Abstract
Aminoglycoside antibiotics are “the drug of choice” for treating many bacterial infections, but their administration results in hearing loss in up to one fourth of the patients who receive them. Several biochemical pathways have been implicated in aminoglycoside antibiotic ototoxicity; however, little is known about how hair cells respond to aminoglycoside antibiotics at the transcriptome level. Here we have investigated the genome-wide response to the aminoglycoside antibiotic gentamicin. Using organotypic cultures of the perinatal organ of Corti, we performed RNA sequencing using cDNA libraries obtained from FACS-purified hair cells. Within 3 h of gentamicin treatment, the messenger RNA level of more than three thousand genes in hair cells changed significantly. Bioinformatic analysis of these changes highlighted several known signal transduction pathways, including the JNK pathway and the NF-κB pathway, in addition to genes involved in the stress response, apoptosis, cell cycle control, and DNA damage repair. In contrast, only 698 genes, mainly involved in cell cycle and metabolite biosynthetic processes, were significantly affected in the non-hair cell population. The gene expression profiles of hair cells in response to gentamicin share a considerable similarity with those previously observed in gentamicin-induced nephrotoxicity. Our findings suggest that previously observed early responses to gentamicin in hair cells in specific signaling pathways are reflected in changes in gene expression. Additionally, the observed changes in gene expression of cell cycle regulatory genes indicate a disruption of the postmitotic state, which may suggest an alternate pathway regulating gentamicin-induced apoptotic hair cell death. This work provides a more comprehensive view of aminoglycoside antibiotic ototoxicity, and thus contributes to identifying potential pathways or therapeutic targets to alleviate this important side effect of aminoglycoside antibiotics.
Keywords: sensory hair cells, aminoglycoside antibiotics, gentamicin, ototoxicity, RNA sequencing, differential gene expression
Introduction
Ototoxicity is a well-known side effect limiting the use of aminoglycoside antibiotics, with reported incidence of hearing loss between 2% and 25% of treated patients (Huth et al., 2011). Several biochemical mechanisms of aminoglycoside ototoxicity have been investigated, including production of reactive oxygen species (ROS) (Forge and Schacht, 2000), disruption of intracellular calcium storage (Matsui et al., 2004), and inhibition of cytoplasmic protein synthesis (Francis et al., 2013). Through biochemical assays, some pathways have also been identified as signaling pathways mediating aminoglycoside-induced hair cell death, such as the pro-apoptotic JNK pathway (Ylikoski et al., 2002) and the protective NF-κB pathway (Jiang et al., 2005). Based on observations made in organ cultures, a timeline of critical signaling events has been established. ROS production is induced shortly after aminoglycoside administration; the JNK pathway responds to drug treatment within 3 h; the intracellular calcium level is increased dramatically at around 4 h; cytochrome C is released into cytoplasm at around 12 h and apoptosis execution steps occur at around 18 h (Matsui et al., 2004).
The methods utilizing low-throughput biochemical assays have been instrumental in dissecting some of the sequelae of antibiotic treatment of hair cells. However, cells are complex systems that integrate input from multiple mechanisms and respond by adjusting multiple pathways simultaneously. High-throughput methods may help us gain a better understanding of the ototoxicity associated with aminoglycosides by providing a complex view of the response; among them, DNA microarray and antibody array experiments have been performed to profile the changes in the cochlea after ototoxic drug treatment at the transcriptional level and the protein level, respectively (Nagy et al., 2004; Jamesdaniel et al., 2008). However, whole cochlea samples were used in those studies, which consisted largely of many different non-hair cell populations in addition to hair cells, and the heterogeneity of the samples limited the interpretation of the results due to a low signal to noise ratio.
Here we have utilized RNA sequencing to investigate the early gene expression changes in hair cells induced by the aminoglycoside antibiotic gentamicin. Compared to microarray analysis, RNA sequencing is more accurate for analyzing expression levels, more informative for pathway analysis, and more powerful in exploiting unannotated genes (Wang et al., 2009). We made use of the Atoh1-GFP transgenic mice, whose fluorescent reporter under the control of the Atoh1 enhancer faithfully identifies hair cells (Lumpkin et al., 2003), to purify GFP+ hair cells by fluorescence-activated cell sorting (FACS) after a 3 h gentamicin treatment for a direct analysis of gene expression changes by RNA sequencing. We chose the 3 h time point because it was previously shown that signaling pathways have initiated a response to the drug, but the cell death process did not appear to be initiated (Matsui et al., 2004). We present analyses of the gene changes associated with several major pathways known to be dysregulated in response to aminoglycoside antibiotics.
Genome wide, 3709 genes, including those involved in a variety of pathways and cellular processes, were found to change significantly in hair cells after gentamicin treatment. Genes involved in the JNK pathway and the NF-κB pathway were differentially expressed in gentamicin-treated hair cells, which is consistent with previous findings that these pathways are important for aminoglycoside ototoxicity (Ylikoski et al., 2002; Jiang et al., 2005). Surprisingly, we did not observe a strong stress response at the transcriptional level at this early time point. Comparison of gentamicin-induced gene expression changes in hair cells and data from other cell lines and tissues, revealed a similarity between aminoglycoside-induced hair cell death and neuronal cell death, as well as a resemblance between aminoglycoside ototoxicity and nephrotoxicity. Additionally, genes involved in the cell cycle and DNA damage repair were also strongly affected by gentamicin at the transcriptional level, indicating a gentamicin-induced disruption of the postmitotic state, which may underlie an alternate pathway leading to hair cell death. In contrast, significantly fewer genes (698 genes) with altered expression after gentamicin treatment were present in the non-hair cell population at this time. Interestingly, expression of genes involved in cell cycle and metabolite biosynthesis pathways were significantly affected in this population. Analyses of our RNA sequencing data indicate that prior to severe oxidative stress, multiple events, including previously implicated activation of the JNK and the NF-κB pathways, and newly hypothesized disruption of the postmitotic state, occur in hair cells in response to aminoglycoside antibiotic treatment. Our work provides a comprehensive view of aminoglycoside ototoxicity, and our transcriptome data could be used to analyze other potential signaling pathways or biological processes mediating aminoglycoside-induced hair cell death which are not covered here.
Materials and methods
Organotypic culture, drug treatment, and FACS sorting
Atoh1-GFP transgenic mice on CD-1 background (Lumpkin et al., 2003; available in the Jackson Laboratory but in different genomic background: B6.129S-Atoh1tm4.1Hzo/J) were used for this study. All animal procedures and usage were approved by the IACUC committee in House Research Institute. Inner ears were collected from postnatal day 1 (P1) animals under sterile conditions and then transferred into ice cold Ca2+- and Mg2+-free PBS (Invitrogen), in which cochlea were further dissected under the microscope (Chen et al., 2002). The cochleae were mounted on polycarbonate membranes (13 mm diameter, 1.0 μm pore size, SPI supplies) floating on DMEM/F12 medium (Invitrogen) supplemented with 1% N2 (Gibco) and 100 U/ml penicillin (Sigma). Organ cultures were maintained under low oxygen conditions (37°C, 5% CO2 and 5% O2) overnight, and treated with gentamicin the next day.
To trace the accumulation of gentamicin in hair cells, Texas Red conjugated gentamicin (GTTR) was prepared as described in the literature (Steyger et al., 2003). 4.4 ml of gentamicin sulfate (Sigma; 50 mg/ml in K2CO3, pH 9) was mixed with 0.6 ml succinimidyl ester of Texas Red (Invitrogen; 2 mg/ml in dimethyl formamide), and then agitated overnight to produce a mixture of gentamicin and GTTR with an approximate 300:1 molar ratio. This GTTR mixture solution was diluted 1:100 with culture medium for treatment of organotypic cultures. After 30 min incubation, organs were fixed with 4% paraformaldehyde, cryosectioned and photographed by fluorescence microscopy.
To confirm the level of hair cell death induced by gentamicin, organ cultures were treated with 0.5 mM gentamicin for 3 h, and then gentamicin was washed out and replaced with fresh medium. GFP positive hair cells were photographed at 3 and 24 h timepoints (Figure 1B).
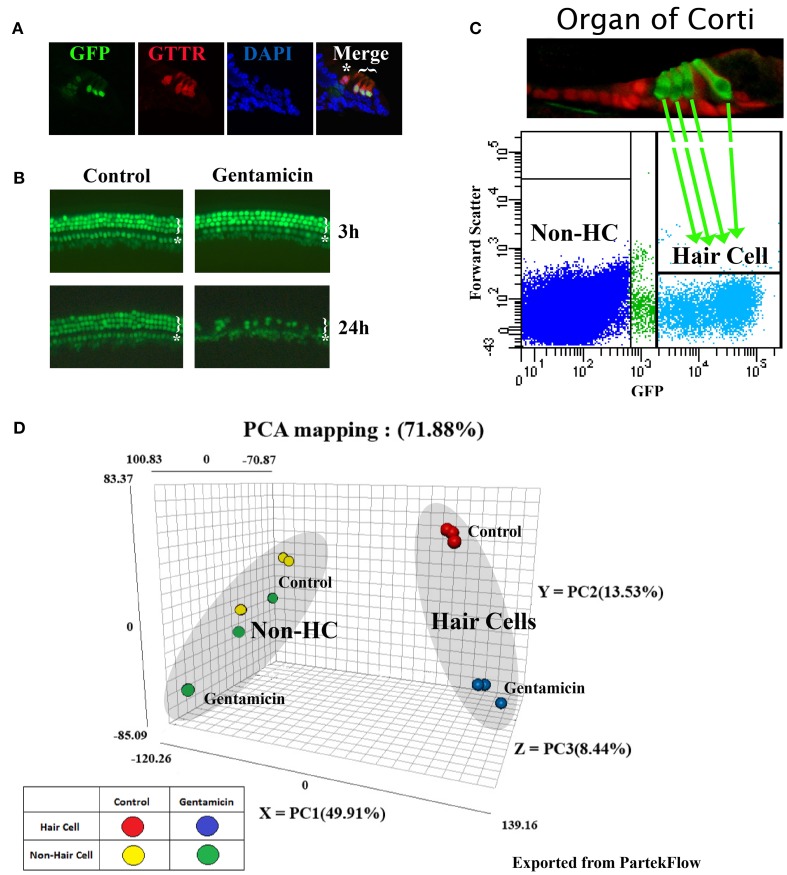
Figure 1.
