Abstract
Strategies to measure functional signaling-associated protein complexes have the potential to augment current molecular biomarker assays, such as genotyping and expression profiling, used to annotate diseases. Aberrant activation of epidermal growth factor receptor (EGFR) signaling contributes to diverse cancers. Here, we used a proximity ligation assay (PLA) to detect EGFR in a complex with growth factor receptor-bound protein 2 (GRB2), the major signaling adaptor for EGFR. We used multiple lung cancer cell lines to develop and characterize EGFR:GRB2 PLA and correlated this assay with established biochemical measures of EGFR signaling. In a panel of patient-derived xenografts in mice, the intensity of EGFR:GRB2 PLA correlated with the reduction in tumor size in response to the EGFR inhibitor cetuximab. In tumor biopsies from three cohorts of lung cancer patients, positive EGFR:GRB2 PLA was observed in patients with and without EGFR mutations and the intensity of EGFR:GRB2 PLA was predictive of overall survival in an EGFR inhibitor-treated cohort. Thus, we established the feasibility of using PLA to measure EGFR signaling-associated protein complexes in patient-based materials, suggesting the potential for similar assays for a broader array of receptor tyrosine kinases and other key signaling molecules.
Introduction
Cellular proteins do not function in isolation, but rather as parts of larger complexes, yet biomarker strategies that identify and measure protein complexes in cancer have not been reported. Current biomarker strategies examine genomic alterations, mRNA expression patterns, and protein levels, which may not reflect underlying biological processes. Furthermore, these approaches cannot evaluate signaling activity driven by protein complexes in tumors and fail to account for contributions of the tumor microenvironment that mediate oncogenic signaling and can be associated with acquired resistance to targeted therapies [1-3], suggesting that the predictive capacity of these assays is often less than ideal.
EGFR is a therapeutic target in non-small cell lung cancer (NSCLC) and other epithelial-derived malignancies. Drugs such as erlotinib, gefitinib and cetuximab are used to treat multiple solid malignancies including tumors of the lung [4], colon [5] and squamous cell cancers of the head and neck (HNSCC) [6]. Erlotinib and gefinitib are structurally-related small molecule inhibitors of EGFR kinase activity [7, 8], whereas cetuximab is a chimeric, monoclonal antibody raised against EGFR that acts by blocking ligand-induced activation [9]. EGFR activation, either through ligand binding or cancer-associated mutations conferring constitutive kinase activity, results in receptor autophosphorylation. This enables SH2 domain-mediated binding of the cytosolic adaptor protein GRB2, a critical mediator of oncogenic EGFR signaling through activation of RAS [10]. GRB2 is required for survival of cells with mutant EGFR [11] and the interaction between EGFR and GRB2 is abrogated by erlotinib, resulting in loss of downstream ERK signaling [12, 13].
Predictive biomarkers for EGFR-directed therapies remain an area of intense investigation, especially in lung cancer. EGFR mutational testing has become a standard of care in lung cancer treatment and presence of activating mutations is clearly associated with response to erlotinib and gefitinib with tumor response rates up to 85% [4]. However, predictive biomarkers for use in cancers with wild-type EGFR are lacking and it remains unclear whether EGFR protein abundance is correlated with response to EGFR-directed therapies. For instance, traditional immunohistochemistry (IHC) has been shown to be positively correlated with response to cetuximab [14], but not correlated with response to erlotinib [15]. In contrast, Automated Quantitative Analysis (AQUA) [16] was used to quantify tumor-specific EGFR, revealing a positive correlation between tumor EGFR protein abundance and response to gefitinib [17].
Previous studies have used the proximity ligation assay (PLA) [18] to measure phosphorylation and dimerization of EGFR in cultured cells and tissues [19-21]. However, these readouts do not capture the intracellular molecular events associated with EGFR activation. Moreover, no PLA studies to date have evaluated EGFR status in tissue samples from large clinical cohorts. We developed a PLA to measure the interaction between EGFR and GRB2. We showed that EGFR:GRB2 PLA correlated with active EGFR signaling and sensitivity to EGFR inhibition using multiple cell lines in culture. Moreover, we demonstrated that EGFR:GRB2 PLA correlated with responsiveness to EGFR inhibitors in 293 patient-derived xenografts (PDX) and 350 tumor specimens from lung cancer patients. Thus, using PLA to measure drug-targetable signaling-associated protein complexes may be an effective way to annotate patient tissues for the purposes of diagnosis, prognosis, and treatment stratification.
Results
Using PLA to measure EGFR signaling activity in cultured cells
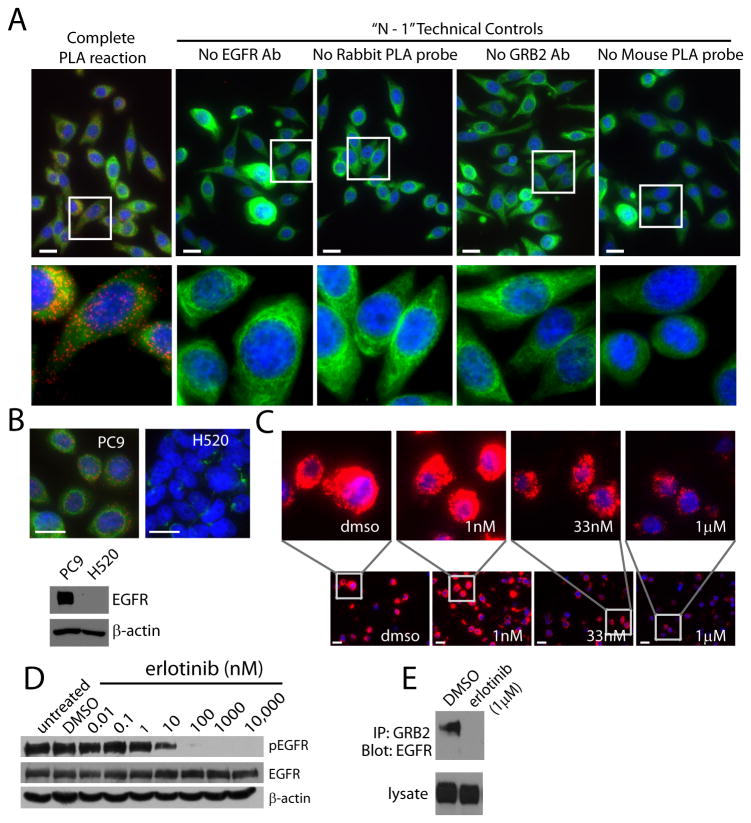
To monitor EGFR signaling, we developed a PLA for EGFR signaling-associated complexes. We performed PLA (fig. S1) [18] using monoclonal antibodies against EGFR and GRB2 and a commercially available PLA kit. We analyzed EGFR:GRB2 PLA in PC9 cells, which are a lung adenocarcinoma cell line with a deletion from Glu746 to Ala750 in the kinase domain of EGFR, which results in ligand-independent constitutive activity [22]. We detected robust EGFR:GRB2 PLA signal in PC9 cells (Fig. 1A), indicative of abundant EGFR:GRB2 complexes. In contrast, there was virtually no PLA signal in the absence of either primary antibody or in the absence of the PLA probe in PC9 cells (Fig. 1A). Moreover, we did not detect PLA signal in H520 lung squamous cell carcinoma cells, which have very low amounts of EGFR (Fig. 1B).
Figure 1. Characterization of EGFR:GRB2 PLA in cultured cell lines.
(A) Images of PC9 NSCLC cells, which have activating mutations in EGFR, fluorescently labeled by EGFR:GRB2 PLA (red), with an antibody targeting cytokeratin (green), and with DAPI (blue). (B) Images of PC9 or H520 cells labeled as in A. Immunoblot shows that H520 cells have little to no detectable EGFR. (C) Images of PC9 cells exposed to the EGFR inhibitor erlotinib and fluorescently labeled by EGFR:GRB2 PLA (red) and DAPI (blue). Images in A-C are representative of three independent experiments. Scale bars = 20μm. (D and E) Western blots of PC9 cells exposed to the indicated concentrations of erlotinib. In D, lysates were probed for activated EGFR with an antibody against Tyr1068 phosphorylated EGFR (pEGFR). β-actin was used as a loading control. In E, lysates were subjected to coimmunoprecipitation with an antibody to GRB2 and blotted for EGFR. Blots in D and E are representative of three independent experiments.
