Abstract
The Tn antigen, which arises from mutation in the Cosmc gene is one of the most common tumor associated carbohydrate antigens. Cosmc resides in X24 encoded by a single gene and functions as a specific molecular chaperone for T-synthase. While the Tn antigen cannot be detected in normal cells, Cosmc mutations inactivate T-synthase and consequently result in Tn antigen expression within certain cancers. In addition to this Cosmc mutation-induced expression, the Tn antigen is also expressed in such cell lines as Jurkat T, LSC and LS174T. Whether the Cosmc mutation is present in the colon cancer cell line HT-29 is still unclear. Here, we isolate HT-29-Tn+ cells from HT-29 cells derived from a female colon cancer patient. These HT-29-Tn+ cells show a loss of the Cosmc gene coding sequence (CDS) leading to an absence of T-synthase activity and Tn antigen expression. Additionally, almost no methylation of Cosmc CpG islands was detected in HT-29-Tn+ as well as in HT-29-Tn- and Tn- tumor cells from male patients. In contrast, the methylation frequency of CpG island of Cosmc in normal female cells was ~50%. Only one active allele of Cosmc existed in HT-29-Tn+ and HT-29-Tn- cells as based upon detection of SNP sites. These results indicate that Tn antigens expression and T-synthase inactivity in HT-29-Tn+ cells can be related to the absence of CDS in Cosmc active alleles, while an inactive allele deletion of Cosmc in HT-29 cells has no influence on Cosmc function.
Keywords: Tn antigen, Cosmc, T-synthase, colon cancer, CpG methylation
Introduction
T-synthase, a key enzyme of the O-glycosylation process, transforms a galactose (Gal) on GalNAc-α-Ser/Thr (Tn antigen) to generate Galβ1-3GalNAc-α-Ser/Thr (T-antigen). These T antigens then extend and branch to form the complex O-glycans [1]. O-glycans in human glycoproteins and mucins as derived from Tn antigen precursors play important roles in vascular biology angiogenesis, lymphangiogenesis and leukocyte trafficking [2-5]. Aberrant alterations of O-glycosylations are generally associated with tumor carbohydrate antigen expression [6-8]. Tn antigen is one of the most common tumor associated carbohydrate antigens. It is expressed in many human carcinomas, including breast carcinomas, cervical, colon, ovarian, stomach and prostate cancers [9-11]. In addition, Tn antigen expression is associated with the potential for metastasis and poor prognosis of tumor cells [12-16]. However, the only currently known genetic basis for controlling Tn antigen expression in humans involves the functional loss of Cosmc, an endoplasmic reticulum-localized molecular chaperone encoded by a single gene on Xq24. As a molecular chaperone of T-synthase, Cosmc promotes the correct folding of T-synthase into unique functional structures that can then specifically interact with denatured T-synthase in vitro [17]. Ablation of the X-linked Cosmc gene completely causes embryonic lethality and Tn antigen expression in many tissues of mice [18]. Dysfunction of Cosmc is associated with T-synthase but not with biosynthesis of other glycosyltransferase [19]. Additionally, transfecting WtCosmc into Tn positive tumor cells restores T-synthase activity and reduces Tn antigen expression [20].
Loss of Cosmc function eliminates T-synthase activity and consequent Tn antigen expression in several human tumors [5], as well as in other diseases, such as Tn syndrome and IgA nephropathy [21,22]. Interestingly, different types of Cosmc mutations are present in Tn+ cells as derived from various diseases associated with Tn antigen expression. The types of Cosmc mutations present in certain tumors and Tn syndrome patients have been summarized in the literature [5]. Recently, Rongjuan Mi, et al. [23] observed an epigenetic silencing of Cosmc due to hypermethylation of its promoter in Tn4 cells, derived from Tn-positive lymphocytes of a male patient with Tn syndrome. However, the mechanisms of Tn antigen expression in IgA nephropathy remain unclear.
We recently discovered that HT-29, a colon cancer cell line derived from a female donor, was mixed with Tn+ and Tn- cells. This two phenotype cells were sorted from HT-29 population by anti-Tn antibody, and designated HT-29-Tn+ and HT-29-Tn- cells. Within HT-29-Tn+ cells, a deletion of the coding region sequence (CDS) in Cosmc resulted in lacking T-synthase activity and Tn antigen expression. Located on X24, Cosmc supposed to have two alleles in normal cells from a female. However, while single nucleotide polymorphism (SNP) sites, which may reflect the numbers of alleles in Cosmc [24], could be detected in cells from a normal female volunteer in our study, it did not exist in HT-29-Tn+ and HT-29-Tn- cells. Clearly, it is unlikely to obtain the normal cells from the female patient who had HT-29 cells. Under the conditions that the normal cells in the female patient from which HT-29 cells derived contained the same SNPs in Cosmc non-coding region as we discovered in normal female volunteer cells, we considerate that the inactive allele of Cosmc in HT-29 cells most likely would have been deleted due to the presence of a tumor, but it would not affect Cosmc function and the absence of CDS only happened in Cosmc active allele in HT-29-Tn+ cells. Alternatively, the deletion CDS of Cosmc in HT-29-Tn+ cells would need to occur on two alleles simultaneously, which is possible but very unlikely. In addition, X-chromosome inactivation (XCI) is an epigenetic mechanism that silences most X-chromosome genes to compensate for gene dosage between males and females [25]. Histone modifications and CpG island methylations are responsible for the majority of gene silencing on inactive X-chromosomes [26]. Findings from previous studies suggest that although differences exist in the CpG islands methylation of X-linked gene between males and females, the overall expression levels were indistinguishable [27]. To identify whether the inactive allele of Cosmc was deleted or not, the CpG methylation in normal female cells and in tumor cells were analyzed by Bisulfite Sequencing PCR (BSP). We found that CpG islands in a normal female cell population showed an ~50% methylation, perhaps due to X-chromosome inactivation, whereas very little methylation was observed in BGC823, HT-29-Tn+ and HT-29-Tn- cells. These results were consistent with our expectation that an inactive X-chromosome in Cosmc as produced by CpG island methylation in HT-29 cells had been deleted. Therefore, only a single DNA sequence was detectable in HT-29-Tn+ and HT-29-Tn- cells. Collectively, these findings suggest that an aberrant Tn antigen expression in HT-29-Tn+ cells results from a CDS deficiency in the active allele of Cosmc. Additionally, the absence of an inactive allele in HT-29 cells, including HT-29-Tn+ and HT-29-Tn- cells, does not affect Cosmc function. Such evidence provides a novel foundation for the study of human diseases associated with abnormal Tn antigen expression and offers new approaches for the diagnosis and therapy of human colorectal cancer.
