Abstract
Lymph node metastasis is recognized as an important mode of liver cancer metastasis. Two hepatocarcinoma cell lines, Hca-F get high (75%) and Hca-P get low (25%) incidences of lymph node metastasis. Forkhead box M1 (FoxM1) is described as a major oncogenic transcription factor in tumor initiation, promotion, and progression. Ezrin is linked to aggressive tumor behavior by involving all stages of tumor metastasis. We firstly compared the expression of FoxM1 and Ezrin between two cells. Then we transiently transfected Hca-P cells with over-FoxM1 plasmid and Hca-F cells with sh-FoxM1 plasmid. We found that both FoxM1 and Ezrin expressed higher in Hca-F than Hca-P. We successfully down-regulated or up-regulated FoxM1 expression in Hca-F or Hca-P cells. And we found that FoxM1 had correlation with proliferation, invasion and migration in mouse hepatocellular carcinoma cell lines.
Keywords: FoxM1, ezrin, lymphatic metastasis, hepatocarcinoma
Introduction
Hepatocellular carcinoma (HCC) is a phenotypically and genetically heterogeneous polyclonal disease and resistant to most conventional chemotherapy. Ninety percent of malignant tumors are carcinomas, and lymph nodes are often the first organ to develop metastasis [1]. HCC is one of the globe’s most common types of cancer and one of the most fatal [2]. At present, HCC is largely a Third World disease, especially Southeast Asia and Africa, China alone accounts for more than 50% of the world HCC cases [3]. Lymphatic metastasis is a complex process involving multiple genes and their products. Lymph node metastasis (LNM) is an important factor resulting in poor prognosis and high mortality. Tumor metastasis includes four steps: proliferation, attachment, migration and invasion. Figuring out HCC lymph node metastasis-related proteins will be of potential benefit to decreasing the relapse and death rates of cancer patients and contribute new ideas to clinical treatment. However, molecular mechanism of metastasis remains poorly understood [4]. In previous studies, Guo et al. established and maintain a pair of syngeneic mouse hepatocarcinoma cell lines, Hca-F and Hca-P in Pathology department of Dalian Medical University. When inoculated subcutaneously in Chinese 615 mice, Hca-P showed a low of LNM rate (< 30%), whereas Hca-F showed a high LNM rate (> 70%) [5,6].
Forkhead box M1 (FoxM1), also previously called HNF-3, HFH-11, WIN, MPP2, or Trident, as a member of Forkhead family of transcription factors, shares homology in Winged Helix/Forkhead box DNA-binding domain [7]. FoxM1 is required for normal G1-S, G2, and M phase cell-cycle transitions. Besides its involvement in cell-cycle transitions, FoxM1 is also a key regulator of mitotic spindle integrity [8], angiogenesis [9], metastasis [9,10], apoptosis [10,11], DNA damage repair [12,13], embryo implantation [14] and tissue regeneration [15]. Several lines of evidence demonstrate that overexpression of FoxM1 occurs in a wide variety of human tumors frequently, including medulloblastoma [16], colorectal cancer [17], hepatocellular carcinoma [18], breast cancer [19], non-small cell lung cancer [20] and so on.
Ezrin is distributed in actin-containing cell surface structures such as microvilli, microspikes, and membrane ruffles in a wide variety of cultured cells and tissues. Ezrin is expressed at high levels in intestine, stomach, lung, pancreas, and kidney; at moderate levels in spleen, lymph nodes, thymus, and bone marrow; at very low levels in heart and brain [21,22]. Ezrin is an integral cytoskeletal linker protein found in numerous cell types but predominantly in polarized epithelial cells [23], where it links filamentous actin to intra-membranous adhesive proteins such as cellular adhesion molecules (CAMs), CD43 and CD44 [24]. In its inactive form, Ezrin is located in the cytoplasm and its C-terminal domain, an F-actin-binding site, is masked by the N-terminal domain of Ezrin or other ERM (ezrin-radixin-moesin) family member proteins. Once Ezrin is activated by threonine and tyrosine phosphorylation, it assumes an active form, in which its N-terminal domain binds the cell membrane and its C-terminal domain binds to F-actin [25,26]. Ezrin is linked to aggressive tumor behavior by involving all stages of tumor metastasis [27,28] including cell adhesion, survival, motility, and signal transduction [29-31].
