Abstract
As with all human herpesviruses, human cytomegalovirus (HCMV) persists for the lifetime of the host by establishing a latent infection, which is broken by periodic reactivation events. One site of HCMV latency is in the progenitor cells of the myeloid lineage such as CD34+ cells and their CD14+ derivatives. The development of experimental techniques to isolate and culture these primary cells in vitro is enabling detailed analysis of the events that occur during virus latency and reactivation. Ex vivo differentiation of latently infected primary myeloid cells to dendritic cells and macrophages results in the reactivation of latent virus and provides model systems in which to analyse the viral and cellular functions involved in latent carriage and reactivation. Such analyses have shown that, in contrast to primary lytic infection or reactivation which is characterised by a regulated cascade of expression of all viral genes, latent infection is associated with a much more restricted viral transcription programme with expression of only a small number of viral genes. Additionally, concomitant changes in the expression of cellular miRNAs and cellular proteins occur, and this includes changes in the expression of a number of secreted cellular proteins and intracellular anti-apoptotic proteins, which all have profound effects on the latently infected cells. In this review, we concentrate on the effects of one of the latency-associated viral proteins, LAcmvIL-10, and describe how it causes a decrease in the cellular miRNA, hsa-miR-92a, and a concomitant upregulation of the GATA2 myeloid transcription factor, which, in turn, drives the expression of cellular IL-10. Taken together, we argue that HCMV latency, rather than a period of viral quiescence, is associated with the virally driven manipulation of host cell functions, perhaps every bit as complex as lytic infection. A full understanding of these changes in cellular and viral gene expression during latent infection could have far-reaching implications for therapeutic intervention.
Keywords: Latency, Human cytomegalovirus, Cell survival
Introduction
Human cytomegalovirus (HCMV) is typical of human herpesviruses in that it has both a latent and a lytic phase of its life cycle, which are bridged by periodic reactivation events. In the immune competent, primary infection is rarely symptomatic, and following the establishment of latency, the virus maintains a persistent infection effectively controlled by the immune system. It is now established that, in vivo, one site of HCMV latency is in bone marrow resident CD34+ myeloid progenitor cells as well as in their derivative CD14+ monocytes present in peripheral blood. Whilst there is little consensus, likely due to differences in cell types analysed and models of latency used, in the spectrum of viral genes expressed during latent infection, it is clear that latency is associated with a much restricted virus gene transcription programme and, in general, an absence of expression of viral major lytic genes [1–5]. This is also true for cytomegalovirus infection in other species, although the mechanisms by which latency is established vary. A number of studies have demonstrated that murine cytomegalovirus (MCMV) establishes latency, which suggests that some parallels may be drawn between the two species [6, 7]. This inhibition of lytic gene expression during latency is likely affected through repressive chromatin marks around the promoter of the viral major immediate early promoter (MIEP). Following differentiation of progenitor myeloid cells into terminally differentiated dendritic cells or macrophages, however, this repressive chromatin structure around the MIEP is relieved, resulting in changes in post-translational modifications of histones around the MIEP associated with transcriptional activation and concomitant induction of viral lytic immediately early (IE) gene expression [5, 8–12]. These data imply that reactivation routinely occurs in vivo, but this is sub-clinical due to robust host immune responses and is supported by the recent observations that macrophages and DCs, in vivo, are sites of virus reactivation in the healthy carrier [13, 14].
Main text
There are numerous effects on the host cell during HCMV latency
It is well established that the numerous HCMV-encoded genes expressed during lytic infection act in concert to exert profound effects on the infected cell, resulting in the modulation of a wide range of cell functions and their downstream effects. This includes modulation of cell metabolism, transcription, translation, cell cycle, cell signalling as well as the inhibition of immune surveillance, cell stress, and cell death [15–30] (Fig. 1a). However, despite a much restricted transcription profile, a number of studies have also shown that latent infection is associated with a profound manipulation of host cell transcription and cell signalling and, again, the inhibition of host immune surveillance, cell stress, and cell death [4, 5, 31–36] (Fig. 1b). Thus, far from being silent, latent infection with HCMV also results in the viral-driven orchestration of cellular gene expression and cell functions, likely, to optimise the cell for latent carriage and reactivation. For example, a number of changes in total cellular mRNAs have been shown to occur upon latent during experimentally latent infection of granulocyte macrophage progenitors (GMPs) [37, 38] resulting in changes in MHC class II expression and secreted monocyte chemoattractant protein-1 (MCP-1) also known as chemokine C–C motif ligand-2 (CCL2) [39]. Consistent with this, latent infection of myeloid progenitors also results in the regulation of MCP-1 as well as a large number of other secreted cell proteins [32]. In addition to the regulation of secreted proteins during latency, an apoptome array shows that there are also a number of changes in levels of anti-apoptotic proteins during latent infection of CD34+ cells with HCMV (Fig. 2).
Fig. 1.

Manipulation of host cell functions during latent HCMV infection, perhaps every bit as complex as lytic infection. Infection of cells results in a wide range of changes to the infected cell. During lytic infection (a), there are reported changes to the modulation of cell metabolism, transcription, translation, cell cycle, cell signalling as well as the inhibition of immune surveillance, cell stress, and cell death [15–25]. Similarly, during latent infection (b), there are reported changes to the manipulation of host cell transcription and cell signalling and, again, the inhibition of host immune surveillance, cell stress, and cell death [4, 5, 31–36]
Fig. 2.

