Abstract
The mortality of colorectal cancer (CRC) is growing due to the unsatisfactory specificity and sensitivity of the existing screening methods. Previous studies have focused on the role of miRNAs as CRC biomarkers. However, few studies have examined the miRNA profiles at each stage. The objective of this study was to identify miRNAs that distinguish CRC patients from normal people to prevent the misdiagnosis of patients with certain stages of CRC. We performed miRNA profiling of 1547 human miRNAs by qRT-PCR in CRC patients with stage II and stage III disease. The statistical analyses showed that there were 96 miRNAs that were significantly dysregulated in CRC relative to normal tissues (P<0.05). There were 28 dysregulated miRNAs associated with separate or combined stages II and III disease. There were 25 downregulated miRNAs, including the following: miR-1, -145, -145*, -137, -363, -143, -4770, -490-5p, -9, -144*, -99a, -99b, -23b, -143*, -100, -768-3p, -24-1*, -125a-5p, -30e*, -574-3p, -126, let-7b, miR-1979, -374b, and -140-3p. We found an upregulation of miR-203, 182, and 96. Our results demonstrated that the expression of miR-1 and miR-374b was significantly decreased in each stage and may function as a biomarker of CRC. Furthermore, 20 miRNAs were dysregulated both in stage II disease without lymph node or distant metastasis and in stage II-III tumors but not in stage III tumors. Only miR-4794 was involved exclusively with stage II tumors, and there were 19 miRNAs that were dysregulated only in stage III disease with lymph node metastasis and in stage II-III disease. There were only 6 miRNAs that were uniquely dysregulated in stage III. Our results indicate that miRNA expression may be valuable in the clinic. However, large prospective studies are required to confirm the role of miRNAs. This study provides a new model for analyzing novel CRC biomarkers by considering more clinical factors.
Keywords: Colorectal, biomarker, miR-1, miR-374b, diagnostic marker
Introduction
Colorectal cancer (CRC) is one of the most serious cancers for human health [1]. It has been estimated that CRC caused more than 50,000 deaths in the United States and more than 500,000 deaths worldwide in 2013 [2]. However, current screening methods for CRC have unsatisfactory sensitivity and specificity. Therefore, novel biomarkers for CRC detection are urgently required. miRNAs may soon represent novel prognostic and diagnostic tools in patients who are at high risk of CRC or have already been diagnosed with CRC.
MiRNAs are a class of small non-coding RNA molecules that regulate gene expression at the post-transcriptional level [3]. Since the identification of miRNAs in 1993 [4], 2578 human miRNA sequences have been collected in the most recent version of miRBase (V20). The discovery of miRNAs has led to a worldwide research effort to establish their roles in carcinogenesis. Previous studies have shown that miRNAs are dysregulated in different human cancers and act as tumor suppressors or oncogenes [5]. miRNA expression signatures provide a more accurate method of cancer subtype classification than the expression profiling of an entire group of known protein-coding RNAs [6]. Thus, miRNAs may be potential biomarkers for diagnosis or prognosis and may act as potential targets in cancer-specific therapies [7,8]. MiR-145 is downregulated in many cancers, including CRC, and is thought to have a tumor-suppressor role [9-11], partly through its targeting of insulin receptor substrate 1 (ISR-1) and type-I insulin-like growth factor receptor (IGF-IR). The loss of miR-145 inhibition increases anti-apoptotic signals in the cell and promote cells growth [12,13]. MiR-155 and miR-21 are increased in the serum of patients with diffuse large B-cell lymphomas. Additionally, the expression of oncogenic miR-155 is upregulated in many other cancer types, and miR-155 alone is sufficient to induce lymphoblastic leukemia [14]. In a recent study, Dinami and colleagues have shown that miR-155 impairs telomere integrity by downregulating the expression level of telomeric repeat binding factor 1 (TERF1) [15]. Furthermore, high miR-155 expression leads to increased genomic instability and correlates with poor clinical outcomes in estrogen-receptor-positive breast cancer. Thus, the downregulation of miR-155 expression improves genomic stability by increasing telomere stability. These findings suggest that miR-155 has multiple functions in causing genomic instability [15]. MiR-21 is overexpressed in most cancers, including breast cancer, glioblastoma, neuroblastoma, leukemia, lymphoma, lung cancer, pancreatic cancer and colorectal cancer [16,17]. In previous studies, investigators have linked the high expression of miR-21 in the serum of patients with improved relapse-free survival [18]. Studies have also shown that miR-21 targets several tumor-suppressor genes, including PTEN, to increase cell proliferation and decrease apoptosis [19]. Many studies have focused on miRNA expression profiling in cancers, including colorectal cancer. Researchers are beginning to uncover the complex role that miRNAs play in malignant disease. However, several issues in miRNA studies of CRC remain to be resolved. These issues include the pattern or signature of differentially expressed miRNAs associated with specific clinical stages [20]. It is important to emphasize the value of identifying the characteristic differential expression levels of miRNAs in different tumor stages because there has been little study of the expression patterns in each tumor stage. These findings might lead to the misdiagnosis of patients with certain stages of CRC. It is also important to identify potential novel biomarkers that can prevent the misdiagnosis of patients with certain stages of CRC. In this study, we profiled 1547 distinct human miRNAs found in tumor and paired adjacent normal mucosa obtained from 28 CRC patients.
Materials and methods
Patients and specimens
The Clinical Research Ethics Committee of Third Affiliated Hospital of Guangzhou Medical University approved the research protocols. All participants provided written informed consent. The patient demographic information was obtained from patient records and registries.
Fifty-six tissue specimens, including 28 tumor tissues and 28 paired adjacent normal mucosal tissues, were selected from 28 cases of CRC. There were 15 cases of stage II disease and 13 cases of stage III disease. All of the CRC patients received surgery at Third Affiliated Hospital of Guangzhou Medical University from June 2012 to October 2014. The stage II and stage III disease cases were selected because 70% of CRC patients seen in the clinic are in these stages [21]. Patients were excluded from the study if they had received preoperative chemotherapy or radiation therapy or had a previous history of malignant tumors. All patients underwent surgical resection in the Department of Pathology, Third Affiliated Hospital of Guangzhou Medical University, and had a final pathological diagnosis of CRC. All tumor specimens were histologically classified and staged according to the seventh edition of the tumor-node-metastasis (TNM) staging system. The tissue samples were flash frozen in liquid nitrogen after resection and stored at -80°C until nucleic acids were extracted.
