Abstract
Introduction: Long non-coding RNAs (lncRNAs) are aberrantly expressed in many diseases including cancer. LncRNA HULC (highly up-regulated in liver cancer) has recently been revealed to be involved in hepatocellular carcinoma development and progression. However, the role and function of HULC in human osteosarcoma remains unknown. Methods: LncRNA HULC expression in osteosarcoma tissues and cell lines was detected by quantitative real-time PCR. Then, the association of HULC level with survival of osteosarcoma patients was performed by the Kaplan-Meier and Cox proportional regression analyses. Furthermore, the effects of HULC on tumorigenicity of osteosarcoma cells were evaluated by in vitro assays. Results: In the present study, we demonstrated that HULC was significantly up-regulated in osteosarcoma tissues and cell lines compared with normal controls, and over-expression of HULC was correlated with clinical stage and distant metastasis. Moreover, higher HULC expression was associated with shorter overall survival of osteosarcoma patients. Furthermore, decreased expression of HULC markedly suppressed osteosarcoma cell proliferation, migration, and invasion. Conclusions: Our results indicated that HULC is a novel molecule involved in osteosarcoma progression, which may provide a new marker of poor prognosis and a potential therapeutic target for osteosarcoma intervention.
Keywords: Osteosarcoma, HULC, long non-coding RNAs, overall survival
Introduction
Osteosarcoma is the most common type of primary sarcoma of the bone and a leading cause of cancer death in adolescents and young adults [1]. Despite the rapid development in therapeutic strategies, such as wide tumor excision, adjuvant chemotherapy and radiotherapy, the cure rate of patients of osteosarcoma is still very low [2,3]. Although recent advances in molecular biology have provided some clues to the molecular pathogenesis of osteosarcoma, the exact molecular mechanisms underlying the histological heterogeneity, drug resistance, and development of metastasis remain unclear [4]. Therefore, it is urgent to develop novel targets for the diagnosis, treatment, and prognosis of osteosarcoma.
Recently, high-throughput transcriptome analysis has revealed that 98% of the human genome can be transcripted into non-coding RNA [5]. Among them, the long non-coding RNA (lncRNA) is an RNA molecular that is longer than 200 nucleotides and cannot be translated into a protein [6]. LncRNAs have been implicated in a large number of cellular processes, such as cell proliferation, cell cycle progression, cell growth and cell apoptosis [7,8]. The deregulation of lncRNAs could play an important role in many types of cancer. For example, Gupta et al showed that lncRNA HOTAIR was over-expressed in breast tumors and could promote metastasis by the interaction with the PRC2 (Polycomb Repressive Complex 2) [9]. Zhang et al indicated that lncRNA ANRIL up-regulated in human gastric cancer tissues and correlated with a higher TNM stage and tumor size. They suggested that ANRIL expression served as an independent predictor for overall survival of gastric cancer patients [10]. Shi et al reported that GAS5 down-regulation was involved in non-small-cell lung carcinoma tumorigenesis and progression, GAS5 over-expression could dramatically induce apoptosis and growth arrest in vitro and reduce tumor growth in vivo, In addition, although they found that p53 and E2F1 are key downstream mediators of GAS5 [11]. However, the role of lncRNAs in osteosarcoma tumor development has only recently been investigated and remains largely unknown.
In the present study, we investigated the role of lncRNA HULC in human osteosarcoma. First, we investigated the expression of HULC in human osteosarcoma tissues and cell lines. Second, HULC correlation with clinicopathologic features and prognosis was analyzed. Finally, cell proliferation, migration and invasion following down-regulation of HULC in osteosarcoma cells were explored.
Materials and methods
Patients and tissue specimens
Osteosarcoma tissues and adjacent non-tumor tissues were obtained between 2004 and 2007 from 78 osteosarcoma patients undergoing surgery at department of orthopedic, The First Affiliated Hospital of Xinxiang Medical University. Tissue samples were cut into two parts, one was fixed with 10% formalin for histopathological diagnosis, and the other was immediately snap-frozen in liquid nitrogen, and stored in liquid nitrogen until RNA extraction. None of the patients received radiotherapy or chemotherapy before surgery. The use of the tissue samples for all experiments was approved by all the patients and by Ethics Committee of The First Affiliated Hospital of Xinxiang Medical University. The characteristics of patients are described in Table 1.
Table 1.
