Abstract
Recent studies have shown that NUF2 (Ndc80 kinetochore complex component) play important roles in multiple human cancers. In our previous report, NUF2 expression was stronger in tumor tissues than in normal pancreatic tissues. However, whether and how NUF2 play a role in pancreatic cancer progression remains largely unknown. The aim of our study is to investigate the expression and functional role of NUF2 in human PC. NUF2 expression was measured in 10 pairs of PC cancerous and noncancerous tissue samples by quantitative real-time PCR. The effects of NUF2 on PC cells were studied by RNA interference. Apoptosis and cell cycle were analyzed by flow cytometry. Cells viability was evaluated using MTT. CDK4/CDK6 activity was measured by Western blot assay. LncRNAs regulated by NUF2 were gained from bioinformatics analysis. The role of LncRNA AF339813, regulated by NUF2, was finally characterized in PC cells by siRNA. Our results showed that NUF2 mRNA and protein were significantly overexpressed in Human PC tissues and several PC cell lines. Through bioinformatics analysis and knockdown NUF2 in PC cells, we found LncRNA AF339813 was positively regulated by NUF2. We further demonstrated that knockdown of AF339813 by siRNA in PC cells significantly reduced cell proliferation and promoted apoptosis. Thus, we conclude that NUF2 is consistently overexpressed in human PC and NUF2 is closely linked with human PC progression through the meditator LncRNA AF339813. Our studies may contribute to understand the molecular mechanism of PC pathogenesis and clinical therapy.
Keywords: NUF2, LncRNA AF339813, pancreatic cancer, RNA interference
Introduction
Pancreatic cancer (PC) is a highly malignant tumor with increasing incidence and mortality in the world and its mortality rate in excess of 95% in China [1,2]. PC is characterized by a highly malignant phenotype that is associated with early metastasis and resistant to chemotherapy and radiation therapy [3,4]. Despite the recent advances in clinical and experimental oncology, the prognosis of PC still remains poor, the five-year survival rates remain at < 5% [5]. Identifying effective therapeutic targets play a key role in illustrating the underlying molecular mechanisms in PC. However, the molecular mechanisms and factors involved PC are still not fully understood.
Recently, high level of NUF2 expression was reported to be associated with poor prognosis for patients with colorectal cancers. DeLuca et al found that in HeLa cells, after knockdown of NUF2 by RNA interference, spindle formation occurred normally, but kinetochores failed to attach to spindle microtubules and cells block in prometaphase, which caused aberrant chromosome segmentation and induced mitotic cells to undergo cell death [6]. Dysregulation of NUF2 has been employed in the development of a series of human cancers including lung cancer, colorectal cancer, gastric cancer, prostate cancer, urinary bladder cancer, renal carcinoma and ovarian cancer, rather than other normal tissues except testis [7-12]. Although extensive studies have been reported, the precise mechanisms leading to PC are only partially understood.
Similarly, the expression levels of certain LncRNAs are associated with recurrence, metastasis and prognosis of cancers. For examples, LncRNA HOTAIR is a strong prognosis marker of patient outcomes and survival in several human cancers [13-15]. Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is not only over expressed in early-stage metastasizing non-small cell lung cancer, but also in breast, pancreas, colon, prostate and liver cancers [16-22]. LncRNA Colon cancer associated transcript 1 (CCAT1) was found a potential marker of colorectal cancer [23]. Besides, LncRNA PCGEM1 gene polymorphisms contribute to prostate cancer risk [24]. However, the relationship between NUF2 and LncRNAs has none reported. Thus, we propose a hypothesis: whether dysregulation of NUF2 operates LncRNAs process in pancreatic cancer cells? The aim of the current studies is to detect the expression of LncRNAs in PC tissues and to explore the relationship between LncRNAs level and NUF2.
In this study, we sought to examine the expression and functional role of NUF2 in human pancreatic cancer. We found that NUF2 was highly expressed in human pancreatic cancer tissues and several PC cell lines. In order to understand the mechanism of NUF2 regulated PC, we further compared the LncRNAs expression profile difference between tumor tissues and adjacent normal tissues. Finally, we also investigated the impact of altered LncRNA AF339813 levels, the most obvious change LncRNA, on the phenotypes of PC cells in vitro. Finally, our results suggested that LncRNA AF339813 may represent a novel indicator of poor prognosis and may be a potential therapeutic target for the diagnosis and gene therapy of PC.
Materials and methods
Patients and tissue specimens
All specimens were handled and made anonymous according to the ethical and legal standards. Paired tissue specimens (tumor and adjacent normal tissues) from 304 patients with PC were obtained and histologically confirmed by a pathologist at Southwest Hospital, Third Military Medical University, China, from January 2010 to December 2010. All samples were derived from patients who had not received adjuvant treatment including chemotherapy or radiotherapy prior to the surgery in order to eliminate potential treatment-induced changes to gene expression profiles. After excision, tissue specimens were immediately frozen in liquid nitrogen for subsequent analysis.