RNA isolation
Frozen tissues (80-100 mg) were used to isolate miRNA using RNAzol reagent (Molecular Research Center) in accordance with the manufacturer’s instructions. Briefly, 100 mg of tissue was homogenized with 1 mL RNAzol. Then, 0.4 mL water was added to the homogenate. After 5-10 min, the mixture was centrifuged at 12 000* g for 15 min for DNA/protein precipitation. One milliliter of the supernatant was mixed with 0.4 mL of 75% ethanol for 10 min and centrifuged at 12 000* g for 8 min to precipitate the miRNAs. The collected miRNA supernatant was mixed with isopropanol (0.8 volumes) for 30 min and then centrifuged at 12000* g for 15 min, washed with 0.4 mL 70% isopropanol, and centrifuged at 88000* g for 3 min. Finally, miRNAs were dissolved in diethylpyrocarbonate-treated water.
The concentration and purity of the miRNAs were determined by electrophoresis and a NanoDrop spectrophotometer (Thermo Scientific, USA). The quality of miRNAs was considered to meet the requirements if the OD260/OD280 was between 1.7 and 2.0. The integrity of small RNAs was evaluated by the determination of robust amplification of small nuclear ubiquitous RNAs (e.g., RNU6b, RNU44, and RNU48) by real-time reverse-transcription PCR (qRT-PCR). These RNAs were chosen because they are commonly used as endogenous controls in miRNA studies.
miRNA profiling
A Universal RT microRNA PCR system (GeneCopoeia, USA) was used for miRNA profiling using 28 tumors and 28 paired normal control tissues. The assay for the profiling included universal reverse transcription (RT) and sequential qRT-PCR amplification with special primers using SYBR Green.
Reaction
The PCR reaction was performed in a 384-well PCR plate with each well containing 20 μL of reaction mix including 1 μL cDNA and 1 μL gene-specific PCR primers. The total volume of the PCR reaction was 50 mL and included 2.5 mL cDNA of each tissue sample for 9×384-well plates. A total of 1547 distinct miRNAs were used for the profiling. The fold changes in the expression of each type of miRNA were determined.
Quantification of miRNAs by qRT-PCR
miRNAs (approximately 500 ng) were reverse transcribed in 25-μL reaction volumes using an All-in-One First-Strand cDNA Synthesis kit (GeneCopoeia, USA). Briefly, 25 μL of RT reaction mix was added to the miRNA sample with 5 μL of reaction buffer, 2.5 U/μL poly (A) polymerase, 10 ng/μL MS2 RNA, and RTase mix. The reaction was performed at 37°C for 60 min and then terminated at 85°C for 5 min.
The cDNA from the RT reaction was diluted ten-fold and then used as the template for the PCR reaction in an Applied Biosystems ViiA 7 Real-Time PCR System (Life Technologies, USA). MS2 RNA was used as an external reference for the quality of extracted miRNAs. The RNAs for RNU6B, RNU44, RNU48, and RNU49 were used for normalization. The PCR system contained the following components within a total 20-μL volume per well: 10 μL 2* All-in-one qPCR Mix, 2 μL PCR forward primer (2 µM), 2 μL PCR reverse primer (2 µM), 1 μL template, 0.2 μL 50* ROX Reference Dye (for calibration) and 4.8 μL ddH2O. A mastermix was prepared that included all components except for the template. If the total volume of the mastermix changed, each component was altered by the proper proportion. The real-time PCR conditions were as follows: hot-start denaturation at 95°C for 10 min; 40 cycles of amplification with denaturation at 95°C for 10 s, annealing at 60°C for 20 s, and extension at 72°C for 15 s. After amplification, the samples were melted at 60°C for 10 min and then cooled at 25°C for 30 s.
A preliminary experiment was performed to ensure that the difference in the cycle threshold (CT) value of MS2 RNA between cancer and paired normal samples did not exceed 1.0. The expression levels of the miRNAs were quantified using SYBR Green-based All-in-One qPCR Mix (GeneCopoeia, USA).
Statistical analyses
The comparative expression of miRNAs was determined using the 2-ΔΔCT method [22]. The differentially expressed miRNAs of tumor tissues relative to paired normal tissues were identified using non-paired t-tests and receiver operating characteristic (ROC) curves. The area under the ROC curve (AUC) was used to evaluate the sensitivity and specificity of miRNAs from tissue specimens as a diagnostic marker for the detection of CRC. In the two-tailed tests, a P-value <0.05 was considered statistically significant. Graph Pad Prism 5.0 (San Diego, CA, USA) and SPSS 16.0 (SPSS, UK) software were used for data analyses.
Results
We performed an analysis using a non-paired t-test by treating normal and tumor samples as two independent groups. We found 96 significantly dysregulated miRNAs. There were 82 downregulated miRNAs and 14 upregulated miRNAs (Table 1). However, miR-26a-2*, miR-338-3p, miR-150, miR-708, miR-503, and miR-21 showed less than a 2-fold change relative to the normal controls. The fold changes of the other 90 dysregulated miRNAs were greater than 2 fold (Table 1). Among the dysregulated miRNAs, the fold changes of miR-1 and miR-145 were the greatest, with changes of more than 15 fold. The other miRNAs with fold changes greater than 5 included the following: miR-145*, miR-137, miR-133a, miR-363, miR-143, miR-4770, miR-490-5p, miR-133b, miR-9, miR-4510, miR-144*, let-7d, and miR-96. Of the upregulated miRNAs, miR-96 was upregulated six-fold (Table 1).
Table 1.