Clinical samples are readily available as formalin-fixed paraffin-embedded (FFPE) specimens. Therefore, we assessed EGFR:GRB2 PLA in FFPE pellets from PC9 cells, which are highly sensitive to EGFR inhibition [22]. We hypothesized that EGFR:GRB2 PLA intensity would be reduced by EGFR inhibition. Therefore, we exposed PC9 cells to erlotinib, a clinically relevant drug that inhibits EGFR signaling [8]. We observed high EGFR:GRB2 PLA intensity in FFPE PC9 cell pellets and this was reduced in cells exposed to erlotinib in a concentration dependent manner (Fig. 1C). Likewise, we found that erlotinib decreased phosphorylation of Tyr1068 in EGFR, which is indicative of activation by EGF or activating mutations and is the major binding site for GRB2 [23], in a similar concentration-dependent manner in PC9 cells (Fig. 1D). Moreover, erlotinib blocked the ability of GRB2 antibodies to coimmunoprecipitate EGFR in PC9 cells (Fig. 1E), consistent with published observations [13]).
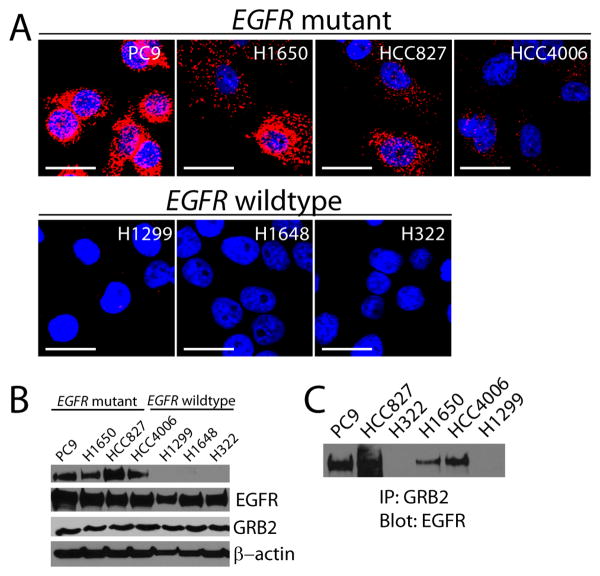
We assessed whether EGFR:GRB2 PLA correlated with activation of EGFR in other cell lines. To test if EGFR:GRB2 PLA correlated with EGFR mutational status and thus reflected activation of EGFR, we examined seven NSCLC cell lines with either mutant (PC9, H1650, HCC827, HCC4006) or wild-type EGFR (H1648, H322, H23). We consistently observed intense EGFR:GRB2 PLA signal in cell lines with mutant EGFR and low to undetectable EGFR:GRB2 PLA signal in cell lines with wild-type EGFR (Fig. 2A). Both EGFR and GRB2 were abundant in all seven cell lines (Fig. 2B), indicating that the absence of EGFR:GRB2 complexes was not due to the low abundance of either protein. Moreover, cell lines with increased EGFR:GRB2 PLA signal had increased abundance of Tyr1068 phosphorylated EGFR (Fig. 2B) and a robust interaction between EGFR and GRB2 as determined by coimmunoprecipitation (Fig. 2C). The intensity of EGFR:GRB2 PLA was lower in EGFR-mutant HCC4006 cells relative to PC9, H1650, or HCC827 cells (Fig. 2A), despite that all four cell lines have identical deletions in exon 19 that impart enhanced sensitivity to EGFR inhibitors [24, 25]. Thus, EGFR:GRB2 PLA intensity was not solely a function of protein abundance or mutational status suggesting that additional signals that modulate EGFR signaling may be reflected by this assay.
Figure 2. EGFR:GRB2 PLA in NSCLC cell lines with known EGFR mutation status.
(A) Images of confocal microscopy (representative z-stacks shown) of seven NSCLC cell lines labeled by EGFR:GRB2 PLA (red) and DAPI (blue). Images are representative of three independent experiments. Scale bars = 20μm. (B) Immunoblots of phosphorylated Tyr1068 EGFR, total EGFR, and GRB2. β-actin was used as aloading control. Blots are representative of three independent experiments. (C) Immunoblot of coimmunoprecipitation of endogenous EGFR using antibodies against GRB2 in lysates of the indicated NSCLC cell lines. Blot is representative of two independent experiments.
Detection of EGFR:GRB2 signaling complexes in mouse xenografts
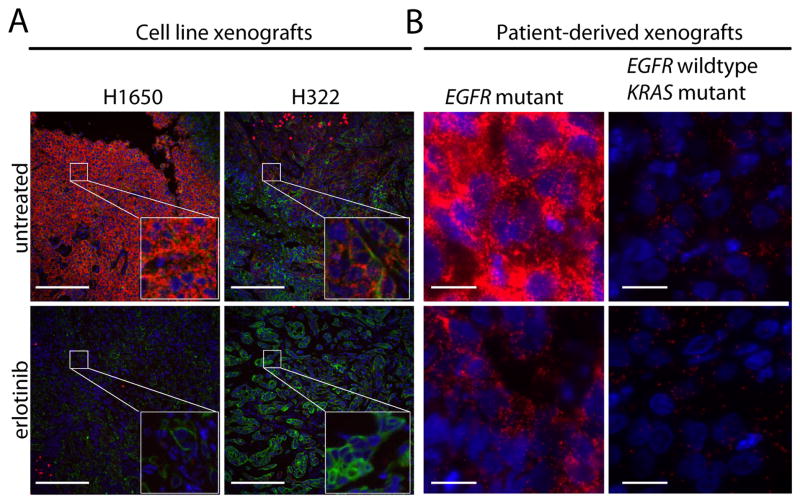
To evaluate the efficacy of EGFR:GRB2 PLA in preclinical models, we examined sections of FFPE tumors from xenografts of NSCLC cell lines in mice. The intensity of the EGFR:GRB2 PLA signal was greater in xenografts made with H1650 (EGFR-mutant) than H322 (EGFR wild-type) cells (Fig. 3A). In addition, treating mice with erlotinib abrogated the EGFR:GRB2 PLA signal in xenografts of H1650 or H322 cells (Fig. 3A), consistent with the observation that erlotinib reduces tumor size in these models [26]. Moreover, we found that the abundance of phosphorylated Tyr1068 EGFR was increased in H1650 xenografts compared to H322 xenografts, and that EGFR Tyr1068 phosphorylation was reduced in xenografts of mice treated with erlotinib (fig. S2A), consistent with the PLA results. EGFR:GRB2 PLA intensity xenografts of H322 cells was higher than that observed in H322 cells in culture, consistent with the response of H322 xenografts to erlotinib [26] and suggesting that EGFR activation in H322 xenografts is driven by EGFR ligands in the tumor microenvironment. EGFR:GRB2 PLA signal localized exclusively to regions of H1650 or H322 xenografts that were positive for cytokeratin (Fig. 3A), which was used to mark the tumor epithelium [27].
Figure 3. EGFR:GRB2 PLA in NSCLC xenografts demonstrates erlotinib-mediated complex dissociation.
(A) Images of sections of FFPE tumors from untreated or erlotinib-treated xenografts from NSCLC cell lines (A) or PDX tumors established from patients harboring an EGFR G179A mutation or a KRAS G12C mutation (B) that were labeled by EGFR:GRB2 PLA (red), an antibody against cytokeratin (green), and with DAPI (blue). Images are representative of three technical replicates. Scale bars = 100μm (A) and 20μm (B).