Association of lncRNA HULC expression with clinicopathologic features in osteosarcoma patients
| Parameter | Total | lncRNA HULC | P | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age (years) | 0.352 | |||
| <25 | 48 | 26 | 22 | |
| ≥25 | 30 | 13 | 17 | |
| Gender | 0.492 | |||
| Female | 33 | 18 | 15 | |
| Male | 45 | 21 | 24 | |
| Tumor size | 0.496 | |||
| <8 cm | 37 | 20 | 17 | |
| ≥8 cm | 41 | 19 | 22 | |
| Location | 0.624 | |||
| Tibia/Femur | 54 | 28 | 26 | |
| Elsewhere | 24 | 11 | 13 | |
| Clinical stage | 0.003 | |||
| IIA | 35 | 24 | 11 | |
| IIB/III | 43 | 15 | 28 | |
| Distant metastasis | 0.005 | |||
| Absent | 57 | 34 | 23 | |
| Present | 21 | 5 | 16 | |
Cell culture and transfection
Human MG-63, U2OS and SAOS-2 osteosarcoma cell lines, and human normal bone cell line hFOB were purchased from the American Type Culture Collection (ATCC, USA). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM)/high glucose supplemented with 10% fetal bovine serum (FBS, Gibco). And cells were incubated in a humidified incubator at 37°C with 5% CO2.
U2OS were transfected with siRNA oligonucleotides using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. The nucleotide sequences of siRNA for HULC (si-HULC) were AACCTCCAGAACTGTGATCCA [12], Negative control siRNA (si-NC) were also purchased from Invitrogen. After transfection, cells were harvested for qRT-PCR analyses.
Cell proliferation assays
Cell proliferation was analyzed by using cell counting assay Kit-8 (CCK-8) (DOJINDO) according to the manufacture’s protocol. Cells were incubated in 10% CCK-8 diluted in normal culture media at 37°C until the visual color conversion occurred. Proliferation rates were determined at 0, 24, 48, 72 and 96 hours after transfection. The absorbance of each well was measured with a microplate reader set at 450 nM.
Cell migration and invasion assay
U2OS cells were grown to confluence in 12-well plastic dishes and were treated with si-HULC or si-NC. Then, 24 hours after transfection, linear scratch wounds (in triplicate) were created on the confluent cell monolayers using a 200 μL pipette tip. To remove cells from the cell cycle prior to wounding, cells were maintained in serum free media. To visualize migrated cells and wound healing, images were taken at 0 and 48 hour.
For these invasion assays, 24 hours after transfection, 1×105 cells in serum free media were seeded in transwell migration chambers (8 μm pore size, Millipore). The upper chamber of these transwell inserts was coated with Matrigel (Sigma). Medium containing 20% FBS was added to the lower chamber. After 24 hours, the non-invading cells were removed with cotton wool. Invasive cells located on the lower surface of the chamber were stained with May-Grunwald-Giemsa stain (Sigma) and then were counted using a microscope (Olympus).
RNA extraction and qRT-PCR analyses
Total RNA was isolated from tissues or cells using TRIzol reagent (Invitrogen), and cDNA was synthesized with PrimeScript reverse transcriptase (TaKaRa) and oligo (dT) following the manufacturer’s instructions. Real-time PCR was performed using SYBR Premix Ex TaqTM II kit (TaKaRa). The conditions of real-time PCR were as follows: 94°C for 10 s, 94°C for 5 s, 52°C for 30 s to anneal, 72°C for 15 s followed by 40 cycles. We used specific primers for HULC (forward primer, 5’-ATCTGCAAGCCAGGAAGAGTC-3’, and reverse primer, 5’-CTTGCTTGATGCTTTGGTCTGT-3’). As a control, GAPDH was amplified with specific primers (forward primer, 5’-CATCACCATCTTCCAGGAGCG-3’, and reverse primer, 5’-TGACCTTGCCCACAGCCTT-3’).
Statistical analysis
All statistical analysis was performed using SPSS version 18 software. The chi-square and t tests were performed to explore the associations between the HULC expression level and the clinical features. Overall survival was estimated by using Kaplan-Meier method, and univariate analysis was conducted by log-rank test. The Cox proportional hazards model was used in the multivariate analysis. P value less than 0.05 was considered to be statistically significant.
Results
HULC expression is up-regulated in osteosarcoma tissues and cell lines
To evaluate the expression of lncRNA HULC in osteosarcoma, qRT-PCR was performed. Our results showed that HULC expression in human osteosarcoma tissues was significantly higher than in adjacent non-tumor tissues (Figure 1A, P < 0.05). In addition, HULC expression was significantly increased in three osteosarcoma cell lines (MG-63, U2OS and SAOS-2) compared with hFOB cell line (Figure 1B, P < 0.05). The U2OS cell line exhibited the highest HULC expression and was thus chosen for the subsequent in vitro experiments.
