Abstract
The oncogenic phosphatidylinositol 3-kinase-AKT-mammlian target of rapamycin pathway (PI3K-AKT-mTOR) pathway is known to be activated in uterine smooth muscle tumors, and Stathmin 1 (STMN1) expression has been identified as a marker of PI3K-AKT-mTOR pathway activation. We hypothesized that STMN1 may have some diagnostic utility and explored how well STMN1 expression correlated with histologic classifications of uterine smooth muscle tumors into benign and malignant groupings. 84 smooth muscle tumors were assessed for STMN1 expression by immunohistochemistry. These included spindle cell leiomyosarcoma (n = 32), conventional spindle cell leiomyomas (n = 30), atypical (symplastic) leiomyoma (n = 5), cellular leiomyoma (n = 7), smooth muscle tumor of uncertain malignant potential (n = 4), mitotically active leiomyomas (n = 2), benign metastasizing leiomyoma (n = 3), and cotyledonoid dissecting leiomyoma (n = 1). All spindle cell leiomyosarcomas were positive (32/32 positive; 100%) as compared with conventional leiomyomata (11/30; 37%) (P < 0.0001). The average immunohistochemical score (0-12+, reflective of intensity and extent) for leiomyosarcomas was 8.7 (± 1.43) whereas the conventional leiomyomata average score was 1.6 (± 1.07) (P < 0.0001). This difference in scores was reflected in the patterns of expression: leiomyosarcomas were predominantly strongly and diffusely positive whereas leiomyomata were predominantly weakly, albeit diffusely positive when expression was present. The sensitivity of STMN1 expression for leiomyosarcomas was 100%. However, the specificity was found to be only 55% (CI = 43-68%). The negative and positive predictive values for leiomyosarcomas were 100% and 52% respectively. The odds ratio (OR) for any STMN1 expression in predicting a spindle cell leiomyosarcoma diagnosis from this dataset was highly significant (OR = 144, P = 0.0006). Thirteen non-smooth muscle tumors that involved the uterus all showed at least focal STMN1 immunoreactivity. In summary, STMN1 is a highly sensitive marker for leiomyosarcoma but is suboptimally specific for diagnostic purposes. The 100% negative predictive value for leiomyosarcoma may offer some diagnostic utility in a small sample, since the absence of STMN1 immunoreactivity in a putative leiomyosarcoma is a strong argument against this diagnostic possibility.
Keywords: Leiomyosarcoma, stathmin, STMN1, immunohistochemistry, leiomyoma
Introduction
Uterine leiomyosarcomas are rare, representing approximately 1% of uterine malignancies, 40% of all uterine sarcomas, and with an annual incidence of approximately 0.64 per 100,000 women [1-5]. Although these are rare tumors, leiomyosarcomas are clinically aggressive, with 5-year survival rates of between 25-75% [3,6-11]. In general, pathologic classifications of smooth muscle tumors show a strong correlation with patient outcomes. However, it has long been recognized that the diagnosis of some smooth muscle tumors can be problematic, including tumors displaying extrauterine disease but bland morphology and favorable patient outcomes, tumors displaying bland histology and clinical aggressiveness, and other tumors that in general, show a dissonance between pathologic features and patient outcomes [11-15]. Accordingly, immunohistochemical (IHC) markers that may assist in differentiating between a leiomyosarcoma and a uterine leiomyoma will be clinically helpful. A variety of such markers, including p21, p27, p53, p16, IMP3, pan-Akt, Ki-67, and fascin have previously been evaluated [10,16-26]. Although each has shown varying degrees of efficacy for this purpose, they generally lack the specificity to be deployed in isolation.
Stathmin 1 (STMN1), also known as oncoprotein 18, is a cytoplasmic phosphoprotein that is involved in regulating the dynamics of mitotic and interphase microtubules. As such, dysregulation of stathmin expression may result in alterations in cell division, which may eventuate in oncogenesis [33]. In the gynecologic tract, STMN1 overexpression has been identified as a negative prognostic factor in both ovarian and endometrial carcinomas, and may have diagnostic utility in cervical and adnexal intraepithelial neoplasias [34-38]. STMN1 expression has also been identified as a significant marker of activation of the oncogenic phosphatidylinositol 3-kinase-AKT-mammlian target of rapamycin pathway (PI3K-AKT-mTOR) pathway [36-40]. This pathway is known to be highly activated in both uterine and extrauterine leiomyosarcoma [27-32]. Accordingly, we hypothesized that STMN1 has some diagnostic utility in classifying uterine smooth muscle tumors regarding their malignant potential and explored how well STMN1 expression correlated with histologic classifications of uterine smooth muscle tumors into benign and malignant groupings. In summary, STMN1 is a highly sensitive marker for leiomyosarcoma but is suboptimally specific for diagnostic purposes. The 100% negative predictive value for leiomyosarcoma may offer some diagnostic utility in a small sample, since the absence of STMN1 immunoreactivity in a putative leiomyosarcoma is a strong argument against the diagnosis.
