Abstract
We aimed to investigate the role of Notch1/Hes signaling pathway in the pathogenesis of abnormal ossification of hip ligament in patients with ankylosing spondylitis (AS). 22 AS patients scheduled for artificial hip arthroplasty were randomly chosen as AS group. As controls, we used 4 patients diagnosed with transcervical fracture who underwent hip replacement surgery. Notch1 and Hes mRNA expressions were detected by real-time fluorescent quantitative polymerase chain reaction (RFQ-PCR). Immunohistochemistry (IHC) was used to detect Notch1 and Hes protein expression. Correlation analyses of Notch-l and Hes with AS-related clinical factors were conducted with spearman’s correlation analysis and partial correlation analysis. RFQ-PCR results showed significant differences in Notch1 and Hes mRNA expressions between AS group and the control group (all P < 0.05). IHC analysis further indicated positive nuclear signals of Notch1 and Hes protein, indicating functional activation of the Notch1 and Hes pathways. Semi-quantitative IHC showed a higher Notch1 and Hes expression levels in AS group compared to the control group (all P < 0.05). Correlation analysis suggested that Hes protein expression was positively associated with the clinical course of the disease in AS patients. In conclusion, Notch1 and Hes overexpression was clearly detected in hip joint ligaments of AS patients, Hes protein expression was associated with the clinical course of AS. Taken together, we suggest that signaling pathways mediated by Notch1-Hes may contribute to ligament ossification of hip joints in AS patients.
Keywords: Notch1, hairy enhancer of split, hip joint ligament, ankylosing spondylitis, mRNA expressions, protein expressions, clinical course, signaling pathway
Introduction
Ankylosing spondylitis (AS) is a chronic inflammatory disease mainly with sacroiliac joint and spine involvement [1,2]. AS mainly affect young adults between 15 and 30 years. The incidence of AS is approximately 0.5% worldwide, while its prevalence is 0.3% in China [1,3]. Importantly, AS is more frequent in men and the incidence ratio among male to female is (2.5-5):1 [4]. Inflammation and pain in the vertebral column and joints result in decreased physical activity, fatigue, sleep disturbances, depression, anxiety and stress, which affects the quality of life [5,6]. The precise pathogenesis and etiology of AS is not elucidated, but autoimmunity, genetic factors and infections are contributing risk factors [7,8]. Management of AS is difficult due to dynamic changes in the underlying AS pathogenesis and shortage of effective interventions for AS [9-11].
The Notch signaling pathway controls cellular differentiation, proliferation, and apoptosis [12]. Increasing evidence suggests that the Notch signaling pathway may be involved in hematological malignancies and solid tumors, as well as in angiogenesis, neurogenesis and homeostasis [13,14]. Notch1, one of the four Notch receptors, is a ligand-activated transmembrane receptor which governs differentiation stimulated by direct cell-cell contact in many tissues [15-17]. The hairy and enhancer of split-1 (Hes-1), a downstream Notch1 effector, has been demonstrated to directly affect cell fate decisions as a primary target gene of Notch signaling pathway [18]. HES1 is a surrogate for Notch1 activity and therefore, HES1 is used to detect Notch1 pathway activation. Hes-1 expression was observed in 65% of rectal neuroendocrine tumors and 10% of pancreatic neuroendocrine tumors [19]. Notch signaling pathway is associated with AS since it might lead to the formation of articular cartilage and coordinate ossification and extension of the growth plate, as well as osteoblast differentiation [20]. Overexpression of Hes could also accelerate osteogenesis and stimulate the expression of osteogenic marker genes, including osteopontin and type1 collagen [21]. Recent studies show a prominent role of the Notch system in mesenchymal stem cell. Therefore, the Notchl-Hes signaling pathway could play a major role in AS development and could be a viable therapeutic target in AS treatment [22-24]. In this study, we investigate the role of Notch1/Hes signaling pathway in AS pathogenesis and explore the potential of this pathway for design of future prevention strategies.
Materials and methods
Ethics statement
The study was approved by the Ethical Committee of Tongren Hospital affiliated to Shanghai Jiaotong University. All study participants provided written informed consent and the study was carried out under the Declaration of Helsinki [25].
