Abstract
The homeobox transcription factor orthodenticle homolog 2 (otx2) is supposed as an organizer that orchestrates a transcription factor network during photoreceptor development. However, its regulation in the process remains unclear. In this study, we have identified a zebrafish limb bud and heart-like gene (lbh-like), which is expressed initially at 30 hours post fertilization (hpf) in the developing brain and eyes. Lbh-like knockdown by morpholinos specifically inhibits expression of multiple photoreceptor-specific genes, such as opsins, gnat1, gnat2 and irbp. Interestingly, otx2 expression in the morphants is not significantly reduced until 32 hpf when lbh-like begins to express, but its expression level in 72 hpf morphants is higher than that in wild type embryos. Co-injection of otx2 and its downstream target neuroD mRNAs can rescue the faults in eyes of Lbh-like morphants. Combined with the results of promoter-reporter assay, we suggest that lbh-like is a new regulator of photoreceptor differentiation directly through affecting otx2 expression in zebrafish. Furthermore, knockdown of lbh-like increases the activity of Notch pathway and perturbs the balance among proliferation, differentiation and survival of photoreceptor precursors.
Keywords: Lbh-like, retina development, photoreceptor differentiation, otx2, Notch
Introduction
The vertebrate retina, a highly organized neural structure with three major cellular layers, is an excellent model for studying cell fate decision 1-3. During retinogenesis, multipotent retinal progenitor cells (RPCs) give rise to six types of neurons and one type of glial cells in a precise and conserved order 4. Rod and cone photoreceptor cells are light-sensitive neurons in vertebrates 5 and account for over 70% of the cells in mammalian retina. Early in retinogenesis, RPCs proliferate and produce additional multipotent progenitors or lineage-restricted progenitors. After exiting from final mitosis, some of lineage-restricted progenitors are directed to become photoreceptor precursors. Although numerous studies on regulatory networks in which photoreceptor precursors differentiate into rod or cone photoreceptors have been made, there is only a limited knowledge about the decision how RPCs become photoreceptor precursors 6. To date, about 20 transcription factors had been identified as intrinsic factors involved in the cell fate determination 6. Among these factors, a homeobox transcription factor orthodenticle homolog 2 (Otx2) was found to act in the very first step of ocular development and to direct RPCs to differentiate into photoreceptor precursors 6, 7, and the Otx2 conditional knockout mouse was observed to lose photoreceptor, bipolar and horizontal cells 8. Furthermore, Otx2 was revealed to induce ciliary- and iris-derived cells to differentiate into photoreceptor-like cells 9. These findings suggest that Otx2 should be a key director in photoreceptor lineage commitment. However, how Otx2 responds to the induced signals is still unclear.
Limb bud and heart (LBH) is a novel high-conserved transcription cofactor in vertebrates involved in embryonic development 10. It was firstly identified as a novel mouse gene and named by its unique spatiotemporal expression pattern in the developing limb bud and heart 11. LBH family proteins contain a conserved acidic glutamate-rich transcriptional activation domain, but lack the ability to bind DNA directly 11. Recently, LBH was identified as a regulator of mammary stem cell expansion/maintenance 12, synoviocyte growth 13, and cranial neural crest cell migration 14. However, the functions remain largely unknown. In this study, we have identified and characterized a novel zebrafish LBH gene, named lbh-like. Furthermore, we have performed a series of experimental investigations, and demonstrated that lbh-like is required for retina development and photoreceptor differentiation through affecting the expression of otx2.
Materials and Methods
Zebrafish husbandry
Wild type zebrafish (AB strain) was raised and maintained at 28.5℃ as described previously 15. The developmental stages were staged in hours or days post-fertilization according to morphological criteria described by Kimmel et al. 16. The animal protocol for this research was approved by the Institute of Hydrobiology Institutional Animal Care and Use Committee (Approval ID: keshuizhuan 0829).
Body length and eye size measurement
The linear distance from the head epiphysis to the tail, along the anterior-posterior axis, was determined as embryo body length. Embryo eye length was quantified by measuring the diameter of eyes. Scales at the same magnification were employed in the measurement.
RNA isolation, semi-quantitative RT-PCR and real-time PCR
Total RNAs were isolated from zebrafish embryos or adult tissues by using SV Total RNA Isolation System (Promega). The quantity and quality were determined by agarose electrophoresis and spectrophotometer 17. The isolated RNAs were then reverse-transcribed by using the Goldscript cDNA Synthesis Kit (Invitrogen) following the manufacturer's recommendation. β-actin was detected as an internal control. Semi-quantitative RT-PCR and real-time PCR analyses were performed as described previously 18, and the sequences of primers were given in Supplementary Table S1.