A number of pro- and anti-apoptotic factors in the FAS signalling pathway are altered during HCMV latency. Either CD34+ cells were uninfected or HCMV latency was established for 10 days, and the cells were harvested for protein analysis (a–d). Relative levels of proteins involved in FAS-mediated and IL-10 signalling were analysed by apoptosis array (R&D systems) (a) and highlighted are Aa (Bcl2), Ab (p53 phospho-serine-15), and Ac (HSP70). Alternatively, cells were harvested for Western blot analysis of total and phosphorylated STAT3 (antibodies from cell signalling) relative to actin loading control (antibody from Abcam) (b) and quantified by densitometry. Data are represented as fold change during latency from representative Western blots (c). The data are summarised in context with the literature in (d). Extrinsic FAS-mediated apoptosis involves FADD, caspase 8, pro-caspase 3, and caspase 3 and leads to apoptosis [81, 82]. Intrinsic mitochondrial-mediated apoptosis involves Bax, Bid, voltage-dependent anion channel (VDAC), cytochrome C (cyt c), pro-caspase 9, and the apoptome [83, 84]. Additionally, anti-apoptotic IL-10 signalling can involve STAT3 phosphorylation, Bcl2, and HSP70 [59, 61, 62]. These anti-apoptotic factors are all positively regulated during HCMV latency. Finally, the two isoforms of virally induced IL-10, cmvIL-10 and LAcmvIL-10, are shown in (e), where grey boxes represent exons. LAcmvIL-10 is generated from alternative splicing, which does not express exon 3
Given the relative paucity of viral genes expressed during HCMV latency [1–5] compared to lytic infection, it may initially seem surprising that such profound changes in the cell result from latent infection. However, another level of regulation of gene expression is via microRNAs (miRNAs). These are highly conserved small (approximately 21 nucleotides in length) RNA molecules encoded in the genomes of plants and animals, which generally regulate the expression of genes by binding to the 3′-untranslated regions (3′-UTR) of specific mRNAs.
Although the first published description of an miRNA was in 1993 [40], the understanding of the functions of many of these small non-coding RNA molecules is still being elucidated and is complicated by the fact that each miRNA is thought to be able to regulate multiple genes. This, coupled with the fact that there are hundreds of miRNAs transcribed in the cells of higher eukaryotes [41–43], reflects the enormous complexity in levels of regulation of gene expression afforded by miRNAs. Various lines of research suggest that miRNAs may act as key regulators of processes as diverse as early development [44], cell proliferation and cell death [45], apoptosis, and fat metabolism [46], as well as cell differentiation [47–49]. There is also evidence to suggest that miRNA expression is involved in the pathogenesis of a number of diseases, including cancer [50, 51] and viral infection [31, 52–55].
It is interesting, therefore, that during HCMV latency, there are changes to a number of cellular miRNAs ([31] and Table 1). Additionally, HCMV itself encodes a number of miRNAs. Consequently, it is possible that many of the changes in the latency-associated secretome as well as the observed changes in anti-apoptotic proteins during latent infection is due, at least in part, to the HCMV-mediated regulation of cellular miRNAs [31] as well as, possibly, expression of a number of viral miRNAs [56].
Table 1.
HCMV-induced latency leads to changes in a number of cellular miRNAs
| MicroRNA | Fold change during latency compared to mock |
|---|---|
| hsa-miR-let-7a | −2.5 |
| hsa-miR-let-7b | −3.7 |
| hsa-miR-206 | −2 |
| hsa-miR-296 3p | −2.6 |
| hsa-miR-297 | −2.9 |
| hsa-miR-32* | −2 |
| hsa-miR-608 | −2.4 |
| hsa-miR-92a | −2.5 |
Following the establishment of latency for 10 days in CD34+ cells, the cells were harvested for miRNA analysis (nCode, Invitrogen), and data are presented as fold change over mock infected cells with probability values
Downregulation of a cellular miRNA can lead to upregulation of cellular proteins during latency
One of the cellular miRNAs which is known to be downregulated during latent infection of CD34+ progenitor cells is hsa-miR-92a ([31] and Table 1). Predictive algorithms and biochemical analysis have shown that this miRNA can target the myeloid transcription factor GATA2, and as predicted, during experimental HCMV latency, there is an increase in levels of this myeloid cellular transcription factor [31, 34].