Study subjects
Between September 2013-October 2013, 22 AS patients scheduled for artificial hip arthroplasty were randomly selected from the Department of Orthopedic Surgery, Tongren Hospital affiliated to Shanghai Jiaotong University. The 22 patients (18 male and 4 female; average age, 40.6) had ongoing AS for 3-15 years range. The eligible AS patients were diagnosed strictly according to the diagnostic criteria of American College of Rheumatology (ACR) (1984 revised version) [26]. Participants who had taken anti-osteoporosis medication, corticosteroids, TNF-α antagonist or non-steroidal anti-inflammatory drug (NSAID) two months before operation were excluded from the present study. In addition, 4 patients with transcervical fracture who underwent hip replacement surgery were selected at random as control patient group. Peripheral ligament tissues samples from caput femoris was collected from the AS patients and the controls during the operation.
Real-time fluorescent quantitative polymerase chain reaction (RFQ-PCR)
Total RNA from ligament tissue was extracted using Trizol (invitrogen) reagent according to manufacturer’s protocol. Purity of isolated RNA was tested by ultraviolet spectrophotometer optical density (OD) and concentration was recorded for further investigation. The integrity of the total RNA was assessed by agarose gel electrophoresis. The cDNA template was synthesized by reverse transcription with Rever TraAceq PCR RT Kit (Toyobo, Shanghai, China). Sequence of primers for PCR was summarized in Table 1. RT-PCR was performed in a total reaction volume of 20 μl, including 10 μl of 2 × SYB qPCR Mix; 0.4 μl of 50 × ROX reference dye; 0.8 μl of forward primer (10 uM); 0.8 μl of reverse primer (10 μM); 2 μl of cDNA; 20 μl of ddH2O. The conditions for RT-PCR reaction were displayed: predenaturing at 95°C for 3 min, 40 cycles of denaturing at 94°C for 30 s, followed by annealing at 65°C for 31 s. The PCR instrument in present study was ABI 7300 system (American). The CT value (average of 3 parallel holes measurements) was collected for each sample after 3 replicates.
Table 1.
Sequence of primers for polymerase chain reactions
| Gene | Forward | Reverse | TM (°C) |
|---|---|---|---|
| Notch1 | CAACATCCAggACAACATgg | ggACTTgCCCAggTCATCTA | 60 |
| Hes | CTAAACTCCCCAACCCACCT | AggCgCAATCCAATATgAAC | 58 |
| GNPTG | AAggCTgAAAggTTTgCTCA | CCAgCCAgCTTCTCTgTAgg | 58 |
Hes, hairy and enhancer of split; TM, temperature.
Sample preparation and HE staining
The collected tissue samples were embedded in paraffin, and cut into 4 μm sections. The sections were deparaffinized in xylene solution and rehydrated with graded concentrations of ethanol (100%-75%) and stored in the refrigerator at 4°C till further use. The tissue sections were stained for 3-8 min in hematoxylin. Subsequently, sections were treated with 1% hydrochloric acid alcohol for 30 s, and then immersed in 1% ammonia solution for 30 s, followed by washing with a stream of water. Subsequently, the sections were stained with eosin for 1-3 min and sealed with neutral gum.
Immunohistochemistry
Hydrogen peroxide (H2O2, 3%) was added to the prepared tissue samples at room temperature in order to quench endogenous peroxidase (POD). Normal rabbit serum was used as blocking agent to avoid non-specific protein adsorption. After 15 min, samples collected from the AS group were washed with phosphate buffered saline (PBS), and incubated with primary antibody diluted with bovine serum albumin (BSA) (1:1200) at 37°C for an hour. Horseradish peroxidase-conjugated secondary antibodies (Genesail Biotech, 1:5000) were added and the sections were incubated at 37°C for 30 min. Substitution of the primary antibody with PBS was used as negative control. PBS washes were performed three times and the samples were developed with diaminobenzidine (DAB), and subsequently, counterstained with hematoxylin for 1-3 min. Samples were dehydrated by an ethanol gradient and vitrified in xylene and sealed with neutral gum. The IHC staining controls were treated with PBS instead of the primary antibody. Stained tissues were observed under Olympus IX70 microscope.
