Abstract
BACKGROUND
Owing to their minimal size, high production yield, versatility and robustness, the recombinant variable domain (nanobody) of camelid single chain antibodies are valued affinity reagents for research, diagnostic, and therapeutic applications. While their preparation against purified antigens is straightforward, the generation of nanobodies to difficult targets such as multi-pass or complex membrane cell receptors remains challenging. Here we devised a platform for high throughput identification of nanobodies to cell receptor based on the use of a biotin handle.
METHODS
Using a biotin-acceptor peptide tag, the in vivo biotinylation of nanobodies in 96 well culture blocks was optimized allowing their parallel analysis by flow cytometry and ELISA, and their direct used for pull-down/MS target identification.
RESULTS
The potential of this strategy was demonstrated by the selection and characterization of panels of nanobodies to Mac-1 (CD11b/CD18), MHC II and the mouse Ly-5 leukocyte common antigen (CD45) receptors, from a VHH library obtained from a llama immunized with mouse bone marrow derived dendritic cells. By on and off switching of the addition of biotin, the method also allowed the epitope binning of the selected Nbs directly on cells.
CONCLUSIONS
This strategy streamline the selection of potent nanobodies to complex antigens, and the selected nanobodies constitute ready-to-use biotinylated reagents.
GENERAL SIGNIFICANCE
This method will accelerate the discovery of nanobodies to cell membrane receptors which comprise the largest group of drug and analytical targets.
Keywords: in vivo biotinylation, flow cytometry, immunoprecipitation, nanobody, phage display, cell receptor
INTRODUCTION
The generation of potent antibodies for research, diagnosis and therapy has been facilitated by the progress in methods for building and selecting large antibody libraries [1, 2]. In addition to conventional heavy/light chain antibody libraries, there has been a growing interest in single domain antibody (sdAb) libraries. These libraries derived from a special type of antibodies that occur in camelids [3] and some species of sharks [4]. These camelid heavy-chain-only antibodies are devoid of light chain and their variable domain shows a high degree of identity with the human VH3 family [5], which has been regarded as an advantageous property for their application as human therapeutic agents [6]. The sdAb antigen binding site sits entirely in the heavy chain variable domain (VHH) and thus the VHH recombinant protein, also referred as nanobody (Nb), represents the smallest antibody fragment (~15 kDa) that retains the parent functional specificity, being half the size of conventional scFv. Recombinant VHHs can be produced with high expression yields in E. coli, as soluble and highly stable proteins, which has popularized their applications [7].
Despite the fact that the VHH antigen binding site is formed by only three and not six CDRs as in heterotetrameric antibodies, sdAbs bind their cognate antigens with similar affinity than conventional antibodies [8]. Although VHHs with specificity for a great variety of antigens have been isolated, including all type of macromolecules and haptens [7, 9], the convex architecture of the sdAb paratope with an extended CDR3 appears to have evolved mainly to interact with cavities on the antigen surface. These are typically found at the active site of enzymes, and hence several sdAb have been shown to affect the catalytic activity of enzymes [10, 11]. This concave topography is also found in many cell receptors, which together with the salient properties of nanobodies make VHH libraries a very attractive source of cell-receptor agonists/antagonists.
VHH libraries are easy to build, and in fact a comprehensive representation of the animal’s original pool of specificities can be accomplished because, conversely to conventional heterodimeric libraries, there is not shuffling of the heavy and light chains [7]. However, a common scenario during the selection of these complex libraries is how to further select a particular antibody with the desire attributes from the large number of positive clones resulting from the initial enrichment. This is more challenging when the antibody target is part of a complex antigen, such as is the case of a membrane protein on the cell surface. Indeed, due to the fact that VHH antigen recognition is highly sensitive to conformational changes in the target antigen [12], the conventional approach of using recombinant membrane proteins for immunization and selection can often lead to the lack of cross-recognition of the native cell receptor if proper measures to replicate its native conformation are not adopted [12]. In addition, for receptor discovery, whole cells have to be used for immunization and panning and thus there is a need for simple methods that enable the characterization of a large number of clones in a simple and systematic fashion, facilitating the identification of the cognate cell receptor even when this is unknown.
To address these limitations we optimized a methodology for the production of in vivo biotinylated nanobodies (BtNb) that facilitates their characterization by ELISA, flow cytometry and pull-down experiments, which is amenable for high-throughput screening, figure 1. The metabolic biotinylation of nanobodies has previously been used for diagnostic applications [13, 14] or for their oriented immobilization in microarrays [15]. The binding of biotin to avidins (avidin/streptavidin) in solution is regarded as one of the strongest non-covalent interactions (KD of ~10−15 M). Although the conjugation of biotin through its carboxyl group is accompanied by a reduction of this affinity [16], the biotin tag provides readily and strong binding to acceptor avidins which has given rise to a profusion of avidin/streptavidin bioconjugates for countless applications [17, 18]. Our labelling approach makes use of the biotin ligase BirA of E. coli which specifically conjugates biotin to the side chain of a Lys residue within a 15 mer acceptor peptide (BtAP) tag [19]. There is only one natural substrate of BirA, the biotin carboxyl carrier protein (BCCP) of E. coli is a minor component of the bacterial cell extract [20] and does not interfere with the intended use of the BtNb. Besides facilitating the isolation of nanobodies against complex targets, the selected antibodies can straightaway be produced in large amounts as biotin-labelled ready-to-use reagents.
