Abstract
This study aimed to identify the key pathways and to explore the mechanism of sorafenib in inhibiting hepatocellular carcinoma (HCC). The gene expression profile of GSE33621, including 6 sorafenib treated group and 6 control samples, was downloaded from the GEO (Gene Expression Omnibus) database. The differentially expressed genes (DEGs) in HCC samples were screened using the ΔΔCt method with the homogenized internal GAPDH. Also, the functions and pathways of DEGs were analyzed using the DAVID. Moreover, the significant pathways of DEGs that involved in HCC were analyzed based on the Latent pathway identification analysis (LPIA). A total of 44 down-regulated DEGs were selected in HCC samples. Also, there were 84 biological pathways that these 44 DEGs involved in. Also, LPIA showed that Osteoclast differentiation and hsa04664-Fc epsilon RI signaling pathway was the most significant interaction pathways. Moreover, Apoptosis, Toll-like receptor signaling pathway, Chagas disease, and T cell receptor signaling pathway were the significant pathways that interacted with hsa04664. In addition, DEGs such as AKT1 (v-akt murine thymoma viral oncogene homolog 1), TNF (tumor necrosis factor), SYK (spleen tyrosine kinase), and PIK3R1 (phosphoinositide-3-kinase, regulatory subunit 1 (alpha)) were the common genes that involved in the significant pathways. Several pathway interaction pairs that caused by several downregulated genes such as SYK, PI3K, AKT1, and TNF, were identified play curial role in sorafenib treated HCC. Sorafenib played important inhibition roles in HCC by affecting a complicate pathway interaction network.
Keywords: Hepatocellular carcinoma, differentially expressed genes, pathway interaction, sorafenib, latent pathway identification analysis
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignancies worldwide that characterized by powerful invasion ability, easy to metastasis and poor prognosis [1]. About 60,000 million cases will diagnose as HCC very year, in which the majority cases are from Chinese people [2]. Treatment methods such as surgery and liver transplantation are benefit to the HCC patients in early stage with the 5-year survival rate of 60%-70% [3]. However, there were no useful treatment methods on HCC patients in later stage due to the complicate mechanism of HCC metastasis and invasion [4]. Therefore, exploring several therapeutic targets for HCC will drive to improve the understanding of HCC metastasis mechanism.
Previous study revealed that the signaling transduction system played crucial roles in HCC development [5]. It has been demonstrated that there were four molecular pathways that driving crucial roles in HCC metastasis and invasion. For instance, overexpression of Ras in Ras-MAPKK (Ras-mitogen-activated protein kinase) pathway down-regulates the expression of tumor suppressor Sprouty and Spred-1 in HCC [6]. Aberrant activation of PI3K/Akt/mTOR (phosphatidylinositol 3-kinase/protein kinase B/mammalian target of rapamycin) pathway is associated with HCC progression [7] and mutation of PI3K contributes to the Akt hyperactivation that leading to a poor HCC prognosis [8]. The activated Wnt/β-catenin pathway results in the β-catenin phosphorylation and inhibition of β-catenin degradation, and results in the combination between β-catenin and TCF (transcription factor) in cells, so as to stimulate the transcription of downstream target genes in HCC [9]. Xie et al. reported that the genetic polymorphisms of several key molecules in JAK/STAT (Janus kinase/signal transducers and activators of transcription) signaling pathway is associated with HCC susceptibility, such as IL-6, STAT3 and mTOR [10].
Sorafenib is a cancer treated drug that is useful for many cancers, such as thyroid cancer [11] and non-small cell lung cancer [12]. It has been reported that sorafenib is the first peroral multi-kinase inhibitor that functioning as a molecular target drug for HCC treatment in recent years [13]. For example, Liu et al. proved that sorafenib inhibited the cell proliferation and induced the cell apoptosis of HCC via inhibiting the activates of Raf-1, β-Raf kinase and tyrosine kinase receptor to block the Raf/MEK/ERK signaling pathway and VEGF signaling pathway [14]. Also, Gedaly et al. reported that sorafenib inhibits the HCC cell proliferation via blocking Ras/MAPK and PI3K/Akt/mTOR pathways [15]. Although many studies have reported the useful treatment of sorafenib in HCC. However, the mechanism of sorafenib in inhibiting HCC metastasis and invasion remains largely unknown due to the complicate signal transduction system in HCC.