Studying on FoxM1 and Ezrin will help us to reveal the role of FoxM1 and Ezrin in the mechanism of tumorigenesis and their lymphatic metastasis, which could be of therapeutic benefit.
Material and methods
Animals and cell lines
The mouse hepatocarcinoma cell line Hca-P, with lymphatic metastasis rate less than 30%, Hca-F, with lymphatic metastasis rate more than 70% were established and maintained in Dalian Medical University as previously described. Inbred Chinese 615 mice (8-week-old males, weighing 18-22 g) were provided by the Experimental Animal center of Dalian Medical University. Hca-P and Hca-F cells were inoculated (2×106 cells in a 0.2 ml cell suspension) into each mouse and grew in the mice peritoneal cavity for 7 days. These cells were drawn and injected again in other 615 mice and allowed to grow for 5 days. Two passages were done in order to harvest large number of cells in around 2 weeks. The cells were then continuously cultured in 90% RPMI 1640 (Life Technologies Corporation, CA, USA) supplemented with 10% fetal bovine serum (Gibco, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C with 5% CO2.
Transient transfection of cells
The FoxM1 overexpression plasmids, FoxM1 siRNA (5’-GCC GGA ACA UGA CCA UCA ATT-3’) and negative control (NC) sequence (5’-GTT CTC CGA ACG TGT CAC GT-3’) were designed and synthesized by Shanghai GenePharma Co., Ltd (Shanghai, China). Cells were cultured in Petri dishes to 80-90% confluence and then transfected with Lipofectamine 2000 (Invitrogen) in strict accordance with the manufacturer’s instructions. After transfection, the cells were cultured for 48 or 72 h before use in subsequent experiments.
RNA extraction and real-time reverse transcriptase PCR
Total RNA was extracted with the TRIzol (Takara, Dalian, China) reagent according to the manufacturer’s protocol and RNA samples were qualified by the measurement of the optic absorbance at 260 and 280 nm with a resultant A260/A280 ratio that ranged from 1.8 to 2.0, which indicated a high purity of the extracted RNA. The concentration of total RNA was calculated according to A260. Aliquots of total RNA (1.0 g each) from each sample were reverse transcribed into cDNA according to the instructions of PrimeScript® RT Reagent Kit (Takara, Dalian, China). Real-time PCR was performed with an ABI Step One Plus Real-time PCR system according to the manufacturers’ recommendations. Real-time PCR reaction contained 10 μl 2× SYBR Premix Ex Taq, 0.8 μl primer mix, 0.4 μl 50× ROX Reference Dye II, 4 μl cDNA, and 4.8 μl deionized water to make a total volume of 20 μl. The relative amount of specific mRNA was normalized to GAPDH. All PCR reactions were run in triplicate and were performed with 40 cycles. The results analysis was carried out using the 2-ΔΔCt method. The primers used were as follows. FoxM1: 5’-CGT CGG CCA CTG ATT CTC AAA-3’ (forward), and 5’-GGC AGG GGA TCT CTT AGG TTC-3’ (reverse); MMP-2: 5’-TTC TTC GCA GGG AAT GAG-3’ (forward), and 5’-ACG ACA GCA TCC AGG TTA T (reverse); MMP-9: 5’-CTT TGT AGG GTC GGT TCT G-3’ (forward), and 5’-GAG TGG ATA GCT CGG TGG-3’ (reverse); GAPDH: 5’-GTG AAG GTC GGA GTC AAC G-3’ (forward), and 5’-TGA GGT CAA TGA AGG GGT C-3’ (reverse).
Western blot
Cells were washed in PBS before incubation with Lysis Buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris, pH 7.4, 1 mM EDTA, 1 mM EGTA, pH 8.0, 0.2 mM Na3VO4, 0.2 mM phenylmethylsulfonyl fluoride, 0.5% Nonidet P-40) on ice for 10 min. The cell lysates were clarified by centrifugation at 9,000 g for 15 min and the supernatants were collected. Protein concentration was determined with the Coomassie Protein Assay reagent using bovine serum albumin (BSA) as a standard. Equal amounts of protein extracts (30 μg) were separated by 12% sodium dodecyl suphate (SDS)-polyacrtlamide gel electrophoresis (PAGE) and transferred to nitrocellulose filter (NC) membranes. The membranes were blocked in 5% non-fatmilk in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 2 hours at room temperature and probed with primary antibodies overnight at 4°C. The membranes were washed with TBST three times. Then the membranes were incubated with horseradish peroxidase-conjugated antibody for 1 hour at room temperature. After washed with TBST four times, the membranes were detected using ECL and visualized using Bio-Rad Laboratories. Western blots shown are representative of at least three independent experiments. Densitometry of each band for the target protein was quantified by densitometry analysis with Labworks 4.6. The protein ban intensity was quantified by the mean ± SD of three experiments for each group as determined from densitometry relative to GAPDH (1:5000).