GATA2 is a cellular transcription factor known to be important in the proliferation, lineage commitment, and survival of haematopoietic progenitor cells [57–60], and the virus targets this important myeloid transcription factor for a number of now well-established reasons. For instance, GATA2 has also been found to regulate the transcription of the latency-associated viral gene product UL144 and may also play a role in the expression of other latency-associated viral gene products. GATA2 not only regulates viral genes but is also known to regulate the expression of a number of cellular genes, including IL-10. Consistent with this, increases in cellular IL-10 (cIL-10) in the secretome of latently infected CD34+ cells have been shown to occur. Detailed analysis of the mechanism by which latency-associated changes in hsa-miR-92a were linked to GATA2 expression and subsequent regulation of cIL-10 came from studies in KG1 cells, a CD34+ cell line which can be manipulated by transfection and recapitulate some aspects of HCMV latent infection, namely the expression of latency-associated genes such as UL138 in the absence of lytic immediate early gene expression [31]. In these cells, transfection of an antagomir to hsa-miR-92a led to increased GATA2 mRNA expression [31] and increased cIL-10. Importantly, this induction of cIL-10 by hsa-miR-92a antagomir did not occur if GATA2 was depleted by RNAi [31]. Taken together, these studies showed that latency-associated changes in hsa-miR-92a result in increased GATA2, which drives the expression of cIL-10 during latent infection (Fig. 3).
Fig. 3.
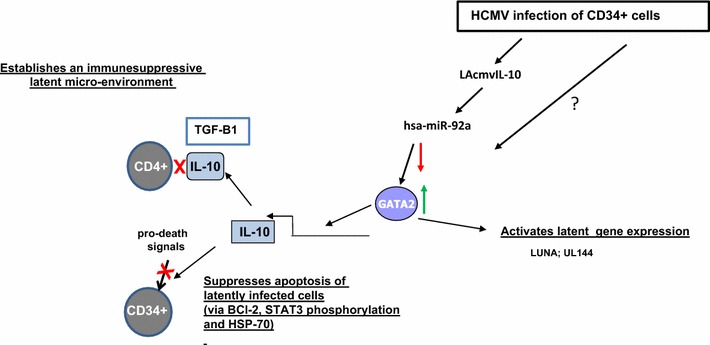
Myeloid transcription factor GATA2 plays multiple roles during HCMV latency. Following the establishment of latency in CD34+ cells for 10 days, there is an induction of cellular hsa-miRNA-92a via LAcmvIL-10 [31, 79]. This leads to a direct upregulation of the cellular transcription factor GATA2 [34]. GATA2 can drive the transcription of the latency-associated viral products LUNA and UL144 [34, 85] as well as driving transcription of the cellular cytokine gene IL-10 [31]. Whether other mechanisms for the upregulation of GATA2 during HCMV latency are also induced is not yet known. IL-10 serves to create an immune suppressive environment [32] as well as to lead to pro-life signalling to the latently infected cell [31]. IL-10 leads to STAT3 phosphorylation and signals to anti-apoptotic factors Bcl2 and HSP70 (see Fig. 2)
Cellular IL-10 is important for the maintenance of latency and immune evasion
Cellular IL-10 is a secreted cytokine known to have immune modulatory properties as well as having pro-life effects on myeloid progenitors, such as CD34+ cells, by driving the expression of the anti-apoptotic factor Bcl2 (Fig. 2a, c, d and [31]). During HCMV latency, cIL-10 is upregulated, and for this reason, the specific functions of cIL-10 in the latent secretome have been analysed [32]. For example, consistent with this observation that cIL-10 is known to play an anti-apoptotic role in CD34+ cells (Fig. 2d), [31, 61], antibody depletion of latency-associated secretion of cIL-10 from the supernatants of latently infected cells results in increased cell death and loss of latent viral genome carriage [31]. Additionally, latency-associated secretion of cIL-10, together with virally induced increases in cellular TGF-beta, results in the establishment of an immune suppressive microenvironment around latently infected cells. This, in turn, inhibits CD4+ cytotoxic T-cell effector functions and thereby suppresses host immune surveillance of the latently infected cell [32].
We know that latent infection of primary CD34+ progenitor cells by HCMV results in their increased survival in the face of pro-apoptotic signals, and this, at least in part, appears to involve the known latency-associated increase in the expression of cIL-10 [31, 59]. However, how cIL-10 mediated this protection was unclear, but the changes in the latent apoptome would suggest that the cIL-10 upregulated during latency modulates cIL-10-mediated suppression of extrinsic and intrinsic pro-apoptotic signals (Fig. 2d).
Engagement of the cIL-10 receptor by cIL-10 is known to induce signalling via STAT3 phosphorylation. This results in positive autoregulation of cIL-10 expression as well as expression of intrinsic death-signalling pathway inhibitors such as Bcl2 [31, 59] and HSP70 [62]. Consistent with this, and the known increase in cIL-10 during HCMV latent infection of CD34+ cells [32], latently infected CD34+ cells also showed extensive increases in STAT3 phosphorylation (Fig. 2b, c) and concomitant increases in the expression of Bcl2 and HSP70 (Fig. 2a, c). HSP70 plays a role in the negative regulation of the intrinsic pathway due to the ability to target pro-caspase 3 [63, 64], present in the FAS-mediated signalling pathway. Thus, during latency, the FAS-mediated signalling pathway is targeted at different stages of FAS-mediated apoptosis via extrinsic apoptosis signalling (Fig. 2d). Interestingly, HSP70 also plays a significant role as a potent inhibitor of the formation of the mitochondrial apoptome [65] and the intrinsic pathway of programmed cell death (Fig. 2d).