Evaluation of immunohistochemical staining results
The results were analyzed by semi-quantitative scoring method: the grading for the percentage of positive cells was carried out: ≤ 5%, 0 point; 6%~25%, 1 point; 26%~50%, 2 point; 51%~75%, 3 point; more than 75%, 4 point. For tinctorial strength, scores were distributed as: no staining, 0 point; light yellow, 1 point; brownish yellow, 2 point; dark brownish yellow, 3 point. Eventually, the result was classified into four grades by multiplying the two scores mentioned above: 0 point, negative (-); 1~4 points, weakly positive (+); 5~8 points, positive (++); 9~12 points, strong positive (+++). Scores for more than 5 point was regarded as positive.
Statistical analysis
Data were tested by the application of homogeneity of variance and two independent samples t-test. Continuous variables with normal distribution are expressed as mean ± standard deviation (SD) and analyzed by Analysis of Variance (ANOVA); skewed distributed continuous variables are expressed as the median value (interquartile range) and tested by nonparametric statistics; while categorical data were presented with frequency counts and was assessed by χ2 tests. Spearman’s correlation analysis and partial correlation analysis were conducted to evaluate correlation between variables. P < 0.05 was considered as significance.
Results
Notch1 and Hes mRNA expression
Dissolution curve analysis was used to determine different reaction products, including non-specific and specific products. The dissolution curves (unimodal) showed specific expression of Notch1 and Hes genes in ligament tissues (Figure 1, SHOW ARROWS TO AREAS OF INTEREST). The independent sample test indicated that mRNA expressions of Notch1 and Hes in AS group was significantly higher than those in the control group (P < 0.05) (Table 2).
Figure 1.
The dissolution curves of (A) Notchl and (B) hairy and enhancer of split (HES) gene in ligament tissues; both Notchl and HES presented with unimodal dissolution curves, showing specific expression of Notchl and HES.
Table 2.
Independent sample test results of mRNA expressions of Notchl and hairy and enhancer of split (Hes) genes in ankylosing spondylitis (AS) group and control group
| Levene’s test | T-test | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| 95% CI | ||||||||||
|
|
|
|||||||||
| F | Sig | t | Df | Sig (Bilateral) | MD | SE | Lower limit | Upper limit | ||
| Notchl | Equal variances assumed | 0.172 | 0.684 | -2.820 | 16 | 0.012 | -1.39583 | 0.49497 | -2.44513 | -0.34654 |
| Unequal variance assumed | -2.710 | 9.128 | 0.024 | -1.39583 | 0.51508 | -2.55854 | -0.23313 | |||
| Hes | Equal variances assumed | 0.20 | 0.889 | -2.740 | 16 | 0.015 | -1.66000 | 0.60587 | -2.94439 | -0.37561 |
| Unequal variance assumed | -2.718 | 9.896 | 0.022 | -1.66000 | 0.61070 | -3.02267 | -0.29733 | |||
Sig, significance; Df, degree of freedom; Hes, hairy and enhancer of split; MD, mean difference; SE, standard error; 95% CI, confidence interval.
HE staining of ligament tissue
HE staining showed the ligament tissue was mainly composed of longitudinal arrangement of elastic fibers (along the long axis), amongst which collagen fibers existed. The main ligament cells were scattered fibroblasts. The cell numbers in the control group appeared to be greater than those in the AS group by visual analysis (Figure 2).
Figure 2.
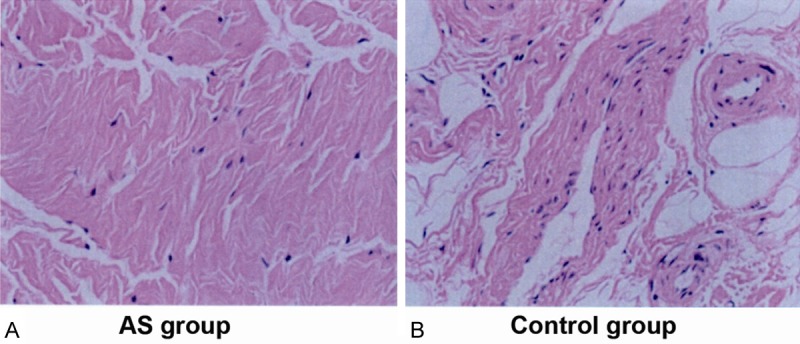
Hematoxylin-eosin (HE) staining of ligament tissue in (A) ankylosing spondylitis (AS) and (B) control group. The ligament tissue was mainly composed of longitudinal arrangement of elastic fibers (along the long axis), amongst which collagen fibers existed. The main ligament cells were scattered fibroblasts. The cell numbers in the control group appeared to be greater than those in the AS group by visual analysis.