Materials and methods
Case selection
84 smooth muscle tumors that had previously been classified based on World Health Organization criteria [41], were selected from the authors’ files. The cases included spindle cell leiomyosarcoma (n = 32), conventional spindle cell leiomyomas (n = 30), atypical (symplastic) leiomyoma (n = 5), cellular leiomyoma (n = 7), smooth muscle tumor of uncertain malignant potential (n = 4), mitotically active leiomyomas (n = 2), benign metastasizing leiomyoma (n = 3), and cotyledonoid dissecting leiomyoma (n = 1). Thirteen non-smooth muscle tumors that involved the uterus were also included, including five low grade endometrial stromal sarcomas, one Ewing sarcoma, one perivascular epithelioid tumor, one pleomorphic rhabdomyosarcomas, one embryonal rhabdomyosarcomas, one alveolar soft part sarcoma, one dedifferentiated liposarcoma, and one osteosarcoma.
Immunohistochemistry
Immunohistochemical analyses were performed on 1 unstained slide from each case. Slides were placed on the Leica Bond Max immunohistochemical stainer. All steps besides dehydration, clearing and coverslipping are performed on the Bond Max. Slides are deparaffinized. Heat induced antigen retrieval was performed on the Bond Max using their Epitope Retrieval 2 solution for 20 minutes. The sections were incubated with a polyclonal antibody to STMN1 (Cell Signaling, Danvers, MA; Catalog #3352) diluted 1:50 for one hour. The Bond Refine Polymer detection system was used for visualization. Slides were then dehydrated, cleared and coverslipped.
Cases were scored using a system that incorporated staining intensity (on a 0-3+) scale and staining extent (0-4+ scale). These values were then multiplied in each case, giving potential scores that ranged from 0-12+. All cases were scored by 2 authors (MMA and OF).
Statistics
Subgroups were compared regarding their STMN1 staining using the Student t-test (comparing mean STMN1 score for each group) or Fisher Exact test (comparing STMN1 rates of positivity in each group). Odds Ratio, Sensitivity, Specificity, Positive Predictive Value, Negative Predictive Value for any STMN1 immunoreactivity in predicting the leiomyosarcoma diagnosis were determined using three thresholds for positivity (≥ 1+, ≥ 4+, and ≥ 6+) using the vassarstats program (http://www.vassarstats.net/clin1.html). A p-value of < 0.05 was considered statistically significant in all analyses.
Results
The distribution of scores for all tumors is reflected in Table 1. The majority of the leiomyosarcomas were strongly and diffusely positive for STMN1 expression (Figures 1 and 2), whereas most leiomyomata were weakly positive or negative for STMN1 expression (Figures 3 and 4). At a ≥ 1+ threshold for positivity, all spindle cell leiomyosarcomas were positive (32/32 positive; 100%) as compared with conventional leiomyomata (11/30; 37%) (P < 0.0001). The average score for leiomyosarcomas was 8.7 (± 1.43) whereas the conventional leiomyomata average score was 1.6 (± 1.07) (P < 0.0001). This difference in scores was reflected in the patterns of expression: leiomyosarcomas were predominantly strongly and diffusely positive whereas leiomyomata were predominantly weakly, albeit diffusely positive when expression was present (Figure 4). The rate of STMN1 positivity in LMS (32/32; 100%) was significantly higher than for all other uterine SM timors when the latter is considered as a group (16/52; 30.8%) (P < 0.0001). All non-smooth muscle tumors were positive.
Table 1.