Differential expressed miRNAs between tumor and paired adjacent normal mucosa in different stages
| miRNAs | II-III | II | III | stages with significant change (P<0.05) | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| fold change | P Value | fold change | P Value | fold change | P Value | ||
| hsa-miR-1 | -16.393 | <0.001 | -17.544 | 0.009 | -15.152 | 0.014 | II, III, II-III |
| hsa-miR-145 | -15.873 | <0.001 | -18.182 | 0.001 | -13.699 | <0.001 | II, III, II-III |
| hsa-miR-145* | -13.158 | <0.001 | -9.615 | <0.001 | -18.868 | <0.001 | II, III, II-III |
| hsa-miR-137 | -10.870 | <0.001 | -13.514 | 0.001 | -8.403 | 0.016 | II, III, II-III |
| hsa-miR-363 | -6.993 | 0.001 | -6.993 | 0.028 | -6.993 | 0.006 | II, III, II-III |
| hsa-miR-143 | -6.944 | <0.001 | -7.407 | 0.022 | -6.452 | 0.005 | II, III, II-III |
| hsa-miR-4770 | -6.803 | <0.001 | -8.333 | 0.001 | -5.263 | 0.022 | II, III, II-III |
| hsa-miR-490-5p | -6.536 | <0.001 | -6.993 | 0.007 | -6.098 | 0.021 | II, III, II-III |
| hsa-miR-9 | -5.848 | <0.001 | -6.494 | 0.014 | -5.208 | 0.014 | II, III, II-III |
| hsa-miR-144* | -5.435 | <0.001 | -3.788 | 0.044 | -8.264 | 0.001 | II, III, II-III |
| hsa-miR-99a | -4.975 | 0.001 | -6.369 | 0.013 | -3.745 | 0.025 | II, III, II-III |
| hsa-miR-99b | -4.739 | <0.001 | -5.556 | 0.010 | -3.953 | 0.004 | II, III, II-III |
| hsa-miR-23b | -4.717 | <0.001 | -3.559 | 0.044 | -6.536 | 0.001 | II, III, II-III |
| hsa-miR-143* | -4.505 | <0.001 | -4.505 | 0.008 | -4.484 | 0.026 | II, III, II-III |
| hsa-miR-100 | -4.386 | 0.002 | -5.495 | 0.022 | -3.356 | 0.047 | II, III, II-III |
| hsa-miR-768-3p | -4.367 | <0.001 | -4.049 | 0.003 | -4.785 | 0.001 | II, III, II-III |
| hsa-miR-24-1* | -4.202 | <0.001 | -2.924 | 0.050 | -6.369 | <0.001 | II, III, II-III |
| hsa-miR-125a-5p | -4.065 | 0.001 | -5.000 | 0.018 | -3.185 | 0.019 | II, III, II-III |
| hsa-miR-30e* | -3.774 | <0.001 | -3.322 | 0.013 | -4.367 | 0.001 | II, III, II-III |
| hsa-miR-574-3p | -3.289 | <0.001 | -2.874 | 0.033 | -3.846 | 0.004 | II, III, II-III |
| hsa-miR-126 | -3.279 | 0.001 | -3.257 | 0.012 | -3.311 | 0.050 | II, III, II-III |
| hsa-let-7b | -3.215 | 0.001 | -3.704 | 0.011 | -2.732 | 0.041 | II, III, II-III |
| hsa-miR-1979 | -2.915 | 0.003 | -2.092 | 0.045 | -4.274 | 0.018 | II, III, II-III |
| hsa-miR-374b | -2.703 | 0.002 | -2.519 | 0.018 | -2.924 | 0.049 | II, III, II-III |
| hsa-miR-140-3p | -2.381 | 0.002 | -2.381 | 0.040 | -2.381 | 0.014 | II, III, II-III |
| hsa-miR-203 | 2.449 | 0.026 | 2.899 | 0.038 | 8.024 | 0.030 | II, III, II-III |
| hsa-miR-182 | 3.290 | 0.002 | 2.769 | 0.022 | 4.015 | 0.040 | II, III, II-III |
| hsa-miR-96 | 6.009 | <0.001 | 5.801 | 0.001 | 6.259 | 0.014 | II, III, II-III |
| hsa-miR-133a | -7.634 | 0.008 | -11.494 | 0.002 | -4.808 | 0.261 | II, II-III |
| hsa-miR-133b | -6.098 | 0.002 | -7.407 | 0.017 | -4.854 | 0.056 | II, II-III |
| hsa-miR-451 | -4.587 | 0.003 | -5.076 | 0.024 | -4.082 | 0.057 | II, II-III |
| hsa-miR-139-5p | -4.184 | 0.005 | -5.376 | 0.026 | -3.135 | 0.111 | II, II-III |
| hsa-miR-218 | -4.167 | 0.007 | -5.435 | 0.028 | -3.067 | 0.138 | II, II-III |
| hsa-miR-193a-5p | -3.472 | 0.002 | -3.788 | 0.013 | -3.155 | 0.065 | II, II-III |
| hsa-miR-365 | -3.472 | 0.002 | -3.717 | 0.010 | 1.239 | 0.786 | II, II-III |
| hsa-miR-204 | -3.344 | 0.004 | -3.509 | 0.039 | -3.165 | 0.056 | II, II-III |
| hsa-miR-328 | -3.311 | 0.009 | -4.831 | 0.012 | -2.137 | 0.285 | II, II-III |
| hsa-miR-193b | -3.257 | 0.009 | -3.861 | 0.048 | -2.681 | 0.094 | II, II-III |
| hsa-miR-3653 | -2.941 | 0.003 | -3.012 | 0.022 | -2.874 | 0.061 | II, II-III |
| hsa-miR-26a | -2.833 | 0.005 | -3.344 | 0.039 | -2.342 | 0.063 | II, II-III |
| hsa-miR-4799-5p | -2.732 | 0.049 | -3.846 | 0.043 | -1.779 | 0.483 | II, II-III |
| hsa-miR-10b | -2.299 | 0.007 | -2.710 | 0.039 | -1.898 | 0.091 | II, II-III |
| hsa-miR-150 | -1.832 | 0.025 | -2.551 | 0.011 | -1.252 | 0.584 | II, II-III |
| hsa-miR-429 | 2.475 | 0.013 | 2.602 | 0.045 | 2.335 | 0.147 | II, II-III |
| hsa-miR-592 | 2.550 | 0.040 | 5.060 | 0.017 | 4.604 | 0.090 | II, II-III |
| hsa-miR-141 | 2.948 | 0.049 | 5.858 | 0.016 | 1.335 | 0.718 | II, II-III |
| hsa-miR-183 | 3.291 | 0.006 | 3.092 | 0.048 | 3.537 | 0.068 | II, II-III |
| hsa-miR-135b | 4.189 | 0.002 | 6.154 | 0.004 | 2.688 | 0.174 | II, II-III |
| hsa-miR-4510 | -5.650 | 0.002 | -4.902 | 0.056 | -6.757 | 0.009 | III, II-III |
| hsa-let-7d | -5.376 | 0.005 | -2.632 | 0.101 | -12.195 | 0.022 | III, II-III |
| hsa-miR-3195 | -4.717 | 0.002 | -4.587 | 0.061 | -4.878 | 0.005 | III, II-III |
| hsa-let-7c | -4.566 | 0.001 | -2.882 | 0.072 | -7.752 | 0.004 | III, II-III |
| hsa-miR-4469 | -4.237 | 0.024 | -2.667 | 0.307 | -7.576 | 0.017 | III, II-III |