Immunohistochemistry analysis
IHC analysis showed intense positive signals of Notch1 and Hes localized in the nucleus (yellow or brown particles). The expression of Notch1 and Hes in ligament tissue of the AS group increased significantly compared with control group (Figure 3). Semi-quantitative IHC showed that the visual score of Notch1 protein expression was 81.2% (13/16) in the AS group compared to 25% (1/4) in the control group, representing statistically significant differences (P < 0.05). The positive rate of Hes protein was 87.5% (14/16) in the AS group, which was statistically higher than that in the control group (25% (1/4)) (P < 0.05) (Figure 4).
Figure 3.
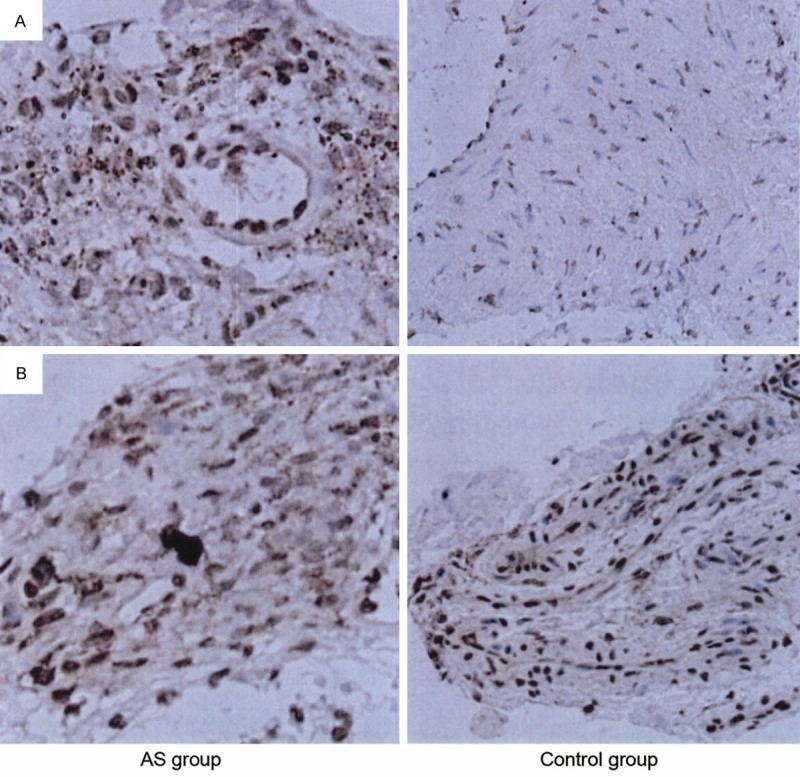
A. Immunohistochemistry staining of Notch1 in ankylosing spondylitis (AS) group and control group; B. Immunohistochemistry staining of hairy and enhancer of split (HES) in ankylosing spondylitis (AS) group and control group. The intense positive signals of Notch1 and Hes localized in the nucleus (yellow or brown particles). The expression of Notch1 and Hes in ligament tissue of the AS group increased significantly compared with control group.
Figure 4.
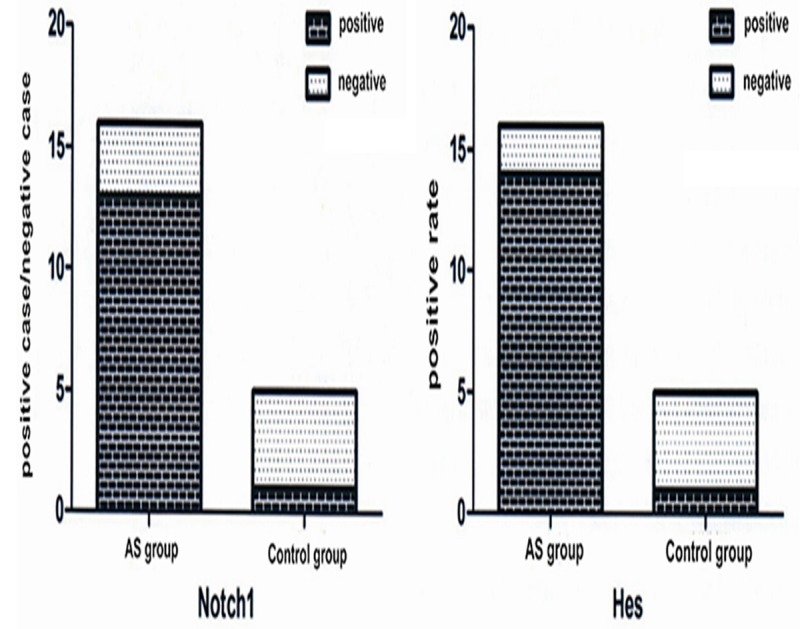
Positive rates of Notch1 and Hes expression in ankylosing spondylitis (AS) group and control group; the positive rate of Notch1 and Hes protein was s statistically higher in the AS group than that in the control group.