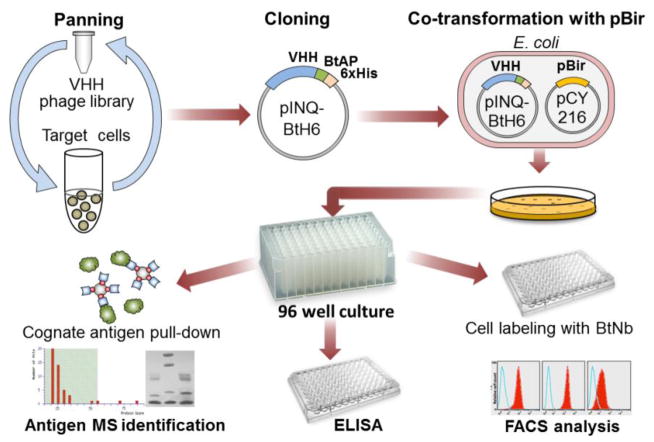
Figure 1. Schematic diagram of the method for the isolation of biotinylated nanobodies to cell receptors.
After panning on cells, the VHH output is cloned en masse in the expression vector pINQ-BtH6 which adds the biotin acceptor peptide. This sub-library of VHH is then co-transformed in E. coli together with pCY216 for over expression of the biotin ligase BirA. Individual clones are then culture in 96 deep well blocks and the soluble cell extract containing the biotinylated Nb can be characterized by different methods in a high throughput fashion.
METHODS
Construction of the pINQ-BtH6 vector
A triclocarban (TCC) specific VHH (T7) cloned between the two SfiI sites of the pComb3 vector [21] was used as template for PCR amplification of the OmpA-SfiI(1)-VHH-SfiI(2) region using the forward aatatctagaaataattttgtttaactttaagaaggagatataccatgaaaaagacagctatcgcgattg and reverse atttctcgagttcgtgccattcgattttctgagcctcgaagatgtcgttcagaccgccaccttggccggcctggcctgaggagacg primers. Upstream of the annealing sequence, the forward primer contained the ribosomal binding site (rbs) of the pET 28a(+) vector (Noavagen) including the XbaI restriction site (underlined). Similarly, the 5′ of the reverse primer contained the coding sequence for the peptide BtAP and the XhoI restriction site (underline). To assemble the pINQ-BtH6 vector, the amplicon was digested with XbaI and XhoI and cloned into the pET 28a(+). The cloning/expression region of pINQ-BtH6 is shown in figure S-2 (supporting information).
In vivo biotinylation of nanobodies
The VHH genes were cloned in the pINQ-BtH6 vector using the SfiI sites and the resulting plasmid was transformed in E. coli BL21(DE3) (Novagen) carrying the plasmid pCY216 [22]. The transformed cells were then seeded in LB (Luria-Bertani) agar plates supplemented with 35 μg/ml of chloramphenicol and 50 μg/ml of kanamycin. Single colonies were grown in 96 deep well culture plates (grainer) in 500 μL of LB supplemented with kanamycin/chloramphenicol in the presence of 0.04 % arabinose and 100 μM D-Biotin at 250 rpm, 37°C. When the OD600 reached 0.6 AU, IPTG was added to a final concentration of 3 μM. After 4 hours the plate was centrifuged and the cell pellets were resuspended in 100 μL of PBS (phosphate buffer saline). To extract the soluble VHHs, the cells were disrupted by three cycles of freezing and thawing or with B-PER Bacterial Protein Extraction reagent (Pierce).
Generation and culture of bone marrow derived-dendritic cells (BMDC)
Murine bone BMDCs were prepared from Balb/c bone marrow precursors by in vitro differentiation in the presence of GM-CSF using standard methods [23]. Briefly, the bone marrow from mouse tibias and femurs was disaggregated by passage through a nylon mesh. Red blood cells were flash lysed with sterile water, the remaining cells were counted and 3 × 106 cells placed in petri dishes (Grainer, Germany) containing 10 mL of DC-media consisting of RPMI supplemented with 10% heat inactivated fetal calf serum (FCS), 2 mM L-glutamine, 500 nM mercapto-ethanol, 100 U/ml penicillin/streptomycin, 20 μg/ml gentamycin, and 2 ng/mL of recombinant murine GM-CSF produced in our lab. Three days later, 10 mL of DC-media was carefully added to the dishes, and on day 6, 10 mL of media was refreshed. On day 7, the purity of the cells was analyzed by flow cytometry as shown in figure S-1 (Supporting information).
Llama immunization and phage display library construction
An adult female llama (Lama glama) from the Montevideo municipal zoo was immunized subcutaneously with 108 BMDC cells in 1mL of PBS (without adjuvant), which was followed by 6 booster injections every 2 weeks. Fifteen days after the last booster 150 mL of blood was collected in blood collection bags with anticoagulant. All activities were performed in accordance with protocols reviewed and approved by the Lecocq Zoo ethical committee. Peripheral blood lymphocytes were isolated by centrifugation on Histopaque 1077 gradients (Sigma) following the manufacturer’s instructions. Ten million cells were then treated with TRIZOL reagent (Invitrogen, Carlsbad) to extract total RNA, and after purification, it was reverser transcribed using the superscript III first-strand synthesis system (Invitrogen) and the primer JH (cca cga ttc tgg ccg gcc tgg cct gag gag acr gtg acc tgg gtc c). The cDNA was then used as template for PCR amplification of the VH and VHH genes using the forward primers, VH1 (catgccatgactcgcggcccaggcggccatggcccaggtgcagctggtgcagtctgg), VH3 (catgccatgactcgcggcccaggcggccatggccgaggtgcagctggtggagtctgg), VH4 (catgccatgactcgcggcccaggcggccatggcccaggtgcagctgcaggagtcggg) and the reverse primer JH [24]. These primers introduce two different SfiI sites (underlined) that are used for subsequent cloning of the fragments into the phagemid pComb3X vector, a kind gift from Dr. Barbas (The Scripps Research Institute, La Jolla, USA). The ligated vector was electroporated in ER2738 E. coli cells (F′proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10(TetR)/fhuA2 glnV Δ(lac-proAB) thi-1 Δ(hsdS-mcrB)5) (Lucigen Corporation, Middleton, WI, USA). The transformed cells were culture in LB containing 0.25% K2HPO4, 0.1% MgSO4, 0.1% glucose and 100 μg/mL ampicillin. After starting the exponential growth, bacterial cells were super-infected with helper phage M13KO7 (Pharmacia Biotech) and cultured overnight in the presence of 40 μg/mL of kanamycin. Phage was precipitated with 0.2 volumes of 20% polyethylene glycol 8000, 2.5M NaCl, on ice during 1 hour, centrifuged and the collected phage resuspended in PBS to a final titer of 1012 cfu/mL.
