Abstract
Background and purpose: MicroRNA-206 (miR-206) acts as a tumor suppressor in melanoma cell lines. However, its clinical significance remains unclear. The aim of this study was to detect the serum level of miR-206 in patients with melanoma and to determine the feasibility of using it as a noninvasive prognostic biomarker. Methods: Expression levels of miR-206 in serum samples from 60 patients with melanoma and 30 healthy controls were detected by real-time quantitative polymerase chain reaction (q-PCR). Results: Expression levels of miR-206 in serum samples from patients with melanoma were significantly lower than those in healthy controls (P < 0.001). In addition, low serum miR-206 level was more frequently observed in patients with two or more metastatic sites (P = 0.02). Its serum level was also significantly associated with the response to treatment (P = 0.01). Moreover, melanoma patients with low serum miR-206 levels had higher clinical stage than those with high serum miR-206 levels (P < 0.001). Furthermore, melanoma patients with low serum miR-206 level had a dramatically shorter 5-year overall and disease-free survival than those with high serum miR-206 level (both P = 0.001). Multivariate analysis also identified the serum miR-206 level as an independent marker for both 5-year overall and disease-free survivals (both P = 0.01) in patients with melanoma. Conclusions: Our results offer the convincing evidence that miR-206 may be implicated in aggressive progression of melanoma. More importantly, the serum level of miR-206 may be a noninvasive prognostic biomarker for the patients with melanoma.
Keywords: MicroRNA-206, melanoma, metastasis, prognosis
Introduction
Melanoma is the most aggressive skin cancer with an increasing incidence worldwide and poor prognosis in the metastatic stage despite recent advances in targeted therapeutic strategies [1]. There are several clinicopathological parameters, such as clinical stage and tumor thickness, used to stratify patients at the time of primary diagnosis. However, clinical stage alone cannot account for the heterogeneity in patient outcomes [2]. Although tumor thickness is the single most important predictor of survival in localized melanoma, the morphologically based staging system only partly explains the variability in the natural history of melanoma [3]. Thus, it is extremely necessary to identify new biomarkers that reflect the underlying melanoma biology. With recent advances in our understanding of melanoma tumorigenesis, growing evidences highlight the interest in the utility of molecular markers that are easily measurable at the time of diagnosis.
MicroRNAs (miRNAs), originally reported in the early 90s of the last century in experiments with Caenorhabditis elegans, are endogenous small non-coding RNAs with 18-23 nucleotides in length [4]. MiRNAs can modulate gene expression by repressing protein translation and inducing messenger RNA (mRNA) degradation via binding to the 3’-untranslated region (3’-UTR) of mRNA [5]. In humans, more than 1000 miRNAs have been identified to date, and they regulate the expression of a third of the human genome [6]. MiRNA alterations have been found to result from genetic mutations, chromosomal aberrations or epigenetic changes, and have been reported to contribute to various developmental defects and diseases. Accumulating studies on individual miRNAs or miRNA patterns have provided strong evidence that miRNAs might play an important role in the development of the malignant phenotype and have supported the role of dysregulated miRNAs as oncogenes or tumor suppressors [7,8]. Particularly, there have been a number of melanoma-associated miRNAs identified by recent studies. For example, let-7 a and b, miR-148, miR-155, miR-182, miR-200c, miR-211, miR-214, miR-221, miR-222, miR-23 a and b, have been reported as differentially expressed miRNAs in melanoma cell lines and clinical tissues compared to benign melanocytes and benign melanocytic lesions, respectively [9,10]; A prognostic signature of six miRNAs consisting of miRNAs miR-150, miR-342-3p, miR-455-3p, miR-145, miR-155, and miR-497 has been identified and overexpression of these miRNAs was shown to be associated with impro-ved long-term survival of metastatic melanoma patients [11]. These findings suggest that miRNA patterns might help to better understand the molecular mechanisms of melanoma development and progression.