Correlation analysis of Notch-l and Hes with AS related clinical factors
As demonstrated in Table 3, there was no correlation between Notchl expression and gender, age, course of disease and onset age (all P > 0.05). Hes expression also had no significant correlation with gender, age and onset age (all P > 0.05). However, the positive expression rate of Hes in patients followed the course of disease at < 5 years and > 15 years being 100% and 50%, respectively, demonstrating a statistical significance (P = 0.01) (Table 4).
Table 3.
Correlation analysis of Notch1 expression with ankylosing spondylitis (AS) related clinical factors, including sex, age, course of disease and onset age
| Parameters | Cases (n) | - | + | ++ | +++ | Positive rate | P value |
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Female | 4 | 0 | 1 | 1 | 2 | 75% | 0.91 |
| Male | 18 | 2 | 2 | 3 | 11 | 77.8% | |
| Age (year) | |||||||
| < 45 | 15 | 1 | 2 | 3 | 9 | 80% | 0.66 |
| ≥ 45 | 7 | 1 | 1 | 1 | 4 | 71.4% | |
| Course of disease (year) | |||||||
| < 5 | 12 | 1 | 2 | 2 | 7 | 75% | 0.78 |
| ≥ 5 | 10 | 1 | 1 | 2 | 6 | 80% | |
| Onset age (year) | |||||||
| < 20 | 9 | 0 | 1 | 3 | 5 | 88.9% | 0.28 |
| ≥ 20 | 13 | 2 | 2 | 1 | 8 | 69.2% | |
Table 4.
Correlation analysis of Hes expression and clinical factors of ankylosing spondylitis (AS) patients, including sex, age, course of disease and onset age
| Parameters | Cases | - | + | ++ | +++ | Positive rate | P value |
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Female | 4 | 0 | 1 | 1 | 2 | 75% | 0.91 |
| Male | 18 | 2 | 2 | 3 | 11 | 77.8% | |
| Age (year) | |||||||
| < 45 | 15 | 1 | 2 | 3 | 9 | 80% | 0.66 |
| ≥ 45 | 7 | 1 | 1 | 1 | 4 | 71.4% | |
| Course of disease (year) | |||||||
| < 5 | 12 | 0 | 0 | 2 | 10 | 100% | 0.01* |
| ≥ 5 | 10 | 2 | 3 | 2 | 3 | 50% | |
| Onset age (year) | |||||||
| < 20 | 9 | 1 | 1 | 2 | 5 | 77.8% | 0.96 |
| ≥ 20 | 13 | 1 | 2 | 2 | 8 | 92.3% | |
Hes, hairy and enhancer of split.
Discussion
Notch signaling pathway participates in many biological processes through maintaining the balance of cell proliferation, differentiation, survival and apoptosis [27,28]. Mutations in Notch family members have an inhibitory effect on several tumors such as cutaneous, chronic myelomonocytic leukemia, lung and head and neck squamous cell carcinomas (HNSCC) tumors [29-31]. Our data showed overexpression of Notch1 and Hes in ligament tissue of hip joints of AS patients and that the positive rate of Hes expression was positively associated with the course of AS. Therefore, the Notchl-Hes signaling pathway may have a prominent role in ligament ossification of hip joints in patients with AS. Notchl-Hes signaling pathway machinery and its cellular consequences are highly conserved [32]. Upon ligand binding, the Notch receptor signaling is activated and the trans-membrane domain cleavage leads to release of the intracellular domain of Notch, which functions as a transcriptional activator for a number of target genes that participate in a variety of cellular activities including cell motility and cell-fate decisions [33,34]. It has been reported that Notch signaling may play a role in bone development and overexpression of ligand or receptors of Notch could potentially be harmful to osteoclast and osteoblast precursors, effecting cell differentiation [35]. Previous research has also revealed that through Jagged1, the Hes-1, can enhance the expression and activity of NFATc1 (nuclear factor of activated T-cells), a transcription factor of osteoclast, and thus promotes osteoclastogenesis [36,37].
