Abstract
Increasing studies have shown that MicroRNAs (miRNAs) play critical roles in the progression of lung carcinoma. In the present study, the expression and functions of miR-570 were investigated. We found that miR-570 was significantly up-regulated in lung cancer tissues, compared with adjacent non-cancerous tissues. In vitro studies further showed that miR-570 mimics could promote, while its antisense oligos inhibit cell proliferation and invasion. At the molecular level, krüppel-like factor 9 (KLF9), a tumor suppressor gene, was identified as a potential target of miR-570 in lung cancer cells. Indeed, miR-570 could negatively regulate protein levels of KLF9 through targeting its 3’-untranslated region. Taken together, our results suggest a previously unknown miR-570/KLF9 molecular network controlling lung carcinoma progression.
Keywords: Lung carcinoma, MicroRNA, miR-570, KLF9
Introduction
Lung cancer, consisting of small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), has become one of the leading causes of cancer deaths worldwide [1,2]. Although great efforts have been made to explore the molecular mechanisms of its initiation and (or) progression, the high mortality rate remains unchanged. Therefore, the identification of novel treatment strategies is crucial and essential for lung cancer management.
In the last decade, microRNAs (miRNAs), a class of small and non-coding RNAs, were identified as key regulators in the tumorigenesis [3,4]. In mammals, miRNAs could post- transcriptionally regulate mRNA and (or) protein levels of target genes through binding to their 3’-untranslated region, and subsequently inducing either mRNA degradation or translational repression [5,6]. Recent studies have demonstrated that several deregulated miRNAs in lung cancer, including miR-30b/c, miR-222, miR-34a and miR-132 [7-11]. Therefore, inhibition of oncogenic miRNAs and restoration of tumor suppressor miRNAs might be effective therapeutic agents in lung cancer.
Previous studies have shown that miR-570 binding site polymorphisms in the B7-H1 or CD274 gene were associated with the risk of gastric cancer [12,13], suggesting it may play an important role in human cancers. In this study, we found that miR-570 was up-regulated in lung cancer tissues and further revealed its oncogenic roles in the tumorigenesis.
Materials and methods
Tissue samples
25 paired NSCLC and adjacent non-cancerous specimens were collected from routine therapeutic surgery with informed consent. All tissue samples were flash-frozen in liquid nitrogen immediately after collection and stored at -80°C until use. The study was approved by the hospital institutional review board.
Cell culture
The human NSCLC cell lines (A549 and NCI-H358) were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Cells were culture in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich, St. Louis, Mo., USA) supplemented with 10% fetal bovine serum (EMD Millipore, Hong Kong, China). Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2.
RNA isolation and real-time PCR
Total RNA from tissues or cell lines was harvested using Trizol according to the manufacturer’s instructions (Invitrogen, Shanghai, China). Expression of mature miRNAs was assayed using Taqman MicroRNA Assay (Applied Biosystems) specific for hsa-miR-570. Small nuclear U6 snRNA was used as an internal control for normalization and quantification of miR-570 expression.
BrdU assays
A cell proliferation enzyme-linked immunosorbent assay (BrdU kit; Beyotime) was used to analyze the incorporation of BrdU during DNA synthesis following the manufacturer’s protocols. All experiments were performed in triplicate. Absorbance was measured at 450 nm in the Spectra Max 190 ELISA reader (Molecular Devices, Sunnyvale, CA).
miR-570 target predictions
The putative targets of miR-570 were predicted using the miRWalk software. The algorithm produced a list of hundreds of target genes for miR-570 by searching for the presence of conserved 7-mer and 8-mer sites matching the seed region of miR-570.
Western blot
Cells were harvested and lysed with ice-cold lysis buffer (50 mM Tris-HCl, pH 6.8, 570 mM 2-ME, 2% w/v SDS, 10% glycerol). After centrifugation at 20000* g for 10 min at 4°C, proteins in the supernatants were quantified and separated by 10% SDS PAGE, transferred to NC membrane (Amersham Bioscience, Buckinghamshire, U.K.). After blocking with 10% nonfat milk in PBS, membranes were immunoblotted with antibodies as indicated, followed by HRP-linked secondary antibodies (Cell Signaling). The signals were detected by SuperSignal West Pico Chemiluminescent Substrate kit (Pierce, Rockford, IL) according to manufacturer’s instructions. Anti-KLF9 and GAPDH antibodies were purchased from Abcam (USA). Protein levels were normalized to GAPDH or Lamin B (Santa Cruz, USA).