Emerging evidence suggests miRNAs may be attractive candidate biomarkers for tumor diagnosis and prognosis for several reasons. First, miRNAs have the ability to classify tumor types more robustly than mRNAs, since they are upstream regulators simultaneously targeting a large number of mRNAs and multiple cancer pathways. Second, miRNAs can modulate classical oncogenes and tumor suppressors, which turn them into candidate tumor suppressors or oncogenes, respectively. Third, the involvement of miRNAs in tumorigenesis is exquisitely context dependent, as the same miRNA can be oncogenic or tumor suppressive in different tumor types. And finally, miRNAs can be extracted both from the initial biopsy specimen and body fluids, and quantified by standardized methods at any point during the patient’s clinical course because of their stability in archival formalin-fixed paraffin-embedded tissues and body fluids.
MiR-206 has been reported to act as a tumor suppressor and play an important role in melanoma tumorigenesis by regulating cell cycle in vitro [12]. However, its clinical implications in human melanoma have not been well defined. Since serum-based biomarkers are attractive because of the ease and minimal invasiveness of collecting multiple serum samples compared with repeat tissue sampling [13], we here focused on the prognostic value of serum miR-206 level in patients with melanoma.
Materials and methods
Patients and tissue samples
The current retrospective study was approved by the Research Ethics Committee of General Hospital of the Air Force & Tangdu Hospital of the Fourth Military Medical University, P. R. China. Written informed consent was obtained from all of the patients enrolled in the current study. All serum samples were handled and made anonymous according to the ethical and legal standards.
A total of 60 patients with melanoma were enrolled from the Department of Dermatology of General Hospital of the Air Force & Tangdu Hospital of the Fourth Military Medical University from 2006 to 2012. The clinicopathological parameters were assessed by two different pathologists who were both unaware of the clinical data, and were summarized in Table 1. All 60 patients were followed clinically for a median of 22 months (range, 1-70 months). In addition, 30 healthy volunteers were enrolled from the Medical center of General Hospital of the Air Force & Tangdu Hospital of the Fourth Military Medical University as a control group. Serum samples were collected from 60 patients with melanoma and 30 healthy controls at the time of primary diagnosis and before starting the chemoimmunotherapy. All serum samples were immediately processed and separated, then aliquoted and stored at -80°C prior to use.
Table 1.
Associations between serum miR-206 level and various clinicopathological parameters
| Clinicopathological features | No. of cases | miR-206 expression | P | |
|---|---|---|---|---|
|
| ||||
| High (n, %) | Low (n, %) | |||
| Age | ||||
| < 60 | 24 | 12 (50.00) | 12 (50.00) | NS |
| ≥ 60 | 36 | 17 (47.22) | 19 (52.78) | |
| Gender | ||||
| Male | 38 | 19 (50.00) | 19 (50.00) | NS |
| Female | 22 | 10 (45.45) | 12 (54.55) | |
| Thickness (mm) | ||||
| < 2.0 | 20 | 11 (55.00) | 9 (45.00) | NS |
| ≥ 2.0 | 40 | 18 (45.00) | 22 (55.00) | |
| Ulceration | ||||
| Negative | 18 | 9 (50.00) | 9 (50.00) | NS |
| Positive | 42 | 20 (47.62) | 22 (52.38) | |
| Differentiation | ||||
| Well | 32 | 19 (59.38) | 13 (40.62) | NS |
| Moderate/poor | 28 | 10 (35.71) | 18 (64.29) | |
| Number of metastatic sites | ||||
| None | 10 | 9 (90.00) | 1 (10.00) | 0.02 |
| One site | 15 | 11 (73.33) | 4 (26.67) | |
| Two or more sites | 35 | 9 (25.71) | 26 (74.29) | |
| Treatment response | ||||
| Complete response | 5 | 5 (100.00) | 0 (0) | 0.01 |
| Partial response | 8 | 6 (75.00) | 2 (25.00) | |
| Stable disease | 20 | 12 (60.00) | 8 (40.00) | |
| Progressive disease | 27 | 6 (22.22) | 21 (77.78) | |
| Clinical stage | ||||
| I/II | 20 | 20 (100.00) | 0 (0) | < 0.001 |
| III/IV | 40 | 9 (22.50) | 31 (77.50) | |
‘NS’ refers to the differences without statistical significance.