Distribution of scores for each uterine tumor
| Tumor | No tested | Distribution of Scores | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| 0 | ≥ 1+ | 1-5+ | 6-9+ | 10-12+ | ||
| Spindle cell leiomyosarcoma | 32 | 0 | 32 | 32 | 25 | 17 |
| Conventional leiomyoma | 30 | 19 | 11 | 10 | 3 | 1 |
| Atypical (symplastic) leiomyoma | 5 | 5 | 0 | 0 | 0 | 0 |
| Cellular leiomyoma | 7 | 7 | 0 | 0 | 0 | 0 |
| Smooth muscle tumor of uncertain malignant potential (STUMP) | 4 | 4 | 0 | 0 | 0 | 0 |
| Cotyledonoid Dissecting leiomyoma | 1 | 0 | 0 | 0 | 0 | 1 |
| Benign metastasizing leiomyoma | 3 | 1 | 2 | 2 | 0 | 0 |
| Mitotically active leiomyoma | 2 | 0 | 2 | 2 | 2 | 2 |
| Non-smooth muscle sarcomas | 13 | 0 | 13 | 3 | 5 | 5 |
| Total | 97 | 37 | 60 | 20 | 15 | 25 |
Figure 1.
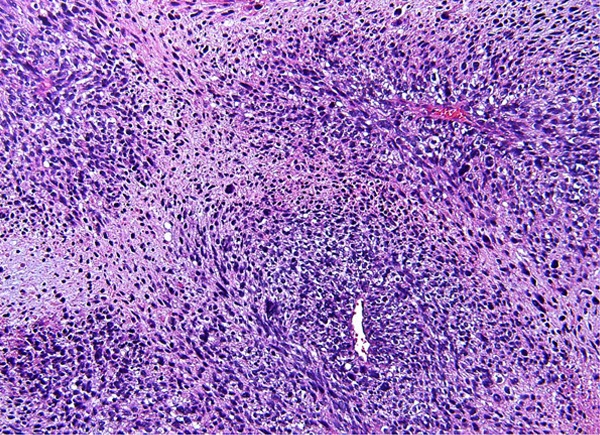
Leiomyosarcoma.
Figure 2.
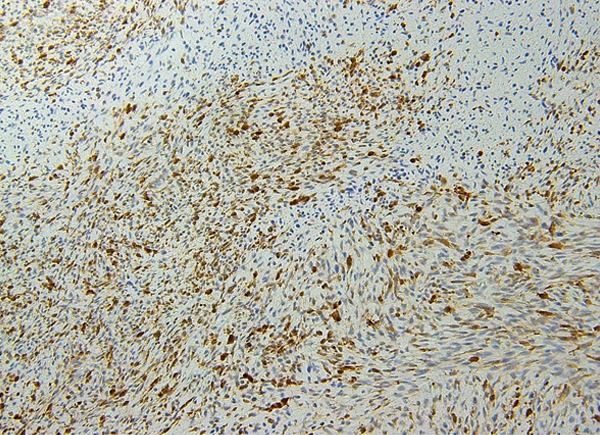
Leiomyosarcoma: diffuse expression of STMN1 in leiomyosarcoma. Necrotic areas show decreased expression.
Figure 3.

Leiomyoma.
Figure 4.
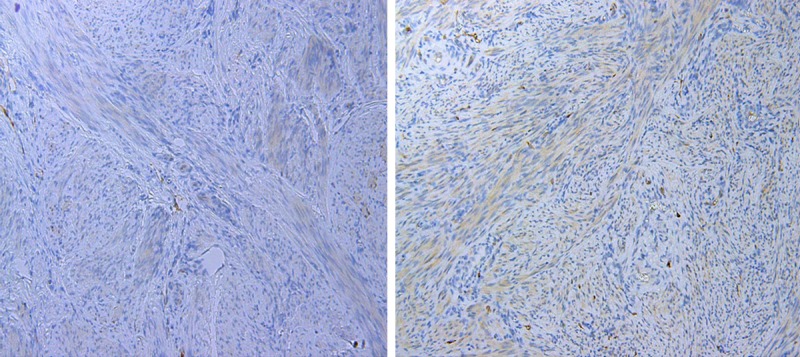
Leiomyoma with foci of no STMN1 expression (left image) and weak STMN1 expression (right image).
For each threshold for positivity, the sensitivity, specificity, positive predictive value, and negative predictive value of the STMN1 as a biomarker for predicting a leiomyosarcoma diagnosis among the uterine smooth muscle tumors was calculated. We repeated the calculations to determine the same parameters for STMN1 as a biomarker for predicting a leiomyosarcoma diagnosis in all uterine tumor cases that were included in this study. Table 2 displays each test parameter for each threshold for positivity for the smooth muscle tumors as well as all tumors.