Gentamicin-induced hair cell loss, and principal component analysis (PCA) of RNA sequencing datasets. (A) Gentamicin accumulates preferentially in hair cells in in vitro organ of Corti culture. Cross sections through cochlear explants from P1, Atoh1-GFP mice (Lumpkin et al., 2003), treated for 30 min with Texas Red conjugated gentamicin (GTTR). Star indicates the inner hair cell, and bracket indicates three outer hair cells. Cells with weak GFP under the inner hair cell are inner phalangeal cells. (B) Atoh1-GFP fluorescence in untreated and 0.5 mM gentamicin-treated organs at 3 and 24 h. There was no detectable hair cell loss at 3 h, but severe hair cell damage caused by gentamicin at 24 h. Star indicates a single row of inner hair cells and bracket indicates three partial rows of outer hair cells. (C) Scatter plot of Atoh1-GFP hair cell purification by FACS shows gate settings and diagram shows a P1 organ of Corti indicating hair cells (green) and supporting cells (red) (Chen and Segil, 1999). (D) PCA map showing the three most significant variances among samples. 78.88% of variance in the combined dataset is captured in the analysis; (49.91% in PC1-X axis, 13.53% in PC2-Y axis, and 8.44% on PC3-Z axis). Each dot represents one biological replicate.
To purify hair cells for RNAseq, organs were digested with 0.05% Trypsin (Invitrogen) and 1 mg/ml Collagenase (Worthington) in PBS at 37°C for 8 min, then incubated with 10% FBS (Life Technologies) in PBS to stop enzymatic digestion. To make single cell suspensions, organs were triturated with a P200 pipette 300 times. The suspension was passed through a cell strainer (40 μm, BD Biosciences) before FACS purification. GFP-positive hair cells, as well as the GFP-negative non-hair cell population (non-hair cell cochlear epithelial cells included Deiters' cells, pillar cells, Hensen cells, cells in the GER, cells in the LER, and other cells constituting surrounding tissues) were purified on a BD FACS Aria II with a 100 μ nozzle. Cells with low-levels of GFP were excluded by stringent gating during FACS purification (Figure 1C). Quality control by FACS-resort, and by immunofluorescence for a hair cell marker (MyosinVI), indicated >95% purity. Sorted cells were collected directly into RNA lysis buffer (Zymo). At least 50,000 cells were collected for each sample, and three replicates were prepared for each condition.
RNA sequencing, reads alignment, PCA and differential gene expression
RNA was extracted from samples using the Zymo Quick-RNA Microprep kit, and then processed for library construction, using the Illumina True-Seq mRNA-seq kit. Six samples were bar-coded, combined into one lane, and sequenced by Illumina Hi-Seq 2000 for single-end 50 cycles (50 bp reads). More than 30 million reads were obtained for each replicate. The reads were trimmed on both ends (quality score ≥25) and aligned against the mouse genome assembly mm10 using TopHat 2 in PartekFlow (Partek Inc.). Normalized read score for each gene was calculated considering total read numbers and gene length (reads per kilobase of transcript per million reads mapped, RPKM). Principal component analysis (PCA) was conducted in PartekFlow based on normalized read numbers for individual genes in each replicate. Differential gene expression was assessed by the embedded gene specific analysis (GSA) module in PartekFlow.
RNA sequence data was deposited into NCBI GEO database (GSE66775).
IPA analysis
Differential gene expression datasets, including gene symbols, fold changes, p-values and total numbers of raw reads, were prepared for Ingenuity pathway analysis software (IPA, Version 21901358, Qiagen Inc.). The gene expression dataset from untreated and gentamicin-treated hair cells, and from untreated and treated non-hair cell samples, were analyzed by the IPA software. A filter of p-value less than 0.01, fold change greater than 1.2 (or less than −1.2), and total number of raw reads greater than 100 was applied, and core pathway analysis was conducted for each dataset. Canonical pathway analysis, diseases and function analysis, upstream regulator analysis, and network analysis were included in the core analysis with default settings.
Q-PCR validation
The cDNA libraries for Q-PCR were made by qScript reverse transcriptase supermix (Quanta Biosciences) using RNA extracted from FACS purified cells as template. Rpl19 was used as internal control for normalization. For validation purpose, four independent biological replicates were collected and analyzed by Q-PCR. Genes were chosen arbitrarily among the list of gentamicin-induced genes in hair cells. SYBR-Green (Applied Biosystems) was used to detect amplified double strand DNA on ViiA 7 machine (Applied Biosystems). Primer pairs used for Q-PCR were listed below. Rpl19 forward 5′-GGTCTGGTTGGATCCCAATG-3′, reverse 5′-CCCGGGAATGGACAGTCA-3′. Atf2 forward 5′-CCGTTGCTATTCCTGCATCAA-3′, reverse 5′-TTGCTTCTGACTGGACTGGTT-3′. Mapk8 forward 5′-AGCAGAAGCAAACGTGACAAC-3′, reverse 5′-GCTGCACACACTATTCCTTGAG-3′. Jun forward 5′-CCTTCTACGACGATGCCCTC-3′, reverse 5′-GGTTCAAGGTCATGCTCTGTTT-3′. Nfkb1 forward 5′-ATGGCAGACGATGATCCCTAC-3′, reverse 5′-TGTTGACAGTGGTATTTCTGGTG-3′. Nfkbib forward 5′-GCGGATGCCGATGAATGGT-3′, reverse 5′-TGACGTAGCCAAAGACTAAGGG-3′. Chuk forward 5′-GTCAGGACCGTGTTCTCAAGG-3′, reverse 5′-GCTTCTTTGATGTTACTGAGGGC-3′. Gadd45g forward 5′-GGGAAAGCACTGCACGAACT-3′, reverse 5′-AGCACGCAAAAGGTCACATTG-3′. Klf11 forward 5′-CATGGACATTTGTGAGTCGATCC-3′, reverse 5′-CCTTTGGTAGATCAGGTGCAG-3′. Ddit3 forward 5′-CTGGAAGCCTGGTATGAGGAT-3′, reverse 5′-CAGGGTCAAGAGTAGTGAAGGT-3′. Bnip1 forward 5′-AGGCTATGCAGACTCTAGTCAG-3′, reverse 5′-CAGTTCTCGGCGGTTGTACT-3′. Casp3 forward 5′-ATGGAGAACAACAAAACCTCAGT-3′, reverse 5′-TTGCTCCCATGTATGGTCTTTAC-3′. Ccar1 forward 5′-AGATGAGTATGACCCAATGGAGG-3′, reverse 5′-CCTTGCAGTACCGGCTGAC-3′. Ccnb2 forward 5′-GCCAAGAGCCATGTGACTATC-3′, reverse 5′-CAGAGCTGGTACTTTGGTGTTC-3′. Ccne2 forward 5′-ATGTCAAGACGCAGCCGTTTA-3′, reverse 5′-GCTGATTCCTCCAGACAGTACA-3′. Ercc8 forward 5′-GAGGAAGATGAAGCTATGGAA-3′, reverse 5′-CTTCAGGGGTTTCTCTTTGTC-3′. Rad52 forward 5′-CTTTGTTGGTGGGAAGTCTGT-3′, reverse 5′-CGGCTGCTAATGTACTCTGGAC-3′. Apex2 forward 5′-GGATGGATGGCTTGCTCAGTA-3′, reverse 5′-ACTTCAGGGAGTAAGAAGGAGG-3′. Alkbh3 forward 5′-GAGCCAGTCTGCTACTCAGC-3′, reverse 5′- AACACAAATTGTCGGTCACATTG-3′.
Results
Perinatal cochleae from Atoh1-GFP transgenic mice, cultured and treated with gentamicin, show that gentamicin accumulated specifically in hair cells, as indicated by the uptake of Texas-Red conjugated gentamicin (Figure 1A), and 91% (SD ± 7%; n = 3) of outer hair cells were killed by gentamicin at 24 h (Figure 1B). To investigate the early transcriptional response of hair cells to gentamicin, cultured cochleae were treated with gentamicin for 3 h, and immediately dissociated and FACS-sorted to obtain purified hair cell and non-hair cell samples (Figure 1C) for RNA sequencing. Since there is a low level of misexpression of GFP in inner phalangeal cells and probably in border cells, cells with low-level GFP expression were excluded by stringently gating GFP during FACS purification to minimize possible contamination. We validated the purity of the hair cell population by immunostaining, as more than 95% of purified GFP-positive cells stained positive for the hair cell marker MyoVI (data not shown). In addition, known hair cell specific genes, such as Atoh1, Pou4f3, Myo6, and Myo7a, have much greater normalized read numbers in hair cell samples than that in non-hair cell samples (Table S1), suggesting the high purity of our hair cell sample. The reliability of the RNA sequencing data was verified by Q-PCR analysis with primers specific for selected genes (Figures 2A, 3A, 4A, 5A, 6A).
