Abstract
Growing evidence suggests a role for the antiapoptotic protein survivin in promotion of cancer cell G1/S transition and proliferation. However, the underlying mechanism is unclear. Further, although upregulation of p21WAF1/CIP1 by p53 plays an important role in p53-mediated cell G1 arrests in response to various distresses, it is unknown whether survivin plays a role in the regulation of p21WAF1/CIP1 expression. Here, we report that exogenous expression of survivin in p53-wild type MCF-7 breast cancer cells inhibits the expression of p21WAF1/CIP1 protein, mRNA and promoter activity, while the survivin C84A mutant and antisense failed to do so. Cotransfection experiments in the p53 mutant H1650 lung cancer cell line showed that survivin neutralizes p53-induced p21WAF1/CIP1 expression and promoter activity. Importantly, genetically silencing of endogenous survivin using lentiviral survivin shRNA also enhances endogenous p21 in p53 wild type cancer cells, suggesting the physiological relevance of the fining. We further demonstrated that both p53 and survivin interacts on the two p53-binding sites in the p21WAF1/CIP1 promoter (−2313 to −2212; −1452 to −1310), and survivin physically interacts with p53 in cancer cells. Together, we propose that survivin may act as a transcription factor or cofactor to interact with p53 on the p21WAF1/CIP1 promoter leading to the inhibition of p21WAF1/CIP1 expression at least in part by neutralizing p53-mediated transcriptional activation of the p21 gene.
Keywords: Survivin, p21WAF1/CIP1, p21 promoter activity, p53, cancer cells
Introduction
Survivin, a unique member in the inhibitor of apoptosis family, is highly expressed in cancers but undetectable in most normal adult tissue. Survivin plays a central role in cancer cell signaling network and is a multi-functional molecule [1,2,3]. Previous studies showed that expression of survivin accelerated S phase entry, resistance to G1 arrest in human hepatoma cells [4]. We demonstrated that vitamin D3-induced cancer cell cycle arrest in the G1 phase requires the downregulation of survivin expression, and in turn, forced expression of survivin blocks cancer cell G1 arrest induced by vitamin D3, and increases S and G2/M populations [5]. Survivin appears to play a role in the regulation of G1-S transition. However, the exact underlying mechanism for this is unclear.
Evidence also suggests a novel role for survivin in regulation of gene expression by potentially acting as a transcription factor (TF) or co-factor [6]. It was reported that survivin regulates the expression of the p53 family gene expression [7]. A report showed that doxorubicin-induced p53-mediated cell cycle inhibitor p21WAF1/CIP1 (p21) expression may transcriptionally inhibit the survivin gene in human hepatocellular carcinoma cell HepG2 [8]. We hypothesized that survivin may act as a TF or cofactor to inhibit p21 transcription in a p53-dependent manner in cancer cells. This hypothesis is consistent with the role for survivin in promoting G1/S transition and for p21 in the inhibition of G1-to-S transition. Additionally, p53 increases p21 gene transcription, and the role of p53 in cell cycle regulation is at least in part via its induction of p21 expression [9,10]. In this report, we have, for the first time, demonstrated that survivin appears to act as a TF or cofactor to inhibit the expression of the p21 gene in a p53-dependent manner, and genetic silencing of survivin increases p21 expression in p53 wild type cancer cells.
Materials and Methods
Cell culture and reagents
MCF-7 and ZR-75 human breast cancer cells expressing wild type p53 were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Cellgro, Herndon, VA, USA) and 100 U/ml of penicillin/0.1μg/ml of streptomycin (Invitrogen, Carlsbad, CA, USA). HCT-8 human colon cancer cells expression wild-type p53 and H1650 human lung cancer cells expressing mutant p53 were maintained in RPMI 1640 medium containing 10% FBS and 100 U/ml of penicillin/0.1μg/ml of streptomycin. Cells were cultured in a humidified atmosphere incubator with 5% CO2 at 37°C and routinely subcultured twice weekly. Survivin antibodies were purchased from Santa Cruz (sc-10811, Santa Cruz, CA) and Epitomics (2565-1, Burlingame, CA). Normal IgG from mouse (sc-2025) and rabbit (sc-2027), Protein A/G plus-agarose bead (sc-2003) were purchased from Santa Cruz. p53 antibodies were purchased from Santa Cruz (DO-1) and CalBiochem (Ab-2, PAb1801). p21 antibodies were purchased from BD Pharmingen (#556430, San Diego, CA). EGFP/GPF A.V. monoclonal antibodies (JL-8) were purchased from Clontech (Mountain View, CA). Actin antibodies (A2066) and horseradish peroxidase-conjugated anti-secondary IgG antibodies were purchased from Sigma (St. Louis, MO).