Using the gene expression profile of GSE33621 [16], Heindryckx et al. proved that inhibition of placental growth factor would be benefit for the therapeutic strategy of HCC metastasis and invasion [17]. In this study, we used microarray analysis to screen the differentially expressed genes (DEGs) in the Sorafenib treated HCC samples. Comprehensive bioinformatics analysis was used to identify the significant pathways that involved in HCC. Our study aimed to identify the significant pathways in HCC metastasis and invasion and explain the role of sorafenib in HCC treatment.
Methods
Microarray data and data preprocessing
The gene expression profile of GSE33621 [16] was downloaded from the GEO (Gene Expression Omnibus) database in NCBI (http://www.ncbi.nlm.nih.gov/geo/) which is the biggest completely public storage, based on the platform of GPL1126 SuperArray GEArray Q series Human Cancer PathwayFinder Gene Array. The platform includes a total of 96 genes that involved in 6 cancer related pathways. The study contains 12 samples which are examined with 6 from sorafenib treated group and 6 from control group.
The single or Multi-Gene qPCR assays in RT2 Profiler PCR Array Data Analysis V3.5 online software [18] was used to preprocess the CEL files.
DEG screening
GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was chosen as the homogenized internal gene. ΔΔCt method [19] was used to screen the DEGs from the normalized profile data with P-value < 0.05 and Fold change = 2-ΔΔCt, in which ΔCt stands for the expression value of normalized GAPDH while ΔΔCt stands for the case group expression value minus control group expression value.
Latent pathway identification analysis (LPIA)
The LPIA, developed by Pham et al. [20], was a method for identification the interactions of pathways associated with DEGs. A significant interaction represented a strong correlation between pathways and disease. The process of LPIA showed as follows:
Step 1: the GO BP (biological process) terms (named G) and KEGG pathways (named P) of DEGs were identified using the clusterProfiler [21] in R; Step 2: a bipartite network was constructed between G and P, one edge of the node was G and the other edge of node was P, edge represents one gene participated in both G and P, the weight of edge was determined by two factors, (1) the relative overlap of G and P was calculated using the Jaccard, (2) mean expression value stand for the expression value of each DEG. The weight formula was shown as follows (Equation 1):
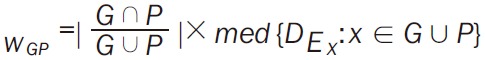 |
Whereaze, |G ∩ P/G ∪ P| stands for the Jaccard similarity coefficient of G and P, DE represents the expression value of DEG. G ∪ P stands for the total DEGs associated with G and P; Step 3: based on the bipartie network, pathways that connected with at least one BP term were chosen to construct the pathway network. The weight formula of edge was Equation 2; Step 4: random walk method [22] was used to calculate the interactive significance of each pathway pair, and the significant interactions was selected. The transfer matrix of random walk method was Equation 3. Whereas Np stands for the total pathways in network, Tij stands for the probability of one pathway from Pi to Pj.
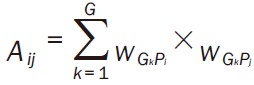 |
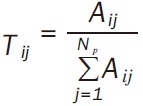 |
Then samples were repeated using the bootstrap method [23] from step 1 to step 4, and then the significant p-value of pathway interaction was obtained.
Results
Data preprocessing and DEGs screening
The Ct value of DEGs was shown in Figure 1. A total of 44 down-regulated DEGs were selected using the ΔΔCt method with P-value < 0.05 (Table 1). There were 84 biological pathways that these 44 DEGs involved in, such as AKT1 (v-akt murine thymoma viral oncogene homolog 1), ERBB2 (v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)), FAS (Fas (TNF receptor superfamily, member 6)), SYK (spleen tyrosine kinase), TNF (tumor necrosis factor), TP53 (tumor protein p53), and PIK3R1 (phosphoinositide-3-kinase, regulatory subunit 1 (alpha)).
Figure 1.
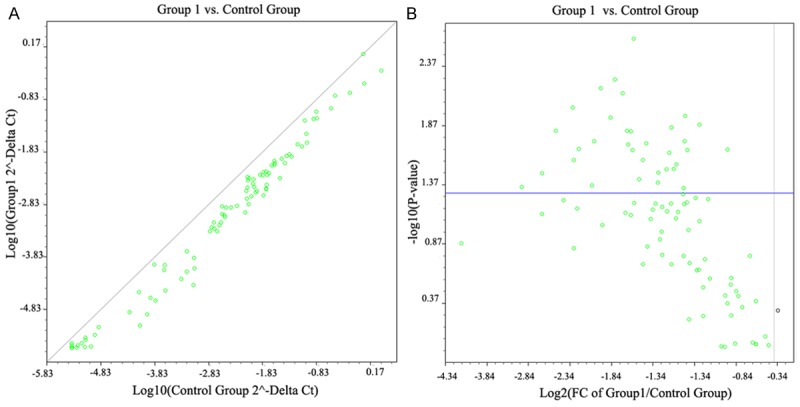
The scatter diagram and volcanic figure of Ct values in case and control group. A: The left figure stands for the Ct of log 10 P-value; B: The right figure stands for the Ct of log 2 fold change.