Table 2.
Test parameters calculated for each threshold for positivity for smooth muscle tumors and all tumors included in this study
| Threshold for positivity | |||
|---|---|---|---|
|
|
|||
| Parameter | ≥ 1+ (95% CI) | ≥ 4+ (95% CI) | ≥ 6+ (95% CI) |
| Smooth Muscle Tumors | |||
| Odds Ratio | 144 (8-2493, P = 0.0006) | 17 (6-51, P < 0.0001) | 33 (9-114, P < 0.0001) |
| Sensitivity | 100 (87-100) | 78 (59-90) | 78 (60-90) |
| Specificity | 69 (55-81) | 83 (69-91) | 90 (77-96) |
| Positive Predictive Value | 67 (51-79) | 73 (55-86) | 83 (65-97) |
| Negative Predictive Value | 100 (87-100) | 86 (73-94) | 87 (74-94) |
| All Tumors | |||
| Odds Ratio | 80 (4-1370, P = 0.0024) | 8.6 (3-23, P < 0.0001) | 11 (4-30, P < 0.0001) |
| Sensitivity | 100 (87-100) | 78.1 (60-90) | 78.1 (60-90) |
| Specificity | 55.4 (43-68) | 70.8 (58-81) | 75.4 (63-85) |
| Positive Predictive Value | 52.5 (39-65) | 56.8 (41-71) | 61 (45-75) |
| Negative Predictive Value | 100 (87-100) | 86.8 (74-94) | 87.5 (75-94) |
Using a threshold for positivity of ≥ 1+, the sensitivity of the STMN1 expression for leiomyosarcomas was 100% (95% confidence interval [CI] = 87-100%). However, the specificity was found to be only 55% (CI = 43-68%). The negative and positive predictive values for leiomyosarcomas were 100% (CI = 87-100%) and 52% (CI = 39-65%) respectively. The odds ratio (OR) for any STMN1 expression in predicting a spindle cell leiomyosarcoma diagnosis from this dataset was highly significant (OR = 144, CI = 8-2493, P = 0.0006, at the ≥ 1+ threshold), see Table 2.
Discussion
The histologic classification of uterine smooth muscle tumors regarding their malignant potential may potentially be problematic [12], and a wide variety of immunohistochemical markers have been assessed as potential diagnostic adjuncts in this classification. Included in this group are p21, p27, p53, p16, IMP3, pan-Akt, Ki-67, progesterone receptor, and fascin [10,16-26,45]. These markers often have a specific drawback that limits their utility. For example, IMP3 is highly specific for leiomyosarcomas but is only moderately sensitive (52% of the leiomyosarcomas were positive, 4.2% of the leiomyomata variants-cellular or symplastic-were positive, all conventional leiomyomata were negative) [16]. Cell cycle regulatory protein expression (such as p16, p21, p27, and p53) is often distinctly heterogeneous in smooth muscle tumors and each cannot be depended upon as a sole discriminator between benign and malignant smooth muscle proliferations [10]. Composite profiles tend to be more useful. For example, Lee et al reported that diffuse expression of p16 and p53 and/or a high Ki-67 proliferation index, yielded a sensitivity of 92% and a specificity of 98% for separating leiomyosarcomas from leiomyomata [20]. Fascin and Pan-AKT also show some potential diagnostic utility, in that most leiomyosarcomas have been found to be positive and most leiomyomata negative, but data are limited and these markers have not been evaluated in multiple laboratories [17,18].
The PI3K-AKT-mTOR pathway appears to be centrally associated with smooth muscle proliferation and neoplasia in the uterus. In animal models, mTOR has been shown to mediate hormone-initiated myometrial hyperplasia in pregnancy [42], and both mTOR and phosphatidylinositol-3 kinase have been reported to be requirements for estrogen-induced proliferation in cell lines derived from uterine leiomyoma and normal myometrium [43]. Mice carrying homologous deletions in an endogenous negative regulator of the P13K-Akt-mTOR pathway developed a variety of benign smooth muscle proliferations as well as rapid onset abdominal leiomyosarcomas [27]. Although none of the resultant malignancies were uterine, this study was noteworthy because constitutive activation of the P13K-Akt-mTOR pathway was only found in the leiomyosarcomas, and not the benign proliferations, suggesting that PI3K-AKT-mTOR pathway activation was a necessary but insufficient event to develop smooth muscle malignancy [27]. The entire PI3K-AKT-mTOR pathway is clearly upregulated in human smooth muscle tumors as assessed by gene expression profiling [44], and constitutive activation of the mTORC2-phospholipase D1 pathway has been demonstrated in uterine leiomyosarcoma (phospholipase D may be an activator of mTOR signaling) [31]. Since STMN1 expression has also been identified as a significant marker of activation of the PI3K-AKT-mTOR pathway [36-40], we explored how well STMN1 expression correlated with histologic classifications of uterine smooth muscle tumors into benign and malignant groupings. The ultimate goal of this study was to explore the diagnostic utility of this marker by establishing its baseline performance in non-problematic cases.