| hsa-miR-27b | -4.115 | 0.006 | -1.927 | 0.222 | -9.901 | 0.011 | III, II-III |
| hsa-miR-214 | -3.333 | 0.003 | -2.695 | 0.099 | -4.237 | 0.006 | III, II-III |
| hsa-miR-9* | -3.135 | 0.020 | -2.119 | 0.279 | -4.902 | 0.020 | III, II-III |
| hsa-miR-378i | -3.115 | 0.002 | -2.632 | 0.097 | -3.861 | 0.003 | III, II-III |
| hsa-let-7g | -3.040 | 0.003 | -2.160 | 0.117 | -4.505 | 0.014 | III, II-III |
| hsa-miR-378* | -2.994 | 0.001 | -2.469 | 0.095 | -3.745 | <0.001 | III, II-III |
| hsa-miR-199a-3p | -2.801 | 0.004 | -2.299 | 0.113 | -3.521 | 0.007 | III, II-III |
| hsa-miR-4684-3p | -2.717 | 0.015 | -2.959 | 0.112 | -2.451 | 0.028 | III, II-III |
| hsa-miR-4443 | -2.445 | 0.008 | -2.092 | 0.076 | -2.967 | 0.047 | III, II-III |
| hsa-miR-214* | -2.421 | 0.009 | -1.869 | 0.189 | -3.257 | 0.009 | III, II-III |
| hsa-miR-199b-3p | -2.364 | 0.012 | -2.309 | 0.107 | -2.427 | 0.048 | III, II-III |
| hsa-miR-422a | -2.364 | 0.014 | -1.821 | 0.201 | -3.185 | 0.029 | III, II-III |
| hsa-miR-26a-2* | -1.984 | 0.038 | -1.511 | 0.344 | -2.717 | 0.038 | III, II-III |
| hsa-miR-1290 | 3.324 | 0.023 | 4.955 | 0.058 | 8.347 | 0.038 | III, II-III |
| hsa-let-7d* | -3.344 | 0.020 | -2.770 | 0.132 | -4.149 | 0.056 | II-III |
| hsa-miR-378d | -3.788 | 0.043 | -3.831 | 0.134 | -3.731 | 0.184 | II-III |
| hsa-miR-4524* | -3.759 | 0.013 | -1.572 | 0.492 | -4.464 | 0.077 | II-III |
| hsa-miR-4423-3p | -3.650 | 0.019 | -2.793 | 0.135 | -5.128 | 0.065 | II-III |
| hsa-miR-151b | -3.571 | 0.021 | -3.509 | 0.122 | -3.650 | 0.093 | II-III |
| hsa-miR-28-5p | -3.165 | 0.004 | -2.967 | 0.051 | 1.168 | 0.843 | II-III |
| hsa-miR-30c-2* | -3.086 | 0.007 | -2.740 | 0.061 | -3.546 | 0.064 | II-III |
| hsa-miR-26b | -3.030 | 0.023 | -3.610 | 0.076 | -2.481 | 0.179 | II-III |
| hsa-miR-195 | -2.976 | 0.015 | -3.155 | 0.083 | -2.786 | 0.097 | II-III |
| hsa-miR-4526 | -2.950 | 0.032 | -2.933 | 0.151 | -2.976 | 0.092 | II-III |
| hsa-miR-30a | -2.857 | 0.031 | -3.030 | 0.134 | -2.674 | 0.120 | II-III |
| hsa-miR-29b-2* | -2.833 | 0.012 | -2.809 | 0.096 | -2.857 | 0.067 | II-III |
| hsa-miR-149 | -2.710 | 0.034 | -2.604 | 0.144 | -2.833 | 0.137 | II-III |
| hsa-miR-628-3p | -2.703 | 0.045 | -2.309 | 0.260 | 1.230 | 0.804 | II-III |
| hsa-miR-490-3p | -2.457 | 0.041 | -1.626 | 0.349 | -3.953 | 0.065 | II-III |
| hsa-miR-130a | -2.347 | 0.033 | -2.674 | 0.110 | -2.016 | 0.152 | II-III |
| hsa-miR-30c | -2.262 | 0.014 | -2.577 | 0.052 | -1.949 | 0.147 | II-III |
| hsa-miR-30d* | -2.212 | 0.034 | -2.114 | 0.126 | -2.331 | 0.157 | II-III |
| hsa-miR-10a | -2.083 | 0.023 | -2.227 | 0.111 | -1.927 | 0.113 | II-III |
| hsa-miR-27b* | -2.083 | 0.048 | -1.748 | 0.311 | -2.551 | 0.069 | II-III |
| hsa-miR-181c | -2.070 | 0.026 | -2.132 | 0.109 | -2.004 | 0.095 | II-III |
| hsa-miR-487b | -2.053 | 0.018 | -1.821 | 0.160 | -2.358 | 0.056 | II-III |
| hsa-miR-4790-5p | -2.020 | 0.027 | -1.645 | 0.112 | -2.604 | 0.123 | II-III |
| hsa-miR-338-3p | -1.961 | 0.041 | -2.375 | 0.051 | -1.575 | 0.371 | II-III |
| hsa-miR-708 | 1.937 | 0.036 | 1.806 | 0.133 | -1.036 | 0.968 | II-III |
| hsa-miR-503 | 1.984 | 0.036 | 2.258 | 0.072 | 1.708 | 0.278 | II-III |
| hsa-miR-21 | 1.997 | 0.040 | 1.985 | 0.171 | 2.010 | 0.125 | II-III |
| hsa-miR-130b | 2.153 | 0.022 | 1.811 | 0.110 | 2.629 | 0.108 | II-III |
| hsa-miR-200a | 2.383 | 0.022 | 2.527 | 0.112 | 2.228 | 0.110 | II-III |
| hsa-miR-4794 | -1.980 | 0.134 | -2.915 | 0.049 | -1.221 | 0.791 | II |
| hsa-miR-155 | -1.053 | 0.862 | -1.107 | 0.736 | 4.010 | 0.032 | III |
| hsa-miR-4489 | -2.252 | 0.078 | -1.389 | 0.599 | -4.115 | 0.040 | III |
| hsa-miR-4634 | -1.340 | 0.493 | 1.212 | 0.791 | -2.457 | 0.012 | III |
| hsa-miR-545 | 1.115 | 0.767 | -1.072 | 0.896 | 5.448 | 0.037 | III |
| hsa-miR-224 | 2.384 | 0.051 | 3.032 | 0.066 | 7.185 | 0.048 | III |
| hsa-miR-7 | 1.762 | 0.089 | 1.577 | 0.281 | 7.968 | 0.019 | III |
Correlation of dysregulated miRNAs and clinical characteristics
We then investigated the expression patterns of the dysregulated miRNAs in stage II and III disease. The analysis showed that there were 49 miRNAs that were significantly dysregulated in stage II tumors compared to the paired normal tissues by non-paired t-test. The results consisted of 41 downregulated and 8 upregulated miRNAs, of which miR-145 had the highest fold-change with 18 fold (Table 1). There were also 53 significantly dysregulated miRNAs identified in stage III tumors. There were 45 downregulated miRNAs, including miR-145* with an 18.87-fold change. Of the 8 upregulated miRNAs, the fold changes of miR-203 and miR-1290 in tumor tissues were the highest, with more than 8-fold induction (Table 1).