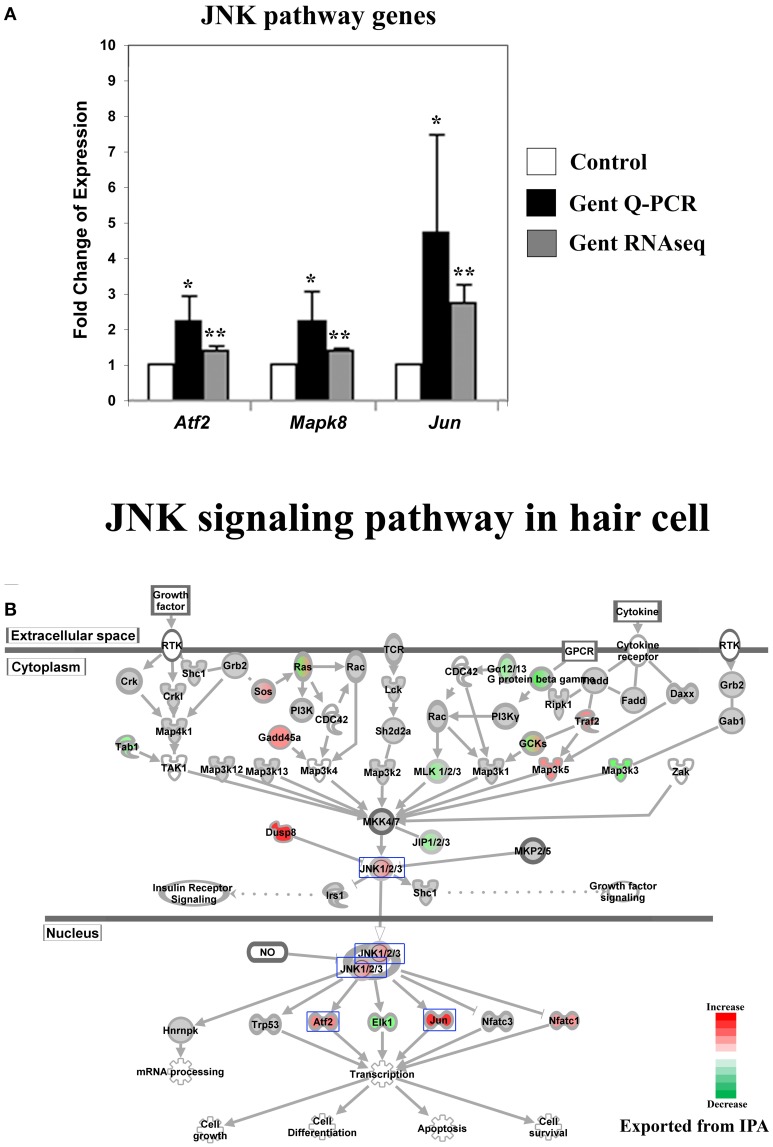
Figure 2.
Gentamicin-induced changes in expression of genes in the Jun-kinase (JNK) pathway. (A) Q-PCR results validating increased messenger RNA levels in hair cells after gentamicin treatment. Error bar, standard deviation: *p < 0.05; **p < 0.01. (B) The JNK signaling pathway with color-coded expression changes indicating significant transcriptional response to gentamicin. Only genes with significant expression changes are colored. Red, increased expression; green, decreased expression relative to control; half red-half green, indicates nodes with multiple genes with some genes upregulated and some genes downregulated. Genes validated and discussed in the text are boxed in the corresponding protein pathway diagrams.
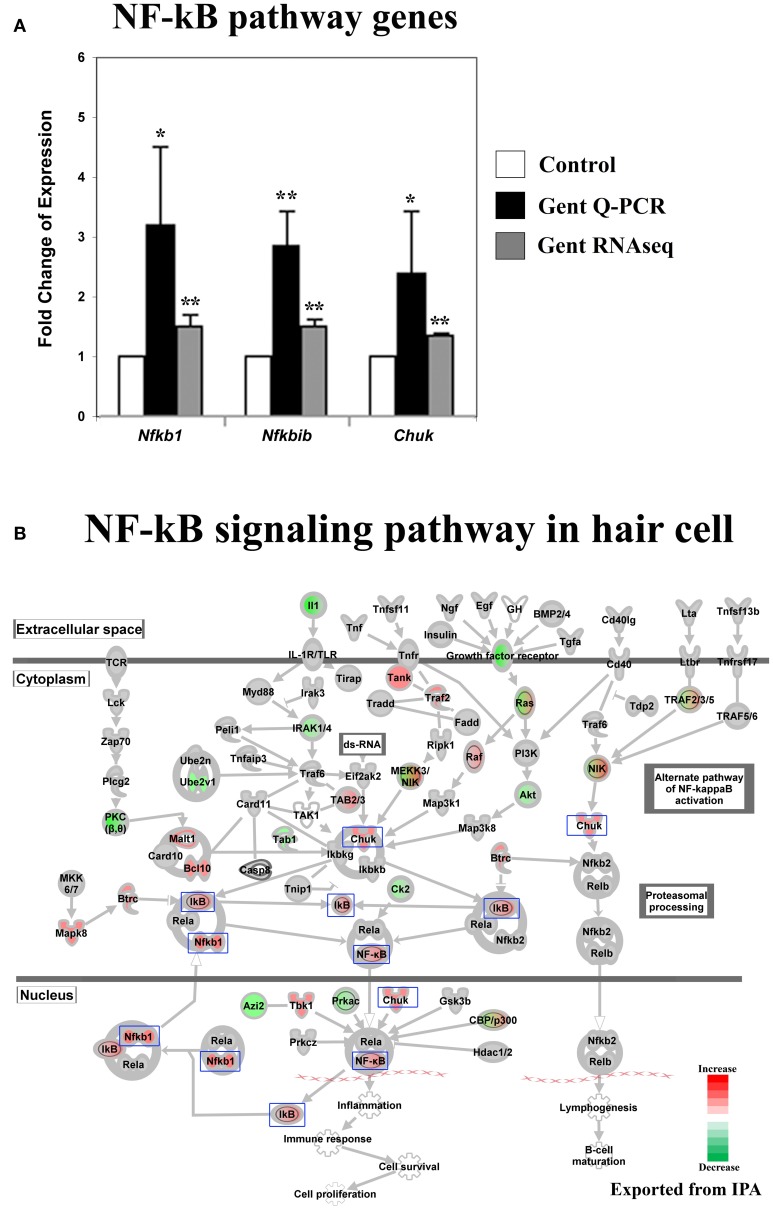
Figure 3.
Gentamicin-induced changes in expression of genes in the NF-κB pathway. (A) Q-PCR results validating increased messenger RNA levels in hair cells after gentamicin treatment. Error bar, standard deviation: *p < 0.05; **p < 0.01. (B) NF-κB pathway with color-coded expression changes suggesting early transcriptional response to gentamicin. Only genes with significant expression changes are colored. Red, increased expression; green, decreased expression relative to control; half red-half green, symbols for nodes consisting of multiple genes with some genes upregulated and some genes downregulated. Genes validated and discussed in the text are boxed in the corresponding protein pathway diagrams.
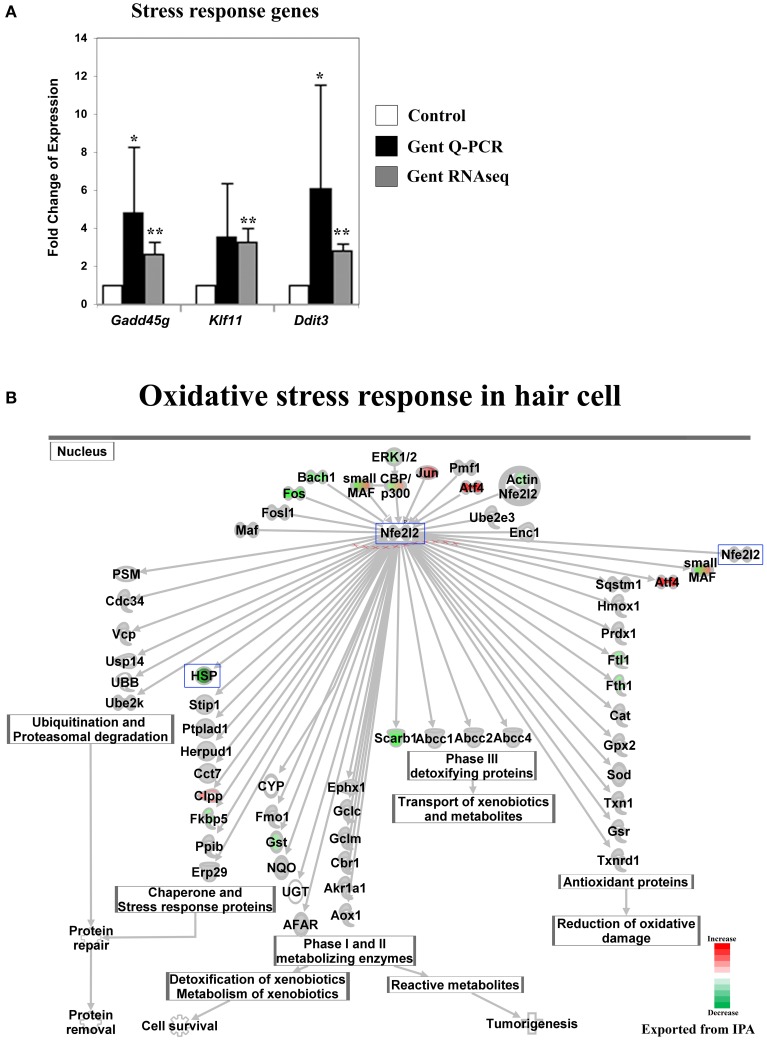
Figure 4.
Gentamicin-induced changes in expression of stress-response genes. (A) Q-PCR results validating increased messenger RNA levels in hair cells after gentamicin treatment. Error bar, standard deviation: *p < 0.1; **p < 0.01. (B) NRF2-mediated oxidative stress response network with color-coded expression shifts showing the absence of significant induction of oxidative stress response at the transcriptional level. Only genes with significant expression changes are color labeled. Red, increased expression; green, decreased expression relative to control; half red-half green, indicates nodes with multiple genes with some genes upregulated and some genes downregulated. Genes validated and discussed in the text are boxed in the corresponding protein pathway diagrams.