We also examined EGFR:GRB2 PLA in tumors from mice with patient-derived xenografts (PDXs), which were created by direct transplantation of primary tumor and stromal tissue from patients.. These PDX models retain phenotypic traits of the original tumor and can be serially passaged in mice, making them attractive models for a variety of preclinical studies [28]. EGFR:GRB2 PLA labeling was robust in PDXs established from a patient tumor with an activating mutation in exon 18 of EGFR (encoding a G719A amino acid substitution), and EGFR:GRB2 PLA was strongly reduced in these mice when treated with erlotinib (Fig. 3B). In contrast, PDXs established from a patient tumor with wild-type EGFR and an activating mutation in exon 1 of KRAS (encoding a G12V amino acid substitution) had markedly less intense EGFR:GRB2 PLA, which was minimally affected by erlotinib treatment (Fig. 3B). The EGFR:GRB2 PLA status in EGFR-mutant or KRAS-mutant PDX samples was similar to IHC staining for Tyr1068 phosphorylated EGFR (fig. S2B).
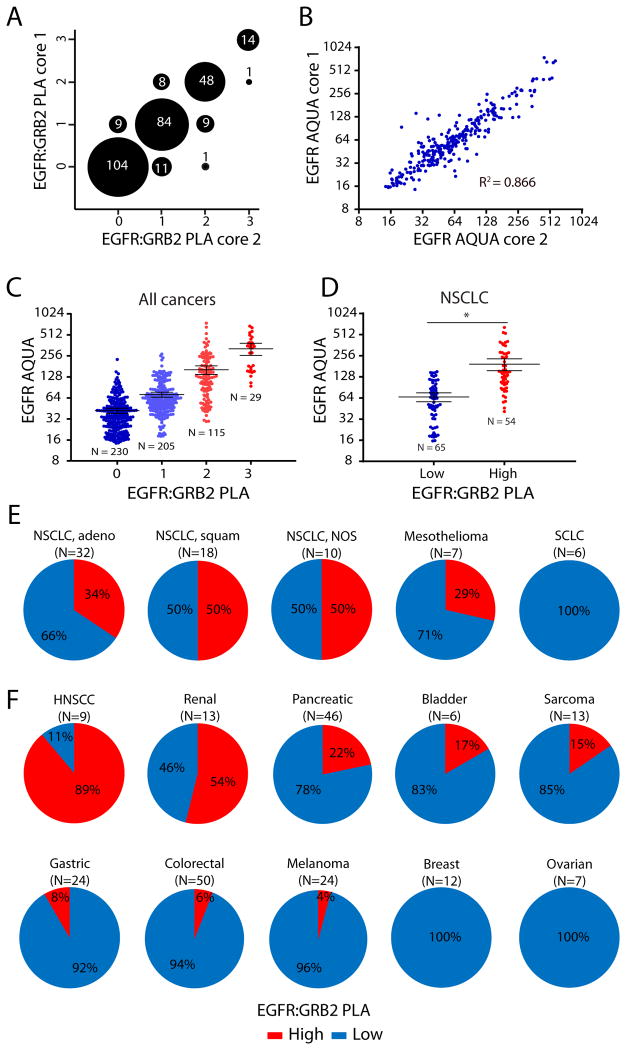
We extended these observations by using EGFR:GRB2 PLA to analyze a tumor microarray containing samples from a panel of 291 PDX tumor types, representing a diverse set of solid tumors from multiple organs of epithelial or non-epithelial origin [29] (fig. S3). We predicted that EGFR:GRB2 PLA could provide additional information related to EGFR activity compared to quantitative immunofluorescence for EGFR abundance. We analyzed the PDX tumor microarray using both EGFR:GRB2 PLA and AQUA for EGFR [30]and observed low variability between duplicate biopsy cores for both assays: the EGFR:GRB2 PLA scores were identical for 86% of PDX tumors (Fig. 4A) and the EGFR AQUA scores were significantly correlated (Fig. 4B). We also observed that PDX tumors with high EGFR:GRB2 PLA scores were more likely to have high EGFR AQUA scores (Fig. 4C). The majority of PDX tumors had intermediate EGFR AQUA scores (32-256), which were widely distributed among EGFR:GRB2 PLA scores (Fig. 4C). Thus, EGFR:GRB2 PLA was not solely a function of the abundance of EGFR and may enable the segregation of specimens with equal amounts of EGFR into different categories based on the degree of EGFR activation.
Fig. 4. EGFR:GRB2 PLA in PDX tumors in mice.
(A and B) Graphs of the variability between replicate cores in a PDX tumor microarray labeled by (A) EGFR:GRB2 PLA or (B) EGFR AQUA. N=289 tumors. For 2 PDX tumors only a single core was available. PLA intensity was scored manually from 0 to 3+ based on the number of foci per cell by blinded observers. For B, P < .0001, Pearson correlation. (C and D) Graphs of the relationship between EGFR:GRB2 PLA and EGFR AQUA for individual cores across all cancer types (C) or in NSCLC PDX tumors (D). Bars represent means ± 95% confidence interval (CI). For C, N=579 cores from 289 tumors. 3 cores were missing either EGFR:GRB2 PLA or EGFR AQUA values. P <.0001, Kruskal-Wallis test. For D, N=119 cores from 60 tumors. Only a single core was available for one tumor. EGFR:GRB2 PLA scores were grouped as Low (0 or 1) or High (2 or 3) . *P < 0.001, Two-tailed Mann-Whitney U-test. (E) Graphs of percent of PDX tumors of the indicated types with EGFR:GRB2 PLA scores grouped as in D. adeno = adenocarcinoma, squam = squamous cell carcinoma, NOS = not otherwise specified. N in parenthesis indicates the number of PDX tumors.
We focused on PDX tumors from lung cancers in more detail because EGFR inhibitors are routinely considered as a therapeutic option in the treatment of NSCLC patients. We segregated EGFR:GRB2 PLA into “low” (0-1) and “high” (2-3) and evaluated the relationship with EGFR protein abundance. Consistent with the analysis of the entire PDX panel, we found that PDX tumors with high EGFR protein abundance were more likely to have high EGFR:GRB2 PLA, but tumors with intermediate EGFR AQUA scores were distributed among both high and low EGFR:GRB2 PLA scoring groups (Fig. 4D). 34% of NSCLC adenocarcinoma PDX tumors, 50% of squamous NSCLC PDX tumors, 50% of NSCLC PDX tumors with poorly defined pathologies, and 29% of pleural mesothelioma PDX tumors had high EGFR:GRB2 PLA scores (Fig. 4E). Moreover, none of the NSCLC adenocarcinoma PDX tumors had activating mutations in EGFR, suggesting that active EGFR signaling is not limited to genomically-encoded aberrant kinase activity. In contrast, high EGFR:GRB2 PLA scores were not observed in small cell lung carcinoma (SCLC) PDX tumors (Fig. 4E), consistent with the low abundance of EGFR in SCLC tumors and lack of clinical efficacy of EGFR-targeted agents in patients with SCLC [31, 32].
We also analyzed additional PDX tumors from other solid tumor types. Some patients with head and neck squamous cell carcinoma (HNSCC) are treated with drugs targeting EGFR [33]. 92% of HNSCC PDX tumors had high EGFR:GRB2 PLA scores (Fig. 4F). High EGFR:GRB2 PLA scores were relatively infrequent in PDX tumors from other solid tumors, except those from renal and pancreatic cancers (Fig. 4F). Despite that colorectal cancer is commonly treated with EGFR-directed therapies [5], only 6% of PDX tumors from colorectal cancers had high EGFR:GRB2 PLA scores (Fig. 4F). Additionally, we observed two sarcoma PDX tumors and a single melanoma PDX tumor with high EGFR:GRB2 PLA scores (Fig. 4F), consistent with the reported involvement of the EGFR pathway in a subset of these malignancies [34-37].