We report that STMN1 is a highly sensitive marker for leiomyosarcoma but is suboptimally specific for diagnostic purposes. It is not a specific marker of malignancy within the smooth muscle group since 37% of leiomyomata showed some STMN1 expression. It is also not a specific marker of smooth muscle neoplasia since non-smooth muscle sarcomas are frequently positive. The 100% negative predictive value for leiomyosarcoma may offer some diagnostic utility in a small sample, in that the absence of STMN1 immunoreactivity in a putative leiomyosarcoma is a strong argument against the diagnosis. However, this marker should be combined with other markers if applied even for this purpose. Irrespective of whether STMN1 can be applied for diagnostic purposes, the patterns of expression may provide valuable insights into the elusive pathogenesis of leiomyosarcomas. For example, 5 of 5 atypical leiomyomas in our dataset were STMN1 negative, indicating that STMN1 expression is not a surrogate indicator for cytologic atypia and suggesting a lack of PI3K-AKT-mTOR pathway activation in this morphologically distinct but biologically benign variant; in contrast, 2 of 2 mitotically active leiomyomata were positive. Since PI3K-AKT-mTOR play such a central role in leiomyosarcomagenesis, STMN1 expression may provide insights into the primary drivers of this process, and/or their morphologic correlates.
Disclosure of conflict of interest
None.
References
- 1.Zacche G, Zanoio L, Sommacampagna P. Uterine sarcoma. Clinical and pathological study of 25 cases. Minerva Ginecologica. 1992;44:355–8. [PubMed] [Google Scholar]
- 2.Nucci MR, Olivia E. Gynecologic Pathology. A Volume in the Series: Foundations in Diagnostic Pathology. 1st edition. China: 2009. Pure Mesenchymal and Mixed Mullerian Tumors of the Uterus; pp. 261–329. [Google Scholar]
- 3.Van Dinh T, Woodruff JD. Leiomyosarcoma of the uterus. Am J Obstet Gynecol. 1982;144:817–23. doi: 10.1016/0002-9378(82)90358-1. [DOI] [PubMed] [Google Scholar]
- 4.Yu KJ, Ho DM, Ng HT, Caho KC, Kan Y. Leiomyosarcoma of uterus: a review of 14 cases. Zhonghua Yi Xue Za Zhi (Taipei) 1989;44:109–14. [PubMed] [Google Scholar]
- 5.Kahanpää KV, Wahlström T, Gröhn P, Heinonen E, Nieminen U, Widholm O. Sarcomas of the uterus: a clinicopathologic study of 119 patients. Obstet Gynecol. 1986;67:417–24. [PubMed] [Google Scholar]
- 6.Mayerhofer K, Obermair A, Windbichler G, Petru E, Kaider A, Hefler L, Czerwenka K, Leodolter S, Kainz C. Leiomyosarcoma of the uterus: a clinicopathologic multicenter study of 71 cases. Gynecol Oncol. 1999;74:196–201. doi: 10.1006/gyno.1999.5436. [DOI] [PubMed] [Google Scholar]
- 7.Barter JF, Smith EB, Szpak CA, Hinshaw W, Clarke-Pearson DL, Creasman WT. Leiomyosarcoma of the uterus: clinicopathologic study of 21 cases. Gynecol Oncol. 1985;21:220–7. doi: 10.1016/0090-8258(85)90256-2. [DOI] [PubMed] [Google Scholar]
- 8.Benoit L, Arnould L, Cheynel N, Goui S, Collin F, Fraisse J, Cuisenier J. The role of surgery and treatment trends in uterine sarcoma. Eur J Surg Oncol. 2005;31:434–42. doi: 10.1016/j.ejso.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Lusby K, Savannah KB, Demicco EG, Zhang Y, Ghadimi MP, Young ED, Colombo C, Lam R, Dogan TE, Hornick JL, Lazar AJ, Hunt KK, Anderson ML, Creighton CJ, Lev D, Pollock RE. Uterine leiomyosarcoma management, outcome, and associated molecular biomarkers: a single institution’s experience. Ann Surg Oncol. 2013;20:2364–72. doi: 10.1245/s10434-012-2834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mills AM, Ly A, Balzer BL, Hendrickson MR, Kempson RL, McKenney JK, Longacre TA. Cell cycle regulatory markers in uterine atypical leiomyoma and leiomyosarcoma: immunohistochemical study of 68 cases with clinical follow-up. Am J Surg Pathol. 2013;37:634–42. doi: 10.1097/PAS.0b013e318287779c. [DOI] [PubMed] [Google Scholar]