We then investigated the intersections of the three sets of significantly differentially expressed miRNAs. There were 28 overlapping miRNAs found, and 25 were downregulated, including the following: miR-1, -145, -145*, -137, -363, -143, -4770, -490-5p, -9, -144*, -99a, -99b, -23b, -143*, -100, -768-3p, -24-1*, -125a-5p, -30e*, -574-3p, -126, let-7b, miR-1979, -374b, -140-3p. We also found that miR-203, -182, and -96 were upregulated (Table 1). These miRNAs are highly likely to be novel biomarkers.
The use of miRNA profiles can contribute to the diagnostic and prognostic classification of human malignancies, which has been reported previously [23-25]. We compared the miRNA expression profile in stage II to that in stage III. Interestingly, the results showed that there were 58 miRNAs with significantly differential expression between stage II and stage III tumors. However, except for miR-768-3p dysregulation with P=0.037, the dysregulation of the other 27 miRNAs in the three sets was not significantly different between stage II and stage III tumors (Table 2).
Table 2.
Differential expressed miRNAs between patients with stage II and stage III
| miRNA | fold change | P Value | miRNA | fold change | P Value | miRNA | fold change | P Value |
|---|---|---|---|---|---|---|---|---|
| hsa-miR-548t | 61.091 | 0.004 | hsa-miR-4648 | 6.147 | 0.029 | hsa-miR-768-3p | 2.795 | 0.037 |
| hsa-miR-3139 | 62.647 | 0.005 | hsa-miR-548ai | 6.261 | 0.030 | hsa-miR-144* | 3.709 | 0.052 |
| hsa-miR-4424 | 11.421 | 0.005 | hsa-miR-4487 | 3.501 | 0.031 | hsa-miR-1979 | 3.029 | 0.055 |
| hsa-miR-4477a | 18.420 | 0.007 | hsa-miR-4439 | 6.380 | 0.032 | hsa-miR-4770 | -2.770 | 0.111 |
| hsa-miR-548x | 11.884 | 0.007 | hsa-miR-4677-3p | 7.092 | 0.034 | hsa-miR-24-1* | 1.786 | 0.199 |
| hsa-miR-3132 | 22.459 | 0.008 | hsa-miR-4793-3p | -5.376 | 0.034 | hsa-miR-99b | 1.747 | 0.347 |
| hsa-miR-23a* | 4.998 | 0.009 | hsa-miR-3144-5p | 9.150 | 0.034 | hsa-miR-145* | 1.964 | 0.373 |
| hsa-miR-4501 | 7.369 | 0.013 | hsa-miR-4468 | 5.848 | 0.035 | hsa-miR-9 | 1.894 | 0.393 |
| hsa-miR-561 | 10.031 | 0.013 | hsa-miR-3689d | 7.680 | 0.036 | hsa-miR-96 | 1.725 | 0.398 |
| hsa-miR-215 | -7.519 | 0.014 | hsa-miR-4473 | 7.847 | 0.037 | hsa-miR-363 | 1.995 | 0.401 |
| hsa-miR-4438 | 9.033 | 0.015 | hsa-miR-452* | 10.969 | 0.037 | hsa-miR-30e* | 1.436 | 0.413 |
| hsa-miR-425 | 5.238 | 0.019 | hsa-miR-768-3p | 2.795 | 0.037 | hsa-miR-374b | 1.474 | 0.414 |
| hsa-miR-4482 | 7.223 | 0.019 | hsa-miR-92a | 2.914 | 0.037 | hsa-miR-145 | 1.895 | 0.419 |
| hsa-miR-4504 | 6.750 | 0.019 | hsa-miR-4633-3p | 5.234 | 0.039 | hsa-miR-125a-5p | 1.536 | 0.451 |
| hsa-miR-4431 | 7.562 | 0.019 | hsa-miR-4645-5p | 6.634 | 0.040 | hsa-miR-137 | 1.847 | 0.451 |
| hsa-miR-4278 | 7.342 | 0.019 | hsa-miR-4513 | 3.144 | 0.040 | hsa-let-7b | 1.521 | 0.480 |
| hsa-miR-3135 | 16.607 | 0.020 | hsa-miR-4644 | 5.853 | 0.041 | hsa-miR-574-3p | 1.383 | 0.502 |
| hsa-miR-3130-5p | 8.219 | 0.021 | hsa-miR-4480 | 5.973 | 0.041 | hsa-miR-143 | 1.332 | 0.709 |
| hsa-miR-4637 | 8.304 | 0.022 | hsa-miR-200b* | 2.600 | 0.042 | hsa-miR-1 | 1.450 | 0.721 |
| hsa-miR-3127 | 8.942 | 0.022 | hsa-miR-4268 | 8.704 | 0.042 | hsa-miR-23b | -1.222 | 0.744 |
| hsa-miR-4503 | 6.484 | 0.023 | hsa-miR-4448 | 5.156 | 0.043 | hsa-miR-143* | -1.229 | 0.748 |
| hsa-miR-3126-5p | 8.438 | 0.025 | hsa-miR-548n | 9.412 | 0.043 | hsa-miR-490-5p | -1.294 | 0.770 |
| hsa-miR-3137 | 69.623 | 0.025 | hsa-miR-4437 | 5.934 | 0.044 | hsa-miR-203 | 1.114 | 0.875 |
| hsa-miR-4464 | 7.620 | 0.026 | hsa-miR-4636 | 4.791 | 0.045 | hsa-miR-140-3p | -1.058 | 0.895 |
| hsa-miR-4427 | 7.179 | 0.026 | hsa-miR-3118 | 6.495 | 0.046 | hsa-miR-182 | -1.054 | 0.918 |
| hsa-miR-146a | 3.447 | 0.027 | hsa-miR-581 | 11.036 | 0.046 | hsa-miR-99a | -1.043 | 0.951 |
| hsa-miR-548ah | 5.448 | 0.027 | hsa-miR-1973 | 3.598 | 0.048 | hsa-miR-100 | -1.018 | 0.978 |
| hsa-miR-4788 | -3.968 | 0.028 | hsa-miR-296-5p | 4.392 | 0.049 | hsa-miR-126 | 1.007 | 0.987 |
| hsa-miR-3179 | 4.303 | 0.028 |
MiR-1 and miR-374b act as CRC biomarkers
The expression of miR-1 and miR-374b were significantly decreased in each stage (Table 1; Figures 1A-C, 2A-C). We then performed ROC analysis to estimate the sensitivity and specificity of miR-1 and miR-374b as diagnosis markers of CRC. The results for miR-1 included the following: AUC 0.806, sensitivity 78.57%, specificity 78.57%, and P<0.001 (Figure 1D). For miR-374b, the results were AUC 0.724, sensitivity 71.43%, specificity 71.43%, and P=0.004 (Figure 2D).