Expression vectors
The expression vectors pcDNA3 (Invitrogen, San Diego, CA) and pEGFPc1 (Clontech, Palo Alto, CA) containing survivin - pcDNA3-survivin [11] pEGFPc1-survivin [12,13], survivin dominant-negative mutants (pcDNA3-survivin C84A [14], pcDNA3-survivin T34A [15]), and survivin antisense cDNA (pcDNA3-AS-survivin [15]), were previously characterized. The wild-type p53 plasmid in pcDNA3 (pc53-SN3) was provided by Dr. Muxiang Zhou (Emory University) via permission from Dr Bert. Vogelstein (Johns Hopkins University). The pAd-GFP and pAd-survivin (pAd-WT) were gifts from Dr. Dario C. Altieri (University of Massachusetts Medical School) and were previously characterized [16]. The −2326p21-Luc plasmid, in which a 2327-bp p21 promoter drives luciferase reporter gene, was provided by Dr. Bert Vogelstein (Johns Hopkins University) and was previously characterized [17].
Adenovirus amplification and infection
Recombinant adenovirus amplification and infection were previously described [18]. Briefly, virus particles for pAd-GFP and pAd-survivin were amplified in HEK293 packaging cells and transfected into MCF-7 cells. Infection efficiency was monitored with GFP marker expression.
Transfection and luciferase activity assay
The pCMV-PRL vector, in which the CMV promoter drives a Renilla luciferase reporter gene, was used as internal transfection efficiency control in a dual luciferase assay system (Promega). Cells were seeded in 48-well plate (5 × 104per well) and grow overnight. Next day, the cultural medium with serum was replaced by serum-free medium before transfection. For transfection, the corresponding plasmid DNA and the liposome solution were separately diluted into 50 μl of optimal-DMEM in 1.5ml tubes using a 1μg:2μl ratio for DNA:Lipofectamine (Invitrogen). After 5 min, the two solutions were gently mixed well. The solution of DNA-liposome complex was added to cells 20-30 min after mixing. The DNA-liposome complexes were stayed with cells for 6 hours and then the medium was replaced with normal culturing medium. Luciferase activity was determined 20 hours after medium replacement as previously described [13] Assays were performed in triplicate.
Reverse transcription (RT)-PCR and real time RT-PCR
The method for semi-quantitative PCR [19] and real time PCR [20] were previously described. The primers used in PCR for determination of p21 mRNA expression are p21-fe1 (5′GCC TTG GCC TGC CCA AGC TC3′, forward primer) and p21-re2 (5′GCC GGC GTT TGG AGT GGT AG3′, reverse primer). The PCR reaction (20μl) containing 0.5 μM of each primer, 20 nM of each dNTPs, and 1 U FideliTaq DNA polymerase (Usb) was performed with an initial step of 94°C for 5 min, followed by 35 cycles of 94°C for 30s, 61°C for 30S, 72°C for 1 min, and a final elongation step of 72°C for 10 min. The PCR products were separated on 1% agarose gels containing ethidium bromide. Rea-time RT-PCR was performed using a standard TaqMan PCR kit (Fermentas) following the kit booklet protocol on an Applied Biosystems 7300 system. A 20μl reaction containing 1μl RT product, 1xTaqMan Universal PCR Master Mix, 1 μM forward and reverse primers was incubated in a 96-well plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All reactions were run in triplicate. The threshold cycle (CT) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold.
Western blots
Western blots were described previously [21]. A total 40μg protein in lysates was used in the studies. Survivin, p53 and p21 proteins were visualized with Western Lightning® Chemiluminescence Reagent Plus (PerkinElmer, Waltham, MA).