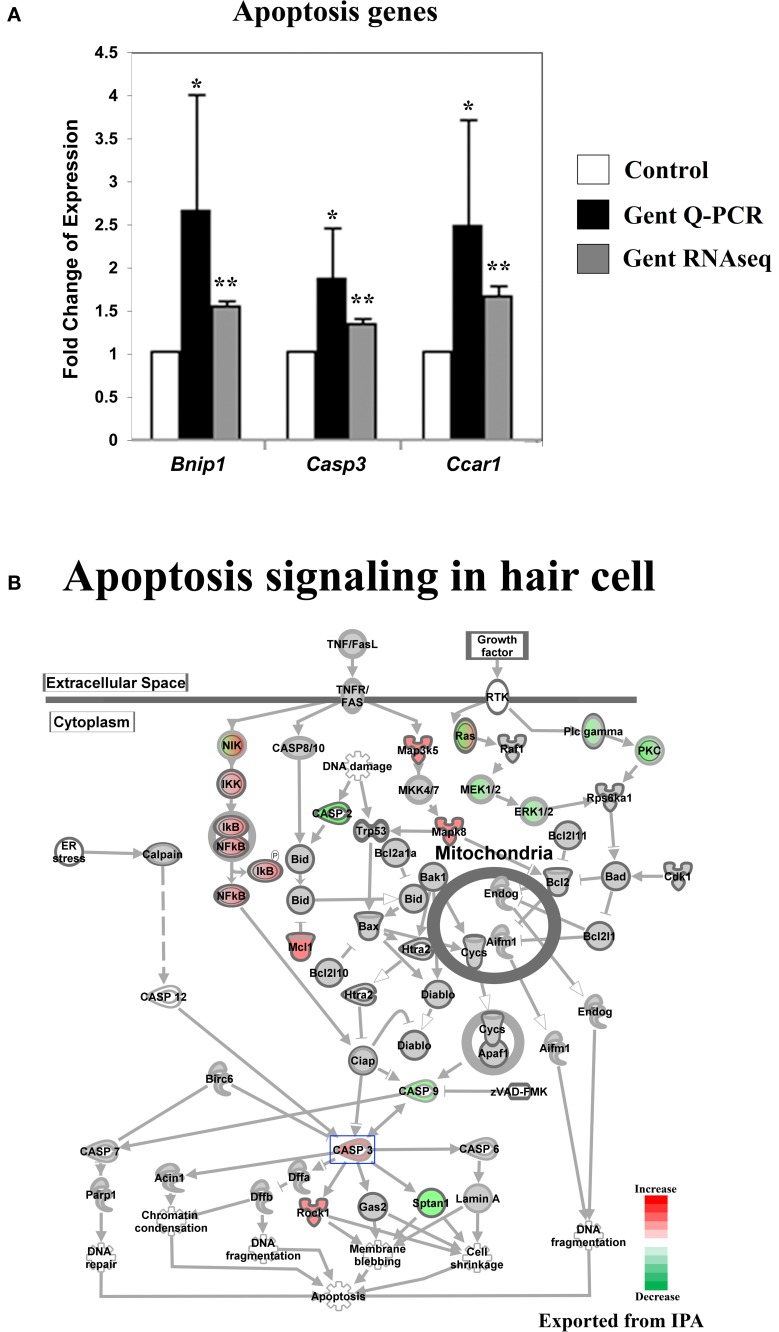
Figure 5.
Gentamicin-induced changes in expression of genes involved in apoptosis. (A) Q-PCR results validating increased messenger RNA levels in hair cells after gentamicin treatment. Error bar, standard deviation: *p < 0.05; **p < 0.01. (B) The apoptosis gene network with color-coded expression changes showing the absence of significant changes for apoptosis initiation and execution genes at transcriptional level. Only genes with significant expression changes are colored. Red, increased expression; green, decreased expression relative to control; half red-half green, indicates nodes with multiple genes with some genes upregulated and some genes downregulated. Genes validated and discussed in the text are boxed in the corresponding protein pathway diagrams.
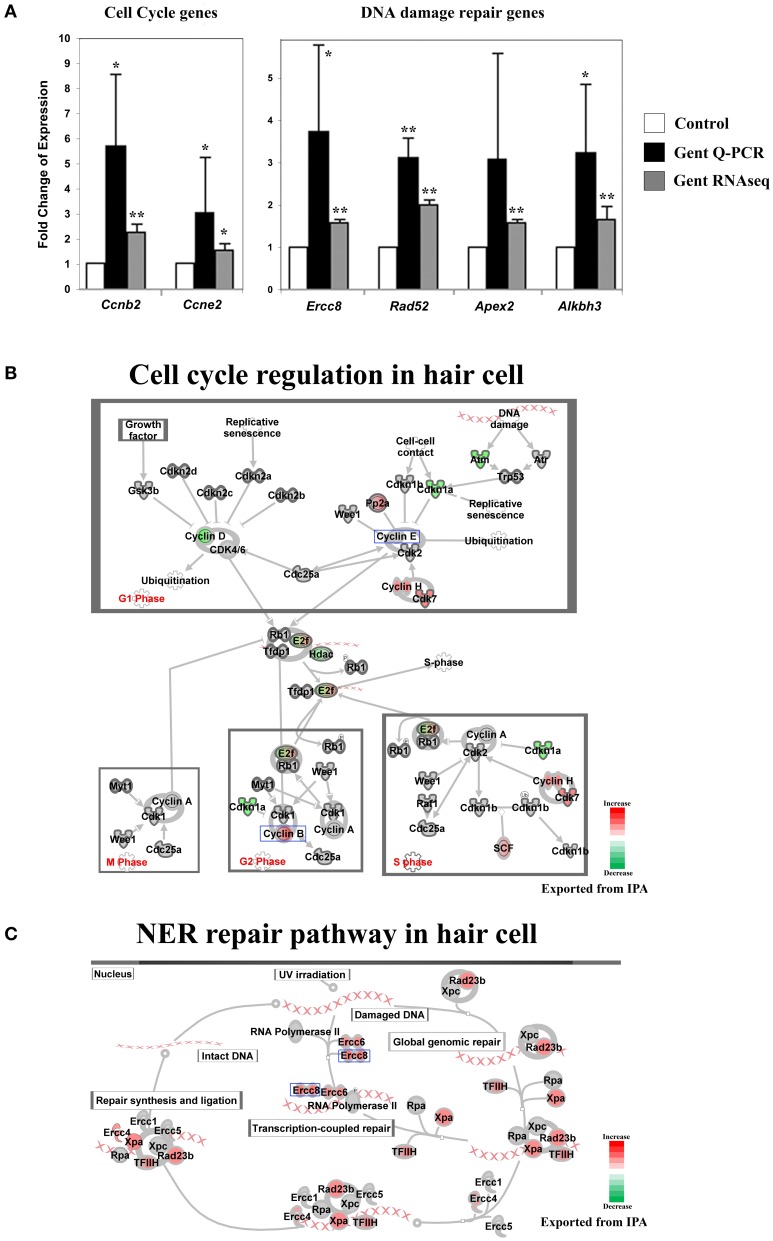
Figure 6.
Gentamicin-induced changes in expression of genes involved in cell cycle and DNA damage repair. (A) Q-PCR results validating increased messenger RNA levels of selected genes in hair cells after gentamicin treatment. These results confirm the up-regulation of cell cycle and DNA damage repair genes in gentamicin-treated hair cells. Error bar, standard deviation: *p < 0.05; **p < 0.01. (B) Cell cycle regulation network with color-coded expression changes suggesting affected cell cycle gene regulation by gentamicin. (C) Nucleotide excision repair (NER) network with color-coded expression changes showing significant induction of NER genes. Only genes with significant expression changes are colored. Red, increased expression; green, decreased expression relative to control; half red-half green, indicates nodes with multiple genes with some genes upregulated and some genes downregulated. Genes validated and discussed in the text are boxed in the corresponding protein pathway diagrams.
Expression of 23420 genes was quantified and analyzed with the Negative Binomial, Poisson or Lognormal Linear distribution model (gene specific analysis algorithm provided by PartekFlow) (Table S1). After 3 h of gentamicin treatment, the mRNA level of 3709 genes in hair cells was significantly altered, with 1917 genes found to be up-regulated and 1792 genes down-regulated (p-value < 0.01, fold change > 1.2 or < −1.2, and raw reads ≥100). In contrast, significantly fewer genes (698) were found to be affected with the same criteria in the non-hair cell population, with 436 genes up-regulated and 262 genes down-regulated.
Principal components analysis
Principal components analysis (PCA) is a method for analyzing the variance in high dimensional datasets, and to present variance in a few dimensions for easier interpretation (Ringnér, 2008). The PCA analysis of our RNA sequencing data shows that 71.88% variance in the dataset was captured within the three most significant principal components (PCs): 49.91% in PC1 (X axis), 13.53% in PC2 (Y axis), and 8.44% in PC3 (Z axis) (Figure 1D). These PCs correspond to different cell types and different treatment conditions.
In the PCA map, each biological replicate of different experimental groups is represented by one dot (Figure 1D). PCA analysis indicates that the variance among the replicates of FACS-purified hair cells is low. Three untreated hair cell replicates cluster tightly in the PCA map (red dots), as do three gentamicin-treated hair cell replicates (blue dots) (Figure 1D). This tight clustering indicates low statistical variance between replicates, and likely stems from the low cellular heterogeneity among FACS-purified replicates. In contrast, both treated (green dots) and untreated (yellow dots) samples from the non-hair cell replicates, which include all other cells in our dissected epithelial preparations, are scattered in the PCA map (Figure 1D), indicating the higher statistical variance among replicates of the non-hair cell samples. Hair cell samples and the non-hair cell samples (Figure 1D, red vs. yellow) lie at the two ends of the most significant variance axis (PC1, X axis), suggesting that the hair cell gene expression profile is significantly different from the non-hair cell population. Along the second variance axis (PC2, Y axis), gentamicin-treated hair cell samples are separated from untreated hair cell samples (Figure 1D, blue vs. red), indicating the significant gene expression shift in hair cells caused by gentamicin treatment at the 3 h time point. In contrast, the non-hair cell samples are not separated by gentamicin treatment (Figure 1D, green vs. yellow), suggesting that the gentamicin-induced gene expression shift in the non-hair cell population was not as consistent as the one in hair cells. Together, the PCA results suggest that gentamicin treatment induced significant and consistent gene expression changes in hair cells.
Signaling pathways and biological processes affected by gentamicin in hair cells
Gene expression in hair cells was affected significantly by gentamicin treatment at 3 h, as shown by the large number of deregulated genes and by the PCA results. To investigate the underlying biological pathways affected in hair cells upon gentamicin treatment, IPA software (Qiagen) was used to analyze affected signaling pathways after gentamicin treatment, and to compare our hair cell datasets with datasets derived from other cell lines and tissues. Pathways affected by gentamicin treatment at 3 h are listed in Table S2. Below we verify and discuss several of these pathways that have been reported to be important in aminoglycoside induced hair cell death, as well as proposing that elements of the cell cycle may provide an additional pathway, signaling apoptosis in hair cells.
Expression of JNK pathway-related genes was significantly affected by gentamicin
The JNK pathway has been strongly implicated in aminoglycoside-induced hair cell death. After aminoglycoside treatment, phosphorylation of JNK and c-Jun was observed in hair cells (Ylikoski et al., 2002), indicating activation of the JNK pathway. Furthermore, inhibition of the JNK pathway by pharmaceutical inhibitors has been shown to protect hair cells from aminoglycoside ototoxicity (Ylikoski et al., 2002), suggesting that JNK activation may be involved in mediating aminoglycoside-induced hair cell death.