Figure 1.
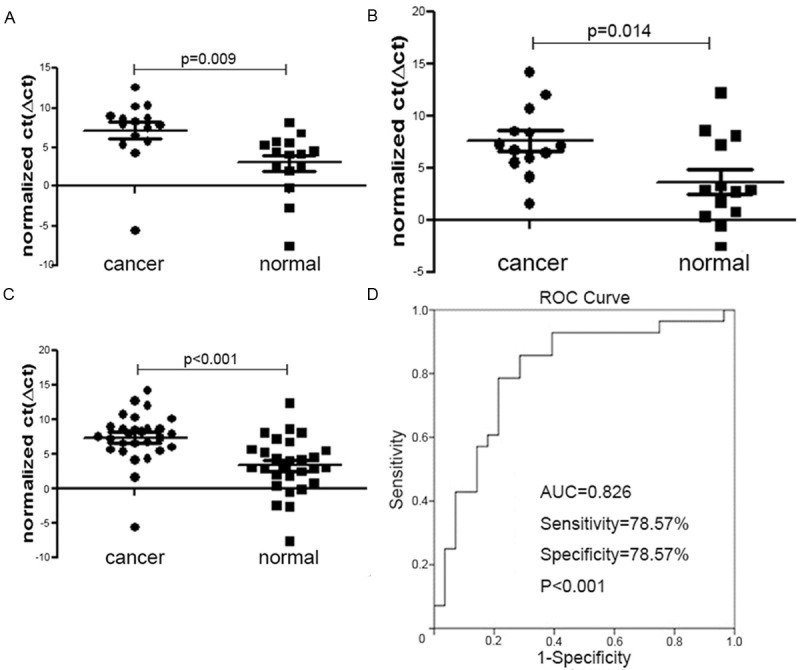
Evaluation of the potential of miR-1 as a diagnostic biomarker in clinical applications. A: Compared to normal tissues, miR-1 was downregulated in stage II; B: Compared to normal tissues, miR-1 was downregulated in stage III; C: Compared to normal tissues, miR-1 was downregulated in stage II-III; D: ROC analysis to estimate the sensitivity and specificity of miR-1 showed AUC 0.806, sensitivity 78.57%, specificity 78.57%, and P<0.001.
Figure 2.
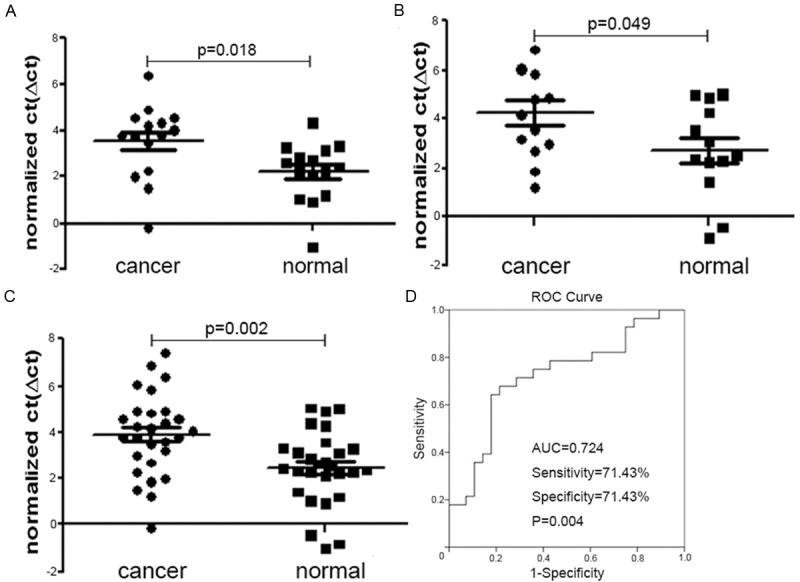
Evaluation of the potential of miR-374b as a diagnostic biomarker in clinical applications. A: Compared to normal tissues, miR-374b was downregulated in stage II; B: Compared to normal tissues, miR-374b was downregulated in stage III; C: Compared to normal tissues, miR-374b was downregulated in stage II-III; D: ROC analysis to estimate the sensitivity and specificity of miR-374b showed AUC 0.724, sensitivity 71.43%, specificity 71.43%, and P=0.004.
The expression levels of miR-1 and miR-374b between stage II and stage III did not show significant differences. The non-paired t-test results were P=0.728 and 0.295 (P>0.05), respectively (Figure 3A, 3B).
Figure 3.
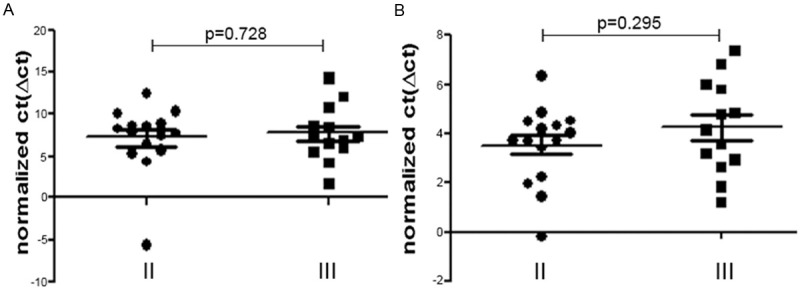
The expression levels of miR-1 and miR-374b between stage II and stage III did not show significant differences. The non-paired t-test results were P=0.728 and 0.295 (P>0.05). A: Compared to stage II tumors, miR-1 was not significantly dysregulated in stage III, with P=0.728; B: Compared to stage II tumors, miR-374b was not significantly dysregulated in stage III, with P=0.295.
Novel biomarkers in CRC without metastasis
It has been reported that the prognosis of cancer patients depends on tumor stage at the time of diagnosis, and many CRC patients have lymph node or distant metastasis at the time of diagnosis [26]. To address this issue, we focused only on miRNAs that were aberrantly expressed in stage II tumors (P<0.05) without lymph node or distant metastasis. Based on this criterion, 20 of the 49 miRNAs were dysregulated in both stage II and stage II-III tumors but not in stage III. Only one miRNA (miR-4794) was involved exclusively in stage II tumors, and it was downregulated 2.915 fold with P=0.049 (Table 1).