Reagents
Antibodies against Caspase-3, Caspase-9, Bax, Bcl-2 and GAPDH were purchased from Cell Signaling technologies (Danvers, MA). Whereas, antibodies against Hec1, Cyclin A2, Cyclin B1, Cyclin D1, Cyclin E1, CDK4 and CDK6 were purchased from Abcam (Cambridge, MA, USA). Rabbit antibodies conjugated with horseradish peroxidase (HRP) and sheep anti-mouse-HRP were purchased from Zhongsan Jinqiao (Beijing, China). All others chemical reagents were purchased from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China).
Cell culture
Human pancreatic cancer cell lines Sw1990, PANC-1, BXPC-3and human embryonic kidney cell line 293 were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China), and cultured in Dulbecco’s modified eagle’s medium (DMEM) (Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (FBS) (Biowest, Nuaillé, France), 100 U/mL penicillin, and 100 mg/mL streptomycin (Hyclone). All cell lines were maintained in a humidified atmosphere of 5% CO2/air at 37°C.
Quantitative real-time RT-PCR analysis
Total mRNA was extracted from pancreatic cancer tissues and cultured cell lines using RNeasy Mini Kit (Qiagen, Valencia, CA, USA). cDNA was then synthesized by RNA reverse transcribing with a Super Script III First-Strand Synthesis System for RT-PCR Kit (Invitrogen). The expression level of NUF2 and LncRNA AF339813 mRNA was measured by RT-PCR with an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). Amplifications were then carried out and the PCR conditions were: Initial denaturation at 95°C for 1 min, 40 cycles of denaturation at 95°C for 5 s and annealing extension at 60°C for 20 s. Relative quantification in RT-PCR was performed using 2-ΔΔCT method [21]. Data were presented as CT values, which were defined as the threshold PCR cycle number at which an amplified product is first detected. ΔCT = Avg.CT (NUF2)-Avg. CT (β-actin). Primers used in qRT-PCR were as follows: NUF2: 5’-TACCATTCAGCAATTTAGTTACT-3’ (forward); and 5’-TAGAATATCAGCAGTCTCAAAG-3’ (reverse). LncRNA AF339813: 5’-GACATACATCAGCCATTT-3’ (forward); and 5’-ACTGACTTAACCAGGAGA-3’. The primers of β-actin, used as internal control, were: 5’-CAGAGCCTCGCCTTTGCCG-3’ (forward); and 5’-ACGCCCTGGTGCCTGGGGCG-3’ (reverse).
Choice of differentially expressed LncRNAslist using heat map analysis
We obtained the microarray date from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/), and the GEO accession number is GSE3325. The date was generated using the genechip Affymetrix Human Genome U133 Plus 2.0 Array GPL570 (HG-U133_Plus_2), which completely coverage Human Genome U133 Set plus 6500 additional genes for analysis of over 47,000 transcripts.
Observations with adjusted p-values ≥ 0.05 were removed, and thus excluded from further analysis. The heat map of the 50 LncRNAs most obvious differences was created using a method of hierarchical clustering by GeneSpring GX, version 7.3 (Agilent Technologies, California, United States).
Construction of the LncRNA AF339813 and NUF2 siRNA vector and transfection
siRNA of human lncRNA AF339813 and NUF2 lentivirus vector carrying GFP sequence was provided by Gene Pharma (Shanghai, China).The recombinant lentivirus of LncRNA AF339813 and NUF2 siRNA and the control lentivirus (GFP lentivirus) were prepared and titered to 108 TU/ml. After lentivirus infection, cells were washed by PBS and collected to perform RT-PCR analysis and western blot analysis. The sequences of siRNAs were as follows: LncRNA AF339813: si-1: 5’-CCCAAAGACATAATCTGGTTATTTG-3’, si-2: 5’-CCTGTGTAAGGTAGTTCATGCATTT-3’, si-3: 5’-GGTAGTTCATGCATTTCCTTCTTCT-3’, si-Scramble: 5’-CCCAGATACTAAGTCTTGTAAATTG-3’; NUF2: Si-1: 5’-GAAGAAACCAGAGCCTGGGAGATTA-3’, Si-2: 5’-CAGAGCCTGGGAGATTAACAGGAAA-3’, Si-3: 5’-CAATAAGATCTTAACAGGAGCTGAT-3’, Si-Scramble: 5’-GAACAAGACCGAGTCAGGAGGATTA-3’.