Luciferase reporter assays
Total cDNA from A549 cells was used to amplify the 3’UTR of KLF9 by PCR. The KLF9 3’UTR was cloned into pMir-Report (Ambion), yielding pMir-Report-KLF9. Mutations were introduced in potential miR-570 binding sites using the QuikChange site-directed mutagenesis Kit (Stratagene). Cells were transfected with the pMir-Report vectors containing the 3’-UTR variants, and miR-570 precursor, control plasmids for 36 hours. The pRL-TK vector (Promega, USA) carrying the Renilla luciferase gene was used as an internal control to normalize the transfection efficiency. Luciferase values were determined using the Dual-Luciferase Reporter Assay System (Promega).
Statistical analysis
Data are expressed as the mean ± SEM from at least three separate experiments. Differences between groups were analyzed using Student’s t-test analysis. A value of P < 0.05 was considered statistically significant.
Results
Up-regulation of miR-570 expression in lung carcinoma tissues
In order to explore the biological significance of miR-570 in NSCLC, its expression level was determined by quantitative real-time PCR analysis in 25 paired NSCLC and adjacent non-cancerous specimens. As a result, we found that its expression was significantly up-regulated in cancer tissues in comparison with the adjacent noncancerous tissues (Figure 1).
Figure 1.
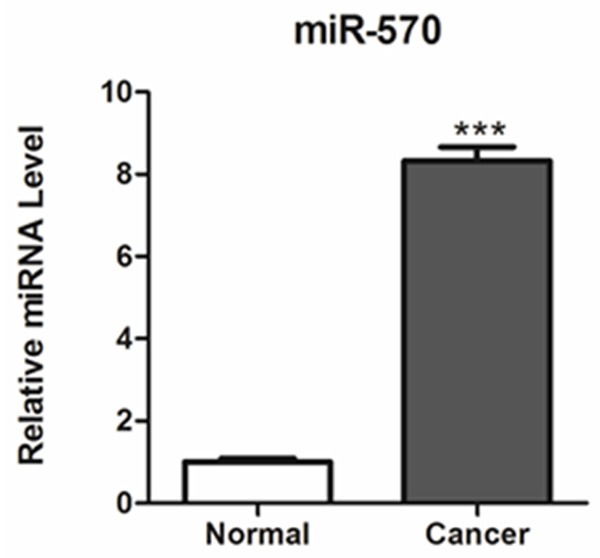
Expression levels of miR-570 in lung cancer tissues. miR-570 expression was determined by TaqMan real-time PCR in human lung cancer tissues (Tumor) and adjacent non-cancerous normal tissues (Normal). ***P < 0.001 compared with normal tissues.
miR-570 overexpression promotes NSCLC cell proliferation
In order to investigate the effect of miR-570 on NSCLC cell proliferation, miR-570 mimics were transfected into A549 and NCI-H358 cells. Real-time PCR results confirmed the overexpression of miR-570 in these two cells (Figure 2A, 2B). As a result, cell viability and proliferation were significantly enhanced by miR-570 overexpression (Figure 2C-F).
Figure 2.