Real-time quantitative polymerase chain reaction (q-PCR)
MiRNAs were isolated from serum samples using miRNeasyTM RNA isolation kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. RNA quality and quantity were assessed using a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Synthesis of cDNA with reverse transcriptase was performed by NCodeTM miRNA quantitative RT-PCR Kits (Applied Biosystems, Foster City, CA, USA). For analysis of miRNA expression, real-time q-PCR analyses were carried out using SYBR Green Reagents (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. All real-time q-RT-PCR were performed on a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). RNA U6 was used as an internal control. The primer sequences used in this study were as follows: miR-206, reverse-transcribed primer 5’-GTCAGAAGGAATGATGCACAGCCAACAACA-3’, forward: 5’-CGTCAGAAGGAATGATGCACAG-3’, reverse: 5’-ACCTGCGTAGGTAGTTTCATGT-3’; U6, reverse transcribed primer: 5’-AACGCTTCACGAATTTGCGT-3’, forward: 5’-CTCGCTTCGGCAGCACA-3’, reverse: 5’-AACGCTTCACGAATTTGCGT-3’. Relative expression of miR-206 to U6 was calculated using the 2-ΔΔCt method [14].
Statistical analysis
Data analyses in the current study were all carried out using the software of SPSS version11.0 for Windows (SPSS Inc, IL, USA). For continuous variables, the data were expressed as means ± standard deviation (S.D.). The Mann-Whitney U test was used to evaluate the associations between serum miR-206 level and clinicopathological parameters. The overall survival (OS) and disease-free survival (DFS) were calculated using the Kaplan-Meier method. The differences between the survival curves were tested by using the log-rank test. The Cox proportional hazards regression model was used to determine the joint effects of several variables on survival. Differences were considered statistically significant when P was less than 0.05.
Results
Decreased serum miR-206 levels in patients with melanoma
Serum miR-206 levels in 60 patients with melanoma and 30 healthy controls were detected by q-PCR. Statistical analysis showed that serum miR-206 levels in patients with melanoma were significantly lower than those in healthy controls (melanoma vs. healthy: 2.05 ± 0.40 vs. 4.02 ± 0.63, P < 0.001, Figure 1). In the current study, melanoma patients with serum miR-206 levels less than the median value of 2.06 were assigned to the low expression Group (n = 31), whereas those with serum miR-206 levels more than the median value of 2.95 were assigned to the high expression group (n = 29).
Figure 1.
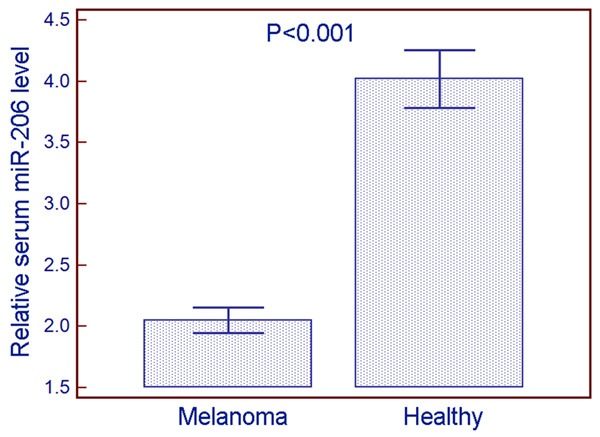
Relative serum miR-206 levels in patients with melanoma and healthy controls. Statistical analysis showed that serum miR-206 levels in patients with melanoma were significantly lower than those in healthy controls (melanoma vs. healthy: 2.05 ± 0.40 vs. 4.02 ± 0.63, P < 0.001).