Table 1.
Information of differentially expressed genes
| Gene Symbol | Description | Fold Change | P-value |
|---|---|---|---|
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 | 0.147624 | 0.02642 |
| APAF1 | apoptotic peptidase activating factor 1 | 0.297302 | 0.005349 |
| BAX | BCL2-associated X protein | 0.386891 | 0.047011 |
| BCL2L1 | BCL2-like 1 | 0.366021 | 0.048627 |
| CCNE1 | cyclin E1 | 0.34151 | 0.036517 |
| CDC25A | cell division cycle 25 homolog A (S. pombe) | 0.251739 | 0.010465 |
| CDK2 | cyclin-dependent kinase 2 | 0.371131 | 0.048012 |
| CDKN2A | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 0.266093 | 0.026237 |
| CFLAR | CASP8 and FADD-like apoptosis regulator | 0.267943 | 0.023552 |
| CHEK2 | protein kinase CHK2-like; CHK2 checkpoint homolog (S. pombe); similar to hCG1983233 | 0.283221 | 0.016826 |
| ERBB2 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | 0.0960547 | 0.026726 |
| ETS2 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | 0.275476 | 0.01593 |
| FAS | Fas (TNF receptor superfamily, member 6) | 0.0960547 | 0.016608 |
| HTATIP2 | Short Chain Dehydrogenase/Reductase Family | 0.147624 | 0.002546 |
| ITGA1 | integrin, alpha 1 | 0.408951 | 0.016446 |
| ITGA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 0.31864 | 0.02997 |
| ITGA3 | integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | 0.34151 | 0.020447 |
| ITGB1 | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | 0.178006 | 0.007464 |
| ITGB3 | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | 0.41466 | 0.00515 |
| ITGB5 | integrin, beta 5 | 0.291183 | 0.027195 |
| JUN | jun oncogene | 0.356013 | 0.026579 |
| MAP2K1 | mitogen-activated protein kinase kinase 1 | 0.251739 | 0.029829 |
| MDM2 | Mdm2 p53 binding protein homolog (mouse) | 0.363493 | 0.021195 |
| MET | met proto-oncogene (hepatocyte growth factor receptor) | 0.332171 | 0.006027 |
| MMP1 | matrix metallopeptidase 1 (interstitial collagenase) | 0.246558 | 0.025416 |
| MMP2 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 0.125869 | 0.003765 |
| MTA2 | P53 Target Protein In Deacetylase | 0.432269 | 0.000882 |
| MTSS1 | Metastasis Suppressor Protein 1 | 0.373712 | 0.019675 |
| NME1 | non-metastatic cells 1, protein (NM23A) expressed in; NME1-NME2 readthrough transcript; non-metastatic cells 2, protein (NM23B) | 0.295248 | 0.042317 |
| PIK3R1 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | 0.222211 | 0.001802 |
| PLAU | plasminogen activator, urokinase | 0.162668 | 0.012227 |
| PLAUR | plasminogen activator, urokinase receptor | 0.233258 | 0.006666 |
| PNN | SR-Like Protein | 0.173139 | 0.011982 |
| RB1 | retinoblastoma 1 | 0.0559391 | 0.002672 |
| S100A4 | Leukemia Multidrug Resistance | 0.307786 | 0.019805 |
| SERPINB5 | serpin peptidase inhibitor, clade B (ovalbumin), member 5 | 0.0595399 | 0.036155 |
| SYK | spleen tyrosine kinase | 0.0871715 | 0.006901 |
| TGFBR1 | transforming growth factor, beta receptor 1 | 0.303549 | 0.02148 |
| TNF | tumor necrosis factor (TNF superfamily, member 2) | 0.126745 | 0.017021 |
| TP53 | tumor protein p53 | 0.192109 | 0.002516 |
| EPDR1 | Ependymin Related Protein 1 | 0.295248 | 0.035598 |
| B2M | beta-2-microglobulin | 0.248273 | 0.000825 |
| HPRT1 | hypoxanthine phosphoribosyltransferase 1 | 0.417544 | 0.032709 |
| HGDC | (R)-2-hydroxyglutaryl-CoA dehydratase subunit alpha | 0.400535 | 0.045343 |
Latent pathway identification analysis (LPIA)
The interaction network of pathways associated with the selected 44 DEGs was shown in Figure 2. There were 2775 interaction pairs in this constructed interaction network. Besides, the significant interaction pairs of pathways with the top 10 weight were shown in Table 2. Interaction pair between hsa04380-Osteoclast differentiation and hsa04664-Fc epsilon RI signaling pathway was the most significant interaction pathway with weight = 10.4131309, which was caused by the down-regulation of SYK (Table 1). Also, there were 6 pathways that interacted with hsa04664 among the pathways with top 10 weights, such as hsa04210-Apoptosis, hsa04620-Toll-like receptor signaling pathway, hsa05142-chagas disease, and hsa04660-T cell receptor signaling pathway (Table 2).