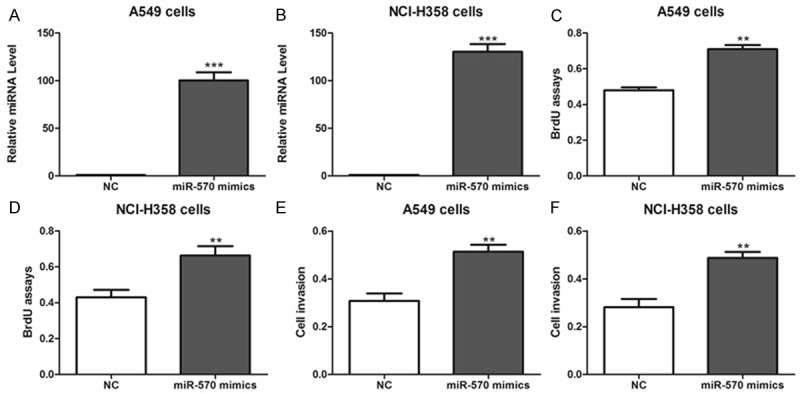
Overexpression of miR-570 promotes lung cancer cell proliferation. (A, B) Expression of miR-570 was determined in A549 (A), NCI-H358 (B) cells after miR-570 mimics or negative control (NC) transfection. ***P < 0.001 compared with NC. (C-F) The cell proliferative potential (BrdU) and viability was determined in A549 and NCI-H358 cells transfected with miR-570 mimics or negative control (NC). A450 absorption was assayed after transfection for 24 hr. *P < 0.05, **P < 0.01 compared with NC.
miR-570 promotes the proliferation of NSCLC cells via down-regulation of KLF9
Through a stringent bioinformatics approach (miRWalk software) [14], we identified the tumor suppressor gene encoding krüppel-like factor 9 (KLF9) harbored a potential miR-570 binding site (Figure 3A). To confirm whether miR-570 regulated the expression of KLF9 gene, we first performed luciferase reporter assays in A549 cells using 3’-untranslated region (3’-UTR) of KLF9. As shown in Figure 3B, overexpression of miR-570 mimics led to a reduction of luciferase activity when the reporter construct contained the KLF9 3’-UTR (Figure 3B). However, mutation of the miR-570 binding site from the KLF9 3’-UTR abolished this effect of miR-570 (Figure 3B). Then, we explored whether the endogenous KLF9 in NSCLC cells was also regulated by miR-570 by Western blot analysis. Our results showed that KLF9 protein levels were substantially down-regulated by miR-570 (Figure 3C, 3D). In agreement, protein levels of KLF9 were down-regulated in clinical NSCLC tissues compared with their adjacent non-tumor tissues (Figure 3E), suggesting that KLF9 was a direct target of miR-570 in NSCLC cells.
Figure 3.
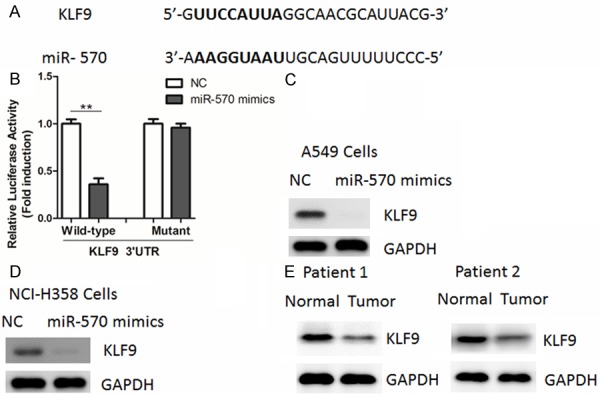
miR-570 represses KLF9 expression through targeting its 3’-UTR. A. Prediction of miR-570 binding sites in the 3’UTRs of human KLF9 gene by miRWalk software. Potential binding site was highlighted in bold. B. Luciferase reporter assays in A549 cells. Cells were transfected with 200 ng of wild-type or mutant 3’-UTR-reporter constructs together with 50 nM of miR-570 precursor or negative control (NC). C, D. Western blot analysis of KLF9 in A549 and NCI-H358 cells transfected with miR-570 mimics or negative control (NC). E. Representative protein levels of KLF9 in human lung cancer tissues (Tumor) and adjacent non-cancerous normal tissues (Normal).
Inhibition of miR-570 inhibits the proliferation of lung cancer cells
As miR-570 overexpression could promote cell proliferation, we speculate that inhibition of endogenous miR-570 might suppress NSCLC cell proliferation. Thus, A549 cells were transfected with miR-570 antisense oligos. As expected, ectopic expression of the miR-570 antisense led to a reduced proliferative ability in A549 and NCI-H358 cells, compared to negative control-transfected cells (Figure 4A-D). Consistently, protein levels of KLF9 were increased in cells transfection of miR-570 (Figure 4E, 4F).