Predictive value of miRNAs for CRC patients with lymph node metastasis
We next investigated the prognostic value of miRNAs to detect CRC and focused on the dysregulated miRNAs that were found only in stage III disease, which is the stage with lymph node metastasis. Among the 53 differentially expressed miRNAs found in stage III, only 6 were uniquely dysregulated in stage III. There were 3 upregulated and 3 downregulated dysregulated miRNAs. Additionally, there were 19 miRNAs that were dysregulated both in stage III and stage II-III tumors (Table 1).
Discussion
CRC is a lethal disease, and current screening methods for CRC are still limited by unsatisfactory sensitivity and specificity. miRNAs have been shown to be important regulators of many gene functions in human cancers [27]. In the present study, we compared the expression level of 1547 miRNAs in CRC tumor tissues to paired normal tissue. We identified 96 miRNAs that were significantly dysregulated in CRC relative to normal tissues (P<0.05). However, verified diagnostic CRC markers should be able to distinguish patients in all stages from normal subjects. The use of such markers might prevent the misdiagnosis of patients with certain stages of CRC [20]. We analyzed the expression profiles of 1547 miRNAs in both stage II and III tumors. Three sets of significantly differentially expressed miRNAs were identified in stages II, III and II-III. Interestingly, there was an intersection among the three sets of miRNAs, and there were 28 dysregulated miRNAs associated with stages II and III separately or in combination (Table 1). With the exception of miR-768-3P, the other 27 miRNAs showed no significant differentiation between stage II and stage III tumors (Table 2). These miRNAs are likely to be novel biomarkers for CRC detection. Previous studies have shown that global changes in miRNA expression are associated with differentiation [28]. Thus, to further investigate the prognostic value of miRNAs for CRC detection, we focused on the miRNAs that were only dysregulated in stage II for screening the stage without lymph node or distant metastasis. We also examined miRNAs that were only uniquely dysregulated in stage III to distinguish the stage with lymph node metastasis but without distant metastasis. In stage II, among the 49 dysregulated miRNAs, there were 20 miRNAs that were dysregulated both in stage II and stage II-III tumors but not in stage III. Only miR-4794 was exclusively involved in stage II tumors, and this miRNA was downregulated 2.915 fold with P=0.049. Among the 53 differentially expressed miRNAs found in stage III, only 6 were uniquely expressed in stage III. There were 3 upregulated and 3 downregulated miRNAs. There were also 19 miRNAs that were dysregulated in both stage III and stage II-III. A previous report demonstrated that miRNA expression profiles are correlated with cancer differentiation. Therefore, we investigated the expression profile differences between stage II and stage III disease. Interestingly, the results showed there were 58 miRNAs with significantly different expression levels between stage II and stage III tumors. However, only miR-768-3p was dysregulated, with P=0.037, and the remaining 27 miRNAs that were dysregulated in the three sets were not significantly different (Table 2). This finding verified that miR-1 and miR-374b might be biomarkers for CRC screening. The ROC curve analysis indicated that miR-1 and miR-374b had a potential use as biomarkers with high sensitivity and specificity for CRC detection.
MiR-1 has previously been shown to have a role in tumorigenesis and is abnormally downregulated in several types of cancers, including lung, prostate, thyroid and colorectal cancers and rhabdomyosarcoma. MiR-1 functions as a tumor suppressor and inhibits cell proliferation and promotes cell differentiation and apoptosis [29]. miR-1 has further been suggested to play a tumor-suppressive role by targeting the transgelin 2 gene (TAGLN2) in bladder cancer [30]. Cristina and colleagues identified a tumor suppressive role for miR-1 in colorectal cancer, controlling MET expression through a feedback loop. The concomitant downregulation of miR-1 and increase of MACC1 contributes to MET overexpression and the metastatic behavior of colon cancer cells [31]. These results are consistent with our findings of miR-1 downregulation in each CRC stage. Additionally, miR-1 expression can be modulated by epigenetic modifications [32]. Data also indicate that miR-1 plays an important role in cancers such as rhabdomyosarcoma, which is the most common soft tissue sarcoma in children [33,34]. Chiyomaru and colleagues showed that miR-1 was barely detectable in primary rhabdomyosarcomas, and its re-expression in tumor cells promoted myogenic differentiation and blocked tumor growth in xenografted mice [35]. Previous observations have suggested that the dysregulation of miR-1 has an oncogenic role via the abolition of the suppressive effect on specific target genes, such as MET, LASP1, IGF-1, IGFR-1, and BCL2 [33,36,37]. James and colleagues showed that miR-1 can have a tumor suppressor function in colorectal cancer of directly downregulating the MET oncogene both at the RNA and protein levels. Furthermore, re-expression of miR-1 leads to a MET-driven reduction of cell proliferation and motility. These findings show that miR-1 reduction enhances colorectal cancer progression [38]. Lu and colleagues observed an inverse correlation between EDN1 and miR-1 expression in HCC patients, and miR-1 was found to inhibit the expression of EDN1. These results suggest that EDN1 plays an important role in HCC progression by activating the PI3K/AKT pathway and is regulated by miR-1 [39]. Moreover, the results of the present study showed that miR-1 is significantly downregulated in each stage and validated the results of previous studies.
Previous reports have shown that miR-374b is downregulated in prostate cancer tissue and have demonstrated that it is an independent predictor of biochemical recurrence-free survival [40]. However, miR-374b has not been previously correlated with CRC. In this study, miR-374b was downregulated in each stage but exhibited no significant difference (P>0.05) between stage II and stage III.
Furthermore, we found that several dysregulated miRNAs may serve as biomarkers for the identification of CRC patients without lymph node or distant metastasis. The value of miRNAs for CRC patients with lymph node disease was also evaluated in our study. These results will be valuable in identifying biomarkers for specific stage tumors.
In conclusion, the present study has a limited number of experimental samples. Therefore, before miRNA testing can be made available in the clinical setting, several large prospective studies are required. Despite this disadvantage, our study supports the concept that alternations in miRNA expression play an important role in CRC carcinogenesis. This study provides a new model to analyze novel CRC biomarkers by considering additional clinical factors.
Acknowledgements
We thank Mr. Rong Su for his help with samples. The miRNA profiling and qRT-PCR in this study were performed at Genecopoeia Guangzhou, China. Huaping Li and his colleagues provided much support during the experiments.
Disclosure of conflict of interest
None.