Figure 2.
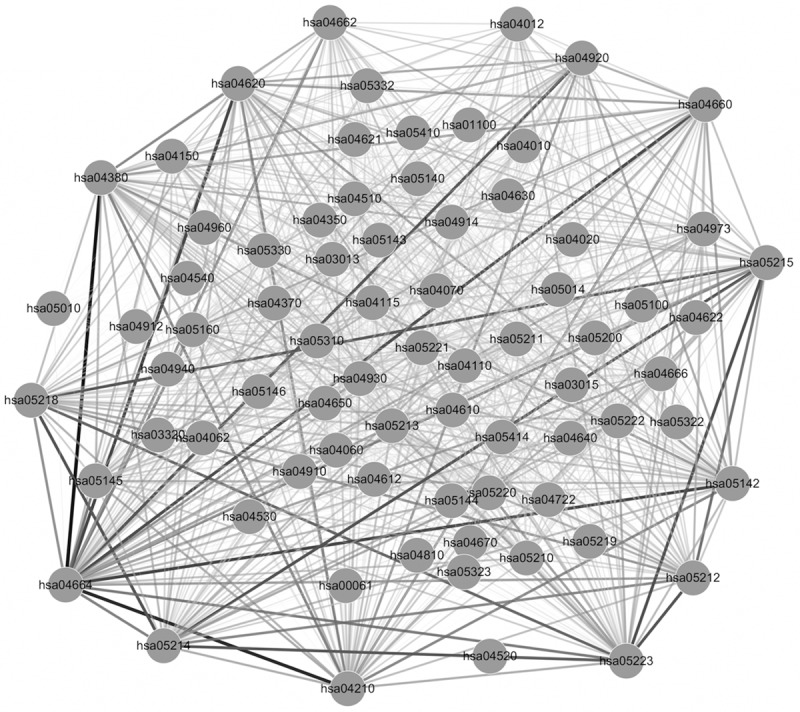
Interaction network of pathways. Edge stands for the interaction between two pathways, the thickness of edge stands for the size of interaction weight.
Table 2.
Pathway interaction pairs with the top 10 weight
| pathway | pathway | weight |
|---|---|---|
| hsa04380~Osteoclast differentiation | hsa04664~Fc epsilon RI signaling pathway | 10.4131309 |
| hsa04210~Apoptosis | hsa04664~Fc epsilon RI signaling pathway | 9.899374049 |
| hsa04620~Toll-like receptor signaling pathway | hsa04664~Fc epsilon RI signaling pathway | 9.151907933 |
| hsa05142~Chagas disease (American trypanosomiasis) | hsa04664~Fc epsilon RI signaling pathway | 9.090325321 |
| hsa04660~T cell receptor signaling pathway | hsa04664~Fc epsilon RI signaling pathway | 8.8833853 |
| hsa05215~Prostate cancer | hsa05223~Non-small cell lung cancer | 8.823397413 |
| hsa05215~Prostate cancer | hsa05214~Glioma | 8.698434482 |
| hsa05218~Melanoma | hsa05214~Glioma | 8.664112555 |
| hsa04920~Adipocytokine signaling pathway | hsa04664~Fc epsilon RI signaling pathway | 8.594524769 |
| hsa05223~Non-small cell lung cancer | hsa05212~Pancreatic cancer | 8.568905067 |
In addition, pathways were interacted via the DEGs involved in the relevant pathways. DEGs such as AKT1, TNF, SYK, and PIK3R1 were the genes involved in hsa04664-Fc epsilon RI signaling pathway, FAS, TNF, SYK, AKT1, TP53, and PIK3R1 were the genes involved in hsa04210-Apoptosis, PIK3R1, AKT1, and TNF were involved in hsa04660-T cell receptor signaling pathway, and TNF and AKT1 were involved in hsa04920-Adipocytokine signaling pathway (Table S1).