Whole mount in situ hybridization
The procedure of whole mount in situ hybridization was performed as described previously 19. The primers with T7 promoter for probe synthesis were shown in the Supplementary Table S1.
Morpholino and mRNA synthesis
Morpholino oligonucleotides were purchased from GeneTools, LLC (Philomath, OR, USA). Sequences of MOs were listed below: lbh-like-MO1 5'-ATCTCCACGCTGCCCATGCCTGGTA-3' (translational blocking morpholino), lbh-like-MO2 5'-AGCACTGCCCCTCCACAGGCCCCAT-3' (translational blocking morpholino), Control MO 5'-CCTCTTACCTCAGTTACAATTTATA-3' (standard control morpholino), otx2-MO 5' - GTTGCTTGAGATACGACATCATGCT - 3' 20, p53 MO: 5'-GCGCCATTGCTTTGCAAGAATTG-3' 21.The primers for rescue experiments were shown in the Supplementary Table S1. Capped RNAs were prepared with the mMESSAGE mMACHINE kit (Ambion). All MOs or mRNAs were injected at the one-cell stage as described previously 21.
Antibody production and Western blot
A peptide (CVSEVESGELRWPPE) was synthesized and conjugated to the KLH peptide (GenScript Company). The production of rabbit polyclonal antibody against zebrafish Lbh-like and analysis of Western blot were performed as described previously 22, 23.
Immunohistochemistry
Section immunohistochemistry and whole-mount immunohistochemistry were performed as described previously 24. Embryos were sectioned at 10 μm in thickness with frozen microtomy (Leica). The following primary antibodies were used: mouse monoclonal antibodies specific for Rhodopsin (1:500 dilution, Abcam), Arrestin3 (1:500 dilution, Abcam), phospho-histone H3 (1:1000 dilution, Cell Signaling), and active caspase3 (1:1000 dilution, BD Biosciences). Fluorescein conjugated goat anti-rabbit/mouse (Thermo) secondary antibodies were used at 1:200 dilution. Nuclei were stained with propidium iodide (PI, 10μg/ml, Sigma). Images were acquired by confocal microscopy (NOL-LSM 710, Carl Zeiss, Germany).
Luciferase assay
A 1.8kb otx2 promoter, containing a 1.2-kb promoter proximal to Otx2 translation start site and a 0.6 kb FM enhancer 25, 26, was subcloned into pGL3-basic luciferase reporter vector (Promega). The fertilized eggs were firstly co-injected with pRL-SV40 and Otx2pro-Luc at a ratio of 1:10. Then, the fertilized eggs were injected with ddH2O, in vitro transcribed lbh-like mRNA or gfp mRNA 27. The luciferase assays in zebrafish embryos were performed as described previously at 36 hpf 28. The primers for luciferase assay were shown in the Supplementary Table S1.
Statistical analyses
Statistical data are presented as Mean ± standard deviation (SD). The body/eye length, numbers of photoreceptor per section, numbers of active caspase3-positive cells and pH3-positive cells in Lbh-like morphants and control groups were statistically analyzed by Independent-Samples T Test with SPSS 13.0 software.
Results
Molecular and expressional characterization of lbh-like in zebrafish
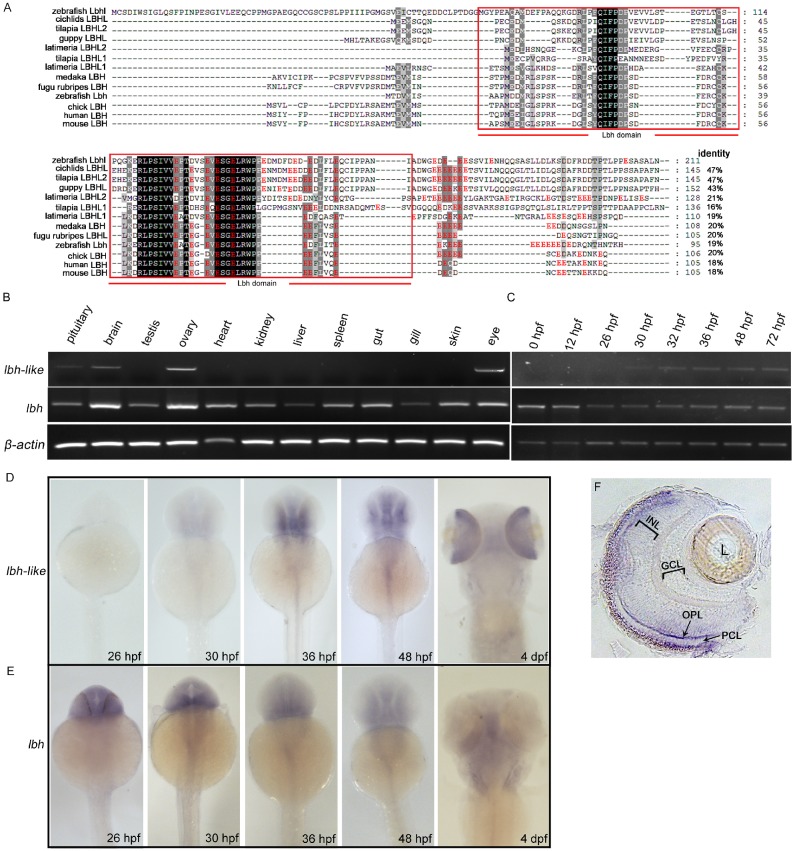