Preparation of lentiviral survivin shRNA infection particles
Plasmid DNA of the control pGIPZ lentiviral vectors (with Puromycin selection and TurboGFP mark) or with survivin shRNA (V2LHS_262484) [22] were first prepared. HEK 293T packaging cells were then seeded in a 100mm cell cultural dish at 80% confluence and incubated for 24 hours at 37°C and 5% C02. Then 250 μl DMEM containing 2.5μg pGIPZ survivin shRNA (or empty pGIPZ vector), 2.5μg psPAX2 (or pCMV-dR8.74), 1.0μg pMD2.G in one tube mixed with 250 μl DMEM containing 9-12 μl lipofectamine and stayed at room temperature for 20 minutes. The cell medium in the 293T cell dish was gently replaced with 500ul DNA/Lipo complex. Three ml 293T media (DMEM with 10%FBS and 1% Pen/Strep) were added after a few minutes at room temperature for 16 hours at 37°C and 5% CO2. The medium in the dish was replaced with new 293T media next day, and the dish was incubated for additional 24 hours at 37°C with 5% CO2. Virus-containing supernatant was harvested and filtered through 0.45 μm cellulose acetate syringe filter, and store virus at 4°C.Check TurboGFP expression before collection of virus in the supernatant. The dish with 293T cells was added another 3.0 - 3.5 ml 293T media and incubated for overnight at 37°C with 5% CO2. The supernatant was collected as above and combined together and stored at 4°C for target cell infection. The 293T cell dish was discarded or were check for gene expression.
Infection of target cells with lentiviral particles
MCF-7 and HCT-8 cancer cells at ~70-80% confluence in 6-well plates were applied with 1 ml lentiviral survivin shRNA particle supernatant harvested above and 4 μg/ml polybrene (infection promoter) directly on to cells. The plate was then spun at 1800 rpm for 45 minutes at room temperature on a microtiter rotor to promote virus infection into cells. The plate was then incubated for 3-6 hours at 37°c with 5% CO2, followed by adding 1 ml cell media for continuous incubation overnight. Next day, cells in each well was split by 1 to 5 and incubated for 24 hours. The infected cells were then selected with purimycin (2 μg/ml) for up to one week. The purimycin-selected cells were used for experiments (e.g. western blots).
Immunoprecipitation (IP) assays
For IP with p53 antibodies, MCF-7 cells (up to 107) were harvested with a scraper in cold PBS buffers and collected by centrifugation at 1000g x 5 min and washed with cold PBS once. Cell pellets were lysed in 1 ml cold RIPA lysis buffer (refer to Western blot) containing proteinase inhibitors. The cell lysates were precleared with centrifuging 14,000 rpm for 15 min. The supernatants were transferred to a new 1.5 ml tube and incubated with up to 3μg p53 antibodies for 1 h at 4°C, followed by incubation with 20-40μl 50% protein A/G-Agarose slurry overnight at 4°C on a rocker platform. After centrifugation at 1000g for 5 min at 4°C, the pellet was washed with RIPA buffer or PBS for 2-4 times and collected by centrifugation as above. The pellets were then resuspended in 20-40 μl 1x sample buffer, boiled for 2-4 min and stored on ice for Western blot analyses.
Chromatin immunoprecipitation (ChIP) assay
MCF-7 and ZR75 cells were washed once with PBS and then crosslinked with 1.5% formaldehyde at 37 °C for 10 minutes. After washed with ice cold PBS twice, the cells were collected in lysis buffer (1% SDS, 10 mM EDTA, 50mM Tris-HCl, pH8.1) and lysed for 30 minutes on ice. Cell lysates were sonicated using Sonicator 3000 (Misonix, NY) and then pre-cleared with salmon sperm DNA/Protein A agarose for 2 hours at 4 °C. Five μg of specific antibodies or normal IgG (control) were used to immunoprecipitate protein-DNA complex from pre-cleared supernatants containing 200 μg of proteins. Immunoprecipitated DNA was amplified using the p21 promoter primers (see below) by PCR with AccuPrime TaqDNA polymerase. PCR products were visualized with ethidium bromide staining after their separation on 2% agarose gels. The following primer sets were used for detecting p53-binding sites in the p21 promoter. Primers for the distal p53-binding site (p53 5′-site) are 5′GCT GTG GCT CTG ATT GGC TTT3′ (forward, −2313 to −2293) and 5′ACA GGC AGC CCA AGG ACA AA3′ (reverse, −2212 to −2231). The primers for the proximal p53-binding site (p53 3′-site) are 5′CAT CCC CAC AGC AGA GGA GAA3′ (forward, −1452 to −1432) and 5′ACC CAG GCT TGG AGC AGC TA3′ (reverse, −1310 to −1329).