Library selection
For negative selection, 600 μl of the phage library (1012 cfu) were added to 4.4 mL DC-media and were incubated with 107 fibroblast (L-929, ATCC) in 75 cm2 culture flasks at 4°C for 1h, with gentle rocking. The supernatant was filtered by 0.2 μm and was further incubated with 2 ×107 BMDC at 4°C for 1h, and were extensively washed with cold DC-media. The cell surface bound phages were stripped by incubation (10 min) with 100 mM glycine, 0.15M NaCl, pH 3.0. After neutralization with 2 M tris base, the eluted phage was amplified and used for additional rounds of panning. After the third round of panning, the output phage was used to infect ER2738 E. coli cells to obtain the phagemid DNA. The purified DNA was digested with SfiI at 50°C for 4 h and the released VHH fragments were gel purified and cloned en masse into the pINQ-BtH6 vector. The ligated plasmid was then electroporated in BL21(DE3) containing the pCY216 vector, and the cells were seeded in kanamycin/chloramphenicol LB agar plates. The next day, single colonies were picked and grown in 96 deep well culture plates (Grainer) as described above.
ELISA Protocol
ELISA plates (Nunc Maxysorp) were coated with 100 μL/well of 1 μg/mL of TCC-BSA, in phosphate-buffered saline (PBS) overnight at 4 °C. After blocking for one hour at room temperature with 1% BSA, dilution of the biotinylated VHH or bacterial soluble extract were incubated for 1 h, followed by washing and incubation with streptavidin HRP for 1 h.
After extensive washing, the peroxidase activity was developed by adding 100 μL of peroxidase substrate ( 0.4 mL of, 6 mg of 3,3′,5,5′-tetramethylbenzidine in 1 mL of DMSO + 0.1 mL of 1% H2O2 in water, in a total of 25 mL of 0.1M acetate buffer, pH 5.5) and incubated for 20 min at room temperature. The enzyme reaction was stopped by the addition of 50 μL of 2 N H2SO4, and the absorbance was read at 450 nm on a Fluostar Optima Reader (BMG, Ortenberg, GE).
Flow Cytometry
Cells (2 × 105 100 μL) were incubated for 1h 4°C with the BtNb present in the soluble fraction of the freeze/thaw extract. Cells were then stained with phycoerythrin- or FITC-streptavidin using standard protocols and flow cytometry parameters were acquired on a FACScalibur (BD Bioscience), followed by analysis with FlowJo (TreeStar). All other antibodies used are from BD Bioscience.
Pull-down experiments and MALDI-TOF identification
A BMDC cell extract was prepared by disrupting the cells with PBS, 1% Triton X100 (PBS-Tx). The clean supernatant was then adsorbed with an excess of streptavidin-agarose (Pierce) and saved for the pull-down experiments. The VHH clones were grown in 96 deep well plates as described and the cell pellet were disrupted with 50 μL of B-PER, diluted to 500 μL with PBS and centrifuged. The soluble fraction was combined with 50 μL of streptavidin-agarose slurry (Pierce) for 30 min with gentle rocking. After washing with PBS-Tx the streptavidin beads were combined with 5 mL of the BMDC cell extract (corresponding to 108 cells) and the mix was kept at 4°C for 4h with gentle rocking. After washing with PBS-Tx, the beads were resuspended in SDS-PAGE loading buffer and were run in 10% or 12.5% gels. After staining with coomassie R-250, the bands were excised and trypsin digested for MALDI-TOF analysis. The tryptic peptides were extracted from the band and analyzed by MALDI-TOF MS with α-cyanocinnamic acid as matrix. A Microflex LR MALDI-TOF (Bruker Daltonics, Billerica, MS, USA) with a 337 nm nitrogen laser was operated in positive ion reflectron mode. Calibration was performed with the peptide calibration standard mix (Bruker Daltonics). The monoisotopic mass list was then searched with Mascot (Matrix Science), with unrestricted proteins mass and taxonomy parameters, allowing one missed cleavage, and using a peptide mass tolerance of ± 0.1 Da or less.
RESULTS AND DISCUSSION
Optimization of the metabolic biotinylation of VHHs
Figure 1 shows the general scheme of the integrated method for the characterization of Nb raised against cell receptors. While some phagemids in combination with amber suppressor bacterial strains allow direct expression of the selected VHHs, it is a common practice to sub-clone them into strong expression vectors to produce them in large amount for their biochemical characterization and final use. When this step is done after the initial selection of clones, each insert has to be sub-cloned individually, which is time consuming and non-compatible with a high throughput method. To overcome this limitation, the initial step in our method consists of the en masse transfer of the pool of panned VHHs from pComb3 to the receptor vector pINQBtH6. The basis for the construction of pINQBtH6 was the pET28a+ vector which allows high levels of expression under the control of the T7 promoter. The pET 28a(+) vector was modify to encode the OmpA signal peptide for periplasmic expression of VHHs, and the BtAP acceptor peptide for site-specific labelling with biotin and a hexa-histidine tag for purification. The conditions for efficient antibody expression were adjusted using VHH T7, a nanobody against the bactericide triclocarban (TCC), as a model VHH [9]. The pINQ-BtH6 vector encoding this nanobody was electroporated into E. coli BL21(DE3) carrying the plasmid pCY216. Initially, the culture conditions (temperature, induction time and IPTG concentration) were adjusted to maximize the expression of soluble VHH (not shown) and were used as starting point for the optimization of the BtNb expression.