Consistent with these findings, we observed changes in the expression of genes involved in JNK signaling pathways upon gentamicin treatment. We verified the expression changes of three genes in this pathway by Q-PCR, namely Atf2, Mapk8 (Jnk1), and Jun (c-Jun) (Figure 2A). IPA pathway analysis revealed that the expression of JNK pathway-related genes was significantly affected in gentamicin-treated hair cells (p = 0.000001). As shown in Figure 2B (color coded, see legend), Mapk8 (Jnk1) itself, in addition to several upstream and downstream factors including Jun, Atf2, Nfatc1, and Elk1, were changed at the transcriptional level in gentamicin-treated hair cells. Interestingly, the induction of Jun expression is regulated by phosphorylation of Jun itself, in an autoregulatory loop, by JNK (Angel et al., 1988); thus the increased mRNA expression of Jun indicates the likely activation of the JNK pathway. Transcriptional changes in other targets of Jun regulation are summarized in Table S3. The pattern of changed expression of JNK pathway-related genes is consistent with the timeline of critical signaling events for aminoglycoside-induced hair cell death, indicating that the JNK pathway is one of the early response pathways that are activated in hair cells after aminoglycoside treatment (Matsui et al., 2004).
Genes involved in the NF-κB pathway were expressed differentially after gentamicin treatment
Activation of the NF-κB pathway has been implicated as protective against aminoglycoside ototoxicity in several biochemical assays (Jiang et al., 2005). Aminoglycoside treatment has been shown to increase DNA binding activity of NF-κB, indicating activation of the NF-κB pathway by aminoglycoside antibiotics (Jiang et al., 2005). Additionally, reagents promoting NF-κB translocation into nuclei reportedly protect hair cells from aminoglycoside ototoxicity (Jiang et al., 2005).
Our RNA sequencing data show that the expression of genes involved in the NF-κB pathway was altered in hair cells after gentamicin treatment. Gentamicin-induced expression changes of Nfkb1 (p50), Nfkbib (Ikbb), and Chuk(Ikk1) were verified by Q-PCR (Figure 3A). Fold changes based on RNA sequencing were smaller but remained highly significant (p < 0.01). In IPA pathway analysis, the NF-κB pathway was identified as a significantly affected pathway in gentamicin-treated hair cells (p = 0.00004). Expression changes of genes involved in the NF-κB pathway are shown in Figure 3B. Among these genes, Nfkb1 and Nfkbib have been identified as target genes of NF-κB (Ten et al., 1992; Schreiber et al., 2006), and their increased expression strongly suggests the activation of the NF-κB pathway in gentamicin-treated hair cells. Transcriptional changes in other NF-κB targets are summarized in Table S3. Our expression data indicate that the NF-κB pathway, like the JNK pathway, is an early response pathway that is activated in hair cells by aminoglycosides.
No significant stress response was observed at the transcriptional level for NRF2 target genes at the 3 h time point
The prevailing hypothesis of aminoglycoside ototoxicity holds that aminoglycoside-induced ROS production causes oxidative stress, which triggers apoptosis in hair cells. Cellular ROS levels are reported to increase within minutes of aminoglycoside administration in vitro (Hirose et al., 1997), and antioxidant reagents are shown to protect hair cells from aminoglyocside ototoxicity (Schacht, 1999). In addition, the oxidative stress response mediated by the transcription factor NRF2 (Nfe2l2) has been implicated in aminoglycoside ototoxicity (Hoshino et al., 2011). Therefore, a response to oxidative stress at the transcriptional level was expected. Surprisingly, although some genes upstream of the Nrf2-dependent oxidative stress pathway were upregulated in response to gentamicin at 3 h, the transcriptional key regulator Nrf2 (Nfe2l2), was not affected, nor were its well-studied transcriptional targets (Ma, 2013) (Figure 4B and Table S3). We interpret this to mean that at this early time point, the Nrf2 oxidative stress pathway has not been fully activated. One explanation for this might be that at the 3 h time point, aminoglycoside-induced ROS production had not increased to a level sufficient to overwhelm the highly-developed antioxidant system in hair cells, and this hypothesis is supported by the observation that after the initial disruption, a new balanced redox state indicated by stable NAD(P)H fluorescence is reached within 15 min post-gentamicin treatment (Schafer and Buettner, 2001; Tiede et al., 2009). Alternatively, the catastrophic oxidative stress may be caused by temporally downstream events such as mitochondrial dysfunction.
Another group of genes, which drew our attention because of a lack of early response, were those encoding heat shock proteins. Expression of these genes was not induced by gentamicin at 3 h, including the HspA group (Hspa1a, Hspa1b, Hspa5, and others), the HspB group (Hspb1, Hspb2, and others), the HspC group (Hsp90aa1, Hsp90ab1, and Hsp90b1), and other members in the HspD, HspE, and HspG groups (Table S1). Among Dnaj family members, which are involved in oxidative stress response, only Dnaja2, Dnajb4, and some Dnajc genes were expressed at slightly higher levels (Table S1). The absence of strong induction of heat shock protein genes suggests that hair cells had not launched a broad stress response at 3 h, and indicates that transcriptional changes in signaling pathways, such as the JNK and the NF-κB pathways, at this time point were not simply caused by dysregulation of general transcription under stress conditions.
Genes involved in the initiation and execution of apoptosis were not significantly affected at the early time point
Aminoglycoside antibiotics are thought to induce hair cell death through an intrinsic mitochondrial apoptosis pathway (Forge and Li, 2000). Apoptosis is an active process that requires gene transcription and protein synthesis (Matsui et al., 2002). In contrast to genes involved in upstream pathways, such as the JNK and the NF-κB pathways, which regulate apoptosis and survival, genes involved in apoptosis initiation and execution were not significantly affected in gentamicin-treated hair cells at 3 h post treatment (Figure 5B). Although we observed up-regulation of Casp3 expression and down-regulation of Casp2 and Casp9 expression, the majority of intrinsic mitochondrial apoptosis pathway-related genes remained unchanged, including pro- and anti-apoptosis Bcl2 family members (Figure 5B). The lack of significant induction of apoptosis factors at this relatively early time point is consistent with the critical events timeline of aminoglycoside-induced hair cell death (Matsui et al., 2004). We verified the expression changes of Bnip1, Casp3 and Ccar1 by Q-PCR (Figure 5A).
We analyzed the cell-type specific response by comparing gentamicin-induced expression changes in hair cells, with gene expression changes during cell death in fibroblasts, neurons, and kidneys (based on published literature as curated in the IPA data base). Like hair cells, neurons are terminally-differentiated, non-dividing cells, but pathways regulating neuronal cell apoptosis are well characterized. We also emphasized the kidney because nephrotoxicity is another well-known sequela to aminoglycoside antibiotics treatment. These comparisons in IPA revealed that gentamicin-induced gene expression changes in hair cells are significantly different from the dataset derived from fibroblasts (p = 0.0000927 and activation z-score −1.81) (activation z-score is a quantitative measure of how similar the hair cell dataset is to the “comparing dataset”: larger positive number indicates there are more genes with expression changes in the same direction in both datasets, while larger negative number means there are more genes with changes in opposite direction between datasets; see Table 1 caption), but similar to datasets of neuronal cell death (p = 0.000280 and activation z-score 0.312), and cell death of kidney cells (p = 0.00576 and activation z-score 0.664). The observed similarity between aminoglycoside-induced hair cell death and neuronal cell death is not surprising, as hair cells and neurons are related types of terminally differentiated, excitable cells. The similarity between hair cells and kidney cells in response to aminoglycoside-induced stress will be discussed in Section: “Similarity between aminoglycoside ototoxicity and nephrotoxicity” found below.
Table 1.
Comparison of the hair cell dataset with known expression changes during cell death in other tissues and cell types.
| Comparing dataset | p-value | Activation z-score | # Genes |
|---|---|---|---|
| Cell death of kidney cells | 5.76E-03 | 0.664 | 73 |
| Cell death of kidney cell lines | 1.09E-03 | 0.579 | 67 |
| Neuronal cell death | 2.80E-04 | 0.312 | 149 |
| Cell death of brain cells | 4.10E-04 | 0.024 | 63 |
| Cell death of central nervous system cells | 6.80E-04 | 0.015 | 66 |
| Cell death of fibroblast cell lines | 2.71E-04 | −0.539 | 98 |
| Cell death of cerebellar cortex cells | 2.77E-05 | −0.574 | 17 |
| Cell death of tumor cell lines | 4.70E-06 | −0.656 | 331 |
| Cell death of lymphoma cell lines | 2.54E-03 | −1.148 | 46 |
| Cell death of fibroblasts | 9.27E-05 | −1.81 | 61 |
Selected entries are shown. The p-value, which is calculated using the right-tailed Fisher Exact Test by IPA, is a measure of the likelihood that the association between a set of focus genes in two datasets is due to random chance. Activation z-score is a quantitative measure of how similar the hair cell dataset is to the “comparing dataset”: larger positive number indicates there are more genes with expression changes in the same direction in both datasets, while larger negative number means there are more genes with changes in opposite direction between datasets. # Genes indicates number of overlapping genes in the comparison.
Gentamicin-induced changes in cell cycle and DNA damage repair genes indicate disruption of the postmitotic state
Another group of early response genes whose expression was significantly affected upon exposure to gentamicin includes those involved in cell cycle regulation and DNA damage repair. We verified several genes in this group, including Ccnb2 (Cyclin B2), Ccne2 (Cyclin E2), Ercc8 (Csa), Rad52, Apex2, and Alkbh3 (Figure 6A). Expression changes of genes involved in cell cycle regulation and nucleotide excision repair are shown in Figures 6B,C, respectively. Comparisons between gentamicin-induced gene expression changes and known transcription regulation of related genes during cell cycle or DNA damage repair are summarized in Tables 2, 3, respectively. These data suggest that the postmitotic state of hair cells was disrupted following gentamicin treatment. Loss of control of the postmitotic state and re-entry into the cell cycle preceding neuronal cell apoptosis has been observed (Arendt, 2000), and inhibition of cell cycle re-entry by blocking CDK activity has been shown to protect neurons from apoptosis induced by certain drugs or stress (Kruman, 2004; Golsteyn, 2005). Consistent with these observations in neuronal cells, aberrant cell cycle re-entry, induced by knocking out Cdkn1a (p21) and Cdkn2d (p19), leads to hair cell apoptosis (Laine et al., 2007). Inhibition of CDK2 activity also protects hair cells from gentamicin ototoxicity (Tao and Segil, unpublished data), suggesting that the cell cycle machinery plays a role in mediating hair cell death. Our RNA sequencing data showing transcriptional changes to cell cycle-related genes indicate a disruption of the postmitotic state, which supports the idea that loss of cell cycle control may underlie an additional pathway leading to hair cell death after gentamicin treatment.