To identify the LBH proteins in zebrafish, the amino acid sequence of LBH domain in mouse LBH was used to search against zebrafish RefSeq DNA database by tblastn with default parameters (NCBI). A significant uncharacterized LOC100000094 (XM_001336435) sequence was observed in zebrafish database. According to the sequence, we cloned the full-length cDNA through RACE PCR. Its full length is 1920 bp, and has an ORF of 633 bp that encodes a protein of 211 amino acids (Supplementary Fig. S1). The encoded protein contains an acidic glutamate-rich region at the carboxy terminus (Fig. 1A), and has 47%, 47% and 43% identities to LBHL of cichlids, tilapia, and guppy, while it only shares 18% to 20% identities to zebrafish Lbh and other mammal LBH (Fig. 1A).
Figure 1.
Molecular and expressional characterization of lbh-like (lbhl) and lbh in zebrafish. (A) Multiple amino acid sequence alignment of zebrafish Lbh-like (Lbhl) and Lbh as well as other vertebrate LBHL and LBH. The black and dark shadings indicate identical sequences. The C-terminal glutamate-rich (red color) conserved LBH domain is labeled by red box. The GenBank IDs for these proteins used in this study are as follows: zebrafish (Danio rerio) Lbhl, XP_001336471; cichlids (Haplochromis burtoni) LBHL, XP_005927379; tilapia (Oreochromis niloticus) LBHL2, XP_003441738; guppy (Poecilia Formosa) LBHL, XP_007575999; latimeria (Latimeria chalumnae) LBHL2, XP_006000505; tilapia (Oreochromis niloticus) LBHL1, XP_005464744; latimeria (Latimeria chalumnae) LBHL1, XP_005988936; medaka (Oryzias latipes) LBH, XP_004082531; fugu rubripes (Takifugu rubripes) LBHL, XP_003962799; zebrafish (Danio rerio) Lbh, NP_956814; chick (Gallus gallus) LBH, NP_001026209; human (Homo sapiens) LBH, NP_112177; mouse (Mus musculus) LBH, NP_084275. (B) Semi-quantitative RT-PCR detection of lbh-like and lbh expression in adult tissues. (C) Semi-quantitative RT-PCR detection of lbh-like and lbh expression during embryogenesis. The β-actin was used as internal control. (D, E) Whole-mount in situ hybridization with lbh-like antisense probe (D) or lbh antisense probe (E). (F) Cryosection of eye from embryo hybridized with lbh-like antisense probe at 4 dpf stage. INL, inner nuclear layer; GCL, ganglion cell layer; OPL, outer plexiform layer; PCL, photoreceptor cell layer; L, lens. Dorsal views.
We further examined the expression patterns of lbh-like and lbh in zebrafish by semi-quantitative RT-PCR analysis and whole-mount in situ hybridization (WISH). As shown in Fig. 1B-E, lbh-like transcript is expressed in pituitary, brain, ovary and eye, whereas lbh is more widely expressed in the analyzed tissues. During embryogenesis, lbh-like message is initially detected in retina and brain at 30 hpf, which increases and maintains a relatively high level at the following stages (Fig. 1C and 1D). At 4 dpf, lbh-like is highly transcribed in retina (Fig. 1D) and mainly distributed in the photoreceptor cell layer (PCL) and outer plexiform layer (OPL) (Fig. 1F). In contrast to lbh-like, lbh is maternally expressed, and zygotic lbh is predominantly expressed in the developing brain and craniofacial structures (Fig. 1C and 1E), as previously shown 14. Therefore, lbh-like and lbh have different expression patterns, suggesting that they might play different functional roles.