Results
Survivin inhibits the expression of p21 mRNA, protein and promoter activity
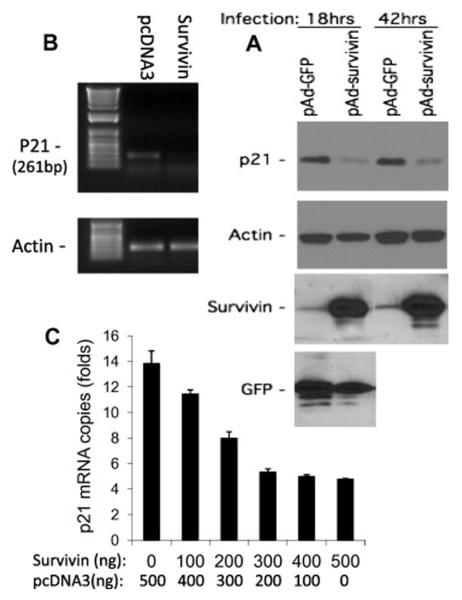
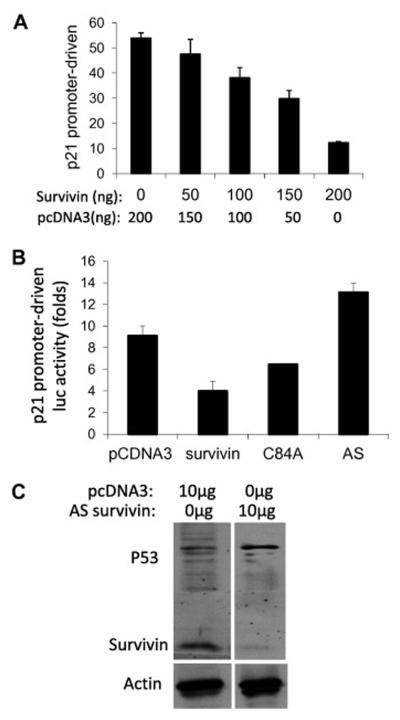
To study the effect of survivin on p21 expression, MCF-7 cells were infected with or without amplification-deficient adenoviral-survivin expression vectors. Cells were lysed and analyzed for p21 expression using western blots 18h and 42h after infection. As shown, survivin significantly inhibited p21 expression compared with empty vector controls (Fig 1A). Next we determined whether survivin-mediated inhibition of p21 protein is associated with the inhibition of p21 mRNA expression. Both semi-quantitative RT-PCR (Fig 1B) and real-time RT-PCR (Fig 1C) indicated that expression of survivin in MCF-7 cells inhibits p21 mRNA expression in a dose-dependent manner. Furthermore, expression of survivin resulted in a dose-dependent inhibition of p21 promoter-driven luciferase construct in MCF-7 cells (Fig 2A). However, survivin C84 mutant or antisense failed to inhibit p21 promoter activity (Fig 2B). Downregulation of survivin by its antisense expression was confirmed in Fig 2C. Interestingly, abrogation of survivin expression increases p53 (Fig 2C), suggesting a role for survivin to control p53 for its potential function in survivin-mediated inhibition of p21 (see results below and discussion).
Fig 1.
Survivin downregulates the p21 gene. A. Survivin inhibits p21 protein. MCF-7 cells were infected with control (pAd-GFP) or survivin expressing (pAd-survivin) adenoviral particles for 18 and 42 hours. The p21 protein was then determined by western blots. Actin, survivin and GFP were also determined as controls. B and C. Survivin decreases p21 mRNA expression. MCF-7 cells were transfected with pcDNA3 (control) and/or pcDNA3-survivin. Total RNAs were isolated and p21 mRNA expression was determined either using semi-QPCR (B) or real time PCR (C, mean ± SE from triplicate).