Cell proliferation assay
Cell proliferation was detected by a Cell Counting Kit-8 assay (Dojindo, Japan) according to the protocol of the manufacturer. cells were suspended in 1640 medium supplemented with 15% heat-inactivated fatal bovine serum and subsequently seeded in 96-well plates. After being incubated, the cultures were added 10 μl CCK-8 solution to each well and incubated at 37°C for another 2 h. Optical density (OD) value of absorbance at 450 nm was measured by Thermo Scientific Fluoroskan Ascent FL. The results were plotted as means ± SD of three independent experiments having three determination per sample for each experiment.
Data analysis
SPSS software version 13.0 for Windows (SPSS Inc, IL, USA). Statistical analysis of the data, expressed as mean ± SEM, was performed using standard one-way analysis of variance (ANOVA) or one-way ANOVA for repeated measures. Statistical significance was set at *P < 0.05, **P < 0.01.
Results
Expression of FoxM1 and Ezrin in Hca-F/Hca-P cells and the ability of proliferation in Hca-F/Hca-P cells
As showed in Figure 1A, FoxM1 and Ezrin expression in mRNA level was detected by qRT-PCR. The expression of FoxM1 and Ezrin were more in Hca-F than Hca-P (P < 0.01). FoxM1 expression at protein level was detected by Western Blot (Figure 1B). The result showed that FoxM1 and Ezrin expressed significantly more in Hca-F than Hca-P (P < 0.01). The CCK-8 cell proliferation test data showed significant promotion of cell proliferation in the Hca-F cells compared to Hca-P cells (Figure 1C, P < 0.05). It indicated that FoxM1 and Ezrin are relevant to tumor lymphatic metastasis.
Figure 1.
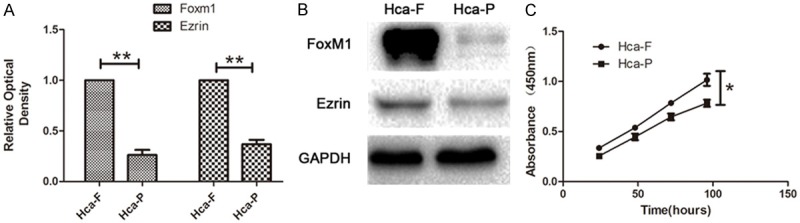
The expression of FoxM1 and Ezrin in Hca-F and Hca-P cells. A. qRT-PCR analysis of FoxM1 and Ezrin in Hca-F and Hca-P cells. B. Western blot analysis of FoxM1 and Ezrin in Hca-F and Hca-P cells. C. Scatter diagram of Hca-F and Hca-P cells proliferation assay using CCK-8 over 96 hours.
Higher expression of FoxM1 was correlated with higher cell proliferation ability and higher migration and invasion ability in Hca-P cells
To evaluate the effect of FoxM1 overexpression on the migration and invasion ability, we performed qRT-PCR and Western Blot. Data showed the mRNA expression of FoxM1, MMP-2, MMP-9 were highly increased in the over-FoxM1 Hca-P cells compared to untreated and control Hca-P cells (Figure 2A, P < 0.05). The protein expression of TIMP-1, TIMP-2 were highly decreased in the over-FoxM1 Hca-P cells compared to untreated and control Hca-P cells, while the expression of Ezrin was increased in the over-FoxM1 Hca-P cells compared to untreated and control Hca-P cells (Figure 2B, P < 0.05).
Figure 2.
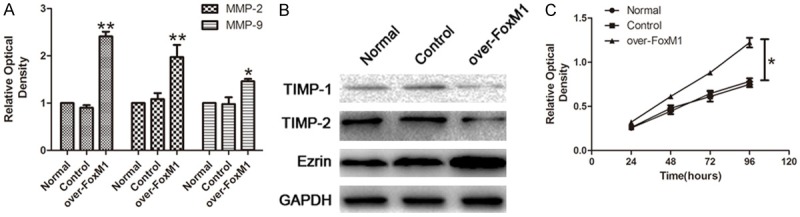
The effects of over-expressed FoxM1 on the proliferation, the expression of invasive proteins of Hca-P cells in vitro. A. qRT-PCR analysis of FoxM1, MMP-2 and MMP-9 in Hca-P cells. B. Western blot analysis of Ezrin, TIMP-1 and TIMP-2 in Hca-P cells. C. After over-expressed FoxM1, Scatter diagram of Hca-P cells proliferation assay using CCK-8 over 96 hours.