Lbh-like knockdown inhibits eye development
To reveal function of Lbh-like during embryogenesis in zebrafish, a translation-blocking morpholino (MO1) was designed to knockdown Lbh-like expression. The lbh-like MO1 effectiveness was firstly evaluated by co-injecting MO1 and plasmid pTag-RFP-N-Lbh-like containing the morpholino binding site. As shown in Supplementary Fig. S2A, MO1 is highly efficient and its activity lasts through 3 dpf. And, Western blot detection verified the down-regulated expression of endogenous Lbh-like in the lbh-like MO1 embryos at 48 hpf (Supplementary Fig. S2B). Moreover, the Lbh-like antibody specificity was also confirmed through Western blot detection of EPC cell extracts with Lbh-like-GST overexpression by pre-immune serum as negative control (NC) and anti-Lbh-like antiserum as positive control (PC) (Supplementary Fig. S2C).
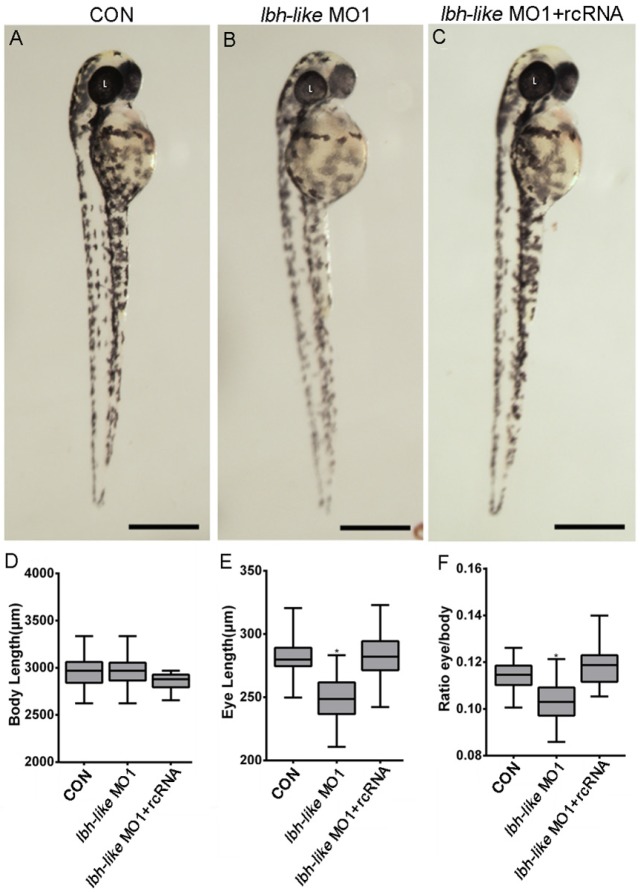
Subsequently, the loss-of-function experiments were performed by MO1 in zebrafish embryos. As shown in Fig. 2, in comparison with wild type embryos or control embryos (CON) injected with control MO (Fig. 2A), 68% embryos (n=31) injected with MO1 show a similar morphant phenotype with smaller eyes at 48 hpf (Fig. 2B). And, the smaller eye defect can be rescued by co-injecting with in vitro transcribed lbh-like mRNA lacking the morpholino binding site (rcRNA) (Fig. 2C). Moreover, we measured their body length and eye length, and calculated the ratio of eye length to body length. As shown in Fig. 2D-F, a significant (9.96%) reduction of eye length/body length is found in the Lbh-like morphants. Another MO (MO2), targeted to a different region around the translational start site, was also tested to be highly efficient and produced similar phenotypes when injected alone or mixed with MO1 (data no shown).
Figure 2.
Smaller eye defect in Lbh-like-deficient embryos. (A-C) Lateral views of control embryos (A), lbh-like deficient embryos (B) and co-injected embryos with lbh-like morpholino and rcRNA (C) at 48 hpf. (D-F) Graphic analysis of body length (D), eye length (E) and the ratio of eye length to body length (F) of embryos shown in A-C. Scale bar=500μm; *p<0.001, t-test; n>20; L, lens.
Morpholino would probably induce development delay that causes small eyes in zebrafish 29. To exclude this possibility, we tested the expression of thymus-specific rag1, since thymus size was usually used as one of morphological markers to distinguish developmental stages of zebrafish larva 30, 31. As shown in Supplementary Fig. S3, the thymus size does not differ between Lbh-like morphants and wild-type embryos at 3 dpf. Therefore, the data indicate that the microphthalmic phenotype in the morphants is specifically resulted from Lbh-like knockdown.