- 11.D’Angelo E, Spagnoli LG, Prat J. Comparative clinicopathologic and immunohistochemical analysis of uterine sarcomas diagnosed using the World Health Organization classification system. Hum Pathol. 2009;40:1571–85. doi: 10.1016/j.humpath.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Bell SW, Kempson RL, Hendrickson MR. Problematic uterine smooth muscle neoplasms. A clinicopathologic study of 213 cases. Am J Surg Pathol. 1994;18:535–58. [PubMed] [Google Scholar]
- 13.Veras E, Zivanovic O, Jacks L, Chiappetta D, Hensley M, Soslow R. “Low-grade leiomyosarcoma” and late-recurring smooth muscle tumors of the uterus: a heterogenous collection of frequently misdiagnosed tumors associated with an overall favorable prognosis relative to conventional uterine leiomyosarcomas. Am J Surg Pathol. 2011;35:1626–37. doi: 10.1097/PAS.0b013e31822b44d2. [DOI] [PubMed] [Google Scholar]
- 14.Posligua L, Silva EG, Deavers MT, Merino MJ, Malpica A. Low-grade smooth muscle tumors of the primary and the secondary mullerian system: a proposed concept of multicentricity. Int J Gynecol Pathol. 2012;31:547–55. doi: 10.1097/PGP.0b013e31824d3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Angelo E, Prat J. Uterine sarcomas: a review. Gynecol Oncol. 2010;116:131–9. doi: 10.1016/j.ygyno.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Cornejo K, Shi M, Jiang Z. Oncofetal protein IMP3: a useful diagnostic biomarker for leiomyosarcoma. Hum Pathol. 2012;43:1567–72. doi: 10.1016/j.humpath.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 17.Kefeli M, Yildiz L, Kaya FC, Aydin O, Kandemir B. Fascin expression in uterine smooth muscle tumors. Int J Gynecol Pathol. 2009;28:328–33. doi: 10.1097/PGP.0b013e318195da9f. [DOI] [PubMed] [Google Scholar]
- 18.Fadare O, Renshaw I, Olson SJ, Liang SX. The phosphatidylinositol 3’ kinase-Akt-mammalian target of rapamycin pathway in smooth muscle tumors of the uterus: selected protein expression patterns and their clinicopathologic implications. Int J Gynecol Pathol. 2011;30:244–51. doi: 10.1097/PGP.0b013e3181fde2ac. [DOI] [PubMed] [Google Scholar]
- 19.Bodner-Adler B, Bodner K, Czerwenka K, Kimberger O, Leodolter S, Mayerhofer K. Expression of p16 protein in patients with uterine smooth muscle tumors: an immunohistochemical analysis. Gynecol Oncol. 2005;96:62–6. doi: 10.1016/j.ygyno.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 20.Lee CH, Turbin DA, Sung YC, Espinosa I, Montgomery K, van de Rijn M, Gilks CB. A panel of antibodies to determine site of origin and malignancy in smooth muscle tumors. Mod Pathol. 2009;22:1519–31. doi: 10.1038/modpathol.2009.122. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Yang B. Immunohistochemical analysis of p16, p53, and Ki-67 expression in uterine smooth muscle tumors. Int J Gynecol Pathol. 2008;27:326–32. doi: 10.1097/PGP.0b013e31815ea7f5. [DOI] [PubMed] [Google Scholar]
- 22.Atkins KA, Arronte N, Darus CJ, Rice LW. The Use of p16 in enhancing the histologic classification of uterine smooth muscle tumors. Am J Surg Pathol. 2008;32:98–102. doi: 10.1097/PAS.0b013e3181574d1e. [DOI] [PubMed] [Google Scholar]