Not all of the changes in proteins we have identified during HCMV latency are in pro-life factors. Figure 2a, c shows that there was a 3.5-fold increase in levels of p53 phosphorylated at serine 15. Ser 15-phosphorylated p53 is known to have pro-apoptotic properties as it is able to upregulate the transcription of the pro-apoptotic factor Bax and concomitantly decrease the expression of anti-apoptotic factor Bcl2 [66]. However, the expression of Bcl2 clearly increased during HCMV latency in our studies (see Fig. 2a, c and [31]) despite elevated levels of phosphorylated p53. Consequently, our view is that other functions associated with latent infection also counter the transcriptional regulation of Bcl2 by p53. One possibility is that HSP70 increased during latent infection and can act to stimulate the expression of Bcl2 via AKT [67]. Therefore, the expression of HSP70 may be strong enough to overcome the repressive effects of phosphorylated p53. Similarly, formation of pro-apoptotic Bax homodimers can be prevented by HSP70 [65]. Therefore, it is likely that HSP70 works at multiple levels to help check and balance the levels of anti-apoptotic factors in the latently infected cell, although this needs to be formally addressed.
Latency-associated viral IL-10 can cause downregulation of the cellular miRNA hsa-miR-92a
It appears, then, that the changes in cellular miRNA expression resulting from latent infection can have important downstream effects on both intrinsic cell survival and host immune evasion. However, until recently, the mechanism by which HCMV latent infection caused such changes in cellular miRNAs expression was far from clear. There are a number of viral genes expressed during latency, which could potentially affect cellular miRNA expression, although the latency-associated functions of many of these latency-associated genes are only just beginning to be unravelled.
One of these is a viral cIL-10 homologue, known as LAcmvIL-10, encoded by the UL111A gene. It is interesting that although latent HCMV infection robustly induces the expression of cIL-10, as discussed above, the virus also expresses LAcmvIL-10 during this life cycle. However, HCMV is not unique amongst the herpesviruses in encoding a cIL-10 homologue [68]. However, HCMV actually encodes two IL-10 homologues. These two isoforms of virus-encoded IL-10 are generated by alternative splicing from the viral UL111A gene as depicted graphically in Fig. 2e. One of these is a protein of 175 amino acids, termed cmvIL-10, which is expressed during lytic infection and has the expression kinetics of a late gene. The second isoform, predicted to consist of 139 amino acids and termed LAcmvIL-10, has a C-terminal truncation and is expressed during both lytic and latent infection [68–70]. The encoding of a cIL-10 homologue is not unique to HCMV. An IL-10 homologue encoded by the UL111A open reading frame (ORF) has also been identified in rhesus macaque CMV (RhCMV). Although it has a slightly different gene structure than cmvIL-10, like cmvIL-10, it shows low amino acid identity to host cIL-10 [69].
The full-length cmvIL-10 gene shares 27 % amino acid homology with cIL-10 [71] and has a number of functions in common with cIL-10: it forms homodimers and binds the cIL-10 receptor [69, 72]; it triggers STAT3 phosphorylation and activation of the JAK/STAT signalling pathway [73, 74]; it signals via the phosphoinositide-3-kinase pathway, contributing to cytokine suppression [74–76] and cIL-10-positive autoregulation [77]; and it shares the ability of cIL-10 to prevent NF-κB activity via inhibition of IKK [75, 78].
In contrast, LAcmvIL-10 appears quite dissimilar to cIL-10 and cmvIL-10. Although, like cIL-10 and cmvIL-10, it can downregulate major histocompatibility complex (MHC) class II in latently infected GMPs [76], either it does not signal through the IL-10 receptor (IL-10R) or it engages the receptor in a different way than cIL-10 and cmvIL-10. Therefore, the mode of action and function during HCMV latency is uncertain.
Interestingly, analysis of viruses lacking the UL111A gene locus was found to be impaired in their ability to induce cIL-10 upon latent infection [79]. Similarly, consistent with these observations, viruses lacking UL111A did not downregulate cellular hsa-miR-92a. Furthermore, recombinant LAcmvIL-10 was found both to induce cIL-10 and to cause the downregulation of hsa-miR-92a [79].
Taken together, then it appears that expression of LAcmvIL-10 during latent infection results in downregulation of cellular hsa-miR-92a, which in turn leads to the upregulation of the myeloid transcription factor GATA2. This increase in GATA2 then drives the expression of cIL-10, which inhibits intrinsic cell death signals and aids immune evasion of the latently infected cell (Fig. 3).
It is also worth emphasising that GATA2 is known to be involved in the hematopoiesis and myeloid cell differentiation [80]. The extent to which latent infection of CD34+ cells, in itself, drives the latently infected progenitor cell down the myeloid lineage, rather than the lymphocyte lineage, is unclear. However, this could, in part, explain the fact that latent viral genomes have not been detected in T and B cells even though these presumably derived from the same CD34+ progenitor cells giving rise to cells of the myeloid lineage.
Future perspectives
The ramifications of latency-associated changes in a number of other cellular miRNAs that have been identified during latent (Table 1 and [31]) infection are unclear. Similarly, latent infection also results in changes in a number of other secreted cellular proteins [32], and the effects of these changes on the latently infected cell will be enlightening. Regardless, it is now clear that latent infection imparts on the latently infected cells a plethora of phenotypic changes through an orchestrated manipulation of cell gene expression and cell functions. These are likely necessary for efficient carriage and reactivation of latent viral genomes, but they may also provide an ‘Achilles heel’ to allow the development of novel therapeutics to target and clear latent infection, at least in some clinical settings.