Fig 2.
Survivin, but not its C84A mutant or antisense (AS), inhibits p21 promoter activity. MCF-7 cells were transfected with various expression vectors alone or in combination. Cells were then lysed to determine luciferase (luc) activity (A and B) or endogenous survivin and p53 expression (C, western blots) 24 hours after transfection. Each bar in A and B is the mean ± SE from triplicate.
Expression of survivin neutralizes p53-mediated upregulation of p21 protein, mRNA and promoter activity
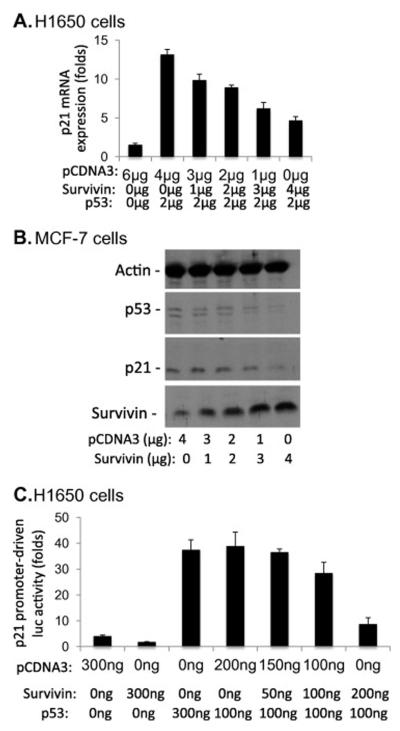
It is known that p53 arrests cells in the G1 phase at least in part through transcriptional upregulation of p21 expression [9,10]. To determine whether downregulation of p21 expression by survivin was at least partially p53-dependant, we transfected p53 and survivin expression vectors alone and in combination into the p53-mutated H1650 lung cancer cells, followed by determination of p21 mRNA using real-time RT-PCR. The results indicated that survivin effectively neutralizes p53-induced p21 mRNA (Fig 3A). Consistently, expression of survivin in MCF-7 breast cancer cells also decreased p21 protein (Fig 3B). Interestingly, increasing survivin expression inhibited p53 protein expression as well (Fig 3B), which is consistent with p53 increase upon abrogation of survivin (Fig 2C), suggesting a potential functional link between survivin and p53. Furthermore, transfection-mediated expression of survivin and p53 in H1650 cells neutralizes the p53-induced p21 promoter activity (Fig 3C).
Fig 3.
Survivin neutralizes p53-induced p21 expression. Cells were transfected as shown. The p21 mRNA (A), protein (B) and promoter activity (C) was determined using real time PCR (A), western blots (B) and luciferase activity assay (C) 24h after transfection. Each bar in A and C is the mean ± SE derived from triplicates. Survivin, p53 and actin were also determined in B as controls.
Genetic silencing of endogenous survivin enhance p21 expression
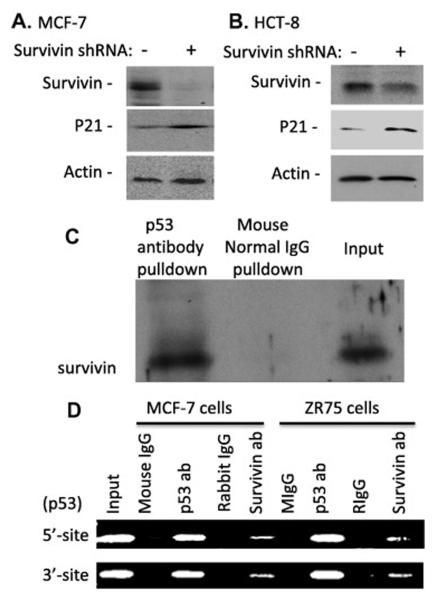
In order to explore the physiological relevance of our finding, we used the lentiviral delivery system carrying the survivin-specific shRNA to silence the endogenous expression of survivin in both MCF-7 and HCT-8 cancer cells with wild type p53. We found that consistent with the data obtained in the figures 1-3, silencing of endogenous survivin enhances the expression of endogenous p21 expression (Fig 4A, B), suggesting that our finding that survivin inhibits p21 expression is highly physiological relevance.