- 23.Gannon BR, Manduch M, Childs TJ. Differential Immunoreactivity of p16 in leiomyosarcomas and leiomyoma variants. Int J Gynecol Pathol. 2008;27:68–73. doi: 10.1097/pgp.0b013e3180ca954f. [DOI] [PubMed] [Google Scholar]
- 24.O’Neill CJ, McBride HA, Connolly LE, McCluggage WG. Uterine leiomyosarcomas are characterized by high p16, p53 and MIB1 expression in comparison with usual leiomyomas, leiomyoma variants and smooth muscle tumours of uncertain malignant potential. Histopathology. 2007;50:851–8. doi: 10.1111/j.1365-2559.2007.02699.x. [DOI] [PubMed] [Google Scholar]
- 25.Hakverdi S, Güngören A, Yaldiz M, Hakverdi AU, Toprak S. Immunohistochemical analysis of p16 expression in uterine smooth muscle tumors. Eur J Gynaecol Oncol. 2011;32:513–5. [PubMed] [Google Scholar]
- 26.Ünver NU, Acikalin MF, Öner Ü, Ciftci E, Ozalp SS, Colak E. Differential expression of P16 and P21 in benign and malignant uterine smooth muscle tumors. Arch Gynecol Obstet. 2011;284:483–90. doi: 10.1007/s00404-010-1690-z. [DOI] [PubMed] [Google Scholar]
- 27.Hernando E, Charytonowicz E, Dudas ME, Menendez S, Matushansky I, Mills J, Socci ND, Behrendt N, Ma L, Maki RG, Pandolfi PP, Cordon-Cardo C. The AKT-mTOR pathway plays a critical role in the development of leiomyosarcomas. Nat Med. 2007;13:748–53. doi: 10.1038/nm1560. [DOI] [PubMed] [Google Scholar]
- 28.Brewer Savannah KJ, Demicco EG, Lusby K, Ghadimi MP, Belousov R, Young E, Zhang Y, Huang KL, Lazar AJ, Hunt KK, Pollock RE, Creighton CJ, Anderson ML, Lev D. Dual Targeting of mTOR and Aurora-A Kinase for the treatment of uterine leiomyosarcoma. Clin Cancer Res. 2012;18:4633–45. doi: 10.1158/1078-0432.CCR-12-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Setsu N, Yamamoto H, Kohashi K, Endo M, Matsuda S, Yokoyama R, Nishiyama K, Iwamoto Y, Dobashi Y, Oda Y. The Akt/mammalian target of rapamycin pathway is activated and associated with adverse prognosis in soft tissue leiomyosarcomas. Cancer. 2012;118:1637–48. doi: 10.1002/cncr.26448. [DOI] [PubMed] [Google Scholar]
- 30.Wong TF, Takeda T, Li B, Tsuiji K, Kitamura M, Kondo A, Yaegashi N. Curcumin disrupts uterine leiomyosarcoma cells through AKT-mTOR pathway inhibition. Gynecol Oncol. 2011;122:141–8. doi: 10.1016/j.ygyno.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Dhingra S, Rodriguez ME, Shen Q, Duan X, Stanton ML, Chen L, Zhang R, Brown RE. Constitutive activation with overexpression of the mTORC2-phospholipase D1 pathway in uterine leiomyosarcoma and STUMP: morphoproteomic analysis with therapeutic implications. Int J Clin Exp Pathol. 2010;4:134–46. [PMC free article] [PubMed] [Google Scholar]
- 32.Gibault L, Ferreira C, Pérot G, Audebourg A, Chibon F, Bonnin S, Lagarde P, Vacher-Lavenu MC, Terrier P, Coindre JM, Aurias A. From PTEN loss of expression to RICTOR role in smooth muscle differentiation: complex involvement of the mTOR pathway in leiomyosarcomas and pleomorphic sarcomas. Mod Pathol. 2012;25:197–211. doi: 10.1038/modpathol.2011.163. [DOI] [PubMed] [Google Scholar]
- 33.Belletti B, Baldassarre G. Stathmin: a protein with many tasks. New biomarker and potential target in cancer. Expert Opin Ther Targets. 2011;15:1249–66. doi: 10.1517/14728222.2011.620951. [DOI] [PubMed] [Google Scholar]