Acknowledgments
We thank Linda Teague, Roy Whiston, and Stuart McGregor Dallas for data and technical support and the MRC for funding, Grant Number G0701279. This research was supported by the Cambridge NIHR BRC cell phenotyping hub.
Footnotes
This article is part of the Special Issue on Cytomegalovirus.
Contributor Information
Emma Poole, Phone: 01223 336850, Email: elp27@cam.ac.uk.
John Sinclair, Email: js152@hermes.cam.ac.uk.
References
- 1.Goodrum FD, Jordan CT, High K, Shenk T. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc Natl Acad Sci USA. 2002;99(25):16255–16260. doi: 10.1073/pnas.252630899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hargett D, Shenk TE. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc Natl Acad Sci USA. 2011;107(46):20039–20044. doi: 10.1073/pnas.1014509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossetto CC, Tarrant-Elorza M, Pari GS. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013;9(5):e1003366. doi: 10.1371/journal.ppat.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poole E, Wills M, Sinclair J. Human cytomegalovirus latency: targeting differences in the latently infected cell with a view to clearing latent infection. New J Sci. 2014;2014:10. doi: 10.1155/2014/313761. [DOI] [Google Scholar]
- 5.Sinclair J, Poole E. Human cytomegalovirus latency and reactivation in and beyond the myeloid lineage. Future Virol. 2014;6:7. doi: 10.1016/j.jcv.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Kurz S, Steffens HP, Mayer A, Harris JR, Reddehase MJ. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71(4):2980–2987. doi: 10.1128/jvi.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seckert CK, Griessl M, Buttner JK, Scheller S, Simon CO, Kropp KA, Renzaho A, Kuhnapfel B, Grzimek NK, Reddehase MJ. Viral latency drives ‘memory inflation’: a unifying hypothesis linking two hallmarks of cytomegalovirus infection. Med Microbiol Immunol. 2012;201(4):551–566. doi: 10.1007/s00430-012-0273-y. [DOI] [PubMed] [Google Scholar]
- 8.Murphy JCFW, Verdin E, Sinclair JH. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 2002;21:1112–1120. doi: 10.1093/emboj/21.5.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves MB, MacAry PA, Lehner PJ, Sissons JG, Sinclair JH. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc Natl Acad Sci USA. 2005;102(11):4140–4145. doi: 10.1073/pnas.0408994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinclair J, Sissons P. Latency and reactivation of human cytomegalovirus. J Gen Virol. 2006;87(Pt 7):1763–1779. doi: 10.1099/vir.0.81891-0. [DOI] [PubMed] [Google Scholar]
- 11.Reeves M, Murphy J, Greaves R, Fairley J, Brehm A, Sinclair J. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J Virol. 2006;80(20):9998–10009. doi: 10.1128/JVI.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groves IJ, Reeves MB, Sinclair JH. Lytic infection of permissive cells with human cytomegalovirus is regulated by an intrinsic ‘pre-immediate-early’ repression of viral gene expression mediated by histone post-translational modification. J Gen Virol. 2009;90(Pt 10):2364–2374. doi: 10.1099/vir.0.012526-0. [DOI] [PubMed] [Google Scholar]
- 13.Reeves MB, Sinclair JH. Circulating dendritic cells isolated from healthy seropositive donors are sites of human cytomegalovirus reactivation in vivo. J Virol. 2013;87(19):10660–10667. doi: 10.1128/JVI.01539-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poole E, Juss JK, Krishna B, Herre J, Chilvers ER, Sinclair J. Alveolar macrophages isolated directly from HCMV seropositive individuals are sites of HCMV reactivation in vivo. J Infect Dis. 2014 doi: 10.1093/infdis/jiu837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poole E, Groves I, MacDonald A, Pang Y, Alcami A, Sinclair J. Identification of TRIM23 as a cofactor involved in the regulation of NF-kappaB by human cytomegalovirus. J Virol. 2009;83(8):3581–3590. doi: 10.1128/JVI.02072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossini G, Cerboni C, Santoni A, Landini MP, Landolfo S, Gatti D, Gribaudo G, Varani S. Interplay between human cytomegalovirus and intrinsic/innate host responses: a complex bidirectional relationship. Mediators Inflamm. 2012;2012:607276. doi: 10.1155/2012/607276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poole E, Bain M, Teague L, Takei Y, Laskey R, Sinclair J. The cellular protein MCM3AP is required for inhibition of cellular DNA synthesis by the IE86 protein of human cytomegalovirus. PLoS One. 2012;7(10):e45686. doi: 10.1371/journal.pone.0045686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun TE, Poole E, Sinclair J. Depletion of cellular pre-replication complex factors results in increased human cytomegalovirus DNA replication. PLoS One. 2012;7(5):e36057. doi: 10.1371/journal.pone.0036057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy EA, Streblow DN, Nelson JA, Stinski MF. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J Virol. 2000;74(15):7108–7118. doi: 10.1128/JVI.74.15.7108-7118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalejta RF. Human cytomegalovirus pp71: a new viral tool to probe the mechanisms of cell cycle progression and oncogenesis controlled by the retinoblastoma family of tumor suppressors. J Cell Biochem. 2004;93(1):37–45. doi: 10.1002/jcb.20177. [DOI] [PubMed] [Google Scholar]