Figure 1.
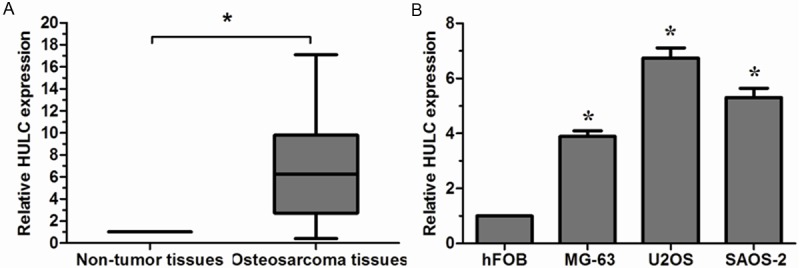
Relative HULC expression in osteosarcoma tissues and cell lines. A. Relative expression of HULC in osteosarcoma tissues in comparison with adjacent non-tumor tissues; B. Relative expression of HULC in osteosarcoma cell lines in comparison with human normal bone cell line hFOB. HULC expression was examined by qRT-PCR and normalized to GAPDH expression. *P < 0.05.
Association of HULC expression with clinicopathologic features of osteosarcoma patients
The 78 patients with osteosarcoma were classified into two groups according to the median expression level of lncRNA HULC. Of the 78 patients with osteosarcoma, 39 were placed in the low HULC expression group and 39 were placed in the high HULC expression group. The associations between clinicopathologic features and HULC expression were summarized in Table 1. High HULC expression level was correlated with clinical stage and distant metastasis (P < 0.05, Table 1). However, high HULC expression was not associated with other clinicopathologic features of osteosarcoma patients, including age, gender, tumor size, as well as tumor location (P > 0.05, Table 1).
High HULC expression levels correlate with osteosarcoma patients’ poor prognosis
The prognostic value of HULC expression was investigated using the Kaplan-Meier method and log-rank test. As shown in Figure 2, there was a significant correlation between HULC expression and overall survival of osteosarcoma patients (P < 0.05, log-rank test). Overall survival rate of osteosarcoma patients with high HULC expression was significantly lower than that of those patients with low HULC expression. Univariate Cox proportional hazards regressions model analysis demonstrated that clinical stage, distant metastasis and HULC expression were statistically significant risk factors affecting the overall survival of osteosarcoma patients (P < 0.05, Table 2). No significant associations were found for age, gender, tumor size, tumor location and patient outcome (P > 0.05, Table 2). Multivariate analysis using the Cox proportional hazard model for all variables that were significant in the univariate analysis confirmed that clinical stage, distant metastasis and HULC expression (P < 0.05) were independent prognostic factors for patients with osteosarcoma (P < 0.05, Table 2). These findings indicated that HULC play key roles in the development and progression of osteosarcoma.
Figure 2.
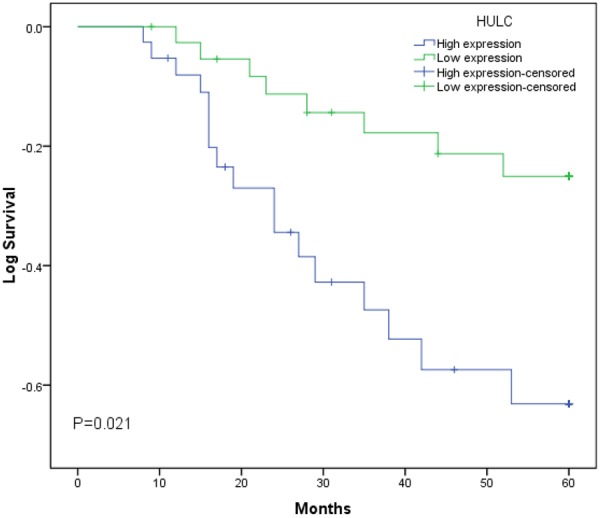
Kaplan-Meier survival curves of patients with osteosarcoma based on HULC expression levels. Patients in the high expression group had significantly poorer prognosis than those in low expression group (P < 0.05, log-rank test).
Table 2.