Decreased serum miR-206 levels associate with aggressive progression of patients with melanoma
Table 1 summarized the associations between serum miR-206 levels with clinicopathological parameters of patients with melanoma. We found that low serum miR-206 level was more frequently observed in patients with two or more metastatic sites (P = 0.02, Table 1). Its serum level was also significantly associated with the response to treatment (P = 0.01, Table 1). Moreover, melanoma patients with low serum miR-206 levels had higher clinical stage than those with high serum miR-206 levels (P < 0.001, Table 1). However, no statistically significant associations of serum miR-206 level with age at diagnosis, gender, tumor thickness, Ulceration status, and differentiation of patients with melanoma were found (all P > 0.05, Table 1).
Decreased serum miR-206 levels predict poor prognosis in patients with melanoma
The Kaplan-Meier plot showed that patients with low serum miR-206 levels had a poorer overall survival than patients with high serum miR-206 levels (log rank, P = 0.001; Figure 2A). On the other hand, the decreased serum miR-206 level was significantly associated with disease-free survival (P = 0.001; Figure 2B).
Figure 2.
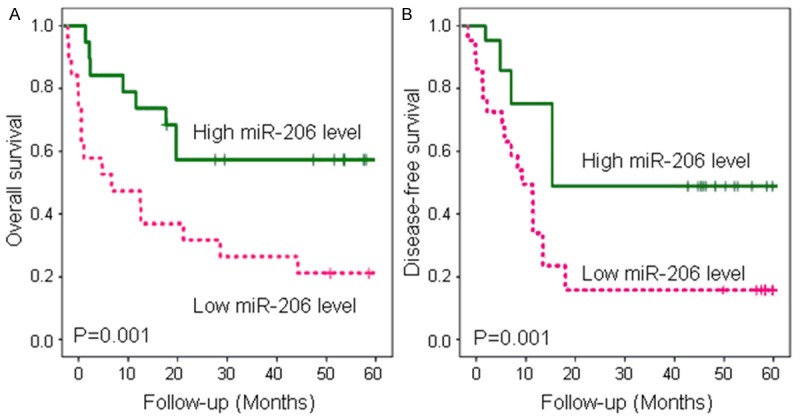
Kaplan-Meier survival curves of melanoma patients. A. The 5-year overall survival of melanoma patients with low serum miR-206 levels was significantly lower than that of melanoma patients with high serum miR-206 levels (P = 0.001). B. The 5-year disease-free survival of melanoma patients with low serum miR-206 levels was significantly lower than that of melanoma patients with high serum miR-206 levels (P = 0.001).
In multivariate analysis, Cox proportional hazards model showed that serum miR-206 level (both P = 0.01), tumor thickness (both P = 0.02), number of metastatic sites (P = 0.008 and 0.01, respectively), response to the treatment response (P = 0.01 and 0.02, respectively) and clinical stage (both P = 0.001), were all independent prognostic factors for both overall survival and disease-free survival in melanoma patients. Statistical values of serum miR-206 level and other clinicopathological parameters derived from Cox stepwise proportional hazards model were shown in Table 2.
Table 2.
Cox multivariate analysis
| Parameter | Overall survival | Disease-free survival | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Risk ratio | 95% confidence interval | P | Risk ratio | 95% confidence interval | P | |
| Age | 0.98 | 0.58-1.96 | NS | 0.72 | 0.36-1.62 | NS |
| Gender | 1.01 | 0.68-2.13 | NS | 0.92 | 0.62-1.88 | NS |
| Thickness | 3.88 | 1.96-8.16 | 0.02 | 3.26 | 1.61-7.01 | 0.02 |
| Ulceration | 1.69 | 0.82-3.59 | NS | 1.21 | 0.68-2.97 | NS |
| Differentiation | 1.92 | 0.97-4.13 | NS | 1.58 | 0.82-3.36 | NS |
| Number of metastatic sites | 5.37 | 2.02-12.24 | 0.008 | 4.66 | 1.92-10.21 | 0.01 |
| Treatment response | 4.23 | 2.01-9.06 | 0.01 | 3.93 | 1.81-8.02 | 0.02 |
| Clinical stage | 7.86 | 2.66-16.66 | 0.001 | 6.89 | 2.09-14.00 | 0.001 |
| Serum miR-206 level | 4.88 | 2.09-10.29 | 0.01 | 4.29 | 1.93-9.63 | 0.01 |
‘NS’ refers to the differences without statistical significance.