Cell proliferation assay
To evaluate the effect of LncRNA AF339813 knockdown on pancreatic cancer cell proliferation, MTT colorimetric assay was performed in PANC-1 cell lines in vitro. Briefly, three days after lentivirus infection, PANC-1 cells (2 × 103/well) was seeded into 96-well plates and cultured in DMEM supplemented with 10% FBS at 37°C under 5% CO2 atmosphere until the cells reached 85% confluence. Then 20 μL of MTT solution (5 mg/mL) was added to each well and incubated for 4 h. Then the medium was then removed, and 150 μL of DMSO was added to dissolve the formazan crystals. Absorbance at 490 nm was measured by using the Model 680 Microplate Reader (Bio-Rad). The measurements for each sample were conducted in triplicate.
Flow cytometric analysis of cell cycle
Transfected cells were harvested after transfection. Cells for cell cycle analysis were stained with propidium oxide by Cell Cycle Analysis Kit (Beyotime, Haimen, China) following the protocol and analyzed by FACScan (BD Biosciences, NY, USA). The percentage of the cells in G1-G0, S and G2-M phase were counted and compared.
Apoptosis analysis
PANC-1 cells were plated into 6-well plates (1 × 106 cells/well) in antibiotic-free medium, after being transfected with siRNA for 48 h. Cells for apoptosis analysis were collected and washed twice and stained with fluorescein isothiocyanate (FITC)-Annexin V and PI (BD Biosciences), using the FITC-Annexin V Apoptosis Detection Kit (BD Biosciences) according to manufacturer’s manual.
Western blotting
Total proteins extracts of each cell treatment group were resolved by 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (Millipore, Bedford, MA, USA) membranes. After blocking, the membranes were washed four times with Tris-buffered saline containing 0.3% Tween-20 (TBST) at room temperature for 15 min and then incubated with primary polyclonal antibodies against Caspase-3 (1:1000), Caspase-9 (1:1000), Bax (1:1500), Bcl-2 (1:1000), Cyclin A2 (1:2000), Cyclin B1 (1:1000), Cyclin D1 (1:5000), Cyclin E1 (1:1500), CDK4 (1:2000) and CDK6 (1:2000). After washing, the membranes were incubated with secondary antibody (1:1000) conjugated with horseradish peroxidase (HRP) at room temperature for 1 h. The protein bands were visualized by enhanced chemiluminescence (ECL Kit; Pierce Biotechnology) and autoradiography. The grey value results of western blotting were subjected to semi-quantitative analysis. The relative amount of target protein could be calculated by dividing the grey value of the target protein band by the grey value of glyceraldehyde 3-phosphate dehydrogenase (GAPDH; internal reference protein) band.
Statistical analysis
Statistical analyses were performed with SPSS 13.0 software. The results were evaluated by χ2 test and the other data were evaluated by Student’s t-test and expressed as the mean ± SD from three independent experiments. A P-value of less than 0.05 was considered statistically significant.
Results
NUF2 was upregulated in pancreatic cancer
To evaluate the functional role of NUF2 in human PC, the expression levels of NUF2 in 304 pairs of PC tissue and adjacent non-tumor samples were determined by qRT-PCR assays. We found that NUF2 expression in cancer tissues from patients with PC was significantly higher than that in adjacent normal tissues (n = 304, P < 0.001, Figure 1A). Subsequently, we also detected the expression of NUF2 in 3 human PC cell lines including PANC-1, SW1990 and BxPC-3 using qRT-PCR and Western Blot. Human embryonic kidney cell line 293T used as a negative control. High expression of NUF2 in PC cell lines were also observed, especially in PANC-1 cells (Figure 1B and 1C) (P < 0.001, Figure 1B).
Figure 1.

Increased expression of NUF2 in human pancreatic cancer tissues and cell lines. A. mRNA levels of NUF2 were detected by real-time quantitative PCR in 10 pairs of PC tissues and adjacent non-cancer tissues. GAPDH gene served as an internal control. Scatter dots represent relative mRNA expression levels and the horizontal lines represent mean with SD. Statistical significance: **P < 0.01, compared to normal tissues. B. qRT-PCR analysis of NUF2 expression in 3 human pancreatic cancer cell lines and HEK293 cell line. GAPDH gene was used as an internal gene. Data represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to HEK293 cells. C. Western blot analysis of NUF2 expression in 3 human pancreatic cancer cell lines Sw1990, PANC-1, BXPC-3 and HEK293 cell line. GAPDH gene served as an internal control. Data represent one of three independent experiments with similar results. *P < 0.05, **P < 0.01, compared to HEK293 cells.