Table 2.
Comparison of the hair cell dataset with known expression changes in datasets from different stages of cell cycle.
| Comparing dataset | p-value | Activation z-score | # Genes |
|---|---|---|---|
| G2 phase | 1.08E-06 | 0.964 | 69 |
| G2/M phase | 1.31E-03 | 0.507 | 46 |
| Cell cycle progression | 3.96E-07 | 0.199 | 237 |
| Mitosis | 2.51E-05 | −0.075 | 112 |
| G1 phase | 6.42E-04 | −0.401 | 86 |
| G1/S phase transition | 1.75E-03 | −0.506 | 46 |
| M phase | 2.37E-04 | −0.891 | 55 |
| S phase | 1.44E-04 | −1.122 | 63 |
| Interphase | 9.91E-08 | −1.334 | 160 |
| Entry into interphase | 4.35E-03 | −1.523 | 35 |
Selected entries are shown.
Table 3.
Comparison of the hair cell dataset with known expression changes in genes associated with the DNA repair process.
| Comparing dataset | p-value | Activation z-score | # Genes |
|---|---|---|---|
| Repair of DNA | 7.05E-06 | 1.953 | 66 |
| DNA damage response of cells | 6.09E-05 | 0.343 | 43 |
| DNA replication | 7.48E-05 | −0.523 | 61 |
| Metabolism of DNA | 6.37E-04 | 0.357 | 90 |
| Excision repair | 1.02E-03 | 2.756 | 20 |
Induction of DNA damage repair genes also supports the hypothesis that the postmitotic state is disrupted in hair cells after gentamicin treatment. It is plausible that the induction of DNA repair genes by DNA damage is caused by elevated ROS levels after gentamicin treatment, but our experimental observations do not support this explanation. First, the absence of significant induction of oxidative stress response genes suggests that the ROS production had not reached a level sufficient to induce the change in known response genes. Second, we failed to detect DNA damage by immunostaining against phosphorylated Chk, or by comet assay (Tao and Segil, unpublished data). Third, we did not observe induction of DNA damage repair genes at the transcriptional level after treating the cochlea with the DNA-damaging agent cisplatin (Ng, Rainey and Segil, unpublished data). Based on these observations, we postulate that up-regulated expression of DNA damage repair genes is not triggered by gentamicin-induced DNA damage and ROS production, but rather is more likely to reflect a change in cell cycle status following gentamicin treatment.
Gene expression changes induced by gentamicin in the non-hair cell population
Hair cells are specifically marked by GFP in Atoh1-GFP transgenic mice (Figure 1A), thus the GFP negative population consists of supporting cells (Deiters' cells, pillar cells and Hensen cells), cells in the greater epithelial ridge, cells in the lesser epithelial ridge, and other cells constituting surrounding tissues (inner phalangeal cells were excluded by FACS, based on their low-level misexpression of GFP, Figures 1A,C). The number of genes whose expression was affected by gentamicin in non-hair cell samples is much smaller than that in hair cells. The PCA results (Figure 1D) also show that the non-hair cell population does not respond to gentamicin uniformly, probably as a result of high heterogeneity. Expression of genes involved in cholesterol synthesis, lipid synthesis, nucleotide metabolism, and other biosynthesis processes, was significantly induced after gentamicin treatment in this population (Table 4). In addition, expression of genes involved in DNA replication and cell cycle was significantly affected (Table 4). The gentamicin-induced gene expression pattern of the non-hair cell population suggests gentamicin-stimulated biosynthesis, and induced cell proliferation in this population.
Table 4.
Comparison of non-hair cell dataset with known expression changes in biosynthesis and proliferation datasets.
| Comparing dataset | p-value | Activation z-score | # Genes |
|---|---|---|---|
| Synthesis of sterol | 4.76E-11 | 1.723 | 16 |
| Synthesis of cholesterol | 1.04E-10 | 1.937 | 14 |
| Metabolism of cholesterol | 3.01E-10 | 1.937 | 17 |
| Steroid metabolism | 1.38E-05 | 1.937 | 20 |
| Steroidogenesis | 2.79E-05 | 1.929 | 17 |
| Synthesis of terpenoid | 3.74E-05 | 2.075 | 20 |
| Metabolism of terpenoid | 5.42E-05 | 1.937 | 21 |
| Metabolism of membrane lipid derivative | 6.59E-05 | 1.66 | 25 |
| Conversion of acyl-coenzyme A | 9.34E-05 | 1.964 | 4 |
| Proliferation of cells | 3.80E-03 | 2.161 | 136 |
Selected entries are shown.
In the presence of Texas-Red conjugated gentamicin (GTTR) in culture medium, hair cells had much higher GTTR fluorescence than other cell types (Figure 1A), suggesting that gentamicin preferentially accumulates in hair cells. However, hair cells and supporting cells form a mosaic linked by tight epithelial junctions and other cell-cell contacts (Gulley and Resse, 1976). Communication between hair cells and supporting cells is continuous as a result of cell-cell contact, and changes in one cell type that result from the gentamicin treatment likely effect physiological change in the other. One such cell-cell signaling pathway significantly affected by gentamicin treatment in non-hair cells is the Notch signaling pathway. Dll1 and Jag2 are highly expressed as Notch ligands in hair cells, and Hes1, Hes5, Hey1, and Hey2 are expressed in supporting cells as Notch target genes (Zine and de Ribaupierre, 2002; Doetzlhofer et al., 2009); after gentamicin treatment, expression of Dll1 and Jag2 were down-regulated in hair cells, and Hes1, Hes5, Hey1, and Hey2 were also expressed at a lower level in the non-hair cell population (p = n.s. for Hes and Hey genes) (Table S1), indicating disrupted cell-cell signaling between hair cells and supporting cells. This observation supports the idea that the expression profile changes that we observe in the non-hair cell population may be brought about by changes in response to gentamicin in hair cells. However, we could not exclude the possibility that non-supporting cells in the GFP-negative population might be responding to gentamicin treatment directly at the transcriptional level.
Similarity between aminoglycoside ototoxicity and nephrotoxicity
Nephrotoxicity is another side effect of aminoglycoside antibiotics, and several publications catalog the changes in response to aminoglycoside antibiotics (Oh et al., 2006; Ozaki et al., 2010). A statistical comparison of gentamicin-induced hair cell gene expression changes and transcriptional profiles after gentamicin treatment in kidney identified a significant overlap between gene expression changes in gentamicin-treated hair cells and kidneys (p = 8.02E-08). In the dataset obtained from kidney, 211 genes were identified by microarray that were significantly changed after gentamicin treatment (Ozaki et al., 2010), and there were 50 genes in our hair cell dataset that were changed in the same direction as they were in kidney (Table 5).
Table 5.
Comparison of gentamicin-induced hair cell gene expression changes with gentamicin-induced expression changes in kidney.