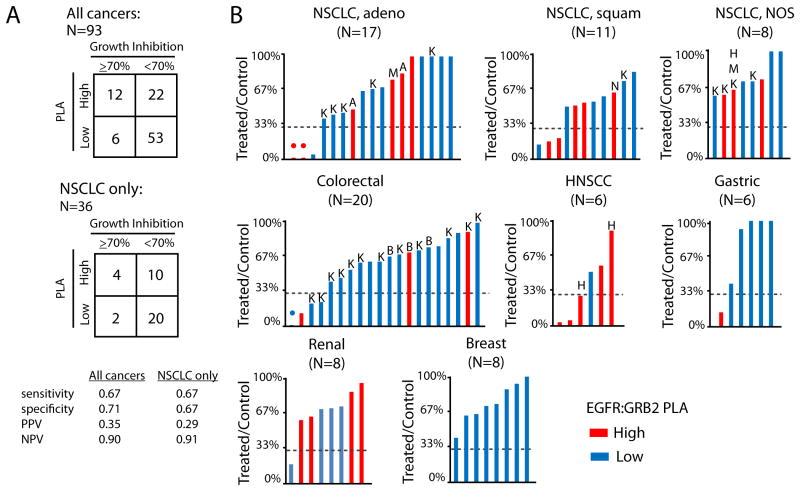
Cetuximab is a monoclonal antibody inhibitor of EGFR, which when used in combination with “standard of care” cytotoxic chemotherapy in patients with NSCLCs with abundant EGFR, improves overall survival relative to chemotherapy alone [14](reference). Cetuximab is approved by the FDA for the treatment of HNSCC [38]) and metastatic colorectal cancers with wild-type KRAS [5]. Cetuximab acts by blocking EGFR and preventing ligand-induced dimerization, thus prevents activation of downstream signaling.). Therefore, we assessed the association of EGFR:GRB2 PLA with tumor response to cetuximab in mice with a diverse library of 93 PDX tumor types (fig. S4). We predicted that tumors with active EGFR signaling (high EGFR:GRB2 PLA signal) would be more likely to respond to inhibition of EGFR than tumors with low EGFR signaling. We performed EGFR:GRB2 PLA on a representative pre-treatment tumor, obtained when mice were randomized to receive either cetuximab or vehicle control. To identify robust responses to cetuximab, we used a threshold of ≥70% inhibition of tumor growth relative to untreated mice, as previously reported with this cohort [29]. We constructed 2×2 contingency tables and calculated assay performance measures (Fig. 5A): Sensitivity of PLA was defined as the proportion of PLA-high responders among all responders, specificity was defined as the proportion of PLA-low non-responders among all non-responders, positive predictive value (PPV) was defined as the proportion of PLA-high-responders among all PLA-high samples, and negative predictive value (NPV) was defined as the proportion of PLA-low non-responders among all PLA-low samples High EGFR:GRB2 PLA scores were associated with cetuximab-response in a combined analysis of all cancer types (Fig. 5A). In a targeted analysis of NSCLC PDX tumors,EGFR:GRB2 PLA did not predict cetuximab-response (Fig. 5A); however, four of six cetuximab-responsive NSCLC PDX tumors, all of which had wild-type EGFR, had high EGFR:GRB2 PLA scores (Fig. 5B). Our cohort of PDX tumors did not include sufficient sample size for other tissues to statistically evaluate other individual cancer types; however, we observed high EGFR:GRB2 PLA scores in three of three cetuximab-responsive HNSCC PDX tumors and one of two cetuximab-responsive colorectal cancer PDX tumors (Fig. 5B). In PDX tumors from gastric cancers (N=6), where cetuximab is not used clinically, we identified a single tumor with a high EGFR:GRB2 PLA score, and this tumor was also the only one with a robust response to cetuximab (Fig. 5B). Furthermore, we did not identify robust responses to cetuximab in PDX tumors from breast cancer where EGFR:GRB2 PLA scores were uniformly low (Fig. 5B).
Figure 5. Predictive capacity of EGFR:GRB2 PLA for cetuximab response in PDX tumors.
(A) 2×2 contingency tables demonstrating EGFR:GRB2 PLA analytical performance to predict robust response to cetuximab (defined as >70% tumor reduction in treated compared to untreated mice). All cancers: P = 0.006; NSCLC only: P = 0.18, Two-tailed Fisher's exact test. (B) Graphs of the % of tumor inhibition in mice treated with cetuximab (50 mg/kg; day 0, 7, 14) compared to untreated mice and sacrificed on day 28. Bars are colored to reflect EGFR:GRB2 PLA score. Lines at 30% (representing 70% reduction in tumor size) demarcate the cutoff between responders (below) and non-responders (above). Drug resistance-associated mutations and molecular events are also annotated. K = KRAS mutant (G12×), B = BRAF mutant, N = NRAS mutant, M = phosphorylated Met, A = phosphorylated AKT, H = phosphorylated HER3.
Detection of EGFR:GRB2 signaling complexes in NSCLC patient specimens
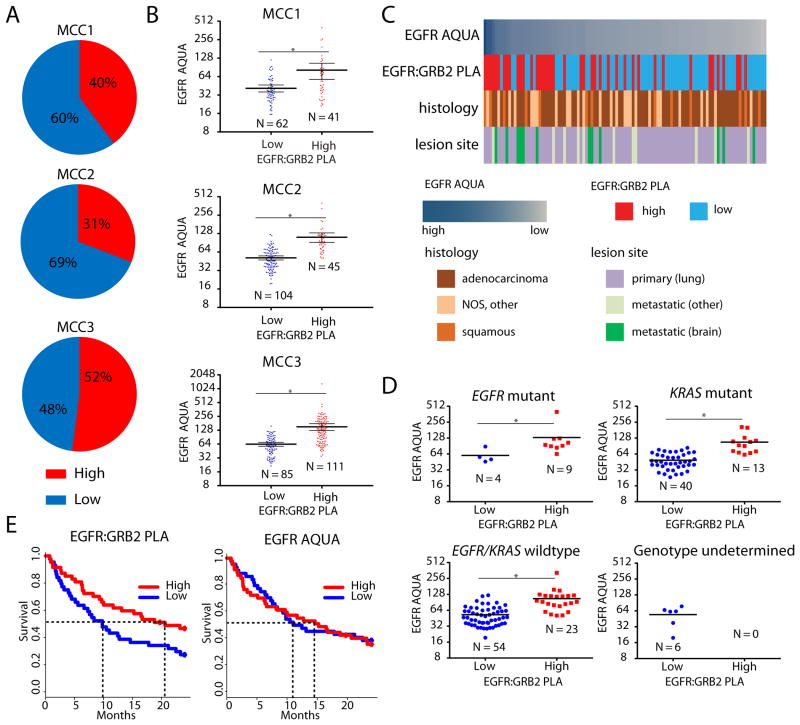
To determine if EGFR:GRB2 PLA could predict clinical outcomes and drug responses in humans, we analyzed tissues from 350 patients with different stages, molecular etiology, and pathological subtypes of NSCLC. We performed EGFR:GRB2 PLA and EGFR AQUA on sequential sections of biopsy cores on tumor microarrays from three patient cohorts [Moffitt Cancer Center (MCC) cohort 1 (MCC1), MCC2, and MCC3]. A panel of blinded observers manually scored the intensity of EGFR:GRB2 PLA across all specimens (fig. S5A), and we found significant concordance among observers (fig. S5B)and a strong correlation between duplicate cores (fig. S5C). We observed increased variability of EGFR AQUA between duplicate cores relative to that observed in PDX models, suggesting that a portion of the PLA core-to-core variability could be driven by spatial variation in EGFR abundance (fig. S7). The percent of tumors with high EGFR:GRB2 PLA scores was similar among the three patient cohorts (Fig 6A). Tumors with high EGFR:GRB2 PLA scores had higher EGFR AQUA scores compared to tumors with low EGFR:GRB2 PLA scores in all three cohorts (Fig. 6B). However, in some individual cases there was discordance between EGFR:GRB2 PLA and EGFR AQUA scores, but this effect did not correlate with histology and was observed in both primary and metastatic tumors (Fig. 6C).
Figure 6. EGFR:GRB2 PLA in NSCLC patient tumor specimens.