Lbh-like knockdown influences expression of marker genes specific to Müller cells and bipolar cells in retina
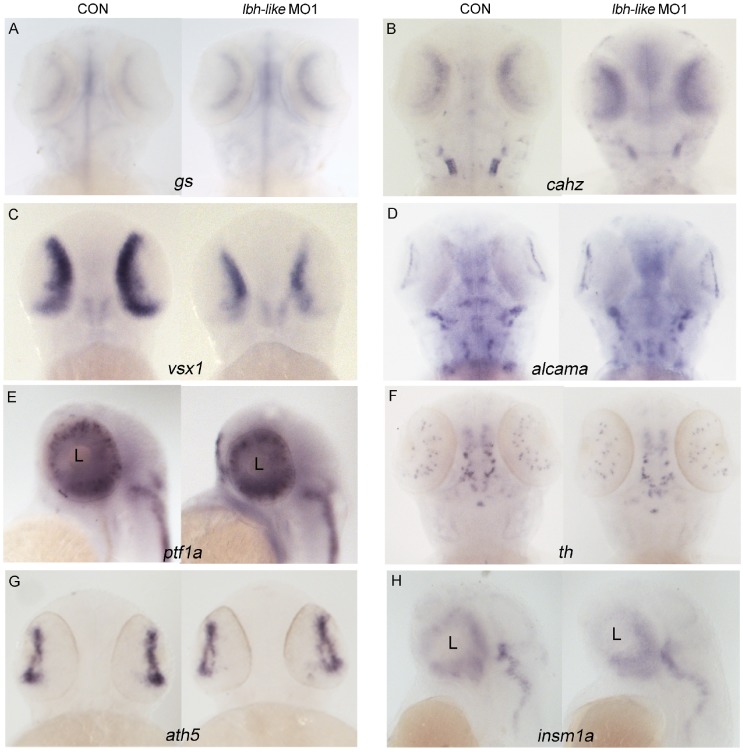
To further understand the effect of lbh-like in eye development, we investigated the expression of retina markers both by WISH and real-time PCR. Compared with that in control embryos, significant expression increases of Müller cell maturation markers glutamine synthetase (gs) and carbonic anhydrase (cahz) 32 were observed in the Lbh-like morphants at 72 hpf (Fig. 3A,B and Supplementary Fig. S4A-B). In contrast, visual system homeobox 1 (vsx1), essential for the proper development of bipolar cells 33-35, was markedly down-regulated in the morphants at 48 hpf (Fig. 3C and Supplementary Fig. S4C). In addition, the expression levels of other type cell marker genes, such as alcama involved in retinal ganglion cell differentiation 36, ptf1a expressed in horizontal and amacrine differentiating cells 37, and amacrine cell's marker gene tyrosine hydroxylase (th) 38, were similar to those in control embryos (Fig. 3D-F and Supplementary Fig. S4D-F). Identical expression patterns between the morphants and control embryos (Fig. 3G, H and Supplementary Fig. S4G, H) were also found in atonal homologue 5 (ath5), a retinal primordium marker involved in RGC specification 39, 40, and insulinoma-associated 1a (insm1a), a developing retina marker required for photoreceptor differentiation 41. Therefore, the data suggest that Lbh-like knockdown might specifically influence expression of marker genes specific to Müller cells and bipolar cells in retina.
Figure 3.
Effects of Lbh-like knockdown on retina marker genes in zebrafish. Gene names are marked in the bottom of panel. In each panel, the left embryo was injected with 4 ng control morpholino and the right one with 4 ng lbh-like MO1. (A, B, D and F) embryos at 72 hpf stage; (C, E, G and H) embryos at 48 hpf stage; (A-D and F, G) Dorsal views; (E and H) Lateral views.
Lbh-like knockdown inhibits photoreceptor differentiation
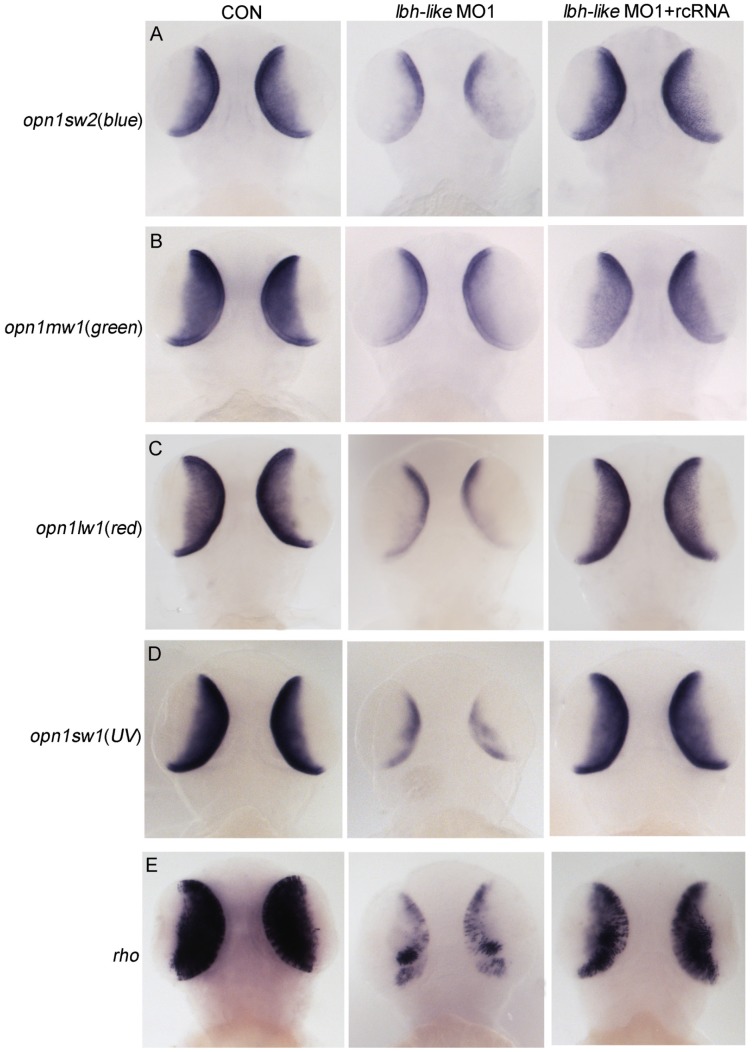