| Gene symbol | Function Annotation | p-value in HC dataset | Fold Change in HC dataset | Findings in nephrotoxicity | Consistent |
|---|---|---|---|---|---|
| Ppp1r10 | Phosphatase | 5.84E-06 | 4.524746 | Upregulates | Yes |
| Ddit3 | Transcription factor | 2.62E-05 | 2.751878 | Upregulates | Yes |
| Jun | Transcription factor | 0.00245 | 2.732988 | Upregulates | Yes |
| Ccnl1 | RNA splicing | 4.46E-06 | 2.482795 | Upregulates | Yes |
| Gnl3 | p53 pathway regulation | 7E-06 | 2.127794 | Upregulates | Yes |
| Chka | Phospholipid synthesis | 6.28E-07 | 2.029311 | Upregulates | Yes |
| Ddx39 | RNA splicing | 4.98E-06 | 1.991224 | Upregulates | Yes |
| Nop58 | Ribosome biogenesis | 7.68E-08 | 1.941254 | Upregulates | Yes |
| Ppp1r15a | Phosphatase | 0.009454 | 1.739694 | Upregulates | Yes |
| Kti12 | Unkown | 0.008232 | 1.649551 | Upregulates | Yes |
| Ptbp1 | RNA splicing | 3.83E-05 | 1.636352 | Upregulates | Yes |
| Eed | Transcription repressor | 8.35E-06 | 1.601639 | Upregulates | Yes |
| Eif2b2 | Translation | 3.21E-06 | 1.572336 | Upregulates | Yes |
| Cul2 | Ubiquitin ligase | 7.77E-06 | 1.510931 | Upregulates | Yes |
| Zfp622 | Transcription regulator | 4.77E-06 | 1.483148 | Upregulates | Yes |
| Naa15 | N-acetyltransferase | 0.000102 | 1.461136 | Upregulates | Yes |
| Smn1 | RNA splicing | 0.000161 | 1.45206 | Upregulates | Yes |
| Phax | RNA export | 0.006074 | 1.438425 | Upregulates | Yes |
| Lig4 | DNA repair | 5.82E-05 | 1.41918 | Upregulates | Yes |
| Azin1 | Metabolism | 0.001597 | 1.388636 | Upregulates | Yes |
| Arf6 | Ras superfamily | 0.000976 | 1.388019 | Upregulates | Yes |
| 2310044G17Rik | Unkown | 0.005972 | 1.37605 | Upregulates | Yes |
| Eaf1 | Transcription factor | 0.000222 | 1.369313 | Upregulates | Yes |
| Cdk13 | Transcription elongation | 0.000244 | 1.365288 | Upregulates | Yes |
| Pafah1b1 | Inactivates platelet-act. factor | 0.000164 | 1.346669 | Upregulates | Yes |
| Gfer | Growth factor | 0.001933 | 1.324081 | Upregulates | Yes |
| Pelo | Mitosis | 0.000148 | 1.319478 | Upregulates | Yes |
| Tsc22d1 | Transcription regulator | 0.000157 | 1.315102 | Upregulates | Yes |
| Ddx50 | RNA helicase | 0.00147 | 1.313465 | Upregulates | Yes |
| Gadd45a | MAPK pathway regulator | 0.007003 | 1.30902 | Upregulates | Yes |
| Tars | tRNA synthesis | 0.001263 | 1.292011 | Upregulates | Yes |
| Mdm2 | p53 pathway regulation | 0.005424 | 1.28435 | Upregulates | Yes |
| Dnaja2 | Chaperone | 0.001429 | 1.282581 | Upregulates | Yes |
| Eif2b4 | Translation | 0.001361 | 1.270756 | Upregulates | Yes |
| Sfpq | RNA splicing | 0.000749 | 1.252767 | Upregulates | Yes |
| Cnksr3 | Sodium transport | 0.00051 | 1.240931 | Upregulates | Yes |
| Cand1 | Ubiquitin ligase | 0.003349 | 1.215329 | Upregulates | Yes |
| Mybbp1a | Transcription regulator | 0.000956 | 1.214954 | Upregulates | Yes |
| Ncl | Ribosome biogenesis | 0.00327 | 1.206809 | Upregulates | Yes |
| Decr2 | Metabolism | 0.003445 | −1.21269 | Downregulates | Yes |
| Asl | Urea cycle | 0.000314 | −1.27227 | Downregulates | Yes |
| Acsl3 | Metabolism | 0.000638 | −1.27564 | Downregulates | Yes |
| Gstm1 | Antioxidant | 0.00249 | −1.32453 | Downregulates | Yes |
| S100a1 | Calcium binding | 0.006299 | −1.37041 | Downregulates | Yes |
| Abat | Metabolism | 0.002967 | −1.38497 | Downregulates | Yes |
| Mrpl23 | Mitochondrial ribosome | 0.003565 | −1.38508 | Downregulates | Yes |
| Hacl1 | Metabolism | 0.002033 | −1.43919 | Downregulates | Yes |
| St6galnac3 | Glycolipid synthesis | 0.005244 | −1.46649 | Downregulates | Yes |
| Pcx | Metabolism | 0.00279 | −1.65712 | Downregulates | Yes |
| Eef1a1 | Translation | 2.35E-05 | −2.0177 | Downregulates | Yes |
| Zfp354a | Unkown | 0.001648 | 1.544937 | Downregulates | No |
| Hyou1 | Stress | 0.002956 | −1.22211 | Upregulates | No |
| Nedd4 | Ubiquitin ligase | 2.45E-06 | −1.27335 | Upregulates | No |
| Ctsd | Protease | 0.00174 | −1.33383 | Upregulates | No |
| Hspa5 | Stress | 0.004463 | −1.3884 | Upregulates | No |
| Calr | Calcium binding | 0.000413 | −1.4266 | Upregulates | No |
| Hsp90b1 | Chaperone | 5.87E-05 | −1.56328 | Upregulates | No |
| Egr1 | Transcription regulator | 0.004467 | −1.61013 | Upregulates | No |
| Zfp36l1 | Growth factor response | 0.001571 | −1.65211 | Upregulates | No |
| Pdia4 | Protein folding | 0.002689 | −1.72559 | Upregulates | No |
| Tubb5 | Cytoskeleton | 0.001836 | −2.0123 | Upregulates | No |
| Spp1 | Cell-matrix interaction | 0.004118 | −4.30391 | Upregulates | No |
We further compared the underlying biological processes and pathways that were affected by gentamicin in kidney and in hair cells, based on the transcriptional changes. We observed similar changes in the expression of genes involved in ribosome biogenesis, RNA processing, and translation in gentamicin-treated kidney and hair cells (Table 5). In addition, up-regulation of expression at the Jun gene, and induction of DNA repair gene Lig4 were observed in both hair cells and kidney. The similarity in transcriptional response to gentamicin between hair cells and kidney cells suggests that there may be some common mechanisms responsible for aminoglycoside ototoxicity and nephrotoxicity, such as translation inhibition.
Despite the aforementioned similarity, the expression of stress response genes differed greatly in gentamicin-treated kidney and hair cells. Expression of oxidative stress response genes and Hsp chaperone genes was collectively increased in gentamicin-treated kidney while expression of these genes was not induced in hair cells. The discrepancy might be based on the different time points of sample collection (3 h post-gentamicin treatment for hair cell sample vs. 7 days after treatment for kidney). It was difficult to directly compare the expression of proliferation genes in kidneys with that in hair cells, because the kidney is an organ consisting of heterogeneous cell populations, with both dividing cells and quiescent cells, whereas hair cells are postmitotic.
Discussion
The causes of hypersensitivity of hair cells to aminoglycoside antibiotics remain unclear, and the prevailing hypothesis of aminoglycoside ototoxicity holds that aminoglycosides induce high levels of ROS leading to apoptotic stimulation. Our purpose in this project was to characterize the hair cell response to aminoglycoside antibiotics at an early time following administration, before the changes associated with apoptosis were apparent. To this end, we analyzed the response of hair cells after 3 h of treatment with gentamicin. As indicated in Figures 2, 3, the JNK pathway and the NF-κB pathway are early responders, with many active components of the pathways showing significant increases. In addition, early changes in cell cycle-related genes, and DNA damage repair genes, signaled the potential disruption of the postmitotic state. In contrast, genes of the apoptosis and stress response pathways remain relatively unchanged at this time point. The transcription profile of hair cells at the early time point suggests that prior to the detection of an ROS response at the transcriptional level, other changes occur that may lead to the stimulation of apoptotic pathways, such as the aforementioned JNK pathway, NF-κB pathway and postmitotic state disruption.
Aminoglycoside-induced ROS production (Hirose et al., 1997), the protective effect of antioxidants (Schacht, 1999), and the NRF2-mediated oxidative stress response (Hoshino et al., 2011), have all been implicated in aminoglycoside ototoxicity. However, we did not observe a significant induction of the transcriptional targets of NRF2 at 3 h in our in vitro system. In addition, expression of heat shock chaperons and other stress response genes were not significantly upregulated. These results suggest that, although ROS changes in hair cells are rapid, at 3 h after gentamicin treatment they have not reached a level sufficient to trigger a broad stress response at the transcriptional level. The catastrophic oxidative stress that triggers apoptosis might be caused by continuous ROS build up, or by subsequent events such as mitochondrial dysfunction.
Expression of genes involved in initiation and execution of apoptosis were not significantly induced at this early time point, which is consistent with the established timeline of hair cell apoptosis (Matsui et al., 2004), and in line with the absence of a strong stress response at the transcriptional level. Together, the data suggest that, at this relatively early time point, hair cells are not yet committed to apoptosis. However, we observed expression changes for genes in upstream signaling pathways regulating apoptosis and cell survival. Not surprisingly, comparison of the observed gentamicin-induced changes in hair cells, with those from cell death studies in other cell lines or tissues, revealed a significant similarity in response to stress.
The disruption of the postmitotic state of hair cells is indicated by expression changes of cell cycle-related genes, suggesting that postmitotic state disruption in dying hair cells is another potential mechanism regulating apoptosis in hair cells, similar to what has been reported in postmitiotic neurons. Cell cycle re-entry has been observed in neurons before neuronal cell death, especially in neurodegenerative diseases (Arendt, 2000). In the central nervous system, mature neurons with mutated genes involved in maintaining the postmitotic state (inactivation of p19Ink4d and p27Kip1) lose control of the postmitotic state, re-enter the cell cycle, and eventually undergo apoptosis (Zindy et al., 1999). Activation of the cell cycle machinery is considered one of the mechanisms controlling programmed cell death in postmitotic neurons, either through direct regulation of apoptosis pathways, or through checkpoint activation (Kruman, 2004; Golsteyn, 2005). In the cochlea, aberrant cell cycle re-entry leads to hair cell apoptosis (Chen et al., 2003; Laine et al., 2007). Inhibition of CDK activity by pharmaceutical inhibitors or genetic mutations of CDK genes protect hair cells from aminoglycoside ototoxicity (Tao and Segil, unpublished data), suggesting that cell cycle machinery regulates hair cell death. This mechanism is also supported by our expression data. Genes involved in cell cycle progression, G2 phase, and G2/M were significantly affected at the transcriptional level (Table 2), indicating a disruption of the quiescent state of hair cells by gentamicin treatment.
Further evidence supporting the loss of cell cycle control mechanisms includes our observation that hair cells up-regulate the expression of DNA repair genes after gentamicin treatment. Although induction of DNA repair genes by gentamicin-induced DNA damage is a plausible explanation, the lack of strong oxidative stress response at the transcriptional level, failure of DNA damage detection in hair cells after gentamicin treatment (Tao and Segil, unpublished data), and unchanged DNA repair gene expression at the transcriptional level in hair cells after DNA damaging agent treatment (Ng, Rainey and Segil, unpublished data) together suggest that gentamicin-induced DNA repair gene expression is not responding to DNA damage, but may be responding to a form of “replicative stress” induced by the disruption of the postmitotic state, similar to that observed in neurons exiting the postmitotic state (Andrusiak et al., 2012). During final differentiation and withdrawal from cell cycle, many postmitotic cells, such as neurons, which need never replicate their genomes again and thus can resort to repairing only essential genes, down-regulate global genomic DNA repair and rely entirely on transcription-coupled repair to maintain actively transcribed genes (Nouspikel and Hanawalt, 2000). Based on the observation that hair cell homeostasis requires transcription-coupled repair for long-term survival (Nagtegaal et al., 2015), we postulate that DNA repair capacity is regulated during hair cell differentiation and cell cycle withdrawal. Thus, up-regulation of the expression of DNA repair genes may indicate disruption of the quiescent state and cell cycle control.
Taken together, our RNA sequencing data suggests that at 3 h, an early time point after gentamicin treatment, activation of the JNK and NF-κB signaling pathways and the disruption of the quiescent state, indicated by significant transcription changes, precede broad stress response and apoptosis initiation, and that these early responses may regulate aminoglycoside-induced hair cell death.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank USC Epigenome Center for library construction and sequencing services, Jane Johnson for Atoh1-GFP transgenic mice, Yibu Chen and Meng Li in USC NML Bioinformatics Service Program for their help with PartekFlow and IPA applications, Juan F. Llamas and Welly Makmura for animal care, and Robert Rainey and Greta Segil for careful reading of the manuscript. This project was supported by funds from NIH Grant R01DC007173, the Sidgmore Family Foundation, and the Hearing Health Foundation.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fncel.2015.00190/abstract
Gentamicin-induced differential gene expression in hair cells and in the non-hair cell population.