To our knowledge, our study is the first to find a correlation between Notchl-Hes signaling pathway and AS. Our data suggests overexpression of Notch1 and Hes occurs in ligament tissue of hip joints of AS patients and the resulting enhanced signaling of Notch1-Hes may contribute to ligament ossification of hip joints in AS patients, through activation of yet unknown processes relevant to the disease. Importantly, we obtain the first correlation between positive rate of Hes expression with the disease course of AS which needs to be further studied.
Acknowledgements
We acknowledge the helpful comments on this paper received from our reviewers.
Disclosure of conflict of interest
None.
References
- 1.Zheng GQ, Zhang YG, Chen JY, Wang Y. Decision making regarding spinal osteotomy and total hip replacement for ankylosing spondylitis: experience with 28 patients. Bone Joint J. 2014;96-B:360–365. doi: 10.1302/0301-620X.96B3.32774. [DOI] [PubMed] [Google Scholar]
- 2.Wang YY, Lu H, Zhao Z, Huang F. The efficacy and safety of Jitongning Capsule in patients with ankylosing spondylitis. Chin J Integr Med. 2013;19:98–103. doi: 10.1007/s11655-012-1212-x. [DOI] [PubMed] [Google Scholar]
- 3.Taylan A, Sari I, Akinci B, Bilge S, Kozaci D, Akar S, Colak A, Yalcin H, Gunay N, Akkoc N. Biomarkers and cytokines of bone turnover: extensive evaluation in a cohort of patients with ankylosing spondylitis. BMC Musculoskelet Disord. 2012;13:191. doi: 10.1186/1471-2474-13-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong X, Zheng Y, Shi TY, Liu HY. Effects of tumor necrosis factor-alpha on sexual activity of male patients with ankylosing spondylitis. Clin Rheumatol. 2015;34:915–20. doi: 10.1007/s10067-014-2751-7. [DOI] [PubMed] [Google Scholar]
- 5.Robinson PC, Brown MA. Genetics of ankylosing spondylitis. Mol Immunol. 2014;57:2–11. doi: 10.1016/j.molimm.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Sommerfleck FA, Schneeberger EE, Buschiazzo EE, Maldonado Cocco JA, Citera G. A simplified version of Ankylosing Spondylitis Disease Activity Score (ASDAS) in patients with ankylosing spondylitis. Clin Rheumatol. 2012;31:1599–1603. doi: 10.1007/s10067-012-2056-7. [DOI] [PubMed] [Google Scholar]
- 7.Ortak H, Inanir A, Demir S, Uysal A, Sahin S, Sagcan M, Onder Y, Alim S, Demir AK. Decreased central corneal thickness in ankylosing spondylitis. Int Ophthalmol. 2014;34:263–268. doi: 10.1007/s10792-013-9827-2. [DOI] [PubMed] [Google Scholar]
- 8.Ma B, Yang B, Guo H, Wang Y, Zhang D, Zhang Y, Xiao Z. The association between tumor necrosis factor alpha promoter polymorphisms and ankylosing spondylitis: a meta-analysis. Hum Immunol. 2013;74:1357–1362. doi: 10.1016/j.humimm.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 9.Song IH, Poddubnyy DA, Rudwaleit M, Sieper J. Benefits and risks of ankylosing spondylitis treatment with nonsteroidal antiinflammatory drugs. Arthritis Rheum. 2008;58:929–938. doi: 10.1002/art.23275. [DOI] [PubMed] [Google Scholar]
- 10.Mebarek S, Hamade E, Thouverey C, Bandorowicz-Pikula J, Pikula S, Magne D, Buchet R. Ankylosing spondylitis, late osteoarthritis, vascular calcification, chondrocalcinosis and pseudo gout: toward a possible drug therapy. Curr Med Chem. 2011;18:2196–2203. doi: 10.2174/092986711795656153. [DOI] [PubMed] [Google Scholar]
- 11.Rajalingham S, Das S. Antagonizing IL-6 in ankylosing spondylitis: a short review. Inflamm Allergy Drug Targets. 2012;11:262–265. doi: 10.2174/187152812800958979. [DOI] [PubMed] [Google Scholar]
