] [PubMed] [Google Scholar]
- 27.Zaltsman A, Krichevsky A, Loyter A, Citovsky V. Agrobacterium induces expression of a plant host F-box protein required for tumorigenicity. Cell Host Microbe. 2010;7:197–209. doi: 10.1016/j.chom.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacroix B, Vaidya M, Tzfira T, Citovsky V. The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J. 2005;24:428–437. doi: 10.1038/sj.emboj.7600524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai H, Roy CR. Show me the substrates: modulation of host cell function by type IV secretion systems. Cell Microbiol. 2003;5:373–383. doi: 10.1046/j.1462-5822.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- 30.Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Trans Res. 1993;2:208–218. [Google Scholar]
- 31.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.van der Linde K, Hemetsberger C, Kastner C, Kaschani F, van der Hoorn RA, Kumlehn J, Doehlemann G. A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell. 2012;24:1285–1300. doi: 10.1105/tpc.111.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimmerman M, Yurewicz E, Patel G. A new fluorogenic substrate for chymotrypsin. Anal Biochem. 1976;70:258–262. doi: 10.1016/s0003-2697(76)80066-8. [DOI] [PubMed] [Google Scholar]