Figure 4.
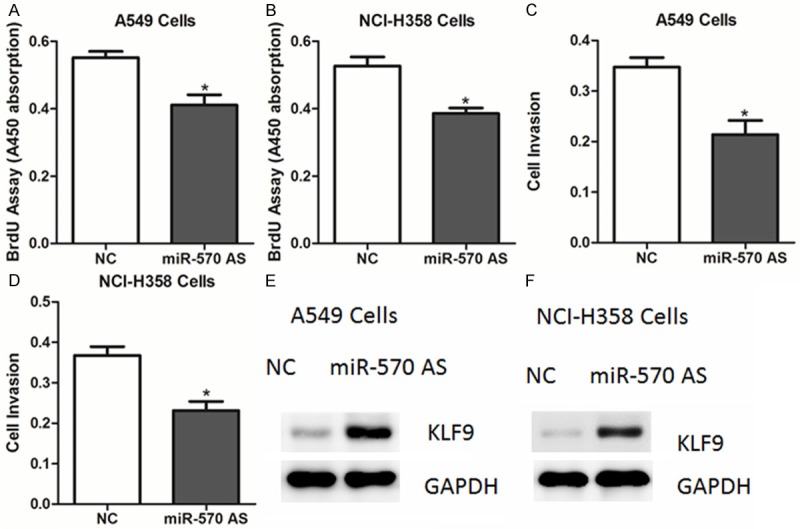
miR-570 antisense inhibits the proliferation of lung cancer cells. A-D. The cell proliferative potential (BrdU) and viability was determined in A549 and NCI-H358 cells transfected with miR-570 antisense or negative control (NC). E, F. Western blot analysis of KLF9 in A549 and NCI-H358 cells transfected with miR-570 antisense or negative control (NC).
Discussion
In the present study, we focused on miR-570 which was up-regulated in NSCLC. Our results showed that KLF9 was a direct functional target of miR-570 in NSCLC cells. Down-regulation of KLF9 protein abundance was also found in NSCLC tissues and cells overexpressing miR-570. Therefore, we demonstrated that down-regulation of KLF9 expression contributed, at least in part, to the oncogenic roles of miR-570. However, the molecular basis for the up-regulation of miR-570 in NSCLC tissues remains elusive. Besides, whether miR-570 has the similar function in other cancer cells remains to be investigated in the future.
The Krüppel-like factor (KLF) family of transcription factors regulates multiple biological events that include cell proliferation, differentiation, apoptosis and metabolism [15,16]. Recent studies have shown that dysregulation of KLFs are involved in human diseases such as cancer, obesity and cardiovascular disease [17,18]. It had been shown that KLF9 was a negative regulator of ligand-dependent estrogen receptor alpha signaling in endometrial adenocarcinoma cells [19]. Besides, KLF9 mRNA abundance was decreased in human endometrial tumors of high tumor grade [20]. KLF9 was also shown to inhibit glioblastoma-initiating stem cells survival by suppression of Notch1 signaling [21]. Moreover, KLF9 could inhibit AKT activation and inhibit tumor growth of prostate cancer cells [22]. Therefore, these reports indicate that KLF9 acts as a tumor suppressor in human cancers. However, the expression of KLF9 is less well known in NSCLC. In this study, we first reported that KLF9 protein levels were reduced in NSCLC tissues compared to matched adjacent non-cancerous tissues. Notably, the biological roles of KLF9 in NSCLC progression remain to be investigated.
Taken together, we demonstrate that miR-570 is up-regulated in NSCLC tissues. Overexpression of miR-570 directly down-regulates KLF9 expression and promotes NSCLC cell proliferation. Our data highlight an oncogenic role for miR-570 in NSCLC pathogenesis.