Materials and methods
Cell culture and PBMC
The human colorectal carcinoma cell line HT-29 and human Gastric adenocarcinoma cell line BGC823 were purchased from the American Type Culture Collection (ATCC), Human Leukemia cells Jurkat [Clone E6-1] were kindly provided by Professor Jiang Nan Xue of Bin Zhou Medical University. All cell lines, including Tn+ and Tn- cells that separated by immune magnetic bead from HT-29 cell lines, and were cultured in RPMI1640 media containing 10% FBS (Hycolon) in 5% CO2 at 37°C.
Normal peripheral blood mononuclear cells (PBMC) as obtained from a healthy female donor, a volunteer from Bin Zhou Medical University Affiliated Hospital, were separated from heparinized blood with Ficoll density gradient centrifugation. This study was approved by the Institutional Review Board of Binzhou Medical University (IRB 2009016). A written informed consent was obtained from the participant.
Flow cytometry and immunofluorescence staining
Cell suspensions were prepared at densities of 1 × 106/100 μL. Cells were divided into two fractions. Initially, 5 μL of mouse anti-Tn mAb (IgM, diluted at 1:10 with PBS) kindly provided by Dr. Tongzhong Ju of Emory University School of Medicine in Altanta, were added to one fraction. The controls were incubated with control mouse IgM. All fractions were incubated at 4°C for 60 min. Cells were then incubated with secondary antibody-FITC-labeled goat anti-mouse IgM (Santa Cruz, Sc-2082). After incubation, cells were washed twice with PBS. Cells were analyzed on flow cytometer (BD FACS Aria III). For immunofluorescence staining, 5 × 105 cells were seeded in 24 well cell culture clusters (Corning Incorporated) and cultured overnight at 37°C. The cells fixed with 4% paraformaldehyde for 30 min, blocked with 1% BSA for 1 h, and then incubated with primary antibody anti-Tn mAb (IgM, diluted at 1:200) for 24 h at 4°C. After three washings with PBS the cells were incubated with FITC-labeled secondary antibody at room temperature for 1 h. Cells were again washed three times with PBS and nuclei were stained with DAPI. Stained cells were then observed under a fluorescence microscope (Olympus, Japan) and the images were recorded at 10 × magnification.
Cell sorting by microBeads
Cells were harvested with Trypsin-EDTA and then washed with Buffer (PBS containing 0.5% BSA and 2 mM EDTA, PH 7.2). The cell pellet was resuspended with Buffer, with approximately 9 × 106 cells and labeled with primary mouse anti-Tn IgM mAb. Cells were washed thrice with the Buffer and then incubated with Anti-mouse MicroBeads (Miltenyi Biotec) for 20 min in 4°C refrigerator. Cells were agitated at 5 min intervals to maintain the suspension. Cells were then washed twice, resuspended in 500 μL Buffer and subjected to magnetic separation according to the manufacture’s recommendation. Tn+ and Tn- cells were sorted and collected.
Preparation of cytoplasmic extraction and enzyme activity assays
Approximately 6-8 × 106 cells were collected to generate a cytosolic fraction using Nuclear and Cytoplasmic Extraction Reagents (Pierce). The cytoplasmic protein concentration was determined using the BCA Protein Assay Kit (Pierce) with bovine serum albumin (BSA) used as a standard according to the manufacture’s instructions. T-synthase activity was measured with a fluorescent assay described previously [28]. Briefly, GalNAc-α-(4-MU) was used as the receptor and UDP-Gal as the donor. 50 μL of reaction system containing 1000 μM GalNAc-α-4-(MU), 500 μM UDP-Gal, 20 mM MnCl2, 0.2% Triton X-100, 800 units of O-glycosidase, 50 mM MES-NaOH buffer (pH 6.8) and an appropriate amount of enzyme was placed in a 96-well black plate (Costar). The fluorescence intensity was assayed with TECAN GENIOS (Austria) after terminating the reaction with the use of Gly-NaOH (pH 10.0).
RT-PCR PCR and sequencing
Total RNA and genomic DNA from cells were prepared using Trizol and a genomic DNA preparation kit (Bioperfectus Technologies), respectively. RT-PCR for human Cosmc and T-synthase were performed with 1 μg total RNA as a template using a transcriptor first strand cDNA synthesis kit (Roche) according to the manufacture’s instructions. PCRs for Cosmc, including non-coding and coding regions and CpG islands, were performed with 100 ng DNA as a template using Kapa 2G Robust HotStart (Kapa Biosystems) according to the manufacture’s protocol. The corresponding primers and band size are listed in Table 1. RT-PCR and PCR products were analyzed on 1% TAE agarose gel. The expected bands were excised, purified and then sequenced (Invitrogen).
Table 1.
PCR primers
| Name | Sequence | Size of Products |
|---|---|---|
| Cosmc cDNA-F | 5’-CGTGAGAGGAAACCCGTG-3’ | 1155 bp |
| Cosmc cDNA-R | 5’-TGTGTGGTTATACCAGTGCC-3’ | |
| T-synthase cDNA-F | 5’-TCTTACAGAAATACACTTTCGG-3’ | 1302 bp |
| T-synthase cDNA-R | 5’-ATTTTATCACACTTCACAGCTC-3’ | |
| DNA in Cosmc coding region-F | 5’-GTCCATAGAGGAGTTGTTGC-3’ | 1218 bp |
| DNA in Cosmc coding region-R | 5’-TCACGCTTTTCTACCACTTC-3’ | |
| DNA in Cosmc no-coding region-F | 5’-AATGGCACAATCTCGGCTC-3’ | 507 bp |
| DNA in Cosmc no-coding region-R | 5’-TGCTCTAACACTCTATGCGGAC-3’ | |
| CpG island of Cosmc-F | 5’-GCTGGCACTGTGGTTAAG-3’ | 1232 bp |
| CpG island of Cosmc-R | 5’-GGAAACAAAACTGCACACG-3’ | |
| GAPDH-F | 5’-TGGGGAAGGTGAAGGTCGG-3’ | 256 bp |
| GAPDH-R | 5’- GGGATCTCGCTCCTGGAAG-3’ |
Analyzing DNA methylation by bisulfite sequencing PCR
Genomic DNA was treated by the MethylCode™ Bisulfite Conversion Kit (Life technology) according to the manufacture’s instructions. Treated DNA was amplified by PCR using the follow primers - Forward primer, 5’-GTAGGTTTATAGTAGTTTTT-3’, and Reverse primer 5’-CTAACCAAACTATTCTAACT-3’. PCR products with 245 bp were analyze by 1.5% agarose gel and were purified using the DNA purification kit (Axygen). The purified PCR fragment was connected to the pMD18-T Vector and transformed into E-coli DH-5α competent cells. Then multiple monoclonal colonies were selected at random for sequencing to detect the methylated frequency of CpG islands (Invitrogen).