Discussion
Improvement in clinical outcomes of patients with melanoma largely depends on increasing the understanding of the molecular mechanisms underlying the tumorigenesis of this malignancy. It is generally recognized that there are several limitations in the traditional staging system, and the molecular biomarker-based prognostic risk stratification will ultimately lead to optimized, personalized cancer treatment. Growing evidence suggest that ideal biomarkers should be easy to measure and strongly correlated with patients’ prognosis [15,16]. In the current study, we confirmed the decreased expression level of miR-206 in sera of patients with melanoma; Then, our statistical analysis found that low serum levels of miR-206 were correlated with aggressive clinicopathological parameters of patients with melanoma, including more metastatic sites, poor response of the treatment and advanced clinical stage; Furthermore, we showed that the serum level of miR-206 was an independent prognostic biomarker for both overall survival and disease-free survival in patients with melanoma. These findings offer a new insight into the relationship between miR-206 and melanoma.
miR-206 belongs to the muscle-specific miR-1 family of muscle-specific miRNAs and plays a role in muscle development [17]. Its activity positively regulates myogenic differentiation [18]. In recent years, accumulating studies have reported that miR-206 may function as a tumor-suppressive miRNA in several cancers. MiR-206 was originally found to be downregulated in ERalpha-positive human breast cancer tissues and introduction of miR-206 into estrogen-dependent MCF-7 breast cancer cells could inhibit cell growth in a dose- and time-dependent manner [19]. Beyond that, the aberrant expression of miR-206 was observed in rhabdomyosarcoma [20], laryngeal cancer [21], lung cancer [22], gastric cancer [23], pancreatic adenocarcinoma [24], hepatocellular carcinoma [25], ovarian cancer [26] and endometrioid adenocarcinoma [27]. In line with these previous reports, we here observed the decreased expression of miR-206 in sera of patients with melanoma. According to its pivotal role in the regulation of tumorigenesis, miR-206 has been indicated to function as a pleiotropic modulator of cell proliferation, invasion and lymphangiogenesis in pancreatic adenocarcinoma by targeting ANXA2 and KRAS genes [24]; miR-206 may inhibit cell migration through direct targeting of the actin-binding protein Coronin 1C in triple-negative breast cancer [28]; miR-206 is also a potential regulator of apoptosis, cell cycle and migration in hepatocellular carcinoma cells [25]; More importantly, miR-206 can induce G1 arrest in melanoma cell lines by inhibition of CDK4 and Cyclin D, supporting miR-206 as a tumor suppressor in melanoma [12]. Consistent with these findings in vitro, our clinical sample-based data in the current study showed the significant associations between low serum miR-206 level and aggressive phenotypes of patients with melanoma. We found that serum levels of miR-206 were predictive for the number of metastatic sites, suggesting it might be a risk factor for metastases. Patients’ response to the treatment may be an important factor for evaluating the subsequent outcome. Our results showed that the decreased expression of miR-206 might confer the poor response to the treatment. Moreover, we also found that low level of miR-206 expression was significantly correlated with advanced clinical stage, suggesting that the loss of miR-206 might be able to play a critical role in the development of melanoma.
The use of serum levels of miRNAs in the clinical setting at the time of diagnosis may determine surveillance strategies by informing clinicians of the risk of disease recurrence. In the current study, we found that the 5-year overall survival and 5-year disease-free survival of melanoma patients with low serum levels of miR-206 were both significantly shorter than those with high serum levels of miR-206, implying that miR-206 might be potential for use as a molecular marker for predicting the prognosis of melanoma patients. Furthermore, we constructed the multivariate regression model to confirm the prognostic value of serum miR-206 level. As a result, miR-206 expression status was identified as an independent prognostic factor for melanoma patients, which was consistent with the previous reports on gastric cancer and breast cancer. Yang et al. [29] found that the gastric cancer patients with a low miR-206 expression had shorter overall survival than those with a high miR-206 expression, and multivariate analysis showed that miR-206 expression was an independent prognostic factor for patients with gastric cancer. Li et al. [30] revealed that a low miR-206 level was an unfavorable prognostic factor for overall survival, in patients with breast cancer.