Acknowledgements
This work was supported by National Nature Science Foundation (30700821). Liaoning BaiQianWan Talents Program (2011921038) and Liaoning S & T Project (2013225585).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 5.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Zhong K, Chen K, Han L, Li B. MicroRNA-30b/c inhibits non-small cell lung cancer cell proliferation by targeting Rab18. BMC Cancer. 2014;14:703. doi: 10.1186/1471-2407-14-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Dai W, Chu X, Yang B, Zhao M, Sun Y. Metformin inhibits lung cancer cells proliferation through repressing microRNA-222. Biotechnol Lett. 2013;35:2013–2019. doi: 10.1007/s10529-013-1309-0. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Liu C, Liu X, Tang DG, Wang J. The microRNA miR-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44hi stem-like NSCLC cells. PLoS One. 2014;9:e90022. doi: 10.1371/journal.pone.0090022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You J, Li Y, Fang N, Liu B, Zu L, Chang R, Li X, Zhou Q. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS One. 2014;9:e91827. doi: 10.1371/journal.pone.0091827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zagryazhskaya A, Zhivotovsky B. miRNAs in lung cancer: a link to aging. Ageing Res Rev. 2014;17:54–67. doi: 10.1016/j.arr.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Wang W, Li F, Mao Y, Zhou H, Sun J, Li R, Liu C, Chen W, Hua D, Zhang X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132:641–648. doi: 10.1007/s00439-013-1275-6. [DOI] [PubMed] [Google Scholar]
- 13.Wang W, Sun J, Li F, Li R, Gu Y, Liu C, Yang P, Zhu M, Chen L, Tian W, Zhou H, Mao Y, Zhang L, Jiang J, Wu C, Hua D, Chen W, Lu B, Ju J, Zhang X. A frequent somatic mutation in CD274 3’-UTR leads to protein over-expression in gastric cancer by disrupting miR-570 binding. Hum Mutat. 2012;33:480–484. doi: 10.1002/humu.22014. [DOI] [PubMed] [Google Scholar]
- 14.Dweep H, Gretz N, Sticht C. miRWalk database for miRNA-target interactions. Methods Mol Biol. 2014;1182:289–305. doi: 10.1007/978-1-4939-1062-5_25. [DOI] [PubMed] [Google Scholar]
- 15.Turner J, Crossley M. Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci. 1999;24:236–240. doi: 10.1016/s0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- 16.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brey CW, Nelder MP, Hailemariam T, Gaugler R, Hashmi S. Kruppel-like family of transcription factors: an emerging new frontier in fat biology. Int J Biol Sci. 2009;5:622–636. doi: 10.7150/ijbs.5.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velarde MC, Zeng Z, McQuown JR, Simmen FA, Simmen RC. Kruppel-like factor 9 is a negative regulator of ligand-dependent estrogen receptor alpha signaling in Ishikawa endometrial adenocarcinoma cells. Mol Endocrinol. 2007;21:2988–3001. doi: 10.1210/me.2007-0242. [DOI] [PubMed] [Google Scholar]
- 20.Simmen FA, Su Y, Xiao R, Zeng Z, Simmen RC. The Kruppel-like factor 9 (KLF9) network in HEC-1-A endometrial carcinoma cells suggests the carcinogenic potential of dys-regulated KLF9 expression. Reprod Biol Endocrinol. 2008;6:41. doi: 10.1186/1477-7827-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ying M, Sang Y, Li Y, Guerrero-Cazares H, Quinones-Hinojosa A, Vescovi AL, Eberhart CG, Xia S, Laterra J. Kruppel-like family of transcription factor 9, a differentiation-associated transcription factor, suppresses Notch1 signaling and inhibits glioblastoma-initiating stem cells. Stem Cells. 2011;29:20–31. doi: 10.1002/stem.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen P, Sun J, Xu G, Zhang L, Yang Z, Xia S, Wang Y, Liu Y, Shi G. KLF9, a transcription factor induced in flutamide-caused cell apoptosis, inhibits AKT activation and suppresses tumor growth of prostate cancer cells. Prostate. 2014;74:946–958. doi: 10.1002/pros.22812. [DOI] [PubMed] [Google Scholar]