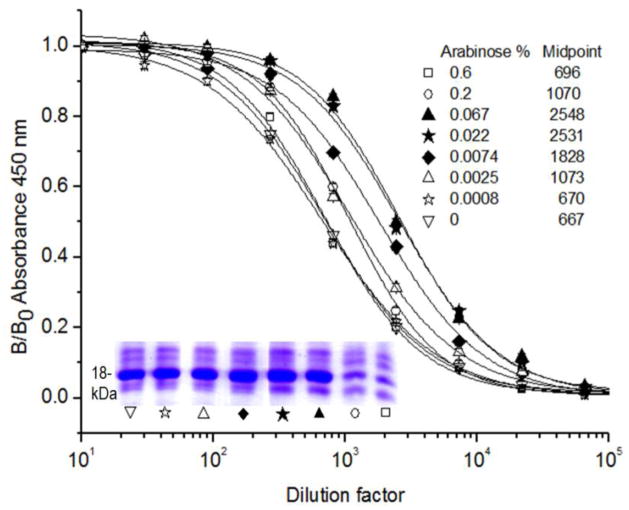
Next, we examined the factors affecting the biotinylation of VHH T7. First the concentration of arabinose used for induction of BirA was tested in the 0.0008–0.6 % range, in the presence of 100 μg/mL of biotin and 3 μM IPTG, figure 2. High concentrations of arabinose affected the expression of VHH T7. The extent of Nb biotinylation was indirectly assessed assuming similar initial concentrations of VHH T7 in all extracts (not true when 0.2 and 0.6% arabinose was used) and analyzing the reactivity of VHH T7 against TCC-BSA by an ELISA developed with streptavidin-HRP. The highest peroxidase activity (higher degree of biotinylation) was found when arabinose concentrations of 0.022 to 0.067% were used, and thus, 0.04% was adopted for all following experiments. Figure S-3a (Supporting information) shows that, in general, there is a good level of expression when randomly picked VHH clones were expressed using these conditions. The expression of VHH T7 (and similarly, for other clones) was not affected by the use of the BtAP tag (figure S-3b), and thus, the yield of the biotinylated version of the VHHs was essentially the same that was obtained when they were expressed with other peptide tags. This yield was typically between 2 and 30 mg/L in shaker flasks, depending on the individual VHH sequence.
Figure 2. Effect of arabinose in the expression and biotinylation of VHH T7.
Different concentrations of arabinose were used to induce the co-expression of VHH T7 and BirA as described in the legend. The graph displays the reactivity of serial dilutions of the VHH T7 soluble extract with its cognate antigen (TCC-BSA) detected with a streptavidin-HRP conjugate. The midpoints of the curves are listed for comparison. The SDS-PAGE analysis of VHH T7 expression in the soluble freeze-and-thaw extract is shown in the insert.
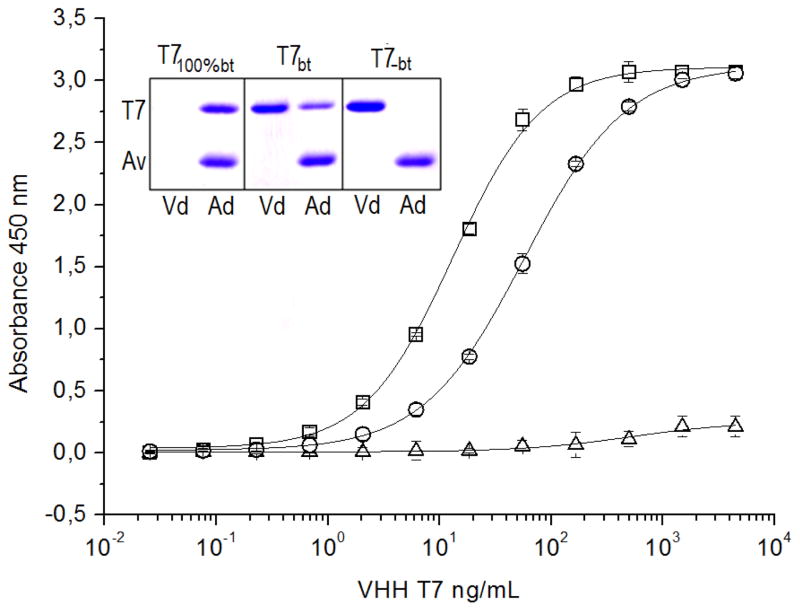
To estimate the degree of biotinylation, VHH T7 was similarly produced and was purified on Ni-NTA agarose (T7bt). Out of this, a 100% biotinylated fraction (T7100%bt) was also prepared using immobilized monomeric-avidin agarose (Pierce, Rockford), which allows gentle elution of biotinylated conjugates. As a negative control VHH T7 was also produced without addition of biotin (T7−bt). The ELISA results shown in figure 3 revealed that only a fraction, ~22% (Δmidpoint), of T7bt is biotinylated, which is in agreement with the SDS-PAGE analysis of the non-bound and bound fractions obtained from avidin-agarose gel. The absence of added biotin resulted in negligible biotinylation.
Figure 3. Analysis of the extent of the in vivo biotinylation of VHH T7.
T7bt: T7 biotinylated in vivo and purified on Ni-NTA-agarose. T7100%bt: fraction of T7bt eluted from monomeric avidin-agarose. T7−bt: VHH T7 expressed without addition of biotin and purified on Ni-NTA-agarose. These three preparation of VHH T7 were titrated against TCC-BSA by ELISA using streptavidin-HRP for developing; T7bt, circles; T7100%bt, squares; T7−bt, triangles. The insert shows the SDS-PAGE analysis of the different preparations of VHH T7 applied to avidin-agarose spin columns; Vd, void; Ad, bound fraction eluted with SDS-PAGE sample buffer. The upper (T7) and lower (Av) bands correspond to the VHH, and the avidin monomer that is released from the avidin-agarose.