Heatmap analysis of the LncRNA
We obtained the microarray date from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/), and the GEO accession number is GSE15471. The Agilent Feature Extraction software (version 7.3) was used to analyze the acquired array images. The quantile normalization and subsequent data processing were performed using the GeneSpring GX v7.3 software package (Agilent Technologies) to analyze differential LncRNAs expression between PC tissue and adjacent non-tumor samples. Firstly, hierarchical clustering analysis was used to compare differential LncRNA expression of the top 50 LncRNAs. The clustered heatmap of 50 LncRNAs for GeneSpring GX is shown in Figure 2A. Then, we further confirmed the selected 6 up-regulation of LncRNAs in PC tissues and PC cell lines through qRT-PCR (Figure 2B). As shown in Figure 2C, comparing to 293T cells, the expression of LncRNA AF339813 was significantly increased about 10-fold in PANC-1 cells, 8-fold in SW1990 cells and 8.9-fold in BxCP-3 cells. The other 5 LncRNAs have a certain degree of up-regulation in PC cell lines. In cancerous tissues, AK022159 expression was also at a level significantly higher than the average level of normal specimens.
Figure 2.
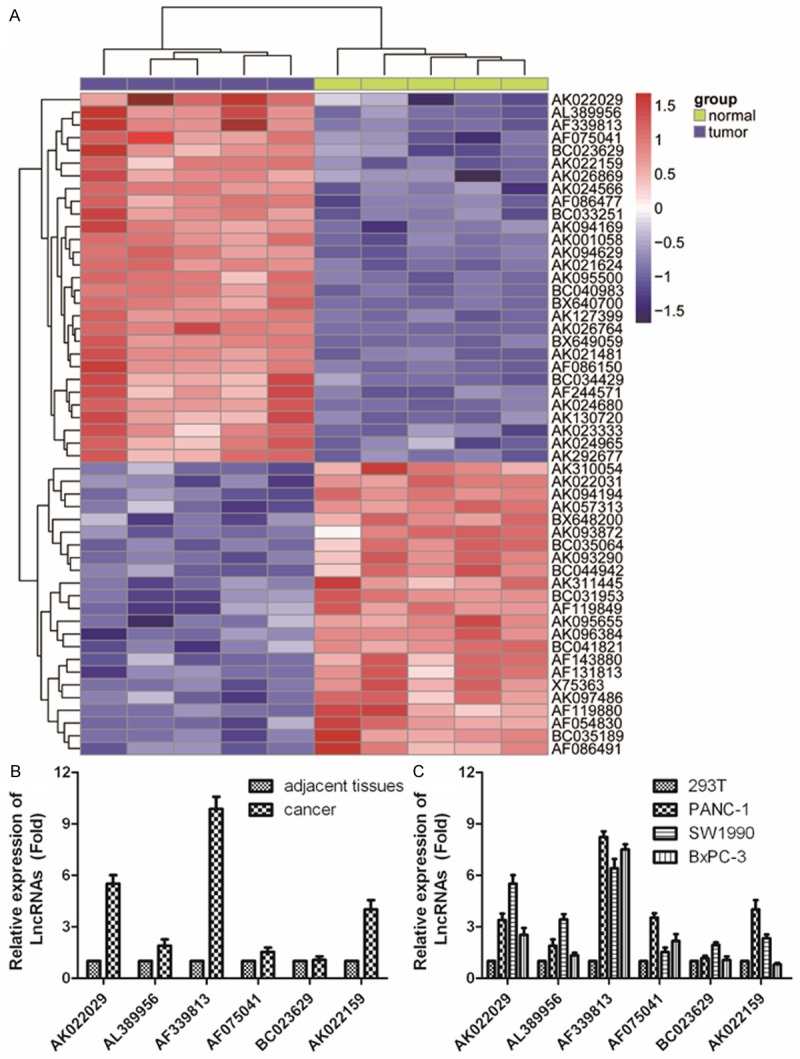
LncRNA expression profiles in PC tissue, adjacent non-tumor samples and cell lines. A. Heatmap analysis of the LncRNAs expression of groups was created using a method of hierarchical clustering by GeneSpring GX, version 7.3. Rows: samples; Columns: LncRNAs; Color key indicates LncRNA expression value, red: highest, blue: lowest. Microarray data obtained from Gene Expression Omnibus (GEO), GSE number isGSE3325. B, C. 6 biggest up-regulated LncRNAs in PC tissue and 4 PC cell lines were further validated for altered transcription level using qRT-PCR, human embryonic kidney cell line 293served as an internal control. The relative amount of each LncRNAs was normalized to GAPDH. Dates in histograms are means ± SD, *P < 0.05, **P < 0.01 compared with normal group (t-test).