Results
Expression of Tn antigen in tumor and normal cells
The Tn antigen was differentially expressed in tumor and normal cells. Normal female cells PBMC sorted from peripheral blood and the BGC823 cells were negative for Tn antigen (Figure 1A). Jurkat and HT-29 cells were stained with anti-Tn mAb (Figure 1B). HT-29-Tn+ and HT-29-Tn- cells were separated from HT-29 cells by microbeads based on their expression or not of Tn antigen (Figure 1C). Consistent with the results of flow cytometry, few HT-29 cells were present and the majority of Jurkat and HT-29-Tn+ cells displayed the Tn (+) phenotype, as detected by fluorescence microscopy (Figure 1D).
Figure 1.
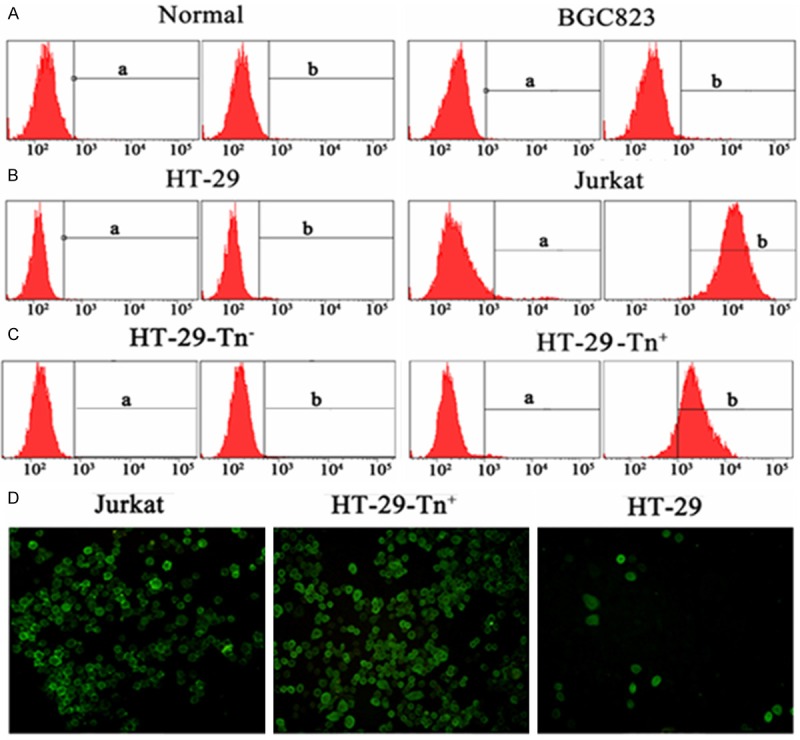
Tn antigen expressed in tumor and normal cells. A and B. Normal cells, BGC823 cells, HT-29 and Jurkat cells were stained with anti-Tn mAb and analyzed with flow cytomerty. a represented isotype control. C. Following separation from HT-29 cells by microbeads, Tn+ and Tn- cells were stained with Tn mAb and analyzed with flow cytometry. Figure a represented negtive control. D. After blocking with BSA, the cells were first incubated with primary anti-Tn mAb and then with the FITC-labeled secondary antibody while cell nuclei were stained with DAPI. Then the Cells were analyzed by fluorescence microscopy. The image presented was at 10x magnification.
Tn antigen positive cells lacking T-synthase activity result from an aberrant alteration of Cosmc expression
To determine whether tumor cells expressing Tn antigen have lower T-synthase activity and Cosmc mutation, we first examined T-synthase activity of the cells. Consistent with previous results [20], Jurkat cells showed lower T-synthase activity due to the T deletion at position 473 of the Cosmc gene, which resulted in an ORF shift and premature stop codon (Figure 2A and 2C). HT-29-Tn- cells showed no difference in T-synthase expression as compared with that in HT-29 cells, whereas HT-29-Tn+ cells lacked T-synthase activity. In addition, T-synthase activity was normal in Tn negative BGC823 cells and normal female cells (Figure 2A). Despite this absence in T-synthase activity within HT-29-Tn+ cells, T-synthase expression was not conspicuously different with regard to cDNA levels when compared with HT-29-Tn- cells. However, Cosmc transcripts were present in all tumor cell lines and normal cells except HT-29-Tn+ cells (Figure 2B and 2C). We also investigated the Cosmc cDNA mutation in all cells which had Cosmc transcripts, with the result being that only Jurkat cells had a position mutation of Cosmc (Figure 2C). These mutations were not apparent in other cells (data not shown). Accordingly, the deletion of Cosmc cDNA can abolish T-synthase activity, while T-synthase expression remains normal in HT-29-Tn+ cells.
Figure 2.