IPA canonical pathways that were affected by gentamicin in hair cells (pathways discussed in text are highlighted in yellow).
Gentamicin-induced expression changes of transcriptional targets of Jun, Nfkb1 and NRF2 (Nfe2l2). Red, significantly upregulated target genes; green, significantly downregulated target genes.
References
- Andrusiak M. G., Vandenbosch R., Park D. S., Slack R. S. (2012). The retinoblastoma protein is essential for survival of postmitotic neurons. J. Neurosci. 32, 14809–14814. 10.1523/JNEUROSCI.1912-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P., Hattori K., Smeal T., Karin M. (1988). The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell 55, 875–885. [DOI] [PubMed] [Google Scholar]
- Arendt T. (2000). Alzheimer's disease as a loss of differentiation control in a subset of neurons that retain immature features in the adult brain. Neurobiol. Aging 21, 783–796. 10.1016/S0197-4580(00)00216-5 [DOI] [PubMed] [Google Scholar]
- Chen P., Johnson J. E., Zoghbi H. Y., Segil N. (2002). The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 129, 2495–2505. [DOI] [PubMed] [Google Scholar]
- Chen P., Segil N. (1999). p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development 126, 1581–1590. [DOI] [PubMed] [Google Scholar]
- Chen P., Zindy F., Abdala C., Liu F., Li X., Roussel M. F., et al. (2003). Progressive hearing loss in mice lacking the cyclin-dependent kinase inhibitor Ink4d. Nat. Cell Biol. 5, 422–426. 10.1038/ncb976 [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A., Basch M. L., Ohyama T., Gessler M., Groves A. K., Segil N., et al. (2009). Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev. Cell 16, 58–69. 10.1016/j.devcel.2008.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forge A., Li L. (2000). Apoptotic death of hair cells in mammalian vestibular sensory epithelia. Hear. Res. 139, 97–115. 10.1016/S0378-5955(99)00177-X [DOI] [PubMed] [Google Scholar]
- Forge A., Schacht J. (2000). Aminoglycoside Antibiotics. Audiol. Neurootol. 5, 3–22. 10.1159/000013861 [DOI] [PubMed] [Google Scholar]
- Francis S. P., Katz J., Fanning K. D., Harris K. A., Nicholas B. D., Lacy M., et al. (2013). A novel role of cytosolic protein synthesis inhibition in aminoglycoside ototoxicity. J. Neurosci. 33, 3079–3093. 10.1523/JNEUROSCI.3430-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golsteyn R. M. (2005). Cdk1 and Cdk2 complexes (cyclin dependent kinases) in apoptosis: a role beyond the cell cycle. Cancer Lett. 217, 129–138. 10.1016/j.canlet.2004.08.005 [DOI] [PubMed] [Google Scholar]
- Gulley R. L., Resse T. S. (1976). Intercellular junctions in the reticular lamina of the organ of Corti. J. Neurocytol. 5, 479–507. 10.1007/BF01181652 [DOI] [PubMed] [Google Scholar]
- Hirose K., Hockenbery D. M., Rubel E. W. (1997). Reactive oxygen species in chick hair cells after gentamicin exposure in vitro. Hear. Res. 104, 1–14. 10.1016/S0378-5955(96)00169-4 [DOI] [PubMed] [Google Scholar]
- Hoshino T., Tabuchi K., Nishimura B., Tanaka S., Nakayama M., Ishii T., et al. (2011). Protective role of Nrf2 in age-related hearing loss and gentamicin ototoxicity. Biochem. Biophys. Res. Commun. 415, 94–98. 10.1016/j.bbrc.2011.10.019 [DOI] [PubMed] [Google Scholar]
- Huth M. E., Ricci A. J., Cheng A. G. (2011). Mechanisms of aminoglycoside ototoxicity and targets of hair cell protection. Int. J. Otolaryngol. 2011:937861. 10.1155/2011/937861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamesdaniel S., Ding D., Kermany M. H., Davidson B. A., Knight P. R., IIIrd., Salvi R., et al. (2008). Proteomic analysis of the balance between survival and cell death responses in cisplatin-mediated ototoxicity. J. Proteome Res. 7, 3516–3524. 10.1021/pr8002479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Sha S. H., Schacht J. (2005). NF-kappaB pathway protects cochlear hair cells from aminoglycoside-induced ototoxicity. J. Neurosci. Res. 79, 644–651. 10.1002/jnr.20392 [DOI] [PubMed] [Google Scholar]
- Kruman I. I. (2004). Why do neurons enter the cell cycle? Cell Cycle 3, 769–773. 10.4161/cc.3.6.901 [DOI] [PubMed] [Google Scholar]
- Laine H., Doetzlhofer A., Mantela J., Ylikoski J., Laiho M., Roussel M. F., et al. (2007). p19(Ink4d) and p21(Cip1) collaborate to maintain the postmitotic state of auditory hair cells, their codeletion leading to DNA damage and p53-mediated apoptosis. J. Neurosci. 27, 1434–1444. 10.1523/JNEUROSCI.4956-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin E. A., Collisson T., Parab P., Omer-Abdalla A., Haeberle H., Chen P., et al. (2003). Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3, 389–395. 10.1016/S1567-133X(03)00089-9 [DOI] [PubMed] [Google Scholar]
- Ma Q. (2013). Role of Nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 53, 401–426. 10.1146/annurev-pharmtox-011112-140320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui J. I., Gale J. E., Warchol M. E. (2004). Critical signaling events during the aminoglycoside-induced death of sensory hair cells in vitro. J. Neurobiol. 61, 250–266. 10.1002/neu.20054 [DOI] [PubMed] [Google Scholar]
- Matsui J. I., Ogilvie J. M., Warchol M. E. (2002). Inhibition of caspases prevents ototoxic and ongoing hair cell death. J. Neurosci. 22, 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagtegaal A. P., Rainey R. N., van der Pluijm I., Brandt R. M. C., van der Horst G. T. J., Borst J. G. G., et al. (2015). Cockayne syndrome group B (Csb) and group A (Csa) deficiencies predispose to hearing loss and cochlear hair cell degeneration in mice. J. Neurosci. 35, 4280–4286. 10.1523/JNEUROSCI.5063-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I., Bodmer M., Brors D., Bodmer D. (2004). Early gene expression in the organ of Corti exposed to gentamicin. Hear. Res. 195, 1–8. 10.1016/j.heares.2004.04.010 [DOI] [PubMed] [Google Scholar]
- Nouspikel T., Hanawalt P. C. (2000). Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol. Cell Biol. 20, 1562–1570. 10.1128/MCB.20.5.1562-1570.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. H., Park H. J., Lim J. S., Jeong S. Y., Hwang J. Y., Kim Y. B., et al. (2006). Gene expression profiling of early renal toxicity induced by gentamicin in mice. Mol. Cell Toxicol. 2, 185–192. 10.1002/jbt.20277 [DOI] [Google Scholar]
- Ozaki N., Matheis K. A., Gamber M., Feidl T., Nolte T., Kalkuhl A., et al. (2010). Identification of genes involved in gentamicin-induced nephrotoxicity in rats – A toxicogenomic investigation. Exp. Toxicol. Pathol. 62, 555–566. 10.1016/j.etp.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Ringnér M. (2008). What is principal component analysis? Nat. Biotechnol. 26, 303–304. 10.1038/nbt0308-303 [DOI] [PubMed] [Google Scholar]
- Schacht J. (1999). Antioxidant therapy attenuates aminoglycoside-induced hearing loss. Ann. N. Y. Acad. Sci. 884, 125–130. [PubMed] [Google Scholar]
- Schafer F. Q., Buettner G. R. (2001). Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 30, 1191–1212. 10.1016/S0891-5849(01)00480-4 [DOI] [PubMed] [Google Scholar]
- Schreiber J., Jenner R. G., Murray H. L., Gerber G. K., Gifford D. K., Young R. A. (2006). Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc. Natl. Acad. Sci. U.S.A. 103, 5899–5904. 10.1073/pnas.0510996103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyger P. S., Peters S. L., Rehling J., Hordichok A., Dai C. F. (2003). Uptake of gentamicin by bullfrog saccular hair cells in vitro. J. Assoc. Res. Otolaryngol. 4, 565–578. 10.1007/s10162-003-4002-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten R. M., Paya C. V., Israël N., Le Bail O., Mattei M. G., Virelizier J. L., et al. (1992). The characterization of the promoter of the gene encoding the p50 subunit of NF-kappa B indicates that it participates in its own regulation. EMBO J. 11, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiede L., Steyger P. S., Nichols M. G., Hallworth R. (2009). Metabolic imaging of the organ of corti–a window on cochlea bioenergetics. Brain Res. 1277, 37–41. 10.1016/j.brainres.2009.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009). RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikoski J., Liang X. Q., Virkkala J., Pirvola U. (2002). Blockade of c-Jun N-terminal kinase pathway attenuates gentamicin-induced cochlear and vestibular hair cell death. Hear. Res. 166, 33–43. 10.1016/S0378-5955(01)00388-4 [DOI] [PubMed] [Google Scholar]
- Zindy F., Cunningham J. J., Sherr C. J., Jogal S., Smeyne R. J., Roussel M. F. (1999). Postnatal neuronal proliferation in mice lacking Ink4d and Kip1 inhibitors of cyclin-dependent kinases. Proc. Natl. Acad. Sci. U.S.A. 96, 13462–13467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zine A., de Ribaupierre F. (2002). Notch/Notch ligands and Math1 expression patterns in the organ of Corti of wild-type and Hes1 and Hes5 mutant mice. Hear. Res. 170, 22–31. 10.1016/S0378-5955(02)00449-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gentamicin-induced differential gene expression in hair cells and in the non-hair cell population.
IPA canonical pathways that were affected by gentamicin in hair cells (pathways discussed in text are highlighted in yellow).
Gentamicin-induced expression changes of transcriptional targets of Jun, Nfkb1 and NRF2 (Nfe2l2). Red, significantly upregulated target genes; green, significantly downregulated target genes.