- 34.Wei SH, Lin F, Wang X, Gao P, Zhang HZ. Prognostic significance of stathmin expression in correlation with metastasis and clinicopathological characteristics in human ovarian carcinoma. Acta Histochem. 2008;110:59–65. doi: 10.1016/j.acthis.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Trovik J, Wik E, Stefansson IM, Marcickiewicz J, Tingulstad S, Staff AC, Njolstad TS MoMaTec Study Group. Vandenput I, Amant F, Akslen LA, Salvesen HB. Stathmin overexpression identifies high-risk patients and lymph node metastasis in endometrial cancer. Clin Cancer Res. 2011;17:3368–77. doi: 10.1158/1078-0432.CCR-10-2412. [DOI] [PubMed] [Google Scholar]
- 36.Trovik J, Wik E, Stefansson I, Carter SL, Beroukhim R, Oyan AM, Kalland KH, Akslen LA, Salvesen HB. Stathmin is superior to AKT and phospho-AKT staining for the detection of phosphoinositide 3-kinase activation and aggressive endometrial cancer. Histopathology. 2010;57:641–6. doi: 10.1111/j.1365-2559.2010.03661.x. [DOI] [PubMed] [Google Scholar]
- 37.Howitt BE, Nucci MR, Drapkin R, Crum CP, Hirsch MS. Stathmin-1 expression as a complement to p16 helps identify high-grade cervical intraepithelial neoplasia with increased specificity. Am J Surg Pathol. 2013;37:89–97. doi: 10.1097/PAS.0b013e3182753f5a. [DOI] [PubMed] [Google Scholar]
- 38.Karst AM, Levanon K, Duraisamy S, Liu JF, Hirsch MS, Hecht JL, Drapkin R. Stathmin 1, a marker of PI3K pathway activation and regulator of microtubule dynamics, is expressed in early pelvic serous carcinomas. Gynecol Oncol. 2011;123:5–12. doi: 10.1016/j.ygyno.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA, Malmström P, Memeo L, Isola J, Bendahl PO, Rosen N, Hibshoosh H, Ringnér M, Borg A, Parsons R. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen JN, Sathyanarayanan S, Di Bacco A, Chi A, Zhang T, Chen AH, Dolinski B, Kraus M, Roberts B, Arthur W, Klinghoffer RA, Gargano D, Li L, Feldman I, Lynch B, Rush J, Hendrickson RC, Blume-Jensen P, Paweletz CP. Pathway-based identification of biomarkers for targeted therapeutics: personalized oncology with PI3K pathway inhibitors. Sci Transl Med. 2010;2:43–55. doi: 10.1126/scitranslmed.3001065. [DOI] [PubMed] [Google Scholar]
- 41.Tavassoli FA, Devilee P. Pathology and Genetics of Tumours of The Breast and Female Genital organs. Lyon: IARC Press; 2003. World Health Organization Classification of Tumours. [Google Scholar]
- 42.Jaffer S, Shynlova O, Lye S. Mammalian target of rapamycin is activated in association with myometrial proliferation during pregnancy. Endocrinology. 2009;150:4672–80. doi: 10.1210/en.2009-0419. [DOI] [PubMed] [Google Scholar]
- 43.Yin XJ, Wang G, Khan-Dawood FS. Requirements of phosphatidylinositol-3 kinase and mammalian target of rapamycin for estrogen-induced proliferation in uterine leiomyoma- and myometrium-derived cell lines. Am J Obstet Gynecol. 2007;196:176, e1–5. doi: 10.1016/j.ajog.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 44.Crabtree JS, Jelinsky SA, Harris HA, Choe SE, Cotreau MM, Kimberland ML, Wilson E, Saraf KA, Liu W, McCampbell AS, Dave B, Broaddus RR, Brown EL, Kao W, Skotnicki JS, Abou-Gharbia M, Winneker RC, Walker CL. Comparison of human and rat uterine leiomyomata: identification of a dysregulated mammalian target of rapamycin pathway. Cancer Res. 2009;69:6171–8. doi: 10.1158/0008-5472.CAN-08-4471. [DOI] [PubMed] [Google Scholar]
- 45.Mittal K, Demopoulos RI. MIB-1 (Ki-67), p53, estrogen receptor, and progesterone receptor expression in uterine smooth muscle tumors. Hum Pathol. 2001;32:984–7. doi: 10.1053/hupa.2001.27113. [DOI] [PubMed] [Google Scholar]