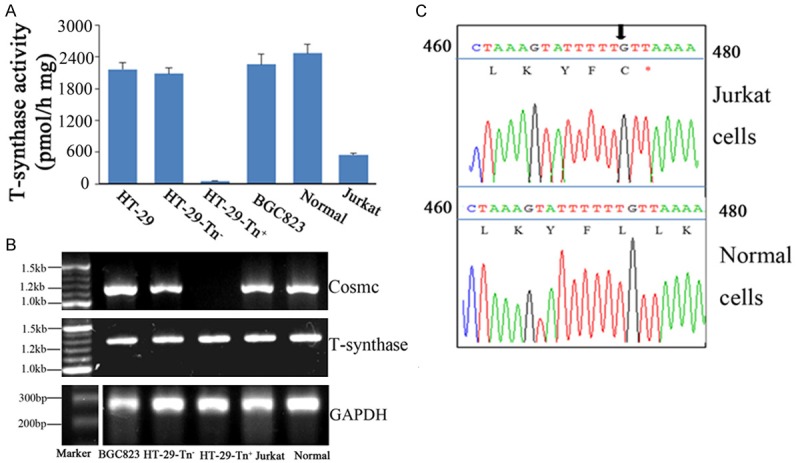
Mutation of the Cosmc gene in Tn+ cells, including Jurkat and HT-29-Tn+ cells, results in lower or a complete absence in T-synthase activity. A. Enzyme activity assay. Cytoplasmic extracts of Tn+ cells, including Jurkat and HT-29-Tn+ cells, and Tn- cells including normal, BGC823 and HT-29-Tn- cells. Which were all generated in triplicates for T-synthase assay using GalNAc-α-4MU as receptor. B. Analysis of Cosmc and T-synthase expression by RT-PCR. mRNA from the cells that we detected was prepared with Trizol, using universal primers to synthesize cDNA. The PCR products were analyzed on 1% agarose gel. C. The mutation of Cosmc in Jurkat cells. PCR products generated using cDNA as template were analyzed on 1% agarose gel, purified and then sequenced. Arrowhead indicates the mutation site.
Deletion of CDS on Cosmc in HT-29-Tn+ cells leads to Cosmc dysfunction
To investigate the mechanisms involved with the lack of Cosmc transcript in HT-29-Tn+ cells, we examined the integrity of the Cosmc gene, including the coding, noncoding regions and the promoter of Cosmc with PCR using DNA as templates. We found that the Cosmc CDS was absent in HT-29-Tn+ cells, while in other cells the Cosmc gene was intact (Figure 3A). The non-coding region bands for Cosmc are not shown. Because there was a very low probability that identical mutations would simultaneously be present in both alleles, we sequenced non-coding regions which contained two SNPs at +1258 bp and +1337 bp (rs591040 and rs 17327439) in HT-29-Tn+ and HT-29-Tn- cells as well as in normal female and BGC823 cells. Besides SNPs, one mosaic sequence at +1266 bp was detected in normal female cells, which was consistent with a previous report [24]. Interestingly, although the DNA from normal female cells contained these two SNPs and one mosaic sequence, the DNA from HT-29-Tn+, HT-29-Tn- and BGC823 cells had only one sequence (Figure 3B). These results suggest that the inactive allele of Cosmc in HT-29-Tn+, HT-29-Tn- cells are absent, but it did not affect Cosmc function. The deletion of CDS in an active allele appears to result in a Cosmc dysfunction in HT-29-Tn+ cells.
Figure 3.
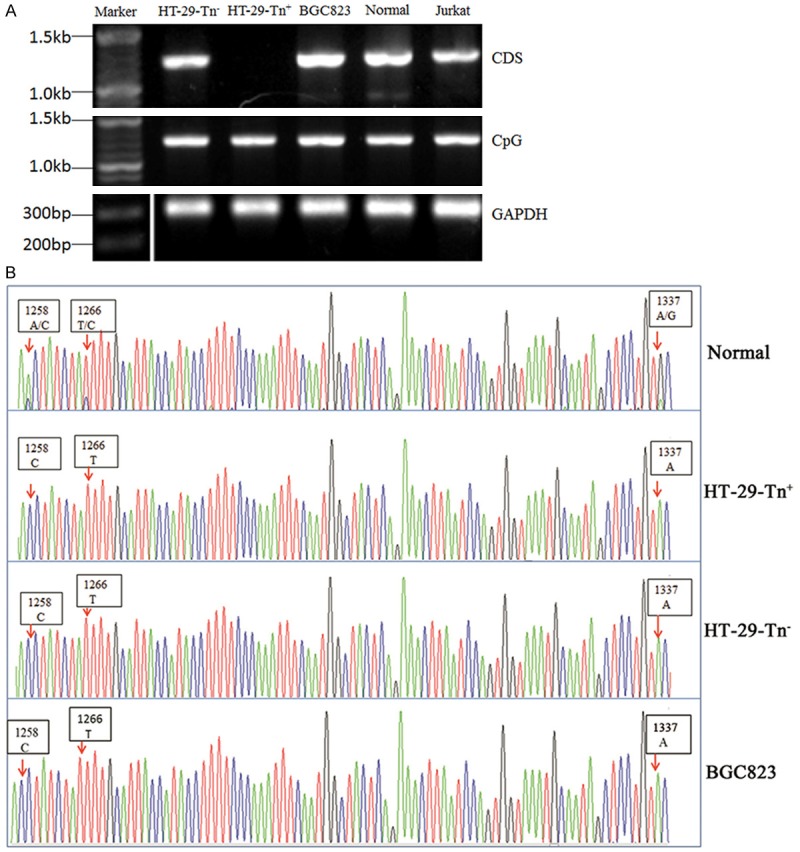
Loss of the active Cosmc allele CDS in HT-29-Tn+ cells. A. PCR analysis of the Cosmc gene. gDNA were extratced from cells and then amplified using corresponding primers. PCR products were analyzed on 1% agarose gel. B. Analysis of SNPs in the non-coding region in Cosmc. Non-coding regions were amplified using gDNA as templates. PCR products were analyzed on 1.5% agarose gel, purified and then sequenced.