Discussion
Hepatocellular carcinoma (HCC) is the sixth most common malignancies worldwide that characterized by powerful invasion ability, easy to metastasis and poor prognosis [1]. The mechanism of sorafenib in inhibiting HCC metastasis and invasion has not been fully reported. In this study, we analyzed the significant pathways that involved in the sorafenib treated HCC to illustrate the mechanism of sorafenib in inhibiting HCC. Hsa04380-Osteoclast differentiation and hsa04664-Fc epsilon RI signaling pathway associated with the down-regulated SYK was the most significant interaction pathway pair. Additionally, hsa04210-Apoptosis, hsa04660-T cell receptor signaling pathway, and hsa04920-Adipocytokine signaling pathway, associated with the DEGs such as AKT1, TNF, and PIK3R1 were the important pathways in HCC.
SYK is a member of the family of non-receptor type Tyr protein kinases that widely expressed in hematopoietic cells and mediates the cellular responses including proliferation, differentiation and phagocytosis [24]. Yuan et al. proved that loss of SYK mRNA was highly correlated with SYK methylation and then contributed to the metastasis of HCC and resulted to the poor treatment [25]. Also, the precious study revealed that Fc epsilon RI signal mediated the tyrosine phosphorylation of SYK in rat tumor mast cells [26], and SYK was a critical factor in immune receptor signaling [27]. Our data showed that SYK involved in Fc epsilon RI signaling pathway was downregulated in sorafenib treated HCC, we speculated that the downregulated SYK enhanced the interaction activity of Fc epsilon RI signaling pathway with other pathways. In addition, Zou et al. reported that osteoclasts with mutation of tyrosine kinase SYK failed to organize the cytoskeleton, suggested its essential role for osteoclast function [28]. Osteoclast differentiation factor is involved in the bone metastasis of cancer [29]. Therefore, SYK may involve in osteoclast differentiation. Also, Ikeda et al. firstly reported that hepatocyte-derived cells from HCC cells had the potential for osteoclastogenesis [30]. In this study, Fc epsilon RI signaling was interacted with osteoclast differentiation pathway, suggesting the important inhibition role of sorafenib in HCC metastasis by downregulating SYK and then affecting the two pathways.
AKT1 is one of 3 closely related serine/threonine-protein kinases of AKT kinase that regulate many processed including metabolism, proliferation, cell survival, and angiogenesis [31], while PIK3R1 is a member of PI3-kinases family of lipid kinases capable of phosphorylating the 3’-OH of phosphoinositides [32]. The downregulated SYK suppressed the Raf-1 expression in the downstream MAPK signaling pathway [33,34] which resulted in the activation of Ras-MAPKK signaling pathway and PI3K/AKT/mTOR pathway [35]. Also, the activated PI3K/AKT/mTOR pathway inhibited the cell growth and proliferation of HCC [36]. Besides, study revealed that the downregulated SYK induced the activation of PI3K [37], and the activated PI3K promoted the AKT/mTOR signaling pathway and NF-κB pathway in HCC [38]. The activated NF-κB weaken the cell proliferation of HCC from the study of Notarbartolo et al. [39]. PI3K negatively regulated the TGF-induced cell apoptosis in HCC [40]. Liu et al. proved that sorafenib induced HCC cell apoptosis via inhibiting the RAF/MEK/ERK signaling pathway [14]. Thus, the interacted two pathways may involve in HCC cell proliferation. In this study, the downregulated PI3K was involved in Apoptosis pathway that interacted with Fc epsilon RI signaling pathway in sorafenib treated HCC samples, implying the important inhibiting role of sorafenib in HCC via affecting the two interactive pathways.
Meanwhile, study reveals that TNF is an endogenous tumor promoter in HCC [41]. Ormandy et al. proved that T cells were gathered in blood of patients with HCC [42]. In this study, TNF that participate in T cell receptor signaling pathway was downregulated in sorafenib treated HCC, indicating that T cell receptor signaling pathway may involve in HCC. On the other hand, Adipose tissue secreted many factors such as leptin and adiponectin [43]. Leptin promotes the invasion and metastasis of HCC by enhancing the cell proliferation and mitosis, which is associated with the activation of PI3K/AKT pathway and ERK pathway [44]. Hence, adipocytokine signaling pathway may be crucial for HCC. Yanawaki et al. said that adipocytokine inhibited the TNF-induced vascular inflammation in human endothelial cells [45] and activated adipocytokine signaling pathway was involved in HCC cell invasion [46]. Based on our study, we speculated that sorafenib may inhibit the HCC metastasis by influencing the interacted Adipocytokine signaling pathway and Fc epsilon RI signaling pathway.