To evaluate the effect of the FoxM1 overexpression on the proliferation potential, we performed CCK-8 cell proliferation test. Data showed significant promotion of cell proliferation in the over-FoxM1 Hca-P cells compared to untreated and control Hca-P cells (Figure 2C, P < 0.05).
Lower expression of FoxM1 was correlated with higher cell proliferation ability and higher migration and invasion ability in Hca-F cells
To evaluate the effect of FoxM1 silencing on the migration and invasion ability, we performed qRT-PCR and Western Blot. Data showed the mRNA expression of FoxM1, MMP-2, MMP-9 were highly decreased in the sh-FoxM1 Hca-F cells compared to untreated and control Hca-F cells (Figure 3A, P < 0.05). The protein expression of TIMP-1, TIMP-2 were highly increased in the sh-FoxM1 Hca-F cells compared to untreated and control Hca-F cells, while the expression of Ezrin was decreased in the sh-FoxM1 Hca-F cells compared to untreated and control Hca-F cells (Figure 3B, P < 0.05).
Figure 3.
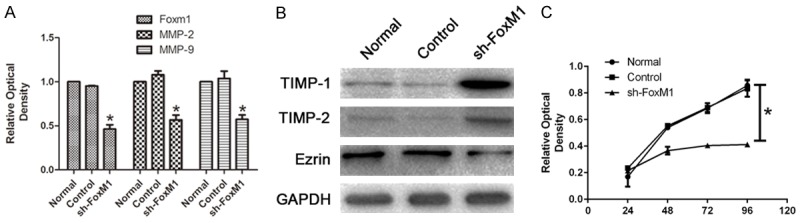
The effects of low-expressed FoxM1 on the proliferation, the expression of invasive proteins of Hca-F cells in vitro. A. qRT-PCR analysis of FoxM1, MMP-2 and MMP-9 in Hca-F cells. B. Western blot analysis of Ezrin, TIMP-1 and TIMP-2 in Hca-F cells. C. After low-expressed FoxM1, Scatter diagram of Hca-F cells proliferation assay using CCK-8 over 96 hours.
To evaluate the effect of the FoxM1 silencing on the proliferation potential, we performed CCK-8 cell proliferation test. Data showed significant inhibition of cell proliferation in the sh-FoxM1 Hca-F cells compared to untreated and control Hca-F cells (Figure 3C, P < 0.05).
Discussions
Tumor metastasis is the major cause of tumor related death. Hca-F and Hca-P cells are hepatocarcinoma cell lines presenting highly and lowly lymph node metastasis separately. We studied found the obviously different expression of FoxM1 between these two cells (Figure 1). It indicated that FoxM1 was a significant factor in lymph node metastasis.
Gene expression profiling revealed that elevated expression of FoxM1 was observed in a multitude of malignancies [32]. It has been reported that FoxM1 promotes tumor invasion, migration and metastasis [33-37]. Furthermore, overexpression of FoxM1 correlated with disease progression and poor prognosis and could serve as an independent predictor of poor survival in various human malignancies [38-41]. In all these processes, MMP-2 and MMP-9 are thought to play a critical role in tumor invasion, migration and metastasis. Among matrix metaloproteases (MMPs), a family of zinc dependent endopeptidases, MMP-2 and MMP-9 have been considered to be critical for tumor growth , invasion and metastasis [42,43]. Cell growth, migration and invasion are important processes involved in tumor progression. In our studies, we explored whether FoxM1 contributed to cell growth, migration and invasion of Hca-P and Hca-F cells in vitro. The results showed that overexpression of FoxM1 by transfection with over-FoxM1 could promote cell growth, invasion and metastasis in Hca-P cells. Similarly, we found that depletion FoxM1 by transfection with shFoxM1 could suppress cell growth, invasion and metastasis in Hca-F cells.