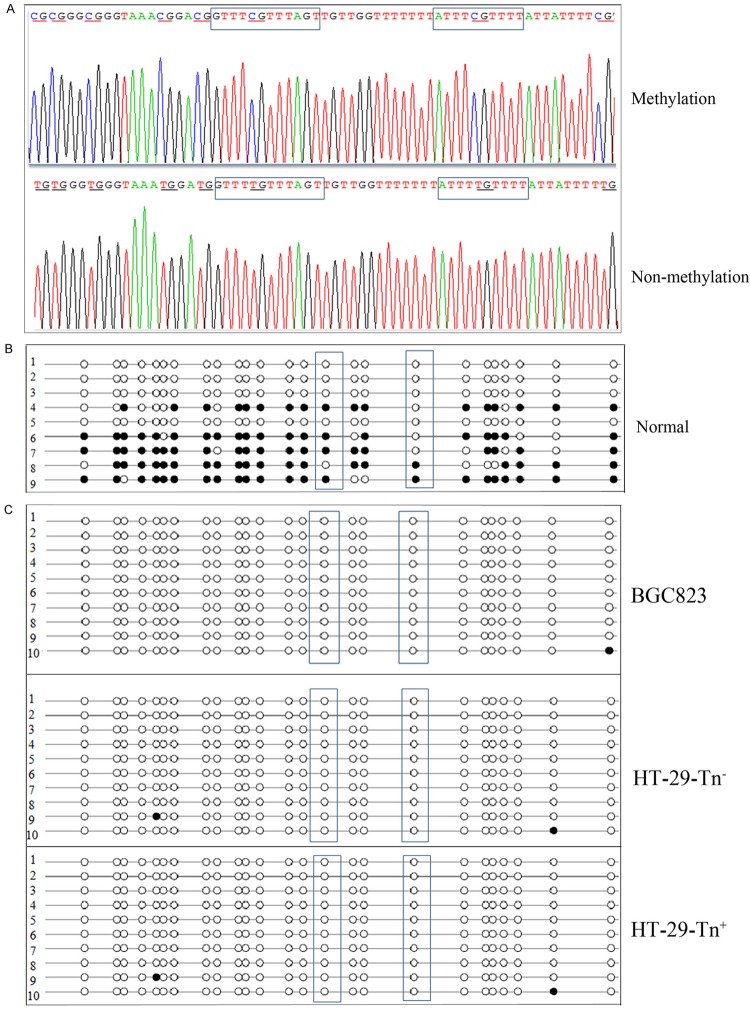
Limited CpG island methylation of Cosmc was obtained in HT-29-Tn+ and HT-29-Tn- cells because of only a single DNA sequence exist in these cells
Methylation status of the CpG island of Cosmc in cells was examined using BSP as a means to determine whether a single active sequence remains in HT-29-Tn+ cells and HT-29-Tn- cells. The CpG island size in Cosmc was 160 bp and contained 25 methylation sites. After treatment with bisulfate, Cytidine (C) was transformed into thymine (T) within non-methylated regions but not within methylated regions (Figure 4A). We selected 9-10 monoclonal colonies for sequencing. As presented in Figure 4B and 4C, all the methylation sites detected were represented by dots, with methylated sites indicated by black dots and non-methylated sites indicated by white dots. With this presentation, the amount of CpG methylation in Cosmc is revealed by the ratio of black: total dots. As expected, in normal female cells the amount of methylation was approximately 50%. The two transfactor SP1/3 binding methylation sites are contained within the boxed region in this presentation. Methylation was close to 50% at one of these sites, but only 20% at the other methylation sites (Figure 4B). By contrast, CpG island methylations of Cosmc in HT-29-Tn+ and HT-29-Tn- cells as well as in BGC823 cells were very low and SP1/3 binding sites were not methylated in these cells (Figure 4C). These results reveal that although the inactive allele of Cosmc did not affect its function, an active allele CDS deletion was associated with the absence of Cosmc transcript in HT-29-Tn+ cells.
Figure 4.
The methylation status of CpG islands in Cosmc. A. The difference between methylated and non-methylated sites after treatment with bisulfate. gDNA from cells were treated with bisulfate and amplified by PCR. CpG islands of Cosmc were identified using specific primers. PCR products were analyzed on 1.5% agarose gel, purified and then subjected to thymine-adenine (TA) colone sequencing. Portions of the sequencing results are shown including methylation and non-methylation sites. Methylation sites detected were underlined and SP1/3 binding sites of Cosmc were boxed. B. CpG methylation status of Cosmc in normal female cells. The amount of CpG island methylation containing 25 methylation sites were examined in normal fenmale cells using BSA. Nine monoclonal colonies were selected for sequencing. SP1/3 binding sites of Cosmc were boxed. C. The amount of CpG island methylation in BGC823, HT-29-Tn+ and HT-29-Tn- cells. Ten monoclonal colonies were selected for sequencing. SP1/3 binding sites in Cosmc were boxed.
Discussion
Our results show that the deletion of Cosmc CDS gene result in T-synthase inactivity and Tn antigen expression in HT-29-Tn+ cells, a subpopulation of Tn positive cells sorted from human colon cancer cell line HT-29. SNP analysis suggested that despite deriving from a female patient, HT-29 cells have only a single allele of the Cosmc gene. Methylation analysis revealed a methylation percentage much lower than normal female Cosmc. Suggesting that perhaps the methylated, inactive copy of Cosmc has been deleted in HT-29, but it does not affect Cosmc function. The absence of CDS gene just occurs in Cosmc active allele of HT-29-Tn+ cells. This is the first example that shed light on some of the potential mechanisms of abnormal Tn antigen expression in HT-29-Tn+ cells.