In conclusion, our study attempt to identify several interactive pathway pairs associated with sorafenib treated HCC. Sorafenib inhibited HCC progression via downregulating the SYK expression and then affecting the interaction pair of Fc epsilon RI signaling and osteoclast differentiation pathway while inducing HCC cell apoptosis by downregulating PI3K and then influencing apoptosis and Fc epsilon RI signaling pathway. This study may provide theoretic basis for the future exploration of drug target therapy in HCC. However, further experimental studies are still needed to confirm our predicted results.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Rahbari NN, Mehrabi A, Mollberg NM, Müller SA, Koch M, Büchler MW, Weitz J. Hepatocellular carcinoma: current management and perspectives for the future. Ann Surg. 2011;253:453–469. doi: 10.1097/SLA.0b013e31820d944f. [DOI] [PubMed] [Google Scholar]
- 2.Yang T, Zhang J, Lu JH, Yang GS, Wu MC, Yu WF. Risk factors influencing postoperative outcomes of major hepatic resection of hepatocellular carcinoma for patients with underlying liver diseases. World J Surg. 2011;35:2073–2082. doi: 10.1007/s00268-011-1161-0. [DOI] [PubMed] [Google Scholar]
- 3.Villanueva A, Llovet JM. Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011;140:1410–1426. doi: 10.1053/j.gastro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singal AG, Nehra M, Adams-Huet B, Yopp AC, Tiro JA, Marrero JA, Lok AS, Lee WM. Detection of hepatocellular carcinoma at advanced stages among patients in the halt-c trial: where did surveillance fail & quest. Am J Gastroenterol. 2013;108:425–432. doi: 10.1038/ajg.2012.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remouchamps C, Boutaffala L, Ganeff C, Dejardin E. Biology and signal transduction pathways of the Lymphotoxin-αβ/LTβR system. Cytokine Growth Factor Rev. 2011;22:301–310. doi: 10.1016/j.cytogfr.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Tarkowski B. Interplay between C-Raf kinase and Rassf1a putative tumour suppressor in liver. Cancer. 2011 uniwien. [Google Scholar]
- 7.Gedaly R, Angulo P, Hundley J, Daily MF, Chen C, Evers BM. PKI-587 and sorafenib targeting PI3K/AKT/mTOR and Ras/Raf/MAPK pathways synergistically inhibit HCC cell proliferation. J Surg Res. 2012;176:542–548. doi: 10.1016/j.jss.2011.10.045. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Q, Lui VW, Yeo W. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Future Oncol. 2011;7:1149–1167. doi: 10.2217/fon.11.95. [DOI] [PubMed] [Google Scholar]
- 9.Dahmani R, Just PA, Perret C. The Wnt/β-catenin pathway as a therapeutic target in human hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2011;35:709–713. doi: 10.1016/j.clinre.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Xie J, Yin J, Zhang Q, Pu R, Zhang Y, Lu W, Cao G. Association of genetic polymorphisms of key molecules in JAK/STAT signaling pathway with susceptibility of hepatocellular carcinoma. Zhonghua Liu Xing Bing Xue Za Zhi. 2012;33:215–219. [PubMed] [Google Scholar]
- 11.Gupta-Abramson V, Troxel AB, Nellore A, Puttaswamy K, Redlinger M, Ransone K, Mandel SJ, Flaherty KT, Loevner LA, O’dwyer PJ. Phase II trial of sorafenib in advanced thyroid cancer. J. Clin. Oncol. 2008;26:4714–4719. doi: 10.1200/JCO.2008.16.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scagliotti G, Novello S, Von Pawel J, Reck M, Pereira JR, Thomas M, Miziara JEA, Balint B, De Marinis F, Keller A. Phase III study of carboplatin and paclitaxel alone or with sorafenib in advanced non-small-cell lung cancer. J. Clin. Oncol. 2010;28:1835–1842. doi: 10.1200/JCO.2009.26.1321. [DOI] [PubMed] [Google Scholar]
- 13.Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, Schwartz B, Simantov R, Kelley S. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5:835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Cao Y, Chen C, Zhang X, Mcnabola A, Wilkie D, Wilhelm S, Lynch M, Carter C. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66:11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