(A) Distributions of EGFR:GRB2 PLA status in 3 NSCLC patient cohorts (MCC1 N=103; MCC2 N=149; MCC3 N= 98). (B) Relationship between EGFR:GRB2 PLA status and EGFR protein abundance determined by AQUA across individual cores in each cohort (MCC3 had 2 cores per patient). Bars represent means ± 95% CI. * P < 0.001, Two-tailed Mann-Whitney U-test. (C) Examples of EGFR:GRB2 PLA status across multiple histologies. Each column represents a single patient specimen. The frequency of high EGFR:GRB2 scores was higher in metastatic brain lesions (P<0.0001, 2-tailed Fisher's exact test). (D) Relationship of EGFR:GRB2 PLA status with the indicated genotypes and relationship to EGFR AQUA in the MCC2 cohort. * indicates P = 0.0112 (EGFR mutant), P < 0.0001 (KRAS mutant), P < 0.0001 (EGFR wild-type) Two-tailed Mann-Whitney U-tests. (E) Survival curves for the MCC3 cohort stratified according to EGFR:GRB2 PLA or EGFR AQUA. P = 0.045 (EGFR:GRB2 PLA), P = 0.81 (EGFR AQUA), Log-rank test.
We investigated the relationship between EGFR:GRB2 PLA and tumor characteristics using tumors from MCC1, which consisted of 103 patients with primary or metastatic NSCLC of multiple histologies, including both adenocarcinoma and squamous cell carcinoma. We found high EGFR:GRB2 PLA scores in 40% of tumors from patients from MCC1 (Fig. 6A-C), and these tumors were enriched for metastatic brain lesions, consistent with recent results with reverse phase protein microarrays showing increased EGFR pathway activation in NSCLC brain metastases [39].
We investigated the relationship between EGFR:GRB2 PLA and the underlying genetic etiology using tumor from MCC2, which consisted of 149 patients with early-stage NSCLC with known EGFR and KRAS mutation status. Whereas 31% of all tumors from this cohort had high EGFR:GRB2 PLA scores (Fig. 6A), 69% (9 of 13) of tumors with mutant EGFR had high EGFR:GRB2 PLA scores (Fig. 6D and fig. S6). Moreover, 25% (13 of 53) of KRAS-mutant tumors and 30% (23 of 77) of tumors without EGFR and KRAS mutations had high EGFR:GRB2 PLA scores (Fig. 6D). Some patients with wild-type EGFR benefit from therapies involving EGFR inhibitors [40, 41]. Thus, using EGFR:GRB2 PLA to annotate such tumors with increased EGFR signaling in the absence of known driver mutations could potentially identify patients responsive to therapeutic EGFR inhibition.
We evaluated the relationship between EGFR:GRB2 PLA scores and response to EGFR-targeted therapies using tumors from MCC3, which included 91 patients with known clinical outcome data and who received the EGFR inhibitors erlotinib or gefitnib. Therapeutic benefits of EGFR inhibitors are observed almost exclusively within the first year due to the emergence of resistance [42]. Thus, we evaluated overall survival using an endpoint of two years of follow-up.. High EGFR:GRB2 PLA was associated with improved overall survival and a doubling in median overall survival (Fig. 6E). In contrast, EGFR AQUA did not effectively separate survival outcomes in the same cohort (Fig. 6F). Analysis of survival in all patients with available outcome data revealed a similar, but not statistically significant, trend of the association between high EGFR:GRB2 PLA scores, but not EGFR AQUA scores, and improved overall survival (fig. S7). Thus, annotation of tumors with EGFR signaling-associated complex abundance by EGFR:GRB2 PLA may be able to identify patients who are more likely to receive benefit from EGFR inhibitor-based therapies.
Discussion
Here, we used EGFR:GRB2 PLA in cell lines, xenografts and patient tumors to visualize signaling-associated protein complexes as measures of targetable signaling activity in human tumors and provide initial evidence that annotation of tumors with knowledge of EGFR signaling-associated protein complexes has the potential to guide therapeutic decisions. Using EGFR:GRB2 PLA in patient specimens and PDX tumors revealed that highly active EGFR signaling was not limited to tumors with activating mutations in EGFR. We found that PDX tumorswith high EGFR:GRB2 PLA were more likely to respond to cetuximab, but EGFR:GRB2 PLA was not entirely predictive of response. The relatively high NPV (.90) in PDX tumors, suggests that patients with little or no detectable EGFR:GRB2 PLA signal would not respond to EGFR-targeted therapies. However, the relatively low specificity (0.71) in PDX tumors may suggest that annotation of tumors with active EGFR:GRB2 signaling alone may not be sufficient to predict response.
Several molecular mechanisms enable tumors to activate compensatory survival pathways enabling drug resistance [43]. Mutations in genes encoding proteins downstream of EGFR or aberrant phosphorylation and activation of Met, AKT, and ERBB3, occur in the PDX tumors used in the current study[29]. In particular, colorectal cancer-derived PDX tumors in cetuximab-treated mice comprise 60% with KRAS mutations, 15% with BRAF mutations, both of which are downstream of EGFR and can be drivers in colorectal cancer [5]. The 25% of colorectal cancer-derived PDX tumors without known oncogenic mutations in either of these two drivers likely represents the population where EGFR:GRB2 PLA would have the most utility as a predictive marker. In patient samples, one tumor from a patient in MCC2 had both an L858R mutation and a T790M “gatekeeper” mutation in EGFR, and thus would not be expected to respond to erlotinib, but had a high EGFR:GRB2 PLA score. Thus, annotation by EGFR:GRB2 PLA may have variable capacity to predict therapeutic responses depending on the underlying genetic landscapes. Moreover, this approach will likely be most valuable in patient populations where EGFR-directed therapies are not initially considered, such as EGFR wildtype tumors, lung squamous cell carcinoma, and others, by identifying therapeutic targets that would otherwise have been missed. Current efforts are underway to prospectively validate EGFR:GRB2 PLA in well-annotated specimens from clinical trials..
PLA to detect protein complexes is a powerful tool to facilitate the detection of disease-associated molecular events in situ. PLA enables detection of protein complexes in a single paraffin section of tumors or other tissue specimens, thereby providing information about the activation of signal transduction in clinically relevant tissues. In addition, PLA can be used to detect protein complexes in archival specimens. Using PLA in patient samples may help translate new knowledge of protein-protein interactions from mass spectrometry and biochemical studies into patient-based materials. Because PLA is an in situ method, it enables assessment of spatial heterogeneity in signaling, which may be particularly relevant at tumor-stroma interfaces. Unlike PLA, nearly all other methods used to characterize protein complexes in situ require genetic manipulation by transient transfection of plasmid DNA or infection with viral vectors and thus are unsuitable for FFPE material. PLA is a highly sensitive method that generates an easily detectable signal that can be visualized with a standard epifluorescence microscope, obviating the need for confocal microscopy or other advanced optical systems. PLA to detect protein complexes uses antibodies that target total protein as opposed to phosphorylation-specific antibodies, which can be difficult to generate and often cannot be used in FFPE samples. Furthermore, because some post-translational modification events, including phosphorylation, are involved in mediating protein complexes, PLA for protein complexes can directly assess the functional endpoint of these signals.
The method of quantification of PLA-labeled tissues is an important issue to consider. We used a qualitative ordinal scoring system (0 to 3+ rating by blinded observers based on the number of foci per cell) to assess the EGFR:GRB2 PLA signal and observed good inter-rater reliability and agreement with independent quantitative measures (EGFR AQUA). Conversion of PLA signals into continuous variables by algorithms in digital imaging software may be necessary to further develop PLA as an analytical platform. Continuous scoring methods may facilitate determination of thresholds and help elucidate the relationship between the abundance of signaling complexes and the degree of drug response. However, because of the long exposure time needed to visualize the PLA signal, our initial attempts at automated quantification were hampered by high fluorescent background arising from red blood cells, regions of necrotic tissue, or other stromal elements in some specimens. Furthermore, when the PLA signal is high (>20 foci per cell) individual foci merge, preventing accurate automated quantification and suggesting that PLA has a limited dynamic range for analog conversion. Thus, given the current widespread and routine use of ordinal scoring systems like the one used in this study for clinical samples labeled by immunofluorescence or fluorescence in situ hybridization (FISH) [44], we believe that ordinal scoring systems have future utility for the assessment of protein complexes by PLA.