LncRNA AF339813 expression is regulated by NUF2 in vitro
In order to understand the functional role of NUF2 in human PC, siRNA experiment was used to silence NUF2 in PANC-1 cells. As shown in Figure 3A, the mRNA level of NUF2 in siRNAs transfected cells down to 0.2-fold, 0.6-fold and 0.4-flod respectively, comparing with si-scramble group. As shown in Figure 3B, the down-regulated expression of NUF2 also observed using Western Blot. These results indicated that NUF2 was efficiently silenced in PANC-1 cells by siRNA-NUF2-1. Therefore siRNA- NUF2-1 is used for all subsequent NUF2 silencing experiments.
Figure 3.
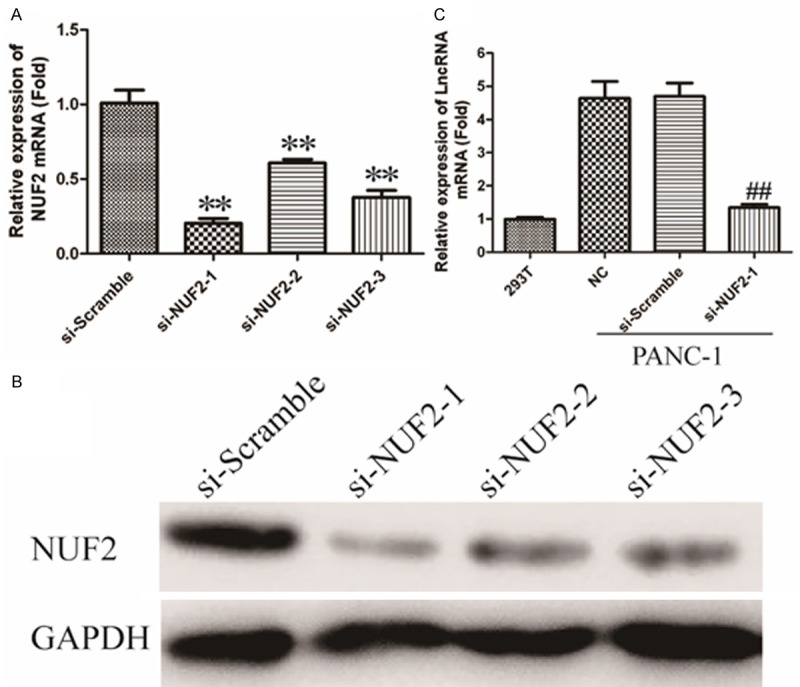
LncRNA AF339813expression is positively regulated by NUF2. A. Silencing efficiency of si-NUF2-1, si-NUF2-2 and si-NUF2-3 were verified by qRT-PCR. *P < 0.05, **P < 0.01 vs. Scramble group. B. Immunoblotting analysis of silencing efficiency of NUF2. GAPDH was used as an internal control. C. LncRNA AF339813 mRNA levels were detected by qRT-PCR in NUF2 knockdown PANC-1 cells. GAPDH gene served as an internal control. The relative gene expression was calculated using 2-ΔΔCt method. Date are expressed as means ± SD from three independent experiments and analyzed by student’s t-test. *P < 0.05, **P < 0.01 vs. non-treated PANC-1 cells.
Subsequently, we determined the relationship between NUF2 and LncRNAs. As shown in Figure 3C, after transfected with siRNA-NUF2-1 in PANC-1 cells, the expression of LncRNA AF339813 was down-regulated about 4.9-fold compared with blank or si-Scramble group. These data imply that LncRNA AF339813 expression is positively regulated by NUF2.
LncRNA AF339813 regulates cell proliferation and apoptosis
We previously proved that silencing of NUF2 inhibits cell growth and induces apoptosis in human PC cell lines [25]. However, the molecular mechanism is unknown. In order to further verify whether the role of NUF2 inregulation of PC cells was mediated via LncRNA AF339813, the expression of LncRNA AF339813 was knockdown via siRNA. The efficacy of siRNA is shown in Figure 4A, more than 70% of the mRNA level was efficiently silenced in PANC-1 cells by siRNA-AF339813-2. As expected, AF339813 knockdown dramatically inhibited cell growth by MTT (Figure 4B). Since cell growth inhibition often due to cell cycle arrest and increased apoptosis, we further analyzed cell cycle and apoptosis by flow cytometry. As shown in Figure 4C, the percentage of apoptotic cells was higher in siRNA-AF339813 transfected cells (10.9%) than that in si-Scramble transfected cells (3.4%) and parental cells (2.86%). To further determine the mechanism of AF339813 on apoptosis, the expression of Caspase-3, Caspase-9, Bax and Bcl-2 was analyzed by Western Blot. As shown in Figure 4D, the expression of Caspase-3, Caspase-9 and Bax were significantly increased and Bcl-2 was significantly decreased with treatment of siRNA-AF339813. Therefore, western blot results provided evidence that silencing AF339813 induced apoptosis through mitochondrial and caspases dependent pathway. Collectively, our data suggest that LncRNA AF339813 expression may play a pivotal role in the proliferation and apoptosis of PC cells in vitro.