We chose to examine the colon cancer cell line HT-29 because it has been well documented that Tn antigen is expressed in HT-29 cells [29,30]. To date, no genetic basis for the expression of Tn antigen in HT-29 cells has been reported. An additional consideration for the use of this model was that, Tn antigen expression is associated with tumor cell metastasis and prognosis of colorectal carcinoma patients [31]. We separated HT-29-Tn+ and HT-29-Tn- cells from the parental stock of HT-29 cells, which derived from a female patient, based on their expression or not of the Tn antigen and found that it was the absence of Cosmc CDS gene result in Tn antigen expression in HT-29-Tn+ cells. However, the likelihood for identical mutation simultaneously occurring was quite remote. To confirm this, we sequenced the non-coding region of Cosmc containing two SNPs in normal female and tumor cells. As expected, were SNPs of Cosmc non-coding regions present, whereas no SNPs of Cosmc were observed in other cell lines. Suggesting that perhaps the active allele of Cosmc had been deleted in HT-29 cells, this deletion did not appear to influence Cosmc function, the absence of CDS just happened in Cosmc active allele in HT-29-Tn+ cells. Moreover, the SNPs and mosaic sequences in the non-coding region of Cosmc that we observed in normal female cells were identical with that reported in the cervical cancer sample DH85, which Tn antigen expression in this sample was caused by loss of the active Cosmc allele [24]. Interestingly, in the SNPs at +1337 bp (rs17327439), thymine (T) remained in the Tn antigen positive cells of DH85, which had been identified only an inactive allele of Cosmc was remained. While adenine (A) was detected at this SNP site in HT-29-Tn+ and HT-29-Tn- cells as well as in BGC823 cells where an active allele of Cosmc was assumed to be present, further confirmed that Cosmc inactive allele had been deleted in HT-29-Tn+ and HT-29-Tn- cells. Furthermore, SNPs located on candidate genes have direct relevance to cancer risk [32,33] and SNPs may affect protein function through altering amino acid sequences [34]. It has been reported that an adenine (A) to quanine (G) mutation in the SNP at position 61 of the 5’ region upstream of the epidermal growth factor (EGF) coding region (rs4444903) was associated with an increased risk in various malignant tumors [35-39]. Recently, Cao et al. [40] demonstrated that the rs3124591 TC genotype of Notch1, one of a family of genes that encode a group of conserved transmembrane receptors, was correlated with high Notch1 expression which had been shown to promote cancer in normal breast tissue, ductal breast carcinomas in situ (DCIS) and other invasive ductal breast carcinomas within the Chinese population. Such a mechanism may provide a theoretical basis for early interventions and treatment of breast cancers. Collating these findings leads to the proposal that Cosmc SNP rs17327439 in the 3’nontranslation region may could serve as a biological marker for estimating the risk of multiple neoplastic lesions. That is, the adenine-adenine (AA) genotype at this site results in increased expression of Cosmc which may modulate (promote or suppress) the process of tumor formation. Additional work will be required to confirm or refute this hypothesis. However, it should be noted that the distribution of SNPs are distinct among individuals or races [32,41]. Therefore, SNPs of Cosmc in normal cells from colon cancer patients who possessed HT-29 cells were not always consistent with that observed in the normal female cells we studied; it is possible that the deletion of CDS may occur simultaneously on two alleles of Cosmc, although the possibility is very small.
Findings from previous reports indicate that most X-linked gene methylations on CpG islands result in XCI methylation in female cells and an absence of methylation in male cells [42]. Methylation sites located around transcription start sites (TSSs) on CpG islands are crucial for gene silencing [43]. To further confirm only one active allele remained in HT-29-Tn+ and HT-29-Tn- cells, the methylation status of the CpG island should be examined since CpG islands are hypermethylated on inactive X chromosomes in somatic cells. In our study almost half of the methylation sites examined were methylated in normal female cells as observed with use of BSP. This may reflect the XCI presence in female cells. In addition, two transcription factor binding sites were included in these methylation sites. Ju et al. [23] reported that methylation of these two sites leads to epigenetic silencing of the Chaperone Cosmc in Tn4 cells, which were derived from a male patient with Tn syndrome. Interestingly, we observed that in one of the SNP binding sites methylation frequency was nearly 50% while in another only 20%. This difference could be due to the limited number of random monoclonal colonies selected. The exact relationship between other methylation sites and Cosmc expression remains unclear. As expected, due to an active allele, very limited amounts of methylation were seen in HT-29-Tn+ and HT-29-Tn- cells. Nor was methylation detected in cells derived from a male patient with a single X chromosome (BGC823 cells). Meissner et al. [44] reported that CpG islands in female cell populations (ES cell-derived and primary astrocytes, respectively), which had XCI, showed an average of ~50% methylation, which is consistent with our results. Up to now, there is so few attention is paid for the CpG methylation, especially for the methylation of transcription factor binding sites in female X-chromosome using BSP, that more work needs to be directed at examining CpG island methylation of Cosmc in normal female samples to identify whether the same methylated status as in our study exists under conditions of coexistence with active and inactive X chromosomes.
Our results established some of the genetic mechanism of Cosmc dysfunction in HT-29-Tn+ cells, which may enable a better understanding of Tn antigen expression in human cancer. Additionally, an alteration in the Cosmc gene abolishes T-synthase activity and consequently an abnormal expression of Tn antigens has been shown in multiple human tumor cells, including colorectal, melanoma cells and cervical cancer samples, as well as in cells from Tn syndrome patients [12,13]. However, Yoo et al. [45] were unable to detect Cosmc mutations after examining the ORF of Cosmc in human colorectal and breast carcinoma samples by SSPS (single strand conformation polymorphism). Perhaps these results might be due to the fact that the colorectal cell line and samples were derived from different tissues. Moreover, it has been confirmed that aberrant Cosmc genes just exist in Tn positive cells, but whether the colorectal and breast carcinoma samples expressed Tn antigen was uncertain in their study. Besides, these investigators did not perform determinations at other regions outside ORF and the CpG island methylation in Cosmc, which may suppress its expression.
In conclusion, we have shown that in HT-29-Tn+ cells, which isolated from colorectal carcinoma-derived cell line HT-29, the inactive allele of Cosmc caused by CpG island methylation was deleted, but this deletion does not affect Cosmc function. T-synthase inactivity and Tn antigen expression, however, are associated with the absence of CDS in Cosmc active allele. Our findings provide new information regarding the mechanisms of Tn antigen expression in tumor cells associated with an abnormal Cosmc gene as well as new directions for future research on DNA methylation and SNP genotype. Our investigation, along with previous studies regarding Tn antigen expression in tumor cells with dysfunctional Cosmc, laid a theoretical basis for the diagnosis and therapy of human cancer using Tn antigens or Cosmc genes as targets.
Acknowledgements
This work was supported by National Natural Science Foundation of China (NSFC 30972778), Shandong Science Foundation (Y2007C143) and Nature Science Foundation from Shandong Province (ZR2014HM020).
Disclosure of conflict of interest
None.