Annotation of signaling-associated protein complexes using PLA may complement currently available genotypic assays, providing a more comprehensive tumor profile. We found that EGFR:GRB2 PLA was a better predictor of clinical response to EGFR inhibitors in patient samples than EGFR AQUA. EGFR:GRB2 PLA could be clinically informative to identify EGFR activity associated with resistance to other targeted therapies. For example, compensatory activation of EGFR signaling is linked to resistance to inhibition of MET [45], BRAF [46], and ALK kinases [47, 48]. Using PLA to monitor other signaling-associated protein complexes may also have clinical utility. For example, PLA could be used to monitor activation of the receptor tyrosine kinases MET and FGFR by either genetic mutation or signals from the tumor microenvironment. Activation of MET or FGFR can play a critical role in bypass signaling associated with cancer drug resistance [1, 2] and several inhibitors of these kinase are in clinical development [49, 50]. Furthermore, recently-described multiplexing strategies could be implemented to develop panels of PLAs for different protein complexes, enabling simultaneous characterization of multiple signaling pathways on a single tissue slide.
Using PLA to detect protein complexes could have utility across a wide spectrum of protein complexes in different types of diseases. PLA could be used to assess rewiring of cellular circuits, by looking directly at the formation or dissolution of protein complexes in different circumstances (for example, drug resistance). Technical hurdles associated with the identification and quantitation of protein complexes across larger numbers of human disease tissues have hampered the study of network rewiring in cancers and other diseases. However, PLAs created using knowledge from signal transduction biology may enable scaling to study hundreds or thousands of tumors. In summary, the use of protein complex annotation by PLA is a novel method for disease annotation and prediction of treatment response with important implications for targeted therapies in a wide variety of diseases.
Methods
Proximity ligation assays
Cells were plated at 25-50 × 104/well in 8-well chamber slides, incubated for 40 hours, fixed with 4% paraformaldehyde, and permeabilized with 0.5% Triton-X 100. For all tissue experiments, slides containing 5 μm sections were rehydrated through xylene and graded alcohols. Heat-induced epitope retrieval was carried out in Tris-EDTA (pH = 9) in a pressure cooker for 20 minutes and then cooled for 20 minutes. Non-specific binding was blocked by incubation with 1.5% BSA at room temperature for 30 min. Primary antibodies were incubated overnight in 1.5% BSA in 0.5% PBS-Tween20 using rabbit antibody targeting EGFR (clone D38B1, Cell Signaling Technology) and mouse antibody targeting GRB2 (clone 81, BD Biosciences). PLA probes were rabbit (-) and mouse (+) and were detected with DuoLink in situ PLA Far Red kit (O-Link Biosciences, Uppsala, Sweden). AlexaFluor488-conjugated anti-cytokeratin was used to demarcate epithelial regions (clone AE1/AE3, eBiosciences).
Confocal images were acquired on a Leica TCS SP5 AOBS laser scanning confocal microscope through a 40× 1.25NA Plan Apochromat oil immersion objective lens (Leica Microsystems CMS GmbH, Germany). 405 diode and HeNe 647 laser lines were applied to excite the samples and tunable emissions were used to minimize crosstalk between fluorochromes. Z stack (0.5μm thick slices) images for each sample were captured with photomultiplier detectors and maximum projections were prepared with the LAS AF software version 2.6 (Leica Microsystems, Germany). Additional fluorescent images were acquired on a fully automated, upright Zeiss Axio- ImagerZ.1 microscope with a 40× 1.25NA oil immersion objective, and DAPI and Cy5 filter cubes. Images were produced using the AxioCam MRm CCD camera and Axiovision version 4.6 software suite (Carl Zeiss Inc, Germany). All tissue-based PLA and AQUA analysis images were acquired using a 20× objective lens (dry) on an AQUA workstation (PM2000, HistoRX) equipped with a fully motorized stage and DAPI, Cy3, FITC and Cy5 filter cubes. Images were saved as individual channels and exported as merged RGB .tiff images.
Immunohistochemistry
Tissue sections were rehydrated and antigens retrieved as described in supplemental methods. Non-specific binding was blocked by incubation with 1.5% BSA, and incubated overnight in BSA in 0.5% PBS-Tween20 using rabbit antibody targeting phospho-EGFR (Tyr1068) (clone D7A5, Cell Signaling Technology). Slides were washed twice with PBST, incubated with Envision+ anti-rabbit (Dako,K4011)) for 1 hr and visualized by DAB. Slides were countered-stained with hematoxylin, rehydrated and hard mounted.
Tissue microarrays
Patient tissue microarrays for MCC1, MCC2, and MCC3 were constructed by the Tissue Procurement Core Facility at Moffitt Cancer Center under an IRB-approved protocol. Patients whom had previously provided informed consent were identified using electronic medical records by an “honest broker.” Donor paraffin blocks were obtained from core biopsies and surgical resection specimens. Core sizes were 1 mm (MCC1) or 0.6 mm (MCC2 and MCC3). For MCC3, two cores per donor were used. Blocks were sectioned at 5 μm, floated onto positively-charged slides, and baked at 60°C for 1 hour. EGFR:GRB2 PLA was manually quantified using a scoring criteria based on foci per cell (0, none detectable; 1+, 1-5 foci per cell; 2+, >5-20 foci per cell; 3+, >20 foci per cell) and annotated as “high” (2+ to 3+ in both cores) or “low” (0 to 1+). Personnel conducting laboratory analyses were blinded to all patient information associated with clinical specimens. PDX tissue microarrays were constructed created at Oncotest GmbH (Freiburg, Germany) using FFPE blocks of PDX tumors. Each core punch was 1mm with two core punches taken from each donor block, spotted in duplicate across 5 slides. For PDX tissues, laboratory analyses were conducted by blinded personnel at a separate location.
Statistical analysis
For EGFR AQUA, non-parametric tests were utilized because EGFR expression values did not follow Gaussian distribution. For PDX specimens treated with cetuximab, no data were available to enable an accurate estimate of effect of size for power calculations; therefore, all available specimens were used to analyze predictive value. For survival analysis in human lung cancer specimens, no data were available to enable an accurate estimate of effect of size for power calculations; therefore, all available specimens were used for analysis (N=91). We used SAS 9.3 (SAS Institute, Cary, NC), R 2.13 and above, Matlab 2009b, and GraphPad Prism 6.02 for the statistical analyses.
Automated quantitative analysis (AQUA)
After heat-induced epitope retrieval and blocking, primary antibodies were incubated overnight in 1.5% BSA-PBST using rabbit anti-EGFR (clone D38B1, Cell Signaling Technology) and detected using AlexaFluor 647-labeled goat targeting rabbit secondary antibodies (Invitrogen). Murine pan-cytokeratin (clone AE1/AE3, Dako) was used to demarcate epithelial regions (tumor mask) and detected using AlexaFluor 555-labeled goat targeting mouse secondary antibodies (Invitrogen). Images were acquired on a PM2000 instrument and AQUA scores were obtained using AQUAnalysis (version 2.3.4, HistoRX).