Univariate and multivariate Cox regression analyses lncRNA HULC for overall survival of osteosarcoma patients
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Risk ratio | 95% CI | P | Risk ratio | 95% CI | P | |
| Age (years) | 0.894 | 0.532-1.287 | 0.413 | |||
| ≥25 vs. <20 | ||||||
| Gender | 1.182 | 0.749-1.523 | 0.296 | |||
| Male vs. Female | ||||||
| Tumor size | 1.371 | 0.668-2.275 | 0.209 | |||
| ≥8 cm vs. <8 cm | ||||||
| Location | 0.942 | 0.687-1.628 | 0.593 | |||
| Tibia/Femur vs. Elsewhere | ||||||
| Clinical stage | 3.176 | 2.175-7.256 | 0.008 | 2.916 | 1.928-6.462 | 0.011 |
| IIB/III vs. IIA | ||||||
| Distant metastasis | 4.703 | 2.881-9.312 | 0.003 | 3.814 | 2.506-8.115 | 0.007 |
| Present vs. Absent | ||||||
| HULC | 2.587 | 1.613-6.058 | 0.004 | 2.277 | 1.481-5.427 | 0.009 |
| High vs. Low | ||||||
Suppressing HULC expression decreases cell proliferation, migration and invasion
To explore the role of HULC in development of osteosarcoma, U2OS cells were transfected with si-HULC or si-NC with high transfection efficiency (Figure 3A, P < 0.05). As shown in Figure 3B, down-regulated expression of HULC resulted in a significantly decreased proliferation of U2OS cells compared to the respective si-NC group (P < 0.05). Furthermore, the effects of HULC lncRNA on the migratory ability and invasiveness of osteosarcoma cells were checked by wound healing assay and transwell invasion assay. As shown in Figure 3C, the migration ability of U2OS cells was significantly reduced after decreased expression of HULC (P < 0.05). In addition, the invasion capacity of U2OS cells transfected with the si-HULC was significantly decreased compared to the respective si-NC group (P < 0.05, Figure 3D).
Figure 3.
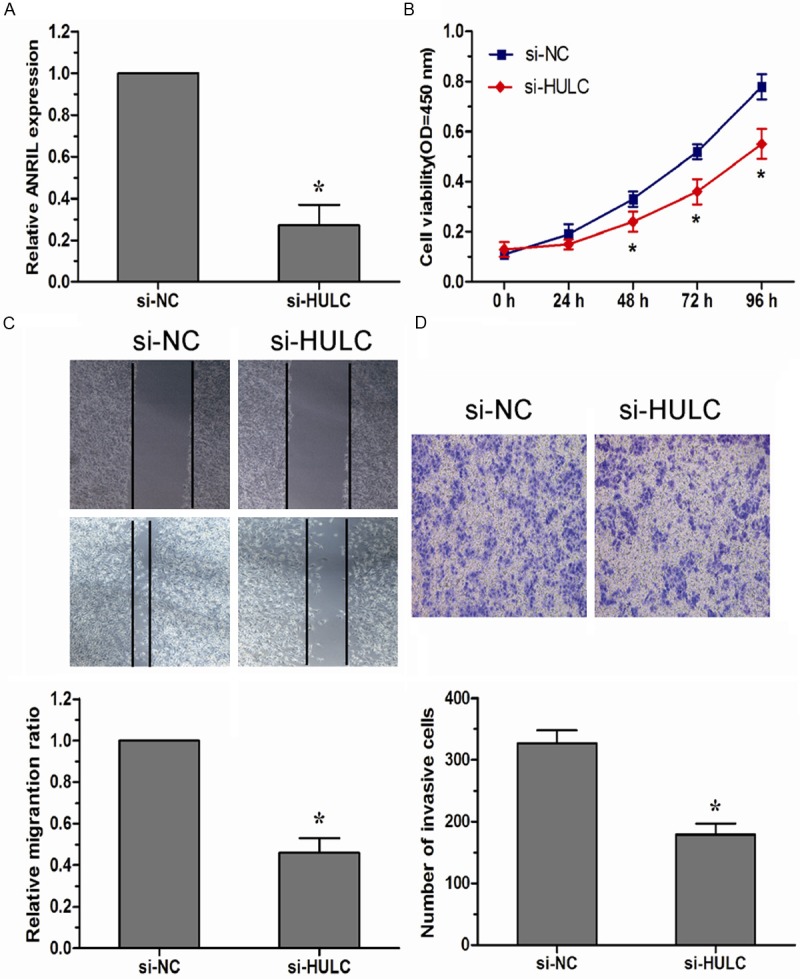
Down-regulated HULC inhibited cell proliferation, migration, and invasion of osteosarcoma cells. A. HULC expression levels were evaluated using qRT-PCR in si-HULC transfected U2OS cells. B. CCK-8 assay was performed to determine the proliferation of U2OS cells transfected with si-HULC. C. Wound healing assay was performed in U2OS cells transfected with si-HULC to investigate changes in cell migration. D. Transwell invasion assay was performed in U2OS cells transfected with si-HULC to investigate changes in cell invasion. Data were presented as mean ± SD from three in dependent experiments. *P < 0.05.