References
- 1.Kleist B, Xu L, Kersten C, Seel V, Li G, Poetsch M. Single nucleotide polymorphisms of the adult intestinal stem cell marker Lgr5 in primary and metastatic colorectal cancer. Am J Transl Res. 2012;4:279–290. [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H, Yu H. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6:391–401. [PMC free article] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans Heterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to &II-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gusev Y, Brackett DJ. MicroRNA expression profiling in cancer from a bioinformatics prospective. Expert Rev Mol Diagn. 2007;7:787–792. doi: 10.1586/14737159.7.6.787. [DOI] [PubMed] [Google Scholar]
- 7.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 8.Cho WC. MicroRNAs in cancer-from research to therapy. Biochim Biophys Acta. 2010;1805:209–217. doi: 10.1016/j.bbcan.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, Kure EH. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One. 2012;7:e34150. doi: 10.1371/journal.pone.0034150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, Chen HY, Sun XF. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009;26:27–34. doi: 10.3233/DMA-2009-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh T, Kojima K, Nakashima R, Kitade Y, Naoe T. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17:398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- 12.La Rocca G, Badin M, Shi B, Xu SQ, Deangelis T, Sepp-Lorenzinoi L, Baserga R. Mechanism of growth inhibition by MicroRNA 145: the role of the IGF-I receptor signaling pathway. J Cell Physiol. 2009;220:485–491. doi: 10.1002/jcp.21796. [DOI] [PubMed] [Google Scholar]
- 13.Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem. 2007;282:32582–32590. doi: 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- 14.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinami R, Ercolani C, Petti E, Piazza S, Ciani Y, Sestito R, Sacconi A, Biagioni F, le Sage C, Agami R, Benetti R, Mottolese M, Schneider C, Blandino G, Schoeftner S. miR-155 Drives Telomere Fragility in Human Breast Cancer by Targeting TRF1. Cancer Res. 2014;74:4145–4156. doi: 10.1158/0008-5472.CAN-13-2038. [DOI] [PubMed] [Google Scholar]
- 16.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 17.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DLW, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 19.Darido C, Georgy SR, Wilanowski T, Dworkin S, Auden A, Zhao Q, Rank G, Srivastava S, Finlay MJ, Papenfuss AT, Pandolfi PP, Pearson RB, Jane SM. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell. 2011;20:635–648. doi: 10.1016/j.ccr.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Xu XH, Wu XB, Wu SB, Liu HB, Chen R, Li Y. Identification of miRNAs differentially expressed in clinical stages of human colorectal carcinoma-an investigation in Guangzhou, China. PLoS One. 2014;9:e94060. doi: 10.1371/journal.pone.0094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki H, Takatsuka S, Akashi H, Yamamoto E, Nojima M, Maruyama R, Kai M, Yamano HO, Sasaki Y, Tokino T, Shinomura Y, Imai K, Toyota M. Genome-wide profiling of chromatin signatures reveals epigenetic regulation of MicroRNA genes in colorectal cancer. Cancer Res. 2011;71:5646–5658. doi: 10.1158/0008-5472.CAN-11-1076. [DOI] [PubMed] [Google Scholar]
- 22.Livaka KJ, Schmittgenb TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Calin GA, Ferracin M, Cimmino A, Leva GD, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 24.Zhu W, Liu X, He J, Chen D, Hunag Y, Zhang YK. Overexpression of members of the microRNA-183 family is a risk factor for lung cancer: a case control study. BMC Cancer. 2011;11:393. doi: 10.1186/1471-2407-11-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosakhani N, Sarhadi VK, Borze I, Karjalainen-Lindsberg ML, Sundstrom J, Ristamaki R, Osterlund P, Knuutila S. MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer. 2012;51:1–9. doi: 10.1002/gcc.20925. [DOI] [PubMed] [Google Scholar]
- 26.Figueredo A, Coombes ME, Mukherjee S. Adjuvant therapy for completely resected stage II colon cancer. Cochrane Database Syst Rev. 2008:CD005390. doi: 10.1002/14651858.CD005390.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 29.Lu JW, Hsia Y, Yang WY, Lin YI, Li CC, Tsai TF, Chang KW, Shieh GS, Tsai SF, Wang HD, Yuh CH. Identification of the common regulators for hepatocellular carcinoma induced by hepatitis B virus X antigen in a mouse model. Carcinogenesis. 2012;33:209–219. doi: 10.1093/carcin/bgr224. [DOI] [PubMed] [Google Scholar]
- 30.Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808–818. doi: 10.1038/bjc.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Migliore C, Martin V, Leoni VP, Restivo A, Atzori L, Petrelli A, Isella C, Zorcolo L, Sarotto I, Casula G, Comoglio PM, Columbano A, Giordano S. MiR-1 downregulation cooperates with MACC1 in promoting MET overexpression in human colon cancer. Clin Cancer Res. 2012;18:737–747. doi: 10.1158/1078-0432.CCR-11-1699. [DOI] [PubMed] [Google Scholar]
- 32.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, Ghoshal K, Jacob ST. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Yan D, Dong Xda E, Chen X, Wang L, Lu C, Wang J, Qu J, Tu L. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Uchida Y, Kawahara K, Nishiyama K, Seki N, Nakagawa M. Functional role of LASP1 in cell viability and its regulation by microRNAs in bladder cancer. Urol Oncol. 2012;30:434–443. doi: 10.1016/j.urolonc.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Ban J, Jug G, Mestdagh P, Schwentner R, Kauer M, Aryee DN, Schaefer KL, Nakatani F, Scotlandi K, Reiter M, Strunk D, Speleman F, Vandesompele J, Kovar H. Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing’s sarcoma. Oncogene. 2011;30:2173–2180. doi: 10.1038/onc.2010.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arlt F, Stein U. Colon cancer metastasis: MACC1 and Met as metastatic pacemakers. Int J Biochem Cell Biol. 2009;41:2356–2359. doi: 10.1016/j.biocel.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Reid JF, Sokolova V, Zoni E, Lampis A, Pizzamiglio S, Bertan C, Zanutto S, Perrone F, Camerini T, Gallino G, Verderio P, Leo E, Pilotti S, Gariboldi M, Pierotti MA. miRNA profiling in colorectal cancer highlights miR-1 involvement in MET-dependent proliferation. Mol Cancer Res. 2012;10:504–515. doi: 10.1158/1541-7786.MCR-11-0342. [DOI] [PubMed] [Google Scholar]
- 39.Lu JW, Liao CY, Yang WY, Lin YM, Jin SL, Wang HD, Yuh CH. Overexpression of endothelin 1 triggers hepatocarcinogenesis in zebrafish and promotes cell proliferation and migration through the AKT pathway. PLoS One. 2014;9:e85318. doi: 10.1371/journal.pone.0085318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin ZY, Deng YH, Qin GQ, Cai C, Chen JH, Jiang FN, Liu XY, Zhong WD. Global analysis of the differentially expressed miRNAs of prostate cancer in Chinese patients. BMC Genomics. 2013;14:757. doi: 10.1186/1471-2164-14-757. [DOI] [PMC free article] [PubMed] [Google Scholar]