- 12.Tamagawa Y, Ishimura N, Uno G, Yuki T, Kazumori H, Ishihara S, Amano Y, Kinoshita Y. Notch signaling pathway and Cdx2 expression in the development of Barrett’s esophagus. Lab Invest. 2012;92:896–909. doi: 10.1038/labinvest.2012.56. [DOI] [PubMed] [Google Scholar]
- 13.Sun YY, Li L, Liu XH, Gu N, Dong HL, Xiong L. The spinal notch signaling pathway plays a pivotal role in the development of neuropathic pain. Mol Brain. 2012;5:23. doi: 10.1186/1756-6606-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takebe N, Nguyen D, Yang SX. Targeting notch signaling pathway in cancer: clinical development advances and challenges. Pharmacol Ther. 2014;141:140–149. doi: 10.1016/j.pharmthera.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y, Wang L, Tang W, Zhang D, Shang T. RNA interference-mediated knockdown of Notch-1 inhibits migration and invasion, down-regulates matrix metalloproteinases and suppresses NF-kappaB signaling pathway in trophoblast cells. Acta Histochem. 2014;116:911–919. doi: 10.1016/j.acthis.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Pupo M, Pisano A, Abonante S, Maggiolini M, Musti AM. GPER activates Notch signaling in breast cancer cells and cancer-associated fibroblasts (CAFs) Int J Biochem Cell Biol. 2014;46:56–67. doi: 10.1016/j.biocel.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Dahan S, Rabinowitz KM, Martin AP, Berin MC, Unkeless JC, Mayer L. Notch-1 signaling regulates intestinal epithelial barrier function, through interaction with CD4+ T cells, in mice and humans. Gastroenterology. 2011;140:550–559. doi: 10.1053/j.gastro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang HP, Sun YY, Chen XM, Yuan LB, Su BX, Ma R, Zhao RN, Dong HL, Xiong L. The neuroprotective effects of isoflurane preconditioning in a murine transient global cerebral ischemia-reperfusion model: the role of the Notch signaling pathway. Neuromolecular Med. 2014;16:191–204. doi: 10.1007/s12017-013-8273-7. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Chen Y, Fernandez-Del Castillo C, Yilmaz O, Deshpande V. Heterogeneity in signaling pathways of gastroenteropancreatic neuroendocrine tumors: a critical look at notch signaling pathway. Mod Pathol. 2013;26:139–147. doi: 10.1038/modpathol.2012.143. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Sun Q, Jiang S, Li J, He C, Xu W. Changes in gene expression profiles of the hip joint ligament of patients with ankylosing spondylitis revealed by DNA chip. Clin Rheumatol. 2012;31:1479–1491. doi: 10.1007/s10067-012-2038-9. [DOI] [PubMed] [Google Scholar]
- 21.Ramasamy SK, Kusumbe AP, Wang L, Adams RH. Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature. 2014;507:376–380. doi: 10.1038/nature13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing L, Jia Y, Lu J, Han R, Li J, Wang S, Peng T, Jia Y. MicroRNA-9 promotes differentiation of mouse bone mesenchymal stem cells into neurons by Notch signaling. Neuroreport. 2011;22:206–211. doi: 10.1097/WNR.0b013e328344a666. [DOI] [PubMed] [Google Scholar]
- 23.Ehninger A, Trumpp A. The bone marrow stem cell niche grows up: mesenchymal stem cells and macrophages move in. J Exp Med. 2011;208:421–428. doi: 10.1084/jem.20110132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moriyama H, Moriyama M, Isshi H, Ishihara S, Okura H, Ichinose A, Ozawa T, Matsuyama A, Hayakawa T. Role of notch signaling in the maintenance of human mesenchymal stem cells under hypoxic conditions. Stem Cells Dev. 2014;23:2211–2224. doi: 10.1089/scd.2013.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunetti G, Oranger A, Carbone C, Mori G, Sardone FR, Mori C, Celi M, Faienza MF, Tarantino U, Zallone A, Grano M, Colucci S. Osteoblasts display different responsiveness to TRAIL-induced apoptosis during their differentiation process. Cell Biochem Biophys. 2013;67:1127–1136. doi: 10.1007/s12013-013-9616-6. [DOI] [PubMed] [Google Scholar]