Figure 4.
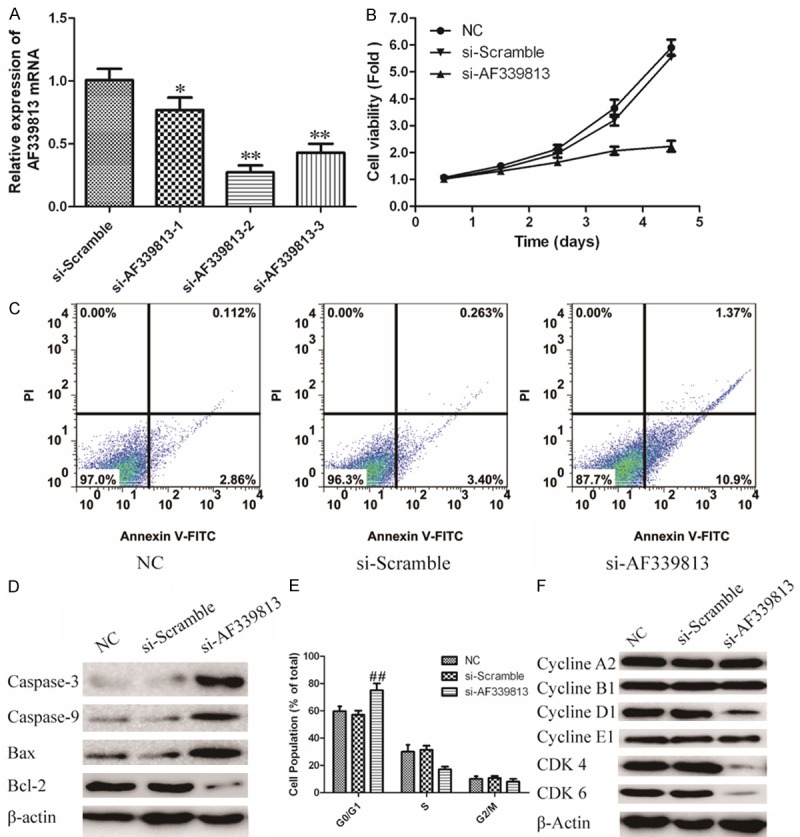
Knockdown BX647187 regulates oncogenic phenotypes of PANC-1 cell. A. Silencing efficiency of siRNAs was verified using qRT-PCR. *P < 0.05, **P < 0.01 vssi-Scramble group. B. Cell viability in AF339813 knockdown cells was measured using MTT. Values represent mean ± SD of three independent experiments. #P < 0.05, ##P < 0.01 vs. NC group. C. Flow cytometry analyze apoptosis of AF339813 knockdown cells stained with AnnexinV-FITC and PI. D. The expression of apoptosis relevant protein: Caspase3, Caspase9, Bax and Bcl-2 were analyzed through Western blot. GAPDH was used as an internal control. E. Cell cycle distribution was analyzed by flow cytometry using PI staining. The percentage of cells in different phases was counted. Values represent mean ± SD of three independent experiments. #P < 0.05, ##P < 0.01 vs. NC group. F. AF339813 silenced PANC-1 cells and controls were subjected to western blot for determining the expression levels of cell cycle related proteins. GAPDH was used as an internal control.
LncRNA AF339813 regulates cell cycle in vitro
To study the role of LncRNA AF339813 in the regulation of cell cycle, the pancreatic cancer cell lines treated with si-LncRNA AF339813 were analyzed. As shown in Figure 4E that the role of LncRNA AF339813 in the regulation of cell cycle. The expression of LncRNA AF339813 is inhibited by specific siRNA and the cell cycle is analyzed by flow cytometry. LncRNA AF339813 inhibition displays a decreased percentage of cells in G1/S phase and more cells in G0/G1 phase compared with si-Scramble.
The G1/S phase transition is primarily regulated by D-typecyclins (D1, D2) in complex with CDK4/CDK6 [26]. Therefore, in order to illuminate the G0/G1 arrest observed, we examined the expression of cyclins A2, B1, D1, E1 as well as CDK4/ CDK6. As shown in Figure 4F, LncRNA AF339813 silencing led to a decrease in cyclins D1 as well as a decrease in protein expression of CDK4/CDK6. However, the expression levels of cyclin A2, cyclin B1 and E1 remained unchanged. These data suggest that G0/G1 arrest is associated with activation of the Cyclin D1 and CDK4/CDK6. Thus, we conclude that LncRNA AF339813 regulates cell cycle arrest at G0/G1 phase via inactivated CDK4/CDK6 complex.