- 21.Wright E, Bain M, Teague L, Murphy J, Sinclair J. Ets-2 repressor factor recruits histone deacetylase to silence human cytomegalovirus immediate-early gene expression in non-permissive cells. J Gen Virol. 2005;86(Pt 3):535–544. doi: 10.1099/vir.0.80352-0. [DOI] [PubMed] [Google Scholar]
- 22.Poole E, King C, Sinclair J, Alcami A. The UL144 gene product of human cytomegalovirus activates NF-kB via a TRAF6-dependent mechanism. EMBO J. 2006;25:4390–4399. doi: 10.1038/sj.emboj.7601287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole E, Atkins E, Nakayama T, Yoshie O, Groves I, Alcami A, Sinclair J. NF-kappaB-mediated activation of the chemokine CCL22 by the product of the human cytomegalovirus gene UL144 escapes regulation by viral IE86. J Virol. 2008;82(9):4250–4256. doi: 10.1128/JVI.02156-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller-Kittrell M, Sparer TE. Feeling manipulated: cytomegalovirus immune manipulation. Virol J. 2009;6:4. doi: 10.1186/1743-422X-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abraham CG, Kulesza CA. Polycomb repressive complex 2 silences human cytomegalovirus transcription in quiescent infection models. J Virol. 2013;87(24):13193–13205. doi: 10.1128/JVI.02420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salvant BS, Fortunato EA, Spector DH. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol. 1998;72(5):3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaumorcel M, Lussignol M, Mouna L, Cavignac Y, Fahie K, Cotte-Laffitte J, Geballe A, Brune W, Beau I, Codogno P, Esclatine A. The human cytomegalovirus protein TRS1 inhibits autophagy via its interaction with Beclin 1. J Virol. 2012;86(5):2571–2584. doi: 10.1128/JVI.05746-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall EE, Bierle CJ, Brune W, Geballe AP. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J Virol. 2009;83(9):4112–4120. doi: 10.1128/JVI.02489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Browne EPST. Evaluation of the host transcriptional response to human cytomegalovirus infection. Proc Natl Acad Sci USA. 2003;100:11439–11444. doi: 10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole E, McGregor Dallas SR, Colston J, Joseph RS, Sinclair J. Virally induced changes in cellular microRNAs maintain latency of human cytomegalovirus in CD34 progenitors. J Gen Virol. 2011;92(Pt 7):1539–1549. doi: 10.1099/vir.0.031377-0. [DOI] [PubMed] [Google Scholar]
- 32.Mason GM, Poole E, Sissons JG, Wills MR, Sinclair JH. Human cytomegalovirus latency alters the cellular secretome, inducing cluster of differentiation (CD)4+ T-cell migration and suppression of effector function. Proc Natl Acad Sci USA. 2012;109(36):14538–14543. doi: 10.1073/pnas.1204836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mason GM, Jackson S, Okecha G, Poole E, Sissons JG, Sinclair J, Wills MR. Human cytomegalovirus latency-associated proteins elicit immune-suppressive IL-10 producing CD4(+) T Cells. PLoS Pathog. 2013;9(10):e1003635. doi: 10.1371/journal.ppat.1003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poole E, Walther A, Raven K, Benedict CA, Mason GM, Sinclair J. The myeloid transcription factor GATA-2 regulates the viral UL144 gene during human cytomegalovirus latency in an isolate-specific manner. J Virol. 2013;87(8):4261–4271. doi: 10.1128/JVI.03497-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weekes MP, Tan SY, Poole E, Talbot S, Antrobus R, Smith DL, Montag C, Gygi SP, Sinclair JH, Lehner PJ. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science. 2013;340(6129):199–202. doi: 10.1126/science.1235047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wills MR, Poole E, Lau B, Krishna B, Sinclair JH. The immunology of human cytomegalovirus latency: Could latent infection be cleared by novel immunotherapeutic strategies? Cell Mol Immunol. 2014 doi: 10.1038/cmi.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slobedman B, Cheung AK. Microarrays for the study of viral gene expression during human cytomegalovirus latent infection. Methods Mol Med. 2008;141:153–175. doi: 10.1007/978-1-60327-148-6_9. [DOI] [PubMed] [Google Scholar]
- 38.Cheung AK, Abendroth A, Cunningham AL, Slobedman B. Viral gene expression during the establishment of human cytomegalovirus latent infection in myeloid progenitor cells. Blood. 2006;108(12):3691–3699. doi: 10.1182/blood-2005-12-026682. [DOI] [PubMed] [Google Scholar]
- 39.Stern JL, Slobedman B. Human cytomegalovirus latent infection of myeloid cells directs monocyte migration by up-regulating monocyte chemotactic protein-1. J Immunol. 2008;180(10):6577–6585. doi: 10.4049/jimmunol.180.10.6577. [DOI] [PubMed] [Google Scholar]