- 15.Gedaly R, Angulo P, Hundley J, Daily MF, Chen C, Koch A, Evers BM. PI-103 and sorafenib inhibit hepatocellular carcinoma cell proliferation by blocking Ras/Raf/MAPK and PI3K/AKT/mTOR pathways. Anticancer Res. 2010;30:4951–4958. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Sun HC, Wang WQ, Zhang QB, Zhuang PY, Xiong YQ, Zhu XD, Xu HX, Kong LQ, Wu WZ. Sorafenib down-regulates expression of HTATIP2 to promote invasiveness and metastasis of orthotopic hepatocellular carcinoma tumors in mice. Gastroenterology. 2012;143:1641–1649. e1645. doi: 10.1053/j.gastro.2012.08.032. [DOI] [PubMed] [Google Scholar]
- 17.Heindryckx F, Coulon S, Terrie E, Casteleyn C, Stassen JM, Geerts A, Libbrecht L, Allemeersch J, Carmeliet P, Colle I. The placental growth factor as a target against hepatocellular carcinoma in a diethylnitrosamine-induced mouse model. J Hepatol. 2013;58:319–328. doi: 10.1016/j.jhep.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Gaj S, Eijssen L, Mensink R, Evelo C. Validating nutrient-related gene expression changes from microarrays using RT2 PCR-arrays. Genes Dev. 2008;3:153–157. doi: 10.1007/s12263-008-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Pham L, Christadore L, Schaus S, Kolaczyk ED. Network-based prediction for sources of transcriptional dysregulation using latent pathway identification analysis. Proc Natl Acad Sci U S A. 2011;108:13347–13352. doi: 10.1073/pnas.1100891108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jamali M, Ester M. TrustWalker: a random walk model for combining trust-based and item-based recommendation in Proceedings of the 15th ACM SIGKDD international conference on Knowledge discovery and data mining. ACM. 2009 [Google Scholar]
- 23.Kim JH. Boostrap Prediction Intervals and Bias-Corrected Forecasting. 2009 [Google Scholar]
- 24.Braselmann S, Taylor V, Zhao H, Wang S, Sylvain C, Baluom M, Qu K, Herlaar E, Lau A, Young C. R406, an orally available spleen tyrosine kinase inhibitor blocks fc receptor signaling and reduces immune complex-mediated inflammation. J Pharmacol Exp Ther. 2006;319:998–1008. doi: 10.1124/jpet.106.109058. [DOI] [PubMed] [Google Scholar]
- 25.Yuan Y, Wang J, Li J, Wang L, Li M, Yang Z, Zhang C, Le Dai J. Frequent epigenetic inactivation of spleen tyrosine kinase gene in human hepatocellular carcinoma. Clin Cancer Res. 2006;12:6687–6695. doi: 10.1158/1078-0432.CCR-06-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchcroft JE, Geahlen RL, Deanin GG, Oliver JM. Fc epsilon RI-mediated tyrosine phosphorylation and activation of the 72-kDa protein-tyrosine kinase, PTK72, in RBL-2H3 rat tumor mast cells. Proc Natl Acad Sci U S A. 1992;89:9107–9111. doi: 10.1073/pnas.89.19.9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner M, Schweighoffer E, Colucci F, Di Santo JP, Tybulewicz VL. Tyrosine kinase SYK: essential functions for immunoreceptor signalling. Immunol Today. 2000;21:148–154. doi: 10.1016/s0167-5699(99)01574-1. [DOI] [PubMed] [Google Scholar]
- 28.Zou W, Kitaura H, Reeve J, Long F, Tybulewicz VL, Shattil SJ, Ginsberg MH, Ross FP, Teitelbaum SL. Syk, c-Src, the αvβ3 integrin, and ITAM immunoreceptors, in concert, regulate osteoclastic bone resorption. J Cell Biol. 2007;176:877–888. doi: 10.1083/jcb.200611083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roodman GD. Biology of osteoclast activation in cancer. J. Clin. Oncol. 2001;19:3562–3571. doi: 10.1200/JCO.2001.19.15.3562. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda T, Seki S, Maki M, Noguchi N, Kawamura T, Arii S, Igari T, Koike M, Hirokawa K. Hepatocellular carcinoma with osteoclast-like giant cells: possibility of osteoclastogenesis by hepatocyte-derived cells. Pathol Int. 2003;53:450–456. doi: 10.1046/j.1440-1827.2003.01503.x. [DOI] [PubMed] [Google Scholar]
- 31.Whiteman EL, Cho H, Birnbaum MJ. Role of Akt/protein kinase B in metabolism. Trends Endocrinol Metab. 2002;13:444–451. doi: 10.1016/s1043-2760(02)00662-8. [DOI] [PubMed] [Google Scholar]