Ezrin, as a membrane-cytoskeleton linker, plays a pivotal role in tumor invasion and metastasis [44-46]. Ezrin can regulate the assembly of cytoskeleton elements to promote cytoskeletal reorganization and phenotypic alternation in cells, and facilitate cell migration and invasion [47,48]. Overexpression of ezrin has been shown to enhance metastatic potential in various types of tumors, while downregulation of ezrin reduced the expression of β-catenin but enhanced the expression of E-cadherin [49,50].
In this paper, we found the expression of Ezrin was increased or decreased as FoxM1 expression changed. So we thought FoxM1 could regulate the expression of Ezrin. We used Ingenuity Pathway Analysis (IPA) software and generated connections between FoxM1 and Ezrin. As showed in Figure 4, FoxM1 could regulate Ezrin by SP1, CTNNB1, SUMO2, etc.
Figure 4.
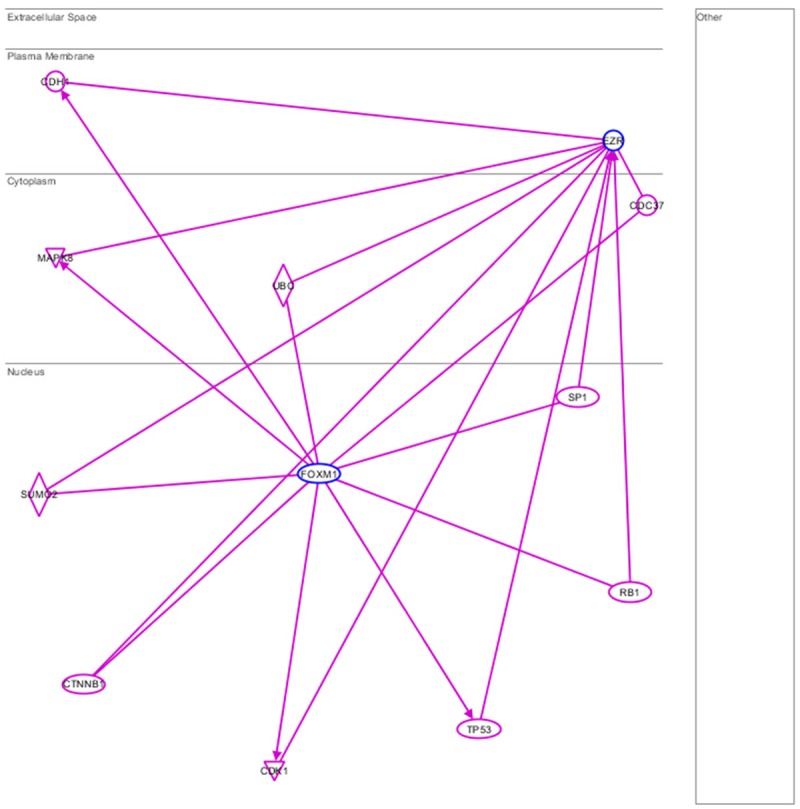
Ingenuity Pathway Analysis (IPA) generated connections between FoxM1 and Ezrin. No direct link between FoxM1 and Ezrin.
In summary, our study is the first to demonstrate the relationship between FoxM1 and Ezrin. FoxM1 could influence tumor growth and metastasis.
Disclosure of conflict of interest
None.