In conclusion, our results offer the convincing evidence that miR-206 may be implicated in the malignant progression of melanoma. More importantly, the serum level of miR-206 may be a noninvasive prognostic biomarker for the patients with melanoma.
Disclosure of conflict of interest
None.
References
- 1.Diepgen TL, Mahler V. The epidemiology of skin cancer. Br J Dermatol. 2002;146(Suppl 61):1–6. doi: 10.1046/j.1365-2133.146.s61.2.x. [DOI] [PubMed] [Google Scholar]
- 2.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, Eggermont AM, Flaherty KT, Gimotty PA, Kirkwood JM, McMasters KM, Mihm MC Jr, Morton DL, Ross MI, Sober AJ, Sondak VK. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009;27:6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mervic L. Prognostic factors in patients with localized primary cutaneous melanoma. Acta Dermatovenerol Alp Pannonica Adriat. 2012;21:27–31. [PubMed] [Google Scholar]
- 4.Palanichamy JK, Rao DS. miRNA dysregulation in cancer: towards a mechanistic understanding. Front Genet. 2014;5:54. doi: 10.3389/fgene.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan JY, Marques AC. The miRNA-mediated cross-talk between transcripts provides a novel layer of posttranscriptional regulation. Adv Genet. 2014;85:149–99. doi: 10.1016/B978-0-12-800271-1.00003-2. [DOI] [PubMed] [Google Scholar]
- 6.Singh TR, Gupta A, Suravajhala P. Challenges in the miRNA research. Int J Bioinform Res Appl. 2013;9:576–83. doi: 10.1504/IJBRA.2013.056620. [DOI] [PubMed] [Google Scholar]
- 7.Rossi M, Amodio N, Di Martino MT, Caracciolo D, Tagliaferri P, Tassone P. From target therapy to miRNA therapeutics of human multiple myeloma: theoretical and technological issues in the evolving scenario. Curr Drug Targets. 2013;14:1144–9. doi: 10.2174/13894501113149990186. [DOI] [PubMed] [Google Scholar]
- 8.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunz M. MicroRNAs in melanoma biology. Adv Exp Med Biol. 2013;774:103–20. doi: 10.1007/978-94-007-5590-1_6. [DOI] [PubMed] [Google Scholar]
- 10.Li P, He QY, Luo CQ, Qian LY. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med Sci Monit. 2014;20:2472–7. doi: 10.12659/MSM.891327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Segura MF, Greenwald HS, Hanniford D, Osman I, Hernando E. MicroRNA and cutaneous melanoma: from discovery to prognosis and therapy. Carcinogenesis. 2012;33:1823–32. doi: 10.1093/carcin/bgs205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgantas RW 3rd, Streicher K, Luo X, Greenlees L, Zhu W, Liu Z, Brohawn P, Morehouse C, Higgs BW, Richman L, Jallal B, Yao Y, Ranade K. MicroRNA-206 induces G1 arrest in melanoma by inhibition of CDK4 and Cyclin D. Pigment Cell Melanoma Res. 2014;27:275–86. doi: 10.1111/pcmr.12200. [DOI] [PubMed] [Google Scholar]
- 13.Shah AA, Leidinger P, Blin N, Meese E. miRNA: small molecules as potential novel biomarkers in cancer. Curr Med Chem. 2010;17:4427–32. doi: 10.2174/092986710794182980. [DOI] [PubMed] [Google Scholar]
- 14.Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol. 2006;15:56–61. doi: 10.1097/00019606-200603000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed FE. Role of miRNA in carcinogenesis and biomarker selection: a methodological view. Expert Rev Mol Diagn. 2007;7:569–603. doi: 10.1586/14737159.7.5.569. [DOI] [PubMed] [Google Scholar]