The fact that only a fraction of the nanobody is coupled to biotin may be related to the expression of the VHH and BirA in different cell compartments. Two strategies were used to investigate this point. To examine their cytoplasmic co-expression, VHH T7 was cloned into pET28a(+) including an initial methionine and this plasmid was transformed into E. coli BL21(DE3) carrying the pCY216 vector. Lacking the oxidative environment of the periplasmic compartment, the expression level of VHH T7 and other VHH clones was significantly affected (figure S-4, Supporting information) and this option was discarded. For periplasmic expression of BirA, its gene was PCR amplified from pCY216, including the pelB signal peptide sequence in the forward primer, and the amplicon was cloned into the pBAD18 (Life Technology), the resulting plasmid was transformed into E. coli BL21(DE3) carrying the pINQ-BtH6 vector. In this case the expression of VHH T7 was normal, but the BirA activity was affected and the degree of biotinylation was greatly reduced, about 10 fold (figure S-5, Supporting information). Based on these results we continued with the co-expression of BirA and VHHs in the cytosol and periplasm, respectively, as the best tradeoff for easy production of biotinylated VHHs. Interestingly, it was possible to reach 100% biotinylation by including a simple step of post-biotinylation in vitro, consisting in a brief incubation with biotin of the freeze-and-thaw extract. In this way BirA and the VHH, properly folded in the cytoplasm and periplasm respectively, get in contact allowing the quantitative conjugation of biotin; this step is highly efficient and does not require biotin during the culture step, figure 4.
Figure 4. SDS-PAGE analysis of VHH T7 biotinylated in vivo and/or in vitro.

T7bt: T7 biotinylated in vivo; T7bt-pbt: prepared as T7bt, but the cell extract was further “post-biotinylated” by incubation with 100 μM biotin, 30 min, 37°C. T7pbt: T7 was co-express with BirA, but biotin (100 μM) was only added during the incubation of the cell extract. In all cases VHH T7 was purified on Ni-NTA-agarose, and was then applied to avidin-agarose spin columns; Vd, void; Ad, bound fraction eluted with SDS-PAGE sample buffer. The upper (T7) and lower (Av) bands correspond to the VHH, and the avidin monomer that is released from the avidin-agarose.
Library construction and initial selection of VHH on BMDC
VHH and VH genes were amplified from 108 peripheral blood leukocytes from a llama immunized with BMDC and cloned into the pComb3X phagemid vector, producing a library of 2 × 109 transformants. For panning experiments, the library was first pre-adsorbed on the mouse fibroblast cell line L-929 (ATCC) to deplete it from ubiquitous cell-receptor binders, and the unbound fraction was then selected on BMDC at 4°C as described. After three rounds of panning the supernatants of 48 individual clones containing soluble VHHs were tested on ELISA plates coated with BMDC membrane cell extracts; more than 95% of clones produced positive results indicating the efficiency of the panning procedure (not shown). The VHHs of this third output were cloned en masse into pINQ-BtH6, and were then transformed into E. coli BL21(DE3) carrying the pCY216 vector. Ninety six individual clones were grown to produce biotinylated VHHs.
The biotin moiety facilitates the large scale screening of nanobodies by flow cytometry
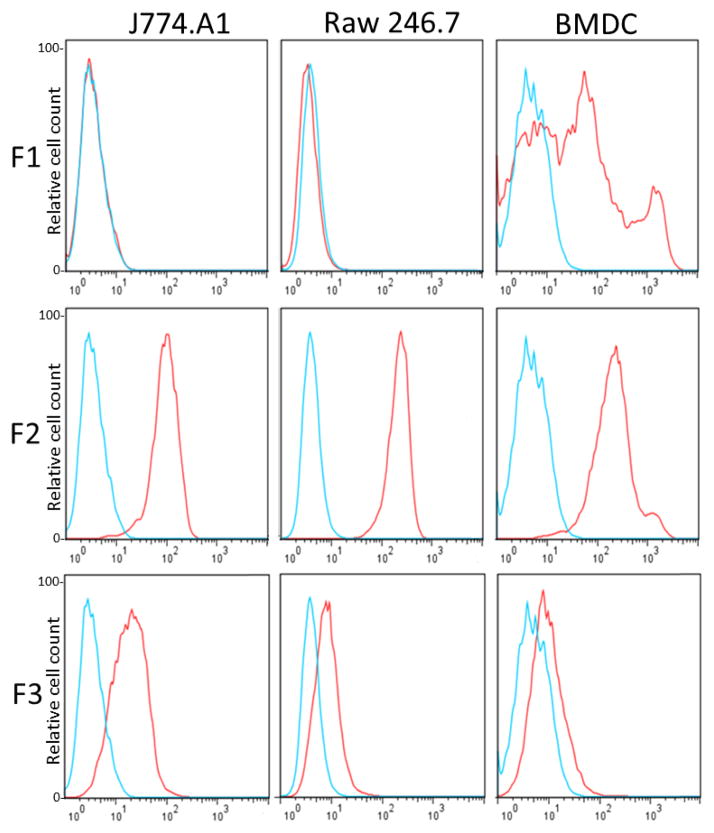
No other method is better for the isolation of antibodies to cell receptors than their direct screening on cells. While this can be accomplished by flow cytometry using fluorescent labeled antibodies against nanobody peptide tags, the use of avidins avoids the worries of unspecific binding of the antibody conjugate (i.e. to Fc receptors), which is a great advantage for high throughput screening. In addition, the broad choice of streptavidin/avidin fluorescent conjugates allows different combinations, which is particularly useful when a multiparametric flow cytometry screening is needed. In our case, to allow the simultaneous analysis of a high numbers of BtNb by flow cytometry, individual clones were grown in 96-deep well plates, the soluble cell extracts were prepared by cycles of freezing and thawing, and the clean supernatants were directly used to label the cells. Initially, positive clones were classified in three families according to their reactivity with BMDC and two mouse macrophage cell lines (ATCC J774.A1 and Raw 246.7); nanobodies that reacted mainly with BMDC (family F1), and nanobodies that reacted with all cell types with high or low geometric mean values (families F2 and F3, respectively), figure 5. The sequences of 12 clones derived from these families are shown in figure S-6, Supporting information. We mainly focused our interest in nanobodies from the F1 and F2 families.