Cell lines and xenograft models
Cell lines were maintained in RPMI 1640 supplemented with 10% fetal bovine serum. All cell lines have been maintained in a central repository at Moffitt Cancer Center since 2008, have been authenticated by STR analysis (ACTG Inc., Wheeling, IL), and are routinely tested for mycoplasma contamination. H1650 and H1299 cells were obtained from ATCC. PC9 cells were provided by Matt Lazarra (MIT, Boston, MA). H1648 cells were provided by John Minna (UTSW, Dallas, TX). HCC827 cells were provided by John Kurie (MD Anderson, Houston, TX). HCC4006 and H322 cells were provided by Paul Bunn (UC Denver, Aurora, CO). H520 cells were provided by Lynn Heasley (UC Denver, Aurora, CO). H1650 and H322 cell line xenografts were established in 4-week old female CD-1 nu/nu mice (Charles River Laboratories) and treated as described [26]. Erlotinib-treated PDXs from NSCLC patients (N=2) were established at Moffitt Cancer Center as described in 4-week old immunodeficient hairless mice Crl:SHO-Prkdc-SCID-Hr-hr[52]. Patients provided written informed consent and all studies were approved by institutional IACUC and IRB. Fresh tissue was obtained from patients undergoing surgical resection; one patient had a KRAS G12V mutation, the other patient had an EGFR G719A, both of which were verified using the OncoCarta Panel. Xenografts from this initial passage were harvested and re-implanted subcutaneously in groups of five mice for each patient sample, with two small pieces per mouse (F2 generation). Tumors were allowed to grow to a size of 1.5 cm, at which point they were harvested, divided into small 3 × 3 × 3 mm pieces, and transplanted to another 18 mice, with two tumors per mouse (F3 generation). Tumors serially transplanted from these mice (F4) were used for experiments. Animals were treated beginning when tumors reached 100mm2 daily for 26 days with 50mg/kg erlotinib by oral gavage.
Multi-histotype PDX panel (N=291) was generated at Oncotest GmbH (Freiburg, Germany) following provisions of German animal welfare acts as described [29]. PDX models were maintained through serial passage and housed at Oncotest. Tumors were serially passaged and animals were randomized into treatment groups when tumors reached approximately 100mm2. A representative tumor was excised before treatment on day 0, fixed in formalin, and embedded into paraffin blocks. For treatment groups, animals were treated either with cetuximab (30 mg/kg) or vehicle control by intraperitoneal injections on days 0, 7, and 14. Animals were euthanized on day 28. Tumor size was measured twice weekly using (width × width × length)/2 measurements.
Quality control (QC) procedures for Tissue Microarrays
We first investigated inter-rater agreement among four raters on the core samples using Kendall's concordance coefficient. The results showed good agreement among raters, and we calculated the average PLA scores among four raters for each core sample. Correlation of the average PLA scores between duplicate samples from the same patient was then examined using Pearson correlation. Ninety-eight patients from the MCC3 cohort each provided two core samples from tumors. Regression models and heat maps was used to detect any potential spatial effects, including row and columns, in tissue microarrays using the average PLA scores of all 226 cores (provided by 102 patients). No significant spatial effects were found in tissue microarrays, and all samples were used in analysis.
Immunoprecipitation and immunoblot analysis
Cells were washed once in cold PBS-1 mM NaVO4, scraped, and lysed in NETN (50 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40) supplemented with phosphatase inhibitor cocktail (Sigma), protease inhibitors (Roche), and 1 mM NaVO4. For immunoprecipitation, anti-GRB2 (sc-255, Santa Cruz) was added to 800 μg lysate overnight. Immune complexes were captured using True-Blot anti-rabbit IP beads (eBiosciences), washed three times in NETN, and resolved by SDS-PAGE. Immunoblotting was performed using antibodies against -EGFR (clone D38B1 Cell Signaling Technology), GRB2 (clone 81, BD), and β-actin (Sigma).
Supplementary Material
Figure S1. PLA assay schematic.
Figure S2. Phosphorylated Tyr1068 EGFR in xenografts.
Figure S3. Distribution of PDX tumors.
Figure S4. Distribution of PDX tumors with cetuximab response data.
Figure S5. Inter-rater reliability and variability between duplicate cores.
Figure S6. Variability of EGFR:GRB2 PLA signal among EGFR-mutant patients.
Figure S7. Overall survival analysis with extended follow-up.
Acknowledgments
This work has been supported in part by the Tissue Procurement, Analytical Microscopy and Biostatistics Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292). The research was made possible through the Total Cancer Care™ Protocol at the Moffitt Cancer Center. We thank Rasa Hamilton (Moffitt Cancer Center) for editorial assistance.
Funding: This work was supported by grants from the National Cancer Institute to E.B.H. (P50-CA119997, R21 CA162178). M.A.S. is partially supported by a fellowship from the Lung Cancer Research Foundation.
Footnotes
Author contributions: MAS designed and performed experiments, analyzed and interpreted data, wrote the manuscript
RH analyzed and interpreted data
KF analyzed and interpreted data, performed statistical analyses
SMH analyzed data
FK annotation of clinical data and patient specimens (Fig 6)
MBS, annotation of clinical data and patient specimens (Fig 6)
VV created patient-derived xenografts (Figs 4, 5)
HHF created patient-derived xenografts (Figs. 4,5)
SA created patient-derived xenografts (Fig 3)
YAC designed experiments, analyzed and interpreted data, performed statistical analyses
EBH designed experiments, analyzed and interpreted data, wrote the manuscript
Data and Materials Availability: All reagents are available upon request.
Competing interests: The authors declare that they have no competing interests. Moffitt Cancer Center has filed a US patent application related to the intellectual property on the use of protein complexes as predictive biomarkers for kinase inhibitor therapies. Soner Altiok is a paid consultant of Pfizer Genetech.
References
- 1.Wilson TR, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487(7408):505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harbinski F, et al. Rescue screens with secreted proteins reveal compensatory potential of receptor tyrosine kinases in driving cancer growth. Cancer Discovery. 2012;2(10):948–59. doi: 10.1158/2159-8290.CD-12-0237. [DOI] [PubMed] [Google Scholar]
- 3.Straussman R, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch FR, et al. Epidermal growth factor receptor inhibition in lung cancer: status 2012. J Thorac Oncol. 2013;8(3):373–84. doi: 10.1097/JTO.0b013e31827ed0ff. [DOI] [PubMed] [Google Scholar]
- 5.Misale S, et al. Resistance to Anti-EGFR Therapy in Colorectal Cancer: From Heterogeneity to Convergent Evolution. Cancer Discov. 2014;4(11):1269–1280. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 6.Boeckx C, et al. Anti-epidermal growth factor receptor therapy in head and neck squamous cell carcinoma: focus on potential molecular mechanisms of drug resistance. Oncologist. 2013;18(7):850–64. doi: 10.1634/theoncologist.2013-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sirotnak FM, et al. Efficacy of cytotoxic agents against human tumor xenografts is markedly enhanced by coadministration of ZD1839 (Iressa), an inhibitor of EGFR tyrosine kinase. Clin Cancer Res. 2000;6(12):4885–92. [PubMed] [Google Scholar]
- 8.Hidalgo M, et al. Phase I and pharmacologic study of OSI-774, an epidermal growth factor receptor tyrosine kinase inhibitor, in patients with advanced solid malignancies. J Clin Oncol. 2001;19(13):3267–79. doi: 10.1200/JCO.2001.19.13.3267. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein NI, et al. Biological efficacy of a chimeric antibody to the epidermal growth factor receptor in a human tumor xenograft model. Clin Cancer Res. 1995;1(11):1311–8. [PubMed] [Google Scholar]
- 10.Lowenstein EJ, et al. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70(3):431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 11.Li J, et al. Perturbation of the mutated EGFR interactome identifies vulnerabilities and resistance mechanisms. Molecular Systems Biology. 2013;9:705. doi: 10.1038/msb.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan PK, et al. Monitoring Interactions between Receptor Tyrosine Kinases and Their Downstream Effector Proteins in Living Cells Using Bioluminescence Resonance Energy Transfer. Molecular Pharmacology. 2007;72(6):1440–1446. doi: 10.1124/mol.107.039636. [DOI] [PubMed] [Google Scholar]