References
- 1.Ju T, Brewer K, D’Souza A, Cummings RD, Canfield WM. Cloning and expression of human core 1 beta1,3-galactosyltransferase. J Biol Chem. 2002;1:178–186. doi: 10.1074/jbc.M109060200. [DOI] [PubMed] [Google Scholar]
- 2.Xia L, Ju T, Westmuckett A, An G, Ivanciu L, McDaniel JM, Lupu F, Cummings RD, McEver RP. Defective angiogenesis and fatal embryonic hemorrhage in mice lacking core 1-derived O-glycans. J Cell Biol. 2004;3:451–459. doi: 10.1083/jcb.200311112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia L, McEver RP. Targeted disruption of the gene encoding core 1 beta1-3-galactosyltransferase (T-synthase) causes embryonic lethality and defective angiogenesis in mice. Methods Enzymol. 2006;416:314–331. doi: 10.1016/S0076-6879(06)16021-8. [DOI] [PubMed] [Google Scholar]
- 4.Homeister JW, Thall AD, Petryniak B, Maly P, Rogers CE, Smith PL, Kelly RJ, Gersten KM, Askari SW, Cheng G, Smithson G, Marks RM, Misra AK, Hindsgaul O, von Andrian UH, Lowe JB. The alpha(1,3)fucosyltransferases FucT-IV and FucT-VII exert collaborative control over selectin-dependent leukocyte recruitment and lymphocyte homing. Immunity. 2001;1:115–126. doi: 10.1016/s1074-7613(01)00166-2. [DOI] [PubMed] [Google Scholar]
- 5.Ju T, Otto VI, Cummings RD. The Tn antigen-structural simplicity and biological complexity. Angew Chem Int Ed Engl. 2011;8:1770–1791. doi: 10.1002/anie.201002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hakomori S. Tumor-associated carbohydrate antigens defining tumor malignancy: basis for development of anti-cancer vaccines. Adv Exp Med Biol. 2001;491:369–402. doi: 10.1007/978-1-4615-1267-7_24. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Sette A, Sidney J, Gendler SJ, Franco A. Tumor-associated carbohydrate antigens: a possible avenue for cancer prevention. Immunol Cell Biol. 2005;4:440–448. doi: 10.1111/j.1440-1711.2005.01347.x. [DOI] [PubMed] [Google Scholar]
- 8.Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;6:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 9.Springer GF. Immunoreactive T and Tn epitopes in cancer diagnosis, prognosis, and immunotherapy. J Mol Med (Berl) 1997;8:594–602. doi: 10.1007/s001090050144. [DOI] [PubMed] [Google Scholar]
- 10.Inoue M, Ogawa H, Nakanishi K, Tanizawa O, Karino K, Endo J. Clinical value of sialyl Tn antigen in patients with gynecologic tumors. Obstet Gynecol. 1990;6:1032–1036. [PubMed] [Google Scholar]
- 11.Desai PR. Immunoreactive T and Tn antigens in malignancy: role in carcinoma diagnosis, prognosis, and immunotherapy. Transfus Med Rev. 2000;4:312–325. doi: 10.1053/tmrv.2000.16229. [DOI] [PubMed] [Google Scholar]
- 12.Laack E, Nikbakht H, Peters A, Kugler C, Jasiewicz Y, Edler L, Hossfeld DK, Schumacher U. Lectin histochemistry of resected adenocarcinoma of the lung: helix pomatia agglutinin binding is an independent prognostic factor. Am J Pathol. 2002;3:1001–1008. doi: 10.1016/S0002-9440(10)64921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konno A, Hoshino Y, Terashima S, Motoki R, Kawaguchi T. Carbohydrate expression profile of colorectal cancer cells is relevant to metastatic pattern and prognosis. Clin Exp Metastasis. 2002;1:61–70. doi: 10.1023/a:1013879702702. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez MF, Tang N, Alansari H, Karvonen RL, Tomkiel JE. Improved approach to identify cancer-associated autoantigens. Autoimmun Rev. 2005;4:230–235. doi: 10.1016/j.autrev.2004.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kakeji Y, Tsujitani S, Mori M, Maehara Y, Sugimachi K. Helix pomatia agglutinin binding activity is a predictor of survival time for patients with gastric carcinoma. Cancer. 1991;11:2438–2442. doi: 10.1002/1097-0142(19911201)68:11<2438::aid-cncr2820681119>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Sewell R, Backstrom M, Dalziel M, Gschmeissner S, Karlsson H, Noll T, Gatgens J, Clausen H, Hansson GC, Burchell J, Taylor-Papadimitriou J. The ST6GalNAc-I sialyltransferase localizes throughout the Golgi and is responsible for the synthesis of the tumor-associated sialyl-Tn O-glycan in human breast cancer. J Biol Chem. 2006;6:3586–3594. doi: 10.1074/jbc.M511826200. [DOI] [PubMed] [Google Scholar]
- 17.Aryal RP, Ju T, Cummings RD. The endoplasmic reticulum chaperone Cosmc directly promotes in vitro folding of T-synthase. J Biol Chem. 2010;4:2456–2462. doi: 10.1074/jbc.M109.065169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Ju T, Ding X, Xia B, Wang W, Xia L, He M, Cummings RD. Cosmc is an essential chaperone for correct protein O-glycosylation. Proc Natl Acad Sci U S A. 2010;20:9228–9233. doi: 10.1073/pnas.0914004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju T, Aryal RP, Stowell CJ, Cummings RD. Regulation of protein O-glycosylation by the endoplasmic reticulum-localized molecular chaperone Cosmc. J Cell Biol. 2008;3:531–542. doi: 10.1083/jcb.200711151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ju T, Cummings RD. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci U S A. 2002;26:16613–16618. doi: 10.1073/pnas.262438199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger EG. Tn-syndrome. Biochim Biophys Acta. 1999;2-3:255–268. doi: 10.1016/s0925-4439(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 22.Mestecky J, Tomana M, Moldoveanu Z, Julian BA, Suzuki H, Matousovic K, Renfrow MB, Novak L, Wyatt RJ, Novak J. Role of aberrant glycosylation of IgA1 molecules in the pathogenesis of IgA nephropathy. Kidney Blood Press Res. 2008;1:29–37. doi: 10.1159/000112922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi R, Song L, Wang Y, Ding X, Zeng J, Lehoux S, Aryal RP, Wang J, Crew VK, van Die I, Chapman AB, Cummings RD, Ju T. Epigenetic silencing of the chaperone Cosmc in human leukocytes expressing tn antigen. J Biol Chem. 2012;49:41523–41533. doi: 10.1074/jbc.M112.371989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ju T, Lanneau GS, Gautam T, Wang Y, Xia B, Stowell SR, Willard MT, Wang W, Xia JY, Zuna RE, Laszik Z, Benbrook DM, Hanigan MH, Cummings RD. Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 2008;6:1636–1646. doi: 10.1158/0008-5472.CAN-07-2345. [DOI] [PubMed] [Google Scholar]