Figure 5. Flow cytometry analysis using three representative nanobodies selected on BMDC.
Each biotinylated nanobody was tested on BMDC and the macrophage cell lines J774.a1 and Raw 246.7 using phycoerythrin-streptavidin, histograms in red. The F1, F2 and F3 representative Nbs were V32, p01 and X6, respectively; VHH T7 nanobody was used as negative control (blue)
The BtNbs can be directly used to pull down their cognate antigen and characterize their reactivity
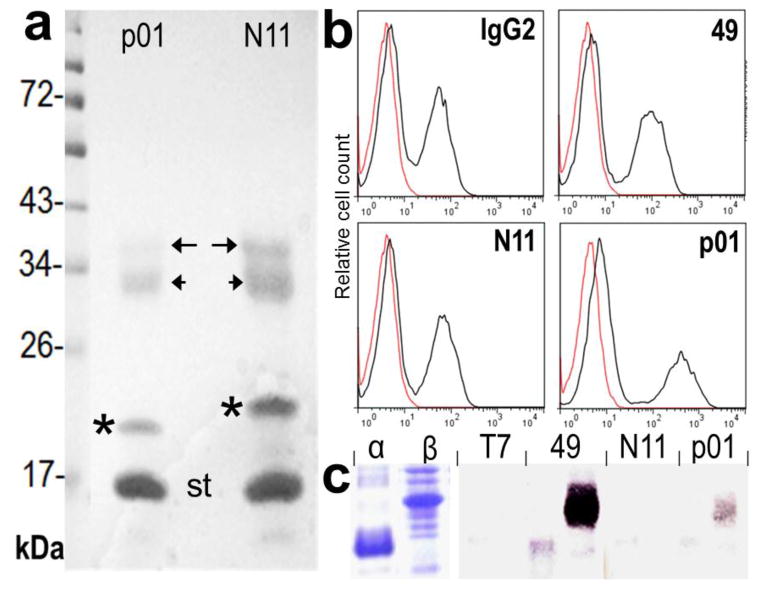
The soluble extracts containing the BtNbs were directly loaded into streptavidin agarose, which was then used for the immunoprecipitation experiments to isolate the VHH target receptor from BMDC triton X-100 extracts. Notice that in this case the post biotinylation step was not included to avoid the competition of a huge excess of biotin. The strong binding of biotin allows for stringent washing conditions helping to reduce unspecific contaminants; a critical point when the antibody target is a minor component of the sample. Moreover, conversely to conventional antibodies, instead of the heavy (~50 kDa) and light (~25 kDa) chain bands, only the VHH (~15–18 kDa) will occur in addition to the avidin/streptavidin monomer (~14 kDa). Figure 6 and 7 show the SDS-PAGE analysis of the immunoprecipitated bands and the MALDI-TOF Mascot analysis of representative bands is displayed in figure S-8, supporting information. As shown in figure 6, the family F1 nanobodies, BtNb p01 and N11 (identical results were obtained with Nb 49), pulled down two components of ~36 kDa (long arrow) and ~30 kDa (short arrow), which were identified as the MHC A-D alpha and beta chain respectively. This is also in agreement with the differential expression of MHC II between BMDC and the macrophages cell lines figure 5, and the actual size of the bands. These antibodies appear to recognize a non-polymorphic epitope on the MHC II molecule because they also reacted with splenocytes of C57bl/6 mice that carry the IAb MHC II haplotype figure 6. To test the specificity of these VHHs, with regard to alpha or beta A-D chain recognition, their ectodomains were amplified from Balb/c mouse cDNA and cloned in the pET28 vector. While expressed at high levels, both proteins were insoluble, and their reactivity could only be tested by western blot, figure 6. There was no reactivity with the denatured proteins, except for Nb 49 which strongly reacted with the MHC beta chain.
Figure 6. Characterization of family F1 nanobodies.
a) The protein bands eluted from the streptavidin-agarose used in the pool-down experiments with Nb p01 and N11 were run on a 12.5% SDS-gel and stained with coomassie blue. The bands are labeled as follows: immunoprecipitated bands (arrows), nanobodies (*), streptavidin monomer (st). b) Flow cytometry analysis of C57bl/6 mouse splenocytes stained with BtNb 49, N11 and p01 or a commercial biotinylated anti-mouse MHC II (I-A/I-E) antibody (IgG2). c) SDS-PAGE and westernblot analysis of the reactivity of the BtNbs with E. coli extracts over expressing the individual chains of the MHC II A-D.
Figure 7. Characterization of family F2 and F3 nanobodies.
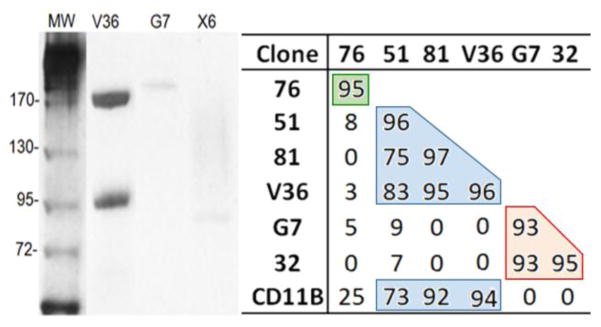
Polled-down components precipitated with Nb V36, G7 and X6 (10% SDS-gel stained with coomassie blue). The table shows the inhibition (%) of the flow cytometry reactivity with BMDC (geometric mean value) of the BtNbs (row header) in the presence of non-biotinylated Nbs (produced in the absence of added biotin) (column header). CD11B, biotinylated Rat IgG to mouse CD11b (BD Pharmingen). Color boxes are used to indicate Nbs that define overlapping epitopes and compete with each other.