Discussion
LncRNAs dysregulation may affect epigenetic information and provide a cellular growth advantage, resulting in progressive and uncontrolled tumor growth [13]. Effective control of both cell growth and cell invasion is critical to the prevention of oncogenesis and to successful cancer therapy [14]. Therefore, identification of cancer associated lncRNAs and investigation of their clinical significance and functions may provide a missing piece of the well-known oncogenic and tumor suppressor network puzzle.
The lncRNA HULC (highly up-regulated in liver cancer) is one of the first strongly over-expressed non-coding transcripts to be identified in human hepatocellular carcinoma (HCC) [15]. The HULC gene is located on chromosome 6p24.3 and is conserved in primates. Transcription of HULC yields an 500 nucleotides long, spliced and polyadenylated non-coding RNA that localizes to the cytoplasm where it has been reported to associate with ribosomes [15]. Wang et al found that HULC was over-expressed in HCC and correlated with tumor stage and tumor grade [16]. Du et al showed that increased HULC levels in HCC cells lead to a higher proliferation rate and tumor growth and induce a down-regulation of the tumor suppressor p18 at a post-transcriptional level [17]. Zhao et al indicated that HULC was increased in gastric cancer, and this over-expression was correlated with advanced clinical features, In vitro assay, over-expression of HULC promoted proliferation and invasion and inhibited cell apoptosis in gastric cancer cells [18]. Peng et al revealed that HULC was up-regulated in human pancreatic cancer tissues and can be considered an independent prognostic factor. Moreover, HULC knockdown could inhibit cell proliferation in vitro [19]. These results suggested that HULC play an essential role in osteosarcoma and its dysregulation may participate in the tumorigenesis. However, the role and function of HULC in osteosarcoma remains unknown.
In the present study, we investigated the clinical significance of lncRNA HULC in osteosarcoma. Our results showed that HULC was increased in osteosarcoma tissues and cell lines. We also proved that the relative expression level of HULC was associated with clinical stage and distance metastasis of osteosarcoma patients. Moreover, HULC high expression was correlated with lower overall survival and could be an independent prognostic factor in patients with osteosarcoma. These findings indicated that HULC could play an important role in development and progression of osteosarcoma and could be used as a candidate prognostic biomarker for osteosarcoma. To explore the possible biological potential of HULC in osteosarcoma progression, we transfected si-HULC into U2OS cells and found that si-HULC markedly inhibit the proliferation, migration and invasion of U2OS cells. It is therefore evident that down-regulated expression of HULC could repress the growth and metastasis of osteosarcoma cells.
In summary, our results suggested that lncRNA HULC expression was up-regulated in osteosarcoma and was associated with the biological aggressiveness and progression of osteosarcoma. Moreover, down-regulated expression of HULC could inhibit the proliferation, migration and invasion of osteosarcoma cells in vitro. These findings demonstrated that HULC could serve as a potential biomarker and therapeutic target for osteosarcoma.
Disclosure of conflict of interest
None.
References
- 1.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayden JB, Hoang BH. Osteosarcoma: basic science and clinical implications. Orthop Clin North Am. 2006;37:1–7. doi: 10.1016/j.ocl.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Marina N, Gebhardt M, Teot L, Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist. 2004;9:422–441. doi: 10.1634/theoncologist.9-4-422. [DOI] [PubMed] [Google Scholar]
- 4.Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- 5.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 6.Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902–906. doi: 10.1042/BST20120020. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Reviews Genetics. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen J. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, Song Y. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2013 doi: 10.1002/mc.22120. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Hammerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, Skawran B, Geffers R, Longerich T, Breuhahn K, Schirmacher P, Stoecklin G, Diederichs S. Posttran scriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1) Hepatology. 2013;58:1703–1712. doi: 10.1002/hep.26537. [DOI] [PubMed] [Google Scholar]
- 13.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 15.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 19.Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31:346. doi: 10.1007/s12032-014-0346-4. [DOI] [PubMed] [Google Scholar]