References
- 1.Sleeman JP. The lymph node as a bridgehead in the metastatic dissemination of tumors. Recent Results Cancer Res. 2000;157:55–81. doi: 10.1007/978-3-642-57151-0_6. [DOI] [PubMed] [Google Scholar]
- 2.Schafer DF, Sorrell MF. Hepatocellular carcinoma. Lancet. 1999;353:1253–1257. doi: 10.1016/S0140-6736(98)09148-X. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 4.Poste G, Fidler IJ. The pathogenesis of cancer metastasis. Nature. 1980;283:139–146. doi: 10.1038/283139a0. [DOI] [PubMed] [Google Scholar]
- 5.Li HF, Ling MY, Xie Y, Xie H. Establishment of a lymph node metastatic model of mouse hepatocellular carcinoma Hca-F cells in C3H/Hej mice. Oncol Res. 1998;10:569–573. [PubMed] [Google Scholar]
- 6.Wu J, Meng J, Du Y, Huang Y, Jin Y, Zhang J, Wang B, Zhang Y, Sun M, Tang J. RACK1 promotes the proliferation, migration and invasion capacity of mouse hepatocellular carcinoma cell line in vitro probably by PI3K/Rac1 signaling pathway. Biomed Pharmacother. 2013;67:313–319. doi: 10.1016/j.biopha.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Ahmad A, Li Y, Banerjee S, Kong D, Sarkar FH. Forkhead box M1 transcription factor: a novel target for cancer therapy. Cancer Treat Rev. 2010;36:151–156. doi: 10.1016/j.ctrv.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 9.Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, Huang S, Tan D, Xie K. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther. 2008;7:2022–2032. doi: 10.1158/1535-7163.MCT-08-0188. [DOI] [PubMed] [Google Scholar]
- 11.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 12.Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, Myatt SS, Lam EW. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res. 2010;8:24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–1016. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Y, Cui D, Kong Y. FoxM1 influences embryo implantation and is regulated by 17 beta-estradiol and progesterone in mouse uteri and endometrium cells. Int J Clin Exp Pathol. 2014;7:6585–6595. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, Kopsombut UG, Costa RH, Gannon M. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol Endocrinol. 2006;20:1853–1866. doi: 10.1210/me.2006-0056. [DOI] [PubMed] [Google Scholar]
- 16.Priller M, Poschl J, Abrao L, von Bueren AO, Cho YJ, Rutkowski S, Kretzschmar HA, Schuller U. Expression of FoxM1 is required for the proliferation of medulloblastoma cells and indicates worse survival of patients. Clin Cancer Res. 2011;17:6791–6801. doi: 10.1158/1078-0432.CCR-11-1214. [DOI] [PubMed] [Google Scholar]
- 17.Uddin S, Ahmed M, Hussain A, Abubaker J, Al-Sanea N, AbdulJabbar A, Ashari LH, Alhomoud S, Al-Dayel F, Jehan Z, Bavi P, Siraj AK, Al-Kuraya KS. Genome-wide expression analysis of Middle Eastern colorectal cancer reveals FOXM1 as a novel target for cancer therapy. Am J Pathol. 2011;178:537–547. doi: 10.1016/j.ajpath.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia L, Huang W, Tian D, Zhu H, Zhang Y, Hu H, Fan D, Nie Y, Wu K. Upregulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinoma. J Hepatol. 2012;57:600–612. doi: 10.1016/j.jhep.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 19.Sanders DA, Ross-Innes CS, Beraldi D, Carroll JS, Balasubramanian S. Genome-wide mapping of FOXM1 binding reveals co-binding with estrogen receptor alpha in breast cancer cells. Genome Biol. 2013;14:R6. doi: 10.1186/gb-2013-14-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu N, Jia D, Chen W, Wang H, Liu F, Ge H, Zhu X, Song Y, Zhang X, Zhang D, Ge D, Bai C. FoxM1 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor metastasis. PLoS One. 2013;8:e59412. doi: 10.1371/journal.pone.0059412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci. 1993;105:1025–1043. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- 22.Gould KL, Bretscher A, Esch FS, Hunter T. cDNA cloning and sequencing of the protein-tyrosine kinase substrate, ezrin, reveals homology to band 4.1. EMBO J. 1989;8:4133–4142. doi: 10.1002/j.1460-2075.1989.tb08598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci. 1993;105:1025–1043. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- 24.Tsukita S, Yonemura S. Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins. J Biol Chem. 1999;274:34507–34510. doi: 10.1074/jbc.274.49.34507. [DOI] [PubMed] [Google Scholar]
- 25.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 26.Turunen O, Wahlstrom T, Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J Cell Biol. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Y, Khan J, Khanna C, Helman L, Meltzer PS, Merlino G. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat Med. 2004;10:175–181. doi: 10.1038/nm966. [DOI] [PubMed] [Google Scholar]