Surprisingly, six out of the eight family F2 nanobodies (V36, 76, 51, 81, B10 and 42) precipitated two bands of about ~170 kDa and ~97kDa (showed for V36 in figure 7), which according to their peptide finger print corresponded to CD11b (170 kDa) and CD18 (95 kDa), the alpha and beta subunits of macrophage-1 antigen (Mac-1) [25]. Mac-1 is a surface integrin receptor expressed on many innate immune cells, which is involved in different immune cell responses, including adhesion, migration, phagocytosis, chemotaxis, cellular activation, and cytotoxicity [26, 27]. Further analysis of their ELISA reactivity with the Mac-1 (CD11b/CD18) or the CR4 (CD11c/CD18) receptors, showed that these antibodies define their epitopes on the CD11b chain of Mac-1, figure S-7, Supporting information.
Nbs G7 and 32b immunoprecipitated a 195 kDa band that was identified as the mouse Ly-5, leukocyte common antigen (CD45), a protein tyrosine phosphatase abundantly expressed on all nucleated hematopoietic cells, an involved in a variety of cellular processes including cell growth, differentiation, activation, and oncogenic transformation [28]. As expected these Nbs stained all leukocyte populations in splenocyte (not shown). Finally, Nb X6, belonging to family F3, precipitated a 92 kDa band. The size of this band and its MALDI-Mascot analysis strongly indicated this as the valosin-containing protein (VCP), a highly abundant (>1%) cytoplasmic protein [29]. VCP is a member of the AAA ATPases family, involved in several cellular functions including cell cycle control, the ubiquitin-proteasome degradation pathway, nuclear trafficking, etc. [30]. It was rather unexpected that the main component pulled-down by Nb X6 was a cytoplasmic protein, because our panning strategy was design to select for antibodies against cell receptors. Since the Nb X6 flow cytometry performance indicates reactivity with a cell receptor, figure 5, the possibility of an artifact due to a fortuitous cross-reactivity with the highly abundant VCP cannot be ruled out.
On and off switching of biotin addition allows binning of the Nb epitopes
A common problem when selecting a panel of antibodies to a particular receptor is how to sort them in terms of their defined epitopes, to avoid functional epitope testing of redundant binders. Interestingly, the versatility of the in vivo biotinylation method also facilitates epitope binning assays for Nbs reacting to a particular target. Indeed, when biotin is not added during the Nb production, the biotinylation of the biotin acceptor peptide tag is negligible (figure 3). This allows to have the labeled and unlabeled version of the same Nb without the need of sub-cloning. In this way, it is possible to perform competitive flow cytometry experiments to study the eventual overlap of their epitopes. Figure 7 shows the inhibition percentage of the flow cytometry signal when pairs of BtNbs and non-biotinylated Nbs to CD11b were tested against each other. The mean fluorescence intensity data is shown in table S-1, Supporting information. Nb 51, 81 and V36, strongly compete with each other, as well as with a commercial anti-CD11b antibody, but not with Nb 76 which appears to react with a non-overlapping epitope on CD11b. A similar analysis showed that the Nbs to CD45, despite bearing dissimilar CDR1 and CDR2, define overlapping epitopes, which is not surprising considering that they share the same CDR3.
CONCLUSIONS
We have optimized a method for efficient production of biotin-tagged nanobodies in 96 deep well plates that allows their rapid characterization without the need of purification of individual clones in a high throughput manner. The more adequate cell compartment for expression of the VHHs and BirA turned out to be the periplasm and cytoplasm, respectively, which allowed to reach ~25% of biotinylation. While this is more than enough for direct use of the cell extracts in flow cytometry or ELISA experiments, fully derivatized Nbs could be easily prepared by a short post-incubation step with biotin, once they got in contact with BirA after cell disruption. In this study a large scale analysis of the reactivity of dozens of Nb clones was carried out against three cell preparations. While still tedious, this task was greatly facilitated by growing and disrupting the E. coli cultures in 96 well blocks, which allowed the simultaneous labeling of BMDC and the macrophage cell lines with BtNbs using a multi-channel micropipette. In addition, since Nbs consist only of the recognition part of the parent antibody, they lack the effector function regions that may cause unspecific binding (i.e. to Fc receptors) making the use of isotypic control antibodies unnecessary. The biotin tag also facilitates the identification/discovery of the Nb cognate antigen, tolerating stringent washing conditions in the pull-down experiments, and thus producing clean peptide finger prints for identification. Using BMDC as a model we demonstrated that it was possible to rapidly identify and characterize the reactivity of a panel of Nb raised against whole cells. Once selected the biotin moiety will be a convenient tag for the intended use of the Nbs, notably in flow cytometry applications, with the additional advantage that typically several tens of mg/L can be obtained in shake flask cultures.
Supplementary Material
Highlights.
Quantitative in vivo biotinylation of nanobodies in 96 culture blocks was optimized.
The biotin moiety allows high throughput FACS analysis of the VHH reactivity.
Pull-down experiments and MS identification are greatly facilitated.
On and off switching of the biotin addition allows binning of VHH epitopes on cells.
Acknowledgments
This work was supported with funds provided by grants FCE 6812 ANII (Agencia Nacional de Investigación e Innovación, Uruguay) and TW05718 Fogarty Center NHI. MR, ST and LA are recipients of scholarships from ANII.