- 32.Jia S, Roberts TM, Zhao JJ. Should individual PI3 kinase isoforms be targeted in cancer? Curr Opin Cell Biol. 2009;21:199–208. doi: 10.1016/j.ceb.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gringhuis SI, Den Dunnen J, Litjens M, Van Der Vlist M, Wevers B, Bruijns SC, Geijtenbeek TB. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-κB activation through Raf-1 and Syk. Nat Immunol. 2009;10:203–213. doi: 10.1038/ni.1692. [DOI] [PubMed] [Google Scholar]
- 34.Zhang P, Wang YZ, Kagan E, Bonner JC. Peroxynitrite targets the epidermal growth factor receptor, Raf-1, and MEK independently to activate MAPK. J Biol Chem. 2000;275:22479–22486. doi: 10.1074/jbc.M910425199. [DOI] [PubMed] [Google Scholar]
- 35.Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J Neurosci. 2005;25:11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen JS, Wang Q, Fu XH, Huang XH, Chen XL, Cao LQ, Chen LZ, Tan HX, Li W, Bi J. Involvement of PI3K/PTEN/AKT/mTOR pathway in invasion and metastasis in hepatocellular carcinoma: Association with MMP-9. Hepatol Res. 2009;39:177–186. doi: 10.1111/j.1872-034X.2008.00449.x. [DOI] [PubMed] [Google Scholar]
- 37.Chiou WF, Don MJ, Liao JF, Wei BL. Psoralidin inhibits LPS-induced iNOS expression via repressing Syk-mediated activation of PI3K-IKK-IκB signaling pathways. Eur J Pharmacol. 2011;650:102–109. doi: 10.1016/j.ejphar.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Yap TA, Garrett MD, Walton MI, Raynaud F, De Bono JS, Workman P. Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol. 2008;8:393–412. doi: 10.1016/j.coph.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Notarbartolo M, Poma P, Perri D, Dusonchet L, Cervello M, D’alessandro N. Antitumor effects of curcumin, alone or in combination with cisplatin or doxorubicin, on human hepatic cancer cells. Analysis of their possible relationship to changes in NF-κB activation levels and in IAP gene expression. Cancer Lett. 2005;224:53–65. doi: 10.1016/j.canlet.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 40.Thorgeirsson SS, Teramoto T, Factor VM. Dysregulation of apoptosis in hepatocellular carcinoma. Semin Liver Dis. 1998;18:115–22. doi: 10.1055/s-2007-1007148. [DOI] [PubMed] [Google Scholar]
- 41.Akkız H, Bayram S, Bekar A, Zdil B, Akgöllü E, Sümbül AT, Demiryürek H, Doran F. G-308A TNF-α polymorphism is associated with an increased risk of hepatocellular carcinoma in the Turkish population: Case-control study. Cancer Epidemiol. 2009;33:261–264. doi: 10.1016/j.canep.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Ormandy LA, Hillemann T, Wedemeyer H, Manns MP, Greten TF, Korangy F. Increased populations of regulatory T cells in peripheral blood of patients with hepatocellular carcinoma. Cancer Res. 2005;65:2457–2464. doi: 10.1158/0008-5472.CAN-04-3232. [DOI] [PubMed] [Google Scholar]
- 43.Fantuzzi G. Adipose tissue, adipokines, and inflammation. J Allergy Clin Immunol. 2005;115:911–919. doi: 10.1016/j.jaci.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 44.Saxena NK, Sharma D, Ding X, Lin S, Marra F, Merlin D, Anania FA. Concomitant activation of the JAK/STAT, PI3K/AKT, and ERK signaling is involved in leptin-mediated promotion of invasion and migration of hepatocellular carcinoma cells. Cancer Res. 2007;67:2497–2507. doi: 10.1158/0008-5472.CAN-06-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamawaki H, Kuramoto J, Kameshima S, Usui T, Okada M, Hara Y. Omentin, a novel adipocytokine inhibits TNF-induced vascular inflammation in human endothelial cells. Biochem Biophys Res Commun. 2011;408:339–343. doi: 10.1016/j.bbrc.2011.04.039. [DOI] [PubMed] [Google Scholar]
- 46.Guo K, Liu Y, Zhou H, Dai Z, Zhang J, Sun R, Chen J, Sun Q, Lu W, Kang X. Involvement of protein kinase C β-extracellular signal-regulating kinase1/2/p38 mitogen-activated protein kinase-heat shock protein 27 activation in hepatocellular carcinoma cell motility and invasion. Cancer Sci. 2008;99:486–496. doi: 10.1111/j.1349-7006.2007.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