We next tested whether Lbh-like knockdown altered photoreceptor-specific gene expression and photoreceptor development in the embryonic retina. As shown in Fig. 4, all extensively expressed opsins including opn1sw2(blue), opn1mw1(green), opn1lw1(red), opn1sw1(UV), and rho in the ONL and ventral patch of control embryos are remarkably reduced in the Lbh-like morphants at 3 dpf (Fig. 4 and Supplementary Fig. S5), and the reduction can be rescued by co-injection with lbh-like rcRNA (Fig. 4), but not with lbh mRNA (Supplementary Fig. S6). And, lbh mRNA co-injection further aggravated the Lbh-like morphant's eye defect rather than rescuing it (Supplementary Fig. S6). This indicates that the defect is specific to the Lbh-like knockdown.
Figure 4.
Effects of Lbh-like knockdown on cone and rod photoreceptor marker genes. Four opsins (A-D) and rho (E) were analyzed by WISH in the embryos at 3 dpf. The left embryos were injected with 4 ng control morpholino, the middle embryos with 4 ng lbh-like MO1, and the right embryos with 4 ng lbh-like MO1 and 0.1ng rcRNA. Dorsal views.
We further examined the expression of several other photoreceptor-specific genes including rod cell-specific gene gnat1 (guanine nucleotide binding protein, alpha transducing activity polypeptide 1), cone cell-specific gene gnat2 42,and interphotoreceptor retinoid-binding protein gene (irbp) expressed in all photoreceptor cells 43. Gnat1 was found to express in the retina and pineal (arrows) of wild type embryos at 72 hpf (Supplementary Fig. S7A). Injection of lbh-like MO1 almost eliminated gnat1 expression in the retina with only a few gnat1-expressing photoreceptors at the initial site of retinal differentiation (arrowhead), while its expression in the pineal was not changed (Supplementary Fig. S7A). Similar to gnat1, the retina expression of gnat2 and irbp was also inhibited in Lbh-like morphants, but the pineal expression remained (Supplementary Fig. S7A, B, C). And, the reduced expression of gnat1, gnat2 and irbp in retina could be recovered by co-injection with lbh-like rcRNA.