- 40.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 41.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedlander MR, Lizano E, Houben AJ, Bezdan D, Banez-Coronel M, Kudla G, Mateu-Huertas E, Kagerbauer B, Gonzalez J, Chen KC, LeProust EM, Marti E, Estivill X. Evidence for the biogenesis of more than 1000 novel human microRNAs. Genome Biol. 2014;15(4):R57. doi: 10.1186/gb-2014-15-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, Blelloch R, Schroth GP, Nusbaum C, Bartel DP. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24(10):992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 45.Brennecke J, Cohen SM. Towards a complete description of the microRNA complement of animal genomes. Genome Biol. 2003;4(9):228. doi: 10.1186/gb-2003-4-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13(9):790–795. doi: 10.1016/S0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 47.Bhushan R, Grunhagen J, Becker J, Robinson PN, Ott CE, Knaus P. miR-181a promotes osteoblastic differentiation through repression of TGF-beta signaling molecules. Int J Biochem Cell Biol. 2013;45(3):696–705. doi: 10.1016/j.biocel.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koga Y, Yasunaga M, Takahashi A, Kuroda J, Moriya Y, Akasu T, Fujita S, Yamamoto S, Baba H, Matsumura Y. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res. 2010;3(11):1435–1442. doi: 10.1158/1940-6207.CAPR-10-0036. [DOI] [PubMed] [Google Scholar]
- 50.Lawrie CH, Ballabio E, Dyar OJ, Jones M, Ventura R, Chi J, Tramonti D, Gooding S, Boultwood J, Wainscoat JS, Hatton CS, Schuh A. MicroRNA expression in chronic lymphocytic leukaemia. Br J Haematol. 2009;147(3):398–402. doi: 10.1111/j.1365-2141.2009.07857.x. [DOI] [PubMed] [Google Scholar]
- 51.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33(6):1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 53.Bennasser Y, Le SY, Yeung ML, Jeang KT. MicroRNAs in human immunodeficiency virus-1 infection. Methods Mol Biol. 2006;342:241–253. doi: 10.1385/1-59745-123-1:241. [DOI] [PubMed] [Google Scholar]
- 54.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315(5818):1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 55.Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol. 2008;82(18):9065–9074. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fannin Rider PJ, Dunn W, Yang E, Liu F. Human cytomegalovirus microRNAs. Curr Top Microbiol Immunol. 2008;325:21–39. doi: 10.1007/978-3-540-77349-8_2. [DOI] [PubMed] [Google Scholar]
- 57.Pan X, Minegishi N, Harigae H, Yamagiwa H, Minegishi M, Akine Y, Yamamoto M. Identification of human GATA-2 gene distal IS exon and its expression in hematopoietic stem cell fractions. J Biochem. 2000;127(1):105–112. doi: 10.1093/oxfordjournals.jbchem.a022570. [DOI] [PubMed] [Google Scholar]
- 58.Hsu AP, Sampaio EP, Khan J, Calvo KR, Lemieux JE, Patel SY, Frucht DM, Vinh DC, Auth RD, Freeman AF, Olivier KN, Uzel G, Zerbe CS, Spalding C, Pittaluga S, Raffeld M, Kuhns DB, Ding L, Paulson ML, Marciano BE, Gea-Banacloche JC, Orange JS, Cuellar-Rodriguez J, Hickstein DD, Holland SM. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118(10):2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weber-Nordt RM, Henschler R, Schott E, Wehinger J, Behringer D, Mertelsmann R, Finke J. Interleukin-10 increases Bcl-2 expression and survival in primary human CD34+ hematopoietic progenitor cells. Blood. 1996;88(7):2549–2558. [PubMed] [Google Scholar]
- 60.Hahn CN, Chong CE, Carmichael CL, Wilkins EJ, Brautigan PJ, Li XC, Babic M, Lin M, Carmagnac A, Lee YK, Kok CH, Gagliardi L, Friend KL, Ekert PG, Butcher CM, Brown AL, Lewis ID, To LB, Timms AE, Storek J, Moore S, Altree M, Escher R, Bardy PG, Suthers GK, D’Andrea RJ, Horwitz MS, Scott HS. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43(10):1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Todaro M, Zerilli M, Ricci-Vitiani L, Bini M, Perez Alea M, Maria Florena A, Miceli L, Condorelli G, Bonventre S, Di Gesu G, De Maria R, Stassi G. Autocrine production of interleukin-4 and interleukin-10 is required for survival and growth of thyroid cancer cells. Cancer Res. 2006;66(3):1491–1499. doi: 10.1158/0008-5472.CAN-05-2514. [DOI] [PubMed] [Google Scholar]
- 62.Zorzi E, Bonvini P. Inducible hsp70 in the regulation of cancer cell survival: analysis of chaperone induction, expression and activity. Cancers. 2011;3(4):3921–3956. doi: 10.3390/cancers3043921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mosser DD, Caron AW, Bourget L, Meriin AB, Sherman MY, Morimoto RI, Massie B. The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Mol Cell Biol. 2000;20(19):7146–7159. doi: 10.1128/MCB.20.19.7146-7159.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sabirzhanov B, Stoica BA, Hanscom M, Piao CS, Faden AI. Over-expression of HSP70 attenuates caspase-dependent and caspase-independent pathways and inhibits neuronal apoptosis. J Neurochem. 2012;123(4):542–554. doi: 10.1111/j.1471-4159.2012.07927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stankiewicz AR, Lachapelle G, Foo CP, Radicioni SM, Mosser DD. Hsp70 inhibits heat-induced apoptosis upstream of mitochondria by preventing Bax translocation. J Biol Chem. 2005;280(46):38729–38739. doi: 10.1074/jbc.M509497200. [DOI] [PubMed] [Google Scholar]