- 26.Martindale J, Goodacre L. The Journey to Diagnosis in AS/Axial SpA: The Impact of Delay. Musculoskeletal Care. 2014;12:221–231. doi: 10.1002/msc.1080. [DOI] [PubMed] [Google Scholar]
- 27.Vartanian A, Gatsina G, Grigorieva I, Solomko E, Dombrovsky V, Baryshnikov A, Stepanova E. The involvement of Notch signaling in melanoma vasculogenic mimicry. Clin Exp Med. 2013;13:201–209. doi: 10.1007/s10238-012-0190-9. [DOI] [PubMed] [Google Scholar]
- 28.Paryan M, Mohammadi-Yeganeh S, Samiee SM, Soleimani M, Arefian E, Azadmanesh K, Poopak B, Mostafavi E, Karimipoor M, Mahdian R. Investigation of deregulated genes of Notch signaling pathway in human T cell acute lymphoblastic leukemia cell lines and clinical samples. Mol Biol Rep. 2013;40:5531–5540. doi: 10.1007/s11033-013-2654-8. [DOI] [PubMed] [Google Scholar]
- 29.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, Bassaganyas L, Baumann T, Juan M, Lopez-Guerra M, Colomer D, Tubio JM, Lopez C, Navarro A, Tornador C, Aymerich M, Rozman M, Hernandez JM, Puente DA, Freije JM, Velasco G, Gutierrez-Fernandez A, Costa D, Carrio A, Guijarro S, Enjuanes A, Hernandez L, Yague J, Nicolas P, Romeo-Casabona CM, Himmelbauer H, Castillo E, Dohm JC, de Sanjose S, Piris MA, de Alava E, San Miguel J, Royo R, Gelpi JL, Torrents D, Orozco M, Pisano DG, Valencia A, Guigo R, Bayes M, Heath S, Gut M, Klatt P, Marshall J, Raine K, Stebbings LA, Futreal PA, Stratton MR, Campbell PJ, Gut I, Lopez-Guillermo A, Estivill X, Montserrat E, Lopez-Otin C, Campo E. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, van De Walle I, Cathelin S, Trimarchi T, Araldi E, Liu C, Ibrahim S, Beran M, Zavadil J, Efstratiadis A, Taghon T, Michor F, Levine RL, Aifantis I. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwanbeck R, Martini S, Bernoth K, Just U. The Notch signaling pathway: molecular basis of cell context dependency. Eur J Cell Biol. 2011;90:572–581. doi: 10.1016/j.ejcb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Eilken HM, Adams RH. Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr Opin Cell Biol. 2010;22:617–625. doi: 10.1016/j.ceb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Lou YL, Guo F, Liu F, Gao FL, Zhang PQ, Niu X, Guo SC, Yin JH, Wang Y, Deng ZF. miR-210 activates notch signaling pathway in angiogenesis induced by cerebral ischemia. Mol Cell Biochem. 2012;370:45–51. doi: 10.1007/s11010-012-1396-6. [DOI] [PubMed] [Google Scholar]
- 35.Won KY, Kim YW, Kim HS, Lee SK, Jung WW, Park YK. MicroRNA-199b-5p is involved in the Notch signaling pathway in osteosarcoma. Hum Pathol. 2013;44:1648–1655. doi: 10.1016/j.humpath.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Schwarzer R, Nickel N, Godau J, Willie BM, Duda GN, Schwarzer R, Cirovic B, Leutz A, Manz R, Bogen B, Dorken B, Jundt F. Notch pathway inhibition controls myeloma bone disease in the murine MOPC315. BM model. Blood Cancer J. 2014;4:e217. doi: 10.1038/bcj.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin GL, Hankenson KD. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J Cell Biochem. 2011;112:3491–3501. doi: 10.1002/jcb.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]