- 13.Koizumi F, et al. Establishment of a human non-small cell lung cancer cell line resistant to gefitinib. Int J Cancer. 2005;116(1):36–44. doi: 10.1002/ijc.20985. [DOI] [PubMed] [Google Scholar]
- 14.Pirker R, et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. The Lancet Oncology. 2012;13(1):33–42. doi: 10.1016/S1470-2045(11)70318-7. [DOI] [PubMed] [Google Scholar]
- 15.Mazières J, et al. Evaluation of EGFR protein expression by immunohistochemistry using H-score and the magnification rule: Re-analysis of the SATURN study. Lung cancer (Amsterdam, Netherlands) 2013 doi: 10.1016/j.lungcan.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 16.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nature Medicine. 2002;8(11):1323–7. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 17.Mascaux C, et al. EGFR protein expression in non-small cell lung cancer predicts response to an EGFR tyrosine kinase inhibitor--a novel antibody for immunohistochemistry or AQUA technology. Clin Cancer Res. 2011;17(24):7796–807. doi: 10.1158/1078-0432.CCR-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature Methods. 2006;3(12):995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 19.Gajadhar AS, et al. In Situ Analysis of Mutant EGFRs Prevalent in Glioblastoma Multiforme Reveals Aberrant Dimerization, Activation, and Differential Response to Anti-EGFR Targeted Therapy. Molecular Cancer Research. 2012;10(3):428–440. doi: 10.1158/1541-7786.MCR-11-0531. [DOI] [PubMed] [Google Scholar]
- 20.Halle C, et al. Phosphorylation of EGFR measured with in situ proximity ligation assay: Relationship to EGFR protein level and gene dosage in cervical cancer. Radiotherapy and Oncology. 2011;101(1):152–157. doi: 10.1016/j.radonc.2011.05.052. [DOI] [PubMed] [Google Scholar]
- 21.Chen TC, et al. Protein Phosphorylation Profiling Using an In Situ Proximity Ligation Assay: Phosphorylation of AURKA-Elicited EGFR-Thr654 and EGFR-Ser1046 in Lung Cancer Cells. PLoS One. 2013;8(3):e55657. doi: 10.1371/journal.pone.0055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukohara T, et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J Natl Cancer Inst. 2005;97(16):1185–94. doi: 10.1093/jnci/dji238. [DOI] [PubMed] [Google Scholar]
- 23.Batzer AG, et al. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Molecular and Cellular Biology. 1994;14(8):5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 26.Song L, et al. Antitumor efficacy of the anti-interleukin-6 (IL-6) antibody siltuximab in mouse xenograft models of lung cancer. Journal of Thoracic Oncology. 2014;9(7):974–82. doi: 10.1097/JTO.0000000000000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng SC, et al. Correlation of specific keratins with different types of epithelial differentiation: monoclonal antibody studies. Cell. 1982;30(2):361–72. doi: 10.1016/0092-8674(82)90234-3. [DOI] [PubMed] [Google Scholar]
- 28.Hidalgo M, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4(9):998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krumbach R, et al. Primary resistance to cetuximab in a panel of patient-derived tumour xenograft models: Activation of MET as one mechanism for drug resistance. European Journal of Cancer. 2011;47(8):1231–1243. doi: 10.1016/j.ejca.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Dimou A, et al. Standardization of epidermal growth factor receptor (EGFR) measurement by quantitative immunofluorescence and impact on antibody-based mutation detection in non-small cell lung cancer. Am J Pathol. 2011;179(2):580–9. doi: 10.1016/j.ajpath.2011.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaseda S, et al. Expression of epidermal growth factor receptors in four histologic cell types of lung cancer. J Surg Oncol. 1989;42(1):16–20. doi: 10.1002/jso.2930420106. [DOI] [PubMed] [Google Scholar]
- 32.Moore AM, et al. Gefitinib in patients with chemo-sensitive and chemo-refractory relapsed small cell cancers: a Hoosier Oncology Group phase II trial. Lung Cancer. 2006;52(1):93–7. doi: 10.1016/j.lungcan.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Sharafinski ME, et al. Epidermal growth factor receptor targeted therapy of squamous cell carcinoma of the head and neck. Head Neck. 2010;32(10):1412–21. doi: 10.1002/hed.21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes DP, et al. Cell surface expression of epidermal growth factor receptor and Her-2 with nuclear expression of Her-4 in primary osteosarcoma. Cancer Res. 2004;64(6):2047–53. doi: 10.1158/0008-5472.can-03-3096. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen TO, et al. Tissue microarray validation of epidermal growth factor receptor and SALL2 in synovial sarcoma with comparison to tumors of similar histology. Am J Pathol. 2003;163(4):1449–56. doi: 10.1016/S0002-9440(10)63502-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boone B, et al. EGFR in melanoma: clinical significance and potential therapeutic target. J Cutan Pathol. 2011;38(6):492–502. doi: 10.1111/j.1600-0560.2011.01673.x. [DOI] [PubMed] [Google Scholar]
- 37.Ivanov VN, Hei TK. Combined treatment with EGFR inhibitors and arsenite upregulated apoptosis in human EGFR-positive melanomas: a role of suppression of the PI3K-AKT pathway. Oncogene. 2005;24(4):616–26. doi: 10.1038/sj.onc.1208125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen MH, et al. Approval summary: Cetuximab in combination with cisplatin or carboplatin and 5-fluorouracil for the first-line treatment of patients with recurrent locoregional or metastatic squamous cell head and neck cancer. Oncologist. 2013;18(4):460–6. doi: 10.1634/theoncologist.2012-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Improta G, et al. Protein pathway activation mapping of brain metastasis from lung and breast cancers reveals organ type specific drug target activation. J Proteome Res. 2011;10(7):3089–97. doi: 10.1021/pr200065t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu CQ, et al. Role of KRAS and EGFR As Biomarkers of Response to Erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. Journal of Clinical Oncology. 2008;26(26):4268–4275. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 41.Ferté C, et al. Durable responses to Erlotinib despite KRAS mutations in two patients with metastatic lung adenocarcinoma. Annals of Oncology. 2010;21(6):1385–1387. doi: 10.1093/annonc/mdq153. [DOI] [PubMed] [Google Scholar]
- 42.Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nature Medicine. 2013;19(11):1389–400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sequist LV, et al. Genotypic and Histological Evolution of Lung Cancers Acquiring Resistance to EGFR Inhibitors. Science Translational Medicine. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 45.Qi J, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res. 2011;71(3):1081–91. doi: 10.1158/0008-5472.CAN-10-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin L, et al. Mapping the molecular determinants of BRAF oncogene dependence in human lung cancer. Proc Natl Acad Sci U S A. 2014;111(7):E748–57. doi: 10.1073/pnas.1320956111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanizaki J, et al. Activation of HER family signaling as a mechanism of acquired resistance to ALK inhibitors in EML4-ALK-positive non-small cell lung cancer. Clin Cancer Res. 2012;18(22):6219–26. doi: 10.1158/1078-0432.CCR-12-0392. [DOI] [PubMed] [Google Scholar]
- 48.Katayama R, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4(120):120ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gavine PR, et al. AZD4547: An Orally Bioavailable, Potent, and Selective Inhibitor of the Fibroblast Growth Factor Receptor Tyrosine Kinase Family. Cancer Research. 2012;72(8):2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 50.Spigel DR, et al. Randomized Phase II Trial of Onartuzumab in Combination With Erlotinib in Patients With Advanced Non-Small-Cell Lung Cancer. Journal of Clinical Oncology. 2013 doi: 10.1200/JCO.2012.47.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leuchowius KJ, et al. Parallel visualization of multiple protein complexes in individual cells in tumor tissue. Molecular & Cellular Proteomics. 2013;12(6):1563–71. doi: 10.1074/mcp.O112.023374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kreahling JM, et al. Wee1 inhibition by MK-1775 leads to tumor inhibition and enhances efficacy of gemcitabine in human sarcomas. PLoS One. 2013;8(3):e57523. doi: 10.1371/journal.pone.0057523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. PLA assay schematic.
Figure S2. Phosphorylated Tyr1068 EGFR in xenografts.
Figure S3. Distribution of PDX tumors.
Figure S4. Distribution of PDX tumors with cetuximab response data.
Figure S5. Inter-rater reliability and variability between duplicate cores.
Figure S6. Variability of EGFR:GRB2 PLA signal among EGFR-mutant patients.
Figure S7. Overall survival analysis with extended follow-up.