Fig 4.
A and B. Genetic silencing of survivin enhances p21 expression. Subconfluent MCF-7 (A) and HCT-8 (B) cancer cells were infected with lentiviral survivin shRNA particles to silence the endogenous survivin expression, followed by western blot analysis of the expression of the p21 and survivin. Actin was used as internal controls for total protein loading. C and D. Survivin and p53 interaction on the p53-binding region of the p21 promoter: C. MCF-7 cell lysates were immunoprecipitated with control or p53 antibodies. The IP pellets were then analyzed for the presence of survivin by western blots. A 5-10% input was used as a positive control. D. MCF-7 and ZR-75 cells were lysed for ChIP assays using normal IgG, p53 antibody or survivin antibody to pull down relevant DNA, followed by PCR using primer sets that span the p53 binding sites in the p21 promoter. The PCR products were analyzed on 2% agarose gel containing ethidium bromide. The input represents PCR product amplified from 1% ChIP assay input material.
Survivin physically interacts with p53 protein
We next determined the possibility that survivin physically interacts with p53 protein in vivo. We performed IP with p53 antibody, followed by western blots using survivin antibody. The result revealed that p53 antibody effectively pulls down survivin protein, while control antibody (normal IgG) failed to do so (Fig 4C), indicating these two proteins (p53 and survivin) physically interact each other in cancer cells.
p53 and survivin bind on the same p53-binding sites-containing DNA region in vivo in the p21 promoter
The physical interaction of survivin with p53 (Fig 4C) provides a possibility that survivin may inhibit p53-mediated upregulation of p21 gene transcription on the p53 binding sites in the p21 promoter. Previous ChIP studies demonstrated that p53 binds to both p53 5′- and 3′-DNA binding sites in the p21 gene promoter [23]. Our ChIP assay experiment revealed that survivin antibodies, showing a similar role to p53 antibodies, pulls down both the p53 5′- and 3′-binding sites, while control antibodies (normal IgG) failed to do so. The physical interaction of survivin with p53 on the p53 binding sites in the p21 gene promoter (Fig 4D) is consistent with the observation that downregulation of p21 by survivin is at least partially p53-dependent (Fig 3).
Discussion
Survivin appears to be a multifunctional molecule and its expression is involved in the inhibition of apoptosis [14] and regulation of mitotic cell division [12] as well as in promotion of G1/S transition [5] and regulation of gene transcription [6]. Therefore, survivin plays a central role in controlling survival and proliferation in cancer cell-signaling networks [3]. However, the mechanism for some of these functions is unclear. For example, the mechanism for survivin to play a role in the promotion of G1/S transition and regulation of gene transcription is unclear nor the potential relationship between its promotion of G1/S transition and its regulation of gene transcription. In the present study, we, for the first time, reported a link between these two functions for survivin via its regulation of the expression of the p21 gene.
There are several issues that should be discussed here. First, why we used H1650 lung cancer cells to determine endogenous p21 expression upon expression of survivin and p53 (Fig 3A), while we used MCF-7 breast cancer cells to determine the p21 and p53 expression after expression of survivin (Fig 3B)? This is because H11650 cells do not have wild type p53, and therefore, we could address whether the effect of survivin on p21 expression is p53-dependent or -independent without interference by endogenous p53. On the other hand, experiments in MCF-7 cells that contain wild type p53 are to look at endogenous p21 and p53 expression upon expression of survivin. So a cancer cell line with a wild type p53 is relevant to the experimental objective.
Expression of survivin also inhibited p53 protein expression (Fig 3B), while downregulation of survivin increases p53 (Fig 2C). In consideration of the interaction of survivin with p53 in cancer cells (Fig 4C), there are two possibilities for survivin to inhibit p53 expression. One is that survivin is able to inhibit p53 gene transcription. The other is that the interaction of survivin with p53 destabilizes p53 protein. In either case, the finding is consistent with the previous report that survivin regulates p53 family protein expression [7].