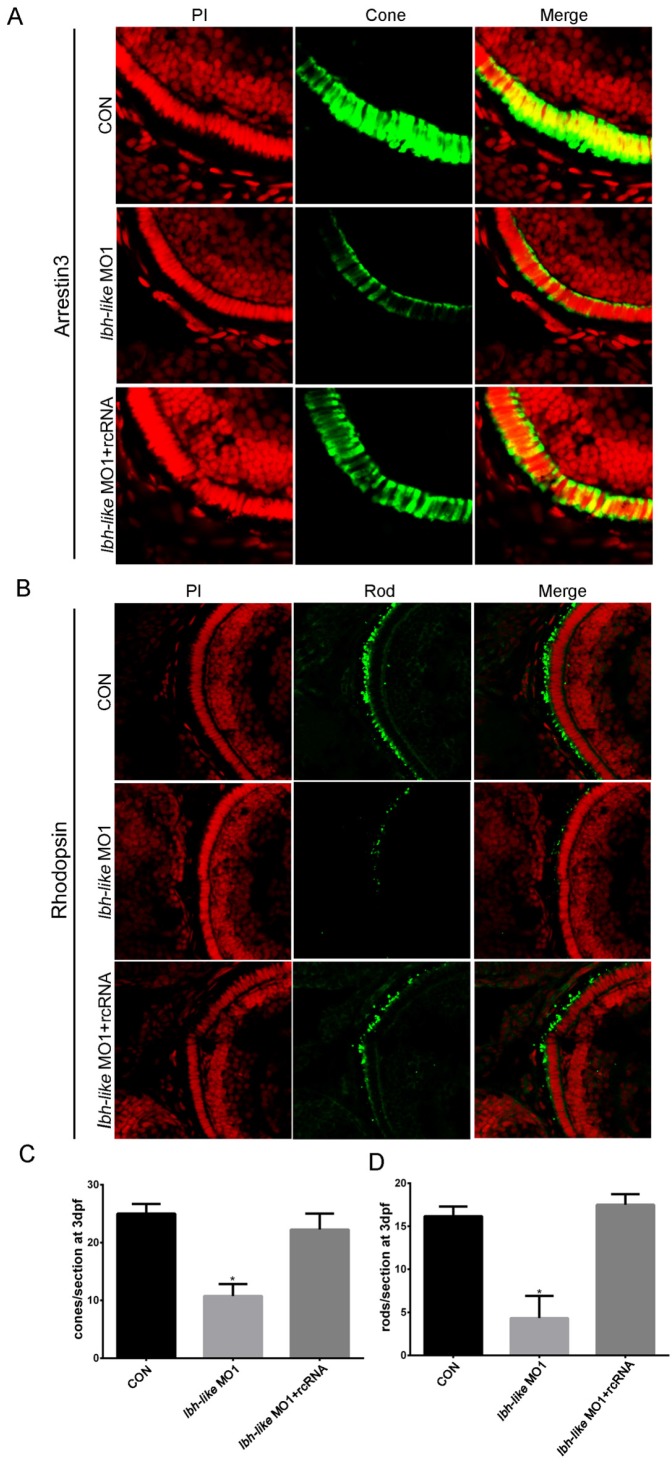
To quantify the decrease of opsin expression, we counted the numbers of cone or rod photoreceptors per section by using immunofluorescence with cone photoreceptor-specific antibody (anti-Arrestin 3) and rod photoreceptor-specific antibody (anti-Rhodopsin). At 3 dpf, an average of 25 cones per section was detected in control embryos (Fig.5A, C). However, the Lbh-like morphants displayed greatly reduced numbers of cone photoreceptors (57% reduction) with an average of 10.75 cones per section (Fig. 5A, C). Significantly, the embryos co-injected with lbh-like MO1 and rcRNA recovered the cone photoreceptor number to an average of 22.25 cones per section (Fig. 5A, C). Similar phenotype of lacking rod photoreceptors (73.2% reduction) was also observed in Lbh-like morphants compared with controls (Fig. 5B, D). Taken together, these results suggest that Lbh-like is required for the formation of photoreceptor cells.
Figure 5.
Reduction of cone or rod photoreceptor cells in Lbh-like morphants. (A-B) Immunofluorescence of zebrafish eye sections. Green fluorescence was stained by the anti-Arrestin 3 antibody (A) or anti-Rhodopsin antibody (B). Red fluorescence was stained by PI. Merge was the overlap of green fluorescence and red fluorescence. (C-D) The numbers of cone (C) or rod (D) photoreceptor cells per section counted by immunofluorescence for anti-Arrestin 3 antibody (A) or anti-Rhodopsin antibody (B), respectively. Cone photoreceptor cells per section, *p<0.01, t-test; n=4 eyes for each treatment; Rod photoreceptor cells per section, *p<0.01, t-test; n=6 eyes for each treatment.