Discussion
In the present study, we report two novel findings (i) expression of NUF2 was significantly enhanced in PC tissues relative to correspondingadjacent normal tissues. Additionally, it was also highly expressed in several kinds of PC cell lines; (ii) down-regulation of NUF2 Inhibits Tumor Growth and Induces apoptosis by regulating LncRNA AF339813.
As a corecomponent of the NDC80 kinetochore complex, NUF2 has been reported to be implicated in tumorigenesis of various kinds of human cancer. Previous studies show that the depletion of NUF2 by specific siRNAs resulted in inhibition of cell proliferation and induction of apoptosis in non-small-cell carcinoma and ovarian cancer [7,10,11]. Likewise, knockdown of NUF2 suppressed tumor growth and induced cell apoptosis in human glioma cells [27]. In previous study, we demonstrated that silencing of NUF2 significantly inhibited the proliferation and colony formation ability of pancreatic cancer cells in vitro through inducing cell cycle arrest at G0/G1 phase [25]. These findings strongly suggested the potential role of NUF2 in tumorigenesis, however, its regulatory mechanisms in human PC is not well characterized.
Recent improvements in high-throughput transcriptome analysis in the last few years , have led to the discovery that > 90% of the total mammalian genome can be transcribed and may yield many short or long noncoding RNAs (lncRNAs) with limited or no protein-coding capacity [28,29]. Increasing numbers of reports have demonstrated that lncRNAs regulate multiple biological processes, including cell growth, cell cycle progression, differentiation, and apoptosis [30-33]. Furthermore, some studies suggest that dysexpression of lncRNAs is in the involvement of a variety of diseases, especially cancer [34,35]. However, the specific role of aberrantly expressed lncRNAs in PC remains largely unknown. In this research, LncRNA AF339813 was found significantly expressed in PC tissue (Figure 2B, 2C) and was positively regulated by NUF2 (Figure 3C). Moreover, silencing AF339813 by siRNA could alter the phenotypes of PC cells in vitro. We found inhibition of AF339813 showed lower cell viability and higher apoptosis compared with the control group in PANC-1 cell lines (Figure 4B, 4C). In addition, flow cytometric analysis showed AF339813 knockdown would lead to cells arrested in G0/G1 phase (Figure 4E). Our data had identified an important role for AF339813 in human PC development and progression.
To further explain the regulatory mechanism of AF339813 in G0/G1 cell cycle arrest, proteins involved in some cycle regulators were analyzed by immunoblotting. Our results indicated that silencing AF339813 markedly decreased the expression of Cyclin D1 and the phosphorylated level of CDK4/CDK6 (Figure 4F). It has been widely accepted that CDK4/CDK6 complex is required for cells transition from G2 to M phase [26]. Thus, we conclude that LncRNA AF339813 regulates cell cycle arrest at G0/G1 phase via inactivated CDK4/CDK6 complex.
However, there are several limitations in our study that should be mentioned. For example, our sample size is relatively small. We do not distinguish whether overexpression of NUF1 was significantly correlated with clinicopathological parameters such as TNM stage, age and gender. Although we found LncRNA AF339813 expression is regulated by NUF2, the detailed regulation mechanisms between NUF2 and AF339813 are not understood. Besides, we detected NUF2 expression pattern and the relationship between NUF2, LncRNA AF339813 and pathophysiology in pancreatic cancer cell lines. Nevertheless, these studies should be pursued by using appropriate animal models in future studies.
In conclusion, we found NUF2 was significantly high expressed in human pancreatic cancer and several PC cell lines for the first time. In addition, NUF2 played a key role in PC phenotypes via regulation of LncRNA AF339813. These findings would be helpful to understand the molecular mechanism of PC pathogenesis and pathophysiology. Our future analysis will focus on whether NUF2 and LncRNA AF339813 is a potential diagnostic even a therapeutic target for PC.
Disclosure of conflict of interest
None.