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The authors declare no competing financial interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Geyer CR, McCafferty J, Dubel S, Bradbury AR, Sidhu SS. Recombinant antibodies and in vitro selection technologies. Methods Mol Biol. 2012;901:11–32. doi: 10.1007/978-1-61779-931-0_2. [DOI] [PubMed] [Google Scholar]
- 2.Hust M, Frenzel A, Schirrmann T, Dubel S. Selection of recombinant antibodies from antibody gene libraries. Methods Mol Biol. 2014;1101:305–320. doi: 10.1007/978-1-62703-721-1_14. [DOI] [PubMed] [Google Scholar]
- 3.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 5.Vu KB, Ghahroudi MA, Wyns L, Muyldermans S. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol Immunol. 1997;34:1121–1131. doi: 10.1016/s0161-5890(97)00146-6. [DOI] [PubMed] [Google Scholar]
- 6.Revets H, De Baetselier P, Muyldermans S. Nanobodies as novel agents for cancer therapy. Expert Opin Biol Ther. 2005;5:111–124. doi: 10.1517/14712598.5.1.111. [DOI] [PubMed] [Google Scholar]
- 7.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 8.van der Linden RH, Frenken LG, de Geus B, Harmsen MM, Ruuls RC, Stok W, de Ron L, Wilson S, Davis P, Verrips CT. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim Biophys Acta. 1999;1431:37–46. doi: 10.1016/s0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- 9.Tabares-da Rosa S, Rossotti M, Carleiza C, Carrion F, Pritsch O, Ahn KC, Last JA, Hammock BD, Gonzalez-Sapienza G. Competitive selection from single domain antibody libraries allows isolation of high-affinity antihapten antibodies that are not favored in the llama immune response. Anal Chem. 2011;83:7213–7220. doi: 10.1021/ac201824z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Transue TR, De Genst E, Ghahroudi MA, Wyns L, Muyldermans S. Camel single-domain antibody inhibits enzyme by mimicking carbohydrate substrate. Proteins. 1998;32:515–522. doi: 10.1002/(sici)1097-0134(19980901)32:4<515::aid-prot9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 11.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardon E, Laeremans T, Triest S, Rasmussen SG, Wohlkonig A, Ruf A, Muyldermans S, Hol WG, Kobilka BK, Steyaert J. A general protocol for the generation of Nanobodies for structural biology. Nature protocols. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saerens D, Frederix F, Reekmans G, Conrath K, Jans K, Brys L, Huang L, Bosmans E, Maes G, Borghs G, Muyldermans S. Engineering camel single-domain antibodies and immobilization chemistry for human prostate-specific antigen sensing. Anal Chem. 2005;77:7547–7555. doi: 10.1021/ac051092j. [DOI] [PubMed] [Google Scholar]
- 14.Sukhanova A, Even-Desrumeaux K, Kisserli A, Tabary T, Reveil B, Millot JM, Chames P, Baty D, Artemyev M, Oleinikov V, Pluot M, Cohen JH, Nabiev I. Oriented conjugates of single-domain antibodies and quantum dots: toward a new generation of ultrasmall diagnostic nanoprobes. Nanomedicine: nanotechnology, biology, and medicine. 2012;8:516–525. doi: 10.1016/j.nano.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Even-Desrumeaux K, Baty D, Chames P. Strong and oriented immobilization of single domain antibodies from crude bacterial lysates for high-throughput compatible cost-effective antibody array generation. Molecular bioSystems. 2010;6:2241–2248. doi: 10.1039/c005279e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anamelechi CC, Clermont EE, Brown MA, Truskey GA, Reichert WM. Streptavidin binding and endothelial cell adhesion to biotinylated fibronectin. Langmuir: the ACS journal of surfaces and colloids. 2007;23:12583–12588. doi: 10.1021/la702322n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diamandis EP, Christopoulos TK. The biotin-(strept)avidin system: principles and applications in biotechnology. Clin Chem. 1991;37:625–636. [PubMed] [Google Scholar]
- 18.Dundas CM, Demonte D, Park S. Streptavidin-biotin technology: improvements and innovations in chemical and biological applications. Appl Microbiol Biotechnol. 2013;97:9343–9353. doi: 10.1007/s00253-013-5232-z. [DOI] [PubMed] [Google Scholar]
- 19.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein science: a publication of the Protein Society. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson BH, Oei J, Saunders M, Gravel R. [3H]biotin-labeled proteins in cultured human skin fibroblasts from patients with pyruvate carboxylase deficiency. J Biol Chem. 1983;258:6660–6664. [PubMed] [Google Scholar]
- 21.Barbas C, III, Burdon D, Scott J, Silverman G. In: Phage Display: A Laboratory Manual. Carlos DRB, Barbas F III, Scott Jamie K, Silverman Gregg J, editors. Cold Spring Harbor Laboratory Press; New York: 2001. pp. 2.1–2.19. [Google Scholar]
- 22.Chapman-Smith A, Turner DL, Cronan JE, Jr, Morris TW, Wallace JC. Expression biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier protein of Escherichia coli acetyl-CoA carboxylase. The Biochemical journal. 1994;302(Pt 3):881–887. doi: 10.1042/bj3020881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zarebski LM, Urrutia M, Goldbaum FA. Llama single domain antibodies as a tool for molecular mimicry. J Mol Biol. 2005;349:814–824. doi: 10.1016/j.jmb.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 25.DuMont AL, Yoong P, Day CJ, Alonzo F, 3rd, McDonald WH, Jennings MP, Torres VJ. Staphylococcus aureus LukAB cytotoxin kills human neutrophils by targeting the CD11b subunit of the integrin Mac-1. Proc Natl Acad Sci U S A. 2013;110:10794–10799. doi: 10.1073/pnas.1305121110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hynes RO. Integrins: bidirectional allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 27.Solovjov DA, Pluskota E, Plow EF. Distinct roles for the alpha and beta subunits in the functions of integrin alphaMbeta2. J Biol Chem. 2005;280:1336–1345. doi: 10.1074/jbc.M406968200. [DOI] [PubMed] [Google Scholar]
- 28.Huntington ND, Tarlinton DM. CD45: direct and indirect government of immune regulation. Immunol Lett. 2004;94:167–174. doi: 10.1016/j.imlet.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Dalal S, Hanson PI. Membrane traffic: what drives the AAA motor? Cell. 2001;104:5–8. doi: 10.1016/s0092-8674(01)00186-6. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT, DeLaBarre B. NSF and p97/VCP: similar at first, different at last. FEBS Lett. 2003;555:126–133. doi: 10.1016/s0014-5793(03)01107-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.