Lbh-like knockdown leads to cell apoptosis and proliferation in retina
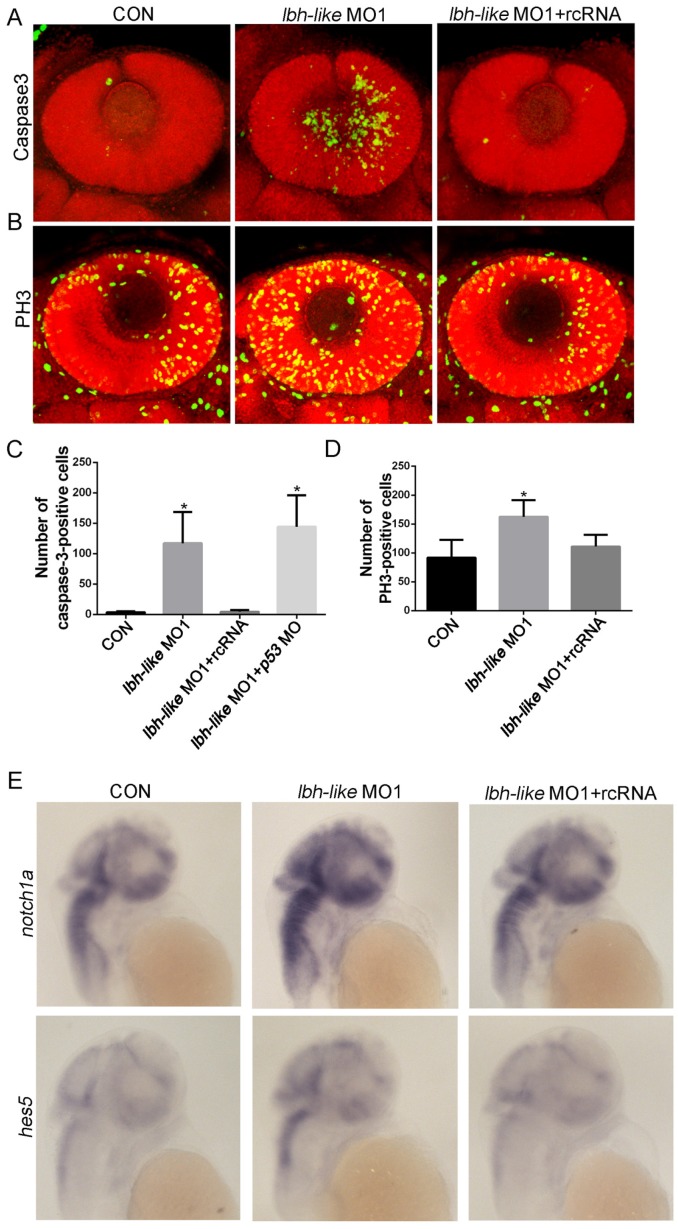
The reduced numbers of photoreceptor cells might be due to the balance change between cell apoptosis and proliferation in the absence of Lbh-like. To address this question, we firstly examined the cell apoptosis in retina by immunostaining assay with antibody against active caspase-3, an apoptosis marker 44. As shown in Fig. 6A, the number of apoptotic cells in retina is much higher in Lbh-like morphants (122.6±55.59 cells) than that in control embryos (3.67±1.63 cells) at 48 hpf, and it can be rescued by co-injecting with lbh-like MO1 and rcRNA (4.43±2.94) (p<0.01; Fig. 6A, C). To avoid unspecific apoptosis due to morpholinos activated by p53 45, we also co-injected lbh-like MO and p53 MO into the fertilized eggs (Fig. 6C). The total number of active caspase-3-positive cells (144.2±51.73) is comparable with that in Lbh-like morphants (Fig. 6C), indicating that the abnormal apoptosis in retina is specifically induced by Lbh-like knockdown. These results suggest that the increased apoptosis might lead to small eye in Lbh-like morphants. On the other hand, we examined cell proliferation through observing PH3-positive cells resided in the retina edge 8, 44 by using anti-phospho-histone H3 antibody. Quantitative counting of positive cells showed that the proliferating cells in retina of the Lbh-like morphants (162.57±28.81 cells) was mildly higher than that in control embryos (91.83±30.72 cells) (Fig. 6B, D), and the increased number of proliferating cells could be reduced by co-injecting with lbh-like MO1 and rcRNA (111.2±20.33). Previous studies have shown that high level of Notch signaling activity preserves a pool of undifferentiated proliferative RPCs and inhibits cell differentiation during retinal development 6, 46, 47. Consistently, Lbh-like morphants also displayed increased expression level of notch1a and its downstream target gene hes5 (Fig. 6E and Supplementary Fig. S8). Therefore, the above results suggest that Lbh-like might have protection against cell apoptosis and balance cell proliferation in retina, and the dual functions might be performed by suppressing Notch signaling.
Figure 6.
Immunostaining assay of cell apoptosis and proliferation as well as WISH detection of Notch pathway in Lbh-like morphants. (A-B) Confocal z-stacks of apoptotic cells (A) or mitotic cells (B) assessed by immunostaining of Caspase-3 or PH3 antibody in embryos at 2 dpf, respectively. Nuclei were visualized by staining with PI. (C-D) Quantification of numbers of Caspase-3-positive cells (C) or PH3-positive cells (D) in embryos, *p<0.01, t-test; n=6 eyes for each treatment. (E) Notch 1 and its downstream target gene hes5 were analyzed by WISH at 2 dpf. Gene names are marked in the left. The left embryos were injected with 4 ng control morpholino, the middle embryos with 4 ng lbh-like MO1, and the right embryos with 4 ng lbh-like MO1 and 0.1ng rcRNA. Lateral views.
Lbh-like affects otx2 expression level during photoreceptor differentiation