In this report, we demonstrated that survivin interacts with p53 and binds to the same p21 promoter region as p53 does (Fig 4C, D). Survivin neutralizes p53-mediated upregulation of p21 gene expression (Fig 3). These observations suggest that while survivin acted as either a TF or a co-factor to inhibit the p21 gene, its function is at least partially p53-dependent. However, a p53-independent pathway may also be involved for survivin to inhibit p21 gene expression. The evidence for the involvement of a p53-independent mechanism for survivin inhibition of the p21 gene is that in the p53 mutant H1650 lung cancer cells, transfection of survivin without cotransfection with wild type p53 also inhibited a basic level of luciferase activity (Fig 3C, lanes 1 versus 2). Therefore, it is likely that survivin may employ both p53-dependent and p53-independent mechanisms to inhibit the expression of the p21 gene. Importantly, these finding is highly physiological relevance, since knockdown of endogenous survivin by infection of lentiviral survivin shRNA particles enhances the expression of the endogenous p21 in p53 wild type cancer cells (Fig. 4A, B).
Finally, the studies in this report also demonstrated that the survivin C84A mutant partially loses survivin function on p21 promoter activity. This finding is consistent with the previous finding that C84A act as a dominant-negative mutant and forced expression of survivin C84A replaces wild type survivin and cause caspase-3 activity in G2/M-synchronized cells [14]. However, although previous studies pointed to an important role for the survivin T34 site in apoptosis inhibition and mitotic regulation [24], we observed that survivin T34A mutant show a similar (if not more) extent of inhibition of p21 gene promoter activity (not shown). This observation may suggest that the role of survivin on G1/S transition is independent of its role in mitosis and apoptosis inhibition.
In summation, we have for the first time linked the rule of survivin in G1/S transition and the regulation of gene expression, and demonstrated that survivin inhibits the expression of p21 protein, mRNA and promoter activity, neutralizes p53-induced p21 expression and promoter activity. Silencing of endogenous survivin enhances p21 expression in p53 wild type cancer cells. Survivin physically interacts with p53 on the two p53-binding sites in the p21 promoter in cancer cells.
- Survivin inhibits the expression of p21, protein, mRNA and promoter activity
- Survivin neutralizes p53-induced p21 expression and promoter activity
- Survivin physically interacts with p53 in cancer cells
- Genetic silencing of endogenous survivin upregulates p21 in p53 wild type cancer cells
- Both p53 and survivin interacts on the two p53-binding sites in the p21 promoter
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- [1].Li F. Survivin study: What Is the Next Wave? J Cell Physiol. 2003;197:8–29. doi: 10.1002/jcp.10327. [DOI] [PubMed] [Google Scholar]
- [2].Fengzhi Li XL. Survivin Study: An Update of “What is the Nex Wave?”. Journal of Cellular Physiology. 2006;208:476–486. doi: 10.1002/jcp.20634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- [4].Suzuki A, Hayashida M, Ito T, Kawano H, Nakano T, Miura M, Akahane K, Shiraki K. Survivin initiates cell cycle entry by the competitive interaction with Cdk4/p16(INK4a) and Cdk2/cyclin E complex activation. Oncogene. 2000;19:3225–3234. doi: 10.1038/sj.onc.1203665. [DOI] [PubMed] [Google Scholar]
- [5].Li F, Ling X, Huang H, Brattain L, Apontes P, Wu J, Binderup L, Brattain MG. Differential regulation of survivin expression and apoptosis by vitamin D(3) compounds in two isogenic MCF-7 breast cancer cell sublines. Oncogene. 2005;24:1385–1395. doi: 10.1038/sj.onc.1208330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang M, Yang J, Li F. Transcriptional and post-transcriptional controls of survivin in cancer cells: novel approaches for cancer treatment. J Exp Clin Cancer Res. 2006;25:391–402. [PMC free article] [PubMed] [Google Scholar]