- 66.Basu A, Haldar S. The relationship between BcI2, Bax and p53: consequences for cell cycle progression and cell death. Mol Hum Reprod. 1998;4(12):1099–1109. doi: 10.1093/molehr/4.12.1099. [DOI] [PubMed] [Google Scholar]
- 67.Liu M, Aneja R, Liu C, Sun L, Gao J, Wang H, Dong JT, Sarli V, Giannis A, Joshi HC, Zhou J. Inhibition of the mitotic kinesin Eg5 up-regulates Hsp70 through the phosphatidylinositol 3-kinase/Akt pathway in multiple myeloma cells. J Biol Chem. 2006;281(26):18090–18097. doi: 10.1074/jbc.M601324200. [DOI] [PubMed] [Google Scholar]
- 68.Slobedman B, Barry PA, Spencer JV, Avdic S, Abendroth A. Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J Virol. 2009;83(19):9618–9629. doi: 10.1128/JVI.01098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jenkins C, Abendroth A, Slobedman B. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J Virol. 2004;78(3):1440–1447. doi: 10.1128/JVI.78.3.1440-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jenkins C, Garcia W, Abendroth A, Slobedman B. Expression of a human cytomegalovirus latency-associated homolog of interleukin-10 during the productive phase of infection. Virology. 2008;370(2):285–294. doi: 10.1016/j.virol.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10) Proc Natl Acad Sci USA. 2000;97(4):1695–1700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ouyang P, Rakus K, van Beurden SJ, Westphal AH, Davison AJ, Gatherer D, Vanderplasschen AF. IL-10 encoded by viruses: a remarkable example of independent acquisition of a cellular gene by viruses and its subsequent evolution in the viral genome. J Gen Virol. 2014;95(Pt 2):245–262. doi: 10.1099/vir.0.058966-0. [DOI] [PubMed] [Google Scholar]
- 73.Wehinger J, Gouilleux F, Groner B, Finke J, Mertelsmann R, Weber-Nordt RM. IL-10 induces DNA binding activity of three STAT proteins (Stat1, Stat3, and Stat5) and their distinct combinatorial assembly in the promoters of selected genes. FEBS Lett. 1996;394(3):365–370. doi: 10.1016/0014-5793(96)00990-8. [DOI] [PubMed] [Google Scholar]
- 74.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 75.Nachtwey J, Spencer JV. HCMV IL-10 suppresses cytokine expression in monocytes through inhibition of nuclear factor-kappaB. Viral Immunol. 2008;21(4):477–482. doi: 10.1089/vim.2008.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spencer JV, Cadaoas J, Castillo PR, Saini V, Slobedman B. Stimulation of B lymphocytes by cmvIL-10 but not LAcmvIL-10. Virology. 2008;374(1):164–169. doi: 10.1016/j.virol.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hedrich CM, Bream JH. Cell type-specific regulation of IL-10 expression in inflammation and disease. Immunol Res. 2010;47(1–3):185–206. doi: 10.1007/s12026-009-8150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang WL, Barry PA. Attenuation of innate immunity by cytomegalovirus IL-10 establishes a long-term deficit of adaptive antiviral immunity. Proc Natl Acad Sci USA. 2010;107(52):22647–22652. doi: 10.1073/pnas.1013794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poole E, Avdic S, Hodkinson J, Jackson S, Wills M, Slobedman B, Sinclair J. Latency-associated viral interleukin-10 (IL-10) encoded by human cytomegalovirus modulates cellular IL-10 and CCL8 secretion during latent infection through changes in the cellular microRNA hsa-miR-92a. J Virol. 2014;88(24):13947–13955. doi: 10.1128/JVI.02424-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vicente C, Conchillo A, Garcia-Sanchez MA, Odero MD. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol Hematol. 2011;82(1):1–17. doi: 10.1016/j.critrevonc.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 81.Kaufmann T, Strasser A, Jost PJ. Fas death receptor signalling: roles of Bid and XIAP. Cell Death Differ. 2012;19(1):42–50. doi: 10.1038/cdd.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lavrik IN. Systems biology of death receptor networks: live and let die. Cell Death Dis. 2014;5:e1259. doi: 10.1038/cddis.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol. 2012;13(12):780–788. doi: 10.1038/nrm3479. [DOI] [PubMed] [Google Scholar]
- 84.Ilmarinen P, Moilanen E, Kankaanranta H. Mitochondria in the center of human eosinophil apoptosis and survival. Int J Mol Sci. 2014;15(3):3952–3969. doi: 10.3390/ijms15033952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reeves M, Woodhall D, Compton T, Sinclair J. Human cytomegalovirus IE72 protein interacts with the transcriptional repressor hDaxx to regulate LUNA gene expression during lytic infection. J Virol. 2010;84(14):7185–7194. doi: 10.1128/JVI.02231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