- 28.Mak H, Naba A, Varma S, Schick C, Day A, SenGupta SK, Arpin M, Elliott BE. Ezrin phosphorylation on tyrosine 477 regulates invasion and metastasis of breast cancer cells. BMC Cancer. 2012;12:82. doi: 10.1186/1471-2407-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gould KL, Bretscher A, Esch FS, Hunter T. cDNA cloning and sequencing of the protein-tyrosine kinase substrate, ezrin, reveals homology to band 4.1. EMBO J. 1989;8:4133–4142. doi: 10.1002/j.1460-2075.1989.tb08598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 31.Wu B, Li J, Huang D, Wang W, Chen Y, Liao Y, Tang X, Xie H, Tang F. Baicalein mediates inhibition of migration and invasiveness of skin carcinoma through Ezrin in A431 cells. Bmc Cancer. 2011;11:527. doi: 10.1186/1471-2407-11-527. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–750. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, Sawaya R, Huang S. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67:8293–8300. doi: 10.1158/0008-5472.CAN-07-1265. [DOI] [PubMed] [Google Scholar]
- 36.Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW, Cheung AN, Ngan HY. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 2008;215:245–252. doi: 10.1002/path.2355. [DOI] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, Huang S, Tan D, Xie K. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia L, Huang W, Tian D, Zhu H, Zhang Y, Hu H, Fan D, Nie Y, Wu K. Upregulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinoma. J Hepatol. 2012;57:600–612. doi: 10.1016/j.jhep.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 40.Okada K, Fujiwara Y, Takahashi T, Nakamura Y, Takiguchi S, Nakajima K, Miyata H, Yamasaki M, Kurokawa Y, Mori M, Doki Y. Overexpression of forkhead box M1 transcription factor (FOXM1) is a potential prognostic marker and enhances chemoresistance for docetaxel in gastric cancer. Ann Surg Oncol. 2013;20:1035–1043. doi: 10.1245/s10434-012-2680-0. [DOI] [PubMed] [Google Scholar]
- 41.Xue YJ, Xiao RH, Long DZ, Zou XF, Wang XN, Zhang GX, Yuan YH, Wu GQ, Yang J, Wu YT, Xu H, Liu FL, Liu M. Overexpression of FoxM1 is associated with tumor progression in patients with clear cell renal cell carcinoma. J Transl Med. 2012;10:200. doi: 10.1186/1479-5876-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hua H, Li M, Luo T, Yin Y, Jiang Y. Matrix metalloproteinases in tumorigenesis: an evolving paradigm. Cell Mol Life Sci. 2011;68:3853–3868. doi: 10.1007/s00018-011-0763-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vihinen P, Kahari VM. Matrix metalloproteinases in cancer: prognostic markers and therapeutic targets. Int J Cancer. 2002;99:157–166. doi: 10.1002/ijc.10329. [DOI] [PubMed] [Google Scholar]
- 44.Ohtani K, Sakamoto H, Rutherford T, Chen Z, Satoh K, Naftolin F. Ezrin, a membrane-cytoskeletal linking protein, is involved in the process of invasion of endometrial cancer cells. Cancer Lett. 1999;147:31–38. doi: 10.1016/s0304-3835(99)00272-4. [DOI] [PubMed] [Google Scholar]
- 45.Elliott BE, Meens JA, SenGupta SK, Louvard D, Arpin M. The membrane cytoskeletal crosslinker ezrin is required for metastasis of breast carcinoma cells. Breast Cancer Res. 2005;7:R365–R373. doi: 10.1186/bcr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osawa H, Smith CA, Ra YS, Kongkham P, Rutka JT. The role of the membrane cytoskeleton cross-linker ezrin in medulloblastoma cells. Neuro Oncol. 2009;11:381–393. doi: 10.1215/15228517-2008-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cant SH, Pitcher JA. G protein-coupled receptor kinase 2-mediated phosphorylation of ezrin is required for G protein-coupled receptor-dependent reorganization of the actin cytoskeleton. Mol Biol Cell. 2005;16:3088–3099. doi: 10.1091/mbc.E04-10-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crepaldi T, Gautreau A, Comoglio PM, Louvard D, Arpin M. Ezrin is an effector of hepatocyte growth factor-mediated migration and morphogenesis in epithelial cells. J Cell Biol. 1997;138:423–434. doi: 10.1083/jcb.138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang HY, Li CF, Fang FM, Tsai JW, Li SH, Lee YT, Wei HM. Prognostic implication of ezrin overexpression in myxofibrosarcomas. Ann Surg Oncol. 2010;17:3212–3219. doi: 10.1245/s10434-010-1185-y. [DOI] [PubMed] [Google Scholar]
- 50.Li Q, Gao H, Xu H, Wang X, Pan Y, Hao F, Qiu X, Stoecker M, Wang E, Wang E. Expression of ezrin correlates with malignant phenotype of lung cancer, and in vitro knockdown of ezrin reverses the aggressive biological behavior of lung cancer cells. Tumour Biol. 2012;33:1493–1504. doi: 10.1007/s13277-012-0400-9. [DOI] [PubMed] [Google Scholar]