- 16.Witwer KW. Circulating MicroRNA Biomarker Studies: Pitfalls and Potential Solutions. Clin Chem. 2015;61:56–63. doi: 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- 17.Elliman SJ, Howley BV, Mehta DS, Fearnhead HO, Kemp DM, Barkley LR. Selective repression of the oncogene cyclin D1 by the tumor suppressor miR-206 in cancers. Oncogenesis. 2014;3:e113. doi: 10.1038/oncsis.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma SB, Lin CC, Farrugia MK, McLaughlin SL, Ellis EJ, Brundage KM, Salkeni MA, Ruppert JM. MicroRNAs 206 and 21 cooperate to promote RAS-extracellular signal-regulated kinase signaling by suppressing the translation of RASA1 and SPRED1. Mol Cell Biol. 2014;34:4143–64. doi: 10.1128/MCB.00480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams BD, Cowee DM, White BA. The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-alpha (ERalpha) signaling and a luminal phenotype in MCF-7 breast cancer cells. Mol Endocrinol. 2009;23:1215–30. doi: 10.1210/me.2009-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–78. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang T, Liu M, Wang C, Lin C, Sun Y, Jin D. Down-regulation of MiR-206 promotes proliferation and invasion of laryngeal cancer by regulating VEGF expression. Anticancer Res. 2011;31:3859–63. [PubMed] [Google Scholar]
- 22.Wang X, Ling C, Bai Y, Zhao J. MicroRNA-206 is associated with invasion and metastasis of lung cancer. Anat Rec (Hoboken) 2011;294:88–92. doi: 10.1002/ar.21287. [DOI] [PubMed] [Google Scholar]
- 23.Ren J, Huang HJ, Gong Y, Yue S, Tang LM, Cheng SY. MicroRNA-206 suppresses gastric cancer cell growth and metastasis. Cell Biosci. 2014;4:26. doi: 10.1186/2045-3701-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keklikoglou I, Hosaka K, Bender C, Bott A, Koerner C, Mitra D, Will R, Woerner A, Muenstermann E, Wilhelm H, Cao Y, Wiemann S. MicroRNA-206 functions as a pleiotropic modulator of cell proliferation, invasion and lymphangiogenesis in pancreatic adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene. 2014 doi: 10.1038/onc.2014.408. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Xu C, Wan H, Liu C, Wen C, Lu H, Wan F. MicroRNA-206 overexpression promotes apoptosis, induces cell cycle arrest and inhibits the migration of human hepatocellular carcinoma HepG2 cells. Int J Mol Med. 2014;34:420–8. doi: 10.3892/ijmm.2014.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Li Y, Wen Z, Kong F, Guan X, Liu W. microRNA-206 overexpression inhibits cellular proliferation and invasion of estrogen receptor α-positive ovarian cancer cells. Mol Med Rep. 2014;9:1703–8. doi: 10.3892/mmr.2014.2021. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y, Kong R, Luo Y, Shi Y, Wang K, Ji G. miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun. 2013;433:207–12. doi: 10.1016/j.bbrc.2013.02.084. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Yan Q, Li S, Zhou L, Yang H, Yang Y, Liu X, Wan X. Expression of the tumor suppressor miR-206 is associated with cellular proliferative inhibition and impairs invasion in ERα-positive endometrioid adenocarcinoma. Cancer Lett. 2012;314:41–53. doi: 10.1016/j.canlet.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Yang Q, Zhang C, Huang B, Li H, Zhang R, Huang Y, Wang J. Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. Eur J Gastroenterol Hepatol. 2013;25:953–7. doi: 10.1097/MEG.0b013e32835ed691. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Hong F, Yu Z. Decreased expression of microRNA-206 in breast cancer and its association with disease characteristics and patient survival. J Int Med Res. 2013;41:596–602. doi: 10.1177/0300060513485856. [DOI] [PubMed] [Google Scholar]