- [7].Wang Z, Fukuda S, Pelus LM. Survivin regulates the p53 tumor suppressor gene family. Oncogene. 2004;23:8146–8153. doi: 10.1038/sj.onc.1207992. [DOI] [PubMed] [Google Scholar]
- [8].Xiong J, Hu L, Li Y, Dou L, Cai P, Tang Z, Wang L. Effect of survivin regulation of transcription level by p21(waf1) overexpression in HepG2 hepatocellular carcinoma cells. J Huazhong Univ Sci Technolog Med Sci. 2008;28:308–313. doi: 10.1007/s11596-008-0318-z. [DOI] [PubMed] [Google Scholar]
- [9].Kim TK. In vitro transcriptional activation of p21 promoter by p53. Biochem Biophys Res Commun. 1997;234:300–302. doi: 10.1006/bbrc.1997.6637. [DOI] [PubMed] [Google Scholar]
- [10].Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- [11].Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- [12].Li F, Ackermann EJ, Bennett CF, Rothermel AL, Plescia J, Tognin S, Villa A, Marchisio PC, Altieri DC. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1:461–466. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- [13].Wu J, Ling X, Pan D, Apontes P, Song L, Liang P, Altieri DC, Beerman T, Li F. Molecular mechanism of inhibition of survivin transcription by the GC-rich sequence selective DNA-binding antitumor agent, hedamycin: evidence of survivin downregulation associated with drug sensitivity. J Biol Chem. 2005;280:9745–9751. doi: 10.1074/jbc.M409350200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- [15].Ambrosini G, Adida C, Sirugo G, Altieri DC. Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem. 1998;273:11177–11182. doi: 10.1074/jbc.273.18.11177. [DOI] [PubMed] [Google Scholar]
- [16].Mesri M, Wall NR, Li J, Kim RW, Altieri DC. Cancer gene therapy using a survivin mutant adenovirus. J Clin Invest. 2001;108:981–990. doi: 10.1172/JCI12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].el-Deiry WS, Tokino T, Waldman T, Oliner JD, Velculescu VE, Burrell M, Hill DE, Healy E, Rees JL, Hamilton SR, et al. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- [18].Ling X, Cheng Q, Black JD, Li F. Forced Expression of Survivin-2B Abrogates Mitotic Cells and Induces Mitochondria-dependent Apoptosis by Blockade of Tubulin Polymerization and Modulation of Bcl-2, Bax, and Survivin. J Biol Chem. 2007;282:27204–27214. doi: 10.1074/jbc.M705161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ling X, Yang J, Tan D, Ramnath N, Younis T, Bundy BN, Slocum HK, Yang L, Zhou M, Li F. Differential expression of survivin-2B and survivin-DeltaEx3 is inversely associated with disease relapse and patient survival in non-small-cell lung cancer (NSCLC) Lung Cancer. 2005;49:353–361. doi: 10.1016/j.lungcan.2005.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wu J, Apontes P, Song L, Liang P, Yang L, Li F. Molecular mechanism of upregulation of survivin transcription by the AT-rich DNA-binding ligand, Hoechst33342: evidence for survivin involvement in drug resistance. Nucleic Acids Res. 2007;35:2390–2402. doi: 10.1093/nar/gkm149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ling X, Bernacki RJ, Brattain MG, Li F. Induction of survivin expression by taxol (paclitaxel) is an early event which is independent on taxol-mediated G2/M arrest. J Biol Chem. 2004;279:15196–15203. doi: 10.1074/jbc.M310947200. [DOI] [PubMed] [Google Scholar]
- [22].Roca H, Varsos Z, Pienta KJ. CCL2 protects prostate cancer PC3 cells from autophagic death via PI3K/AKT-dependent survivin up-regulation. J Biol Chem. 2008;283:25057–25073. doi: 10.1074/jbc.M801073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu W, Konduri SD, Bansal S, Nayak BK, Rajasekaran SA, Karuppayil SM, Rajasekaran AK, Das GM. Estrogen receptor-alpha binds p53 tumor suppressor protein directly and represses its function. J Biol Chem. 2006;281:9837–9840. doi: 10.1074/jbc.C600001200. [DOI] [PubMed] [Google Scholar]
- [24].O’Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, Tognin S, Marchisio PC, Altieri DC. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci U S A. 2000;97:13103–13107. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]