- 25.LYON MF. Gene action in the X-chromosome of the mouse (Mus musculus L. ) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 26.Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, Antonarakis SE. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;10:1592–1600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasukochi Y, Maruyama O, Mahajan MC, Padden C, Euskirchen GM, Schulz V, Hirakawa H, Kuhara S, Pan X H, Newburger PE, Snyder M, Weissman SM. X chromosome-wide analyses of genomic DNA methylation states and gene expression in male and female neutrophils. Proc Natl Acad Sci U S A. 2010;8:3704–3709. doi: 10.1073/pnas.0914812107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju T, Xia B, Aryal RP, Wang W, Wang Y, Ding X, Mi R, He M, Cummings RD. A novel fluorescent assay for T-synthase activity. Glycobiology. 2011;3:352–362. doi: 10.1093/glycob/cwq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh R, Campbell BJ, Yu LG, Fernig DG, Milton JD, Goodlad RA, FitzGerald AJ, Rhodes JM. Cell surface-expressed Thomsen-Friedenreich antigen in colon cancer is predominantly carried on high molecular weight splice variants of CD44. Glycobiology. 2001;7:587–592. doi: 10.1093/glycob/11.7.587. [DOI] [PubMed] [Google Scholar]
- 30.Barrow H, Tam B, Duckworth CA, Rhodes JM, Yu LG. Suppression of core 1 Gal-transferase is associated with reduction of TF and reciprocal increase of Tn, sialyl-Tn and Core 3 glycans in human colon cancer cells. PLoS One. 2013;3:e59792. doi: 10.1371/journal.pone.0059792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konno A, Hoshino Y, Terashima S, Motoki R, Kawaguchi T. Carbohydrate expression profile of colorectal cancer cells is relevant to metastatic pattern and prognosis. Clin Exp Metastasis. 2002;1:61–70. doi: 10.1023/a:1013879702702. [DOI] [PubMed] [Google Scholar]
- 32.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;6788:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 33.Zhong JH, You XM, Gong WF, Ma L, Zhang Y, Mo QG, Wu LC, Xiao J, Li LQ. Epidermal growth factor gene polymorphism and risk of hepatocellular carcinoma: a meta-analysis. PLoS One. 2012;3:e32159. doi: 10.1371/journal.pone.0032159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nuckel H, Frey UH, Bau M, Sellmann L, Stanelle J, Durig J, Jockel KH, Duhrsen U, Siffert W. Association of a novel regulatory polymorphism (-938C>A) in the BCL2 gene promoter with disease progression and survival in chronic lymphocytic leukemia. Blood. 2007;1:290–297. doi: 10.1182/blood-2006-03-007567. [DOI] [PubMed] [Google Scholar]
- 35.Shahbazi M, Pravica V, Nasreen N, Fakhoury H, Fryer AA, Strange RC, Hutchinson PE, Osborne JE, Lear JT, Smith AG, Hutchinson IV. Association between functional polymorphism in EGF gene and malignant melanoma. Lancet. 2002;9304:397–401. doi: 10.1016/S0140-6736(02)07600-6. [DOI] [PubMed] [Google Scholar]
- 36.Lanuti M, Liu G, Goodwin JM, Zhai R, Fuchs BC, Asomaning K, Su L, Nishioka NS, Tanabe KK, Christiani DC. A functional epidermal growth factor (EGF) polymorphism, EGF serum levels, and esophageal adenocarcinoma risk and outcome. Clin Cancer Res. 2008;10:3216–3222. doi: 10.1158/1078-0432.CCR-07-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piao Y, Liu Z, Ding Z, Xu L, Guo F, Sun Q, Xie X. EGF +61A>G polymorphism and gastrointestinal cancer risk: a HuGE review and meta-analysis. Gene. 2013;1:26–33. doi: 10.1016/j.gene.2013.01.057. [DOI] [PubMed] [Google Scholar]
- 38.Yoshiya S, Fujimoto Y, Bekki Y, Konishi H, Yamashita Y, Ikegami T, Yoshizumi T, Shirabe K, Oda Y, Maehara Y. Impact of epidermal growth factor single-nucleotide polymorphism on recurrence of hepatocellular carcinoma after hepatectomy in patients with chronic hepatitis C virus infection. Cancer Sci. 2014;6:646–650. doi: 10.1111/cas.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abu DB, Yang M, Fuchs BC, Karl DL, Yamada S, Sninsky JJ, O’Brien TR, Dienstag JL, Tanabe KK, Chung RT. A functional polymorphism in the epidermal growth factor gene is associated with risk for hepatocellular carcinoma. Gastroenterology. 2011;1:141–149. doi: 10.1053/j.gastro.2011.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao YW, Wan GX, Zhao CX, Hu JM, Li L, Liang WH, Li WQ, Li YC, Li YX, Du XM, Yu SY, Li F. Notch1 single nucleotide polymorphism rs3124591 is associated with the risk of development of invasive ductal breast carcinoma in a Chinese population. Int J Clin Exp Pathol. 2014;7:4286–4294. [PMC free article] [PubMed] [Google Scholar]
- 41.Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M, Wojcik SE, Ferdin J, Kunej T, Xiao L, Manoukian S, Secreto G, Ravagnani F, Wang X, Radice P, Croce CM, Davuluri RV, Calin GA. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;7:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cotton AM, Lam L, Affleck JG, Wilson IM, Penaherrera MS, McFadden DE, Kobor MS, Lam WL, Robinson WP, Brown CJ. Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Hum Genet. 2011;2:187–201. doi: 10.1007/s00439-011-1007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;7:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 44.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;7205:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoo NJ, Kim MS, Lee SH. Absence of COSMC gene mutations in breast and colorectal carcinomas. APMIS. 2008;2:154–155. doi: 10.1111/j.1600-0463.2008.00965.x. [DOI] [PubMed] [Google Scholar]