References
- 1.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2140–2141. doi: 10.1056/NEJMc1412266. [DOI] [PubMed] [Google Scholar]
- 4.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford SM. The importance of primary care for cancer diagnoses. Lancet Oncol. 2014;15:136–137. doi: 10.1016/S1470-2045(14)70013-0. [DOI] [PubMed] [Google Scholar]
- 6.DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon E. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayama S, Daigo Y, Kato T, Ishikawa N, Yamabuki T, Miyamoto M, Ito T, Tsuchiya E, Kondo S, Nakamura Y. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Res. 2006;66:10339–10348. doi: 10.1158/0008-5472.CAN-06-2137. [DOI] [PubMed] [Google Scholar]
- 8.Numnum TM, Makhija S, Lu B, Wang M, Rivera A, Stoff-Khalili M, Alvarez RD, Zhu ZB, Curiel DT. Improved anti-tumor therapy based upon infectivity-enhanced adenoviral delivery of RNA interference in ovarian carcinoma cell lines. Gynecol Oncol. 2008;108:34–41. doi: 10.1016/j.ygyno.2007.08.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurzov EN, Izquierdo M. RNA interference against Hec1 inhibits tumor growth in vivo. Gene Ther. 2006;13:1–7. doi: 10.1038/sj.gt.3302595. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko N, Miura K, Gu Z, Karasawa H, Ohnuma S, Sasaki H, Tsukamoto N, Yokoyama S, Yamamura A, Nagase H, Shibata C, Sasaki I, Horii A. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis. Biochem Biophys Res Commun. 2009;390:1235–1240. doi: 10.1016/j.bbrc.2009.10.127. [DOI] [PubMed] [Google Scholar]
- 11.Sethi G, Pathak HB, Zhang H, Zhou Y, Einarson MB, Vathipadiekal V, Gunewardena S, Birrer MJ, Godwin AK. An RNA interference lethality screen of the human druggable genome to identify molecular vulnerabilities in epithelial ovarian cancer. PLoS One. 2012;7:e47086. doi: 10.1371/journal.pone.0047086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiraishi T, Terada N, Zeng Y, Suyama T, Luo J, Trock B, Kulkarni P, Getzenberg RH. Cancer/Testis Antigens as potential predictors of biochemical recurrence of prostate cancer following radical prostatectomy. J Transl Med. 2011;9:153. doi: 10.1186/1479-5876-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 16.Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L, Cao Y, Bao RF, Mu JS, Tan ZJ, Tao F, Liu YB. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15:806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Su L, Chen X, Li P, Cai Q, Yu B, Liu B, Wu W, Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68:557–564. doi: 10.1016/j.biopha.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, Wei M, Shen J, Hou J, Gao X, Xu C, Huang J, Zhao Y, Sun Y. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190:2278–2287. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie P, Zhou G, Li G. The role of MALAT1 correlates with HPV in cervical cancer. Oncol Lett. 2014;7:2135–2141. doi: 10.3892/ol.2014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, Ren J, Hou F, Li Q. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han Y, Liu Y, Zhang H, Wang T, Diao R, Jiang Z, Gui Y, Cai Z. Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long non-coding RNA MALAT1. FEBS Lett. 2013;587:3875–3882. [PubMed] [Google Scholar]
- 22.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 23.Kam Y, Rubinstein A, Naik S, Djavsarov I, Halle D, Ariel I, Gure AO, Stojadinovic A, Pan H, Tsivin V, Nissan A, Yavin E. Detection of a long non-coding RNA (CCAT1) in living cells and human adenocarcinoma of colon tissues using FIT-PNA molecular beacons. Cancer Lett. 2014;352:90–96. doi: 10.1016/j.canlet.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Crea F, Watahiki A, Quagliata L, Xue H, Pikor L, Parolia A, Wang Y, Lin D, Lam WL, Farrar WL, Isogai T, Morant R, Castori-Eppenberger S, Chi KN, Wang Y, Helgason CD. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5:764–774. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu P, Chen X, Sun J, Bie P, Zhang LD. SiRNA-mediated Knockdown against NUF2 Suppresses Pancreatic Cancer Proliferation in Vitro and in Vivo. Biosci Rep. 2015:35. doi: 10.1042/BSR20140124. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 27.Huang SK, Qian JX, Yuan BQ, Lin YY, Ye ZX, Huang SS. SiRNA-mediated knockdown against NUF2 suppresses tumor growth and induces cell apoptosis in human glioma cells. Cell Mol Biol (Noisy-le-grand) 2014;60:30–36. [PubMed] [Google Scholar]
- 28.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Xu D, Yang F, Yuan JH, Zhang L, Bi HS, Zhou CC, Liu F, Wang F, Sun SH. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology. 2013;58:739–751. doi: 10.1002/hep.26361. [DOI] [PubMed] [Google Scholar]
- 31.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 33.Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5) J Cell Sci. 2008;121:939–946. doi: 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- 34.Tang JY, Lee JC, Chang YT, Hou MF, Huang HW, Liaw CC, Chang HW. Long noncoding RNAs-related diseases, cancers, and drugs. ScientificWorldJournal. 2013;2013:943539. doi: 10.1155/2013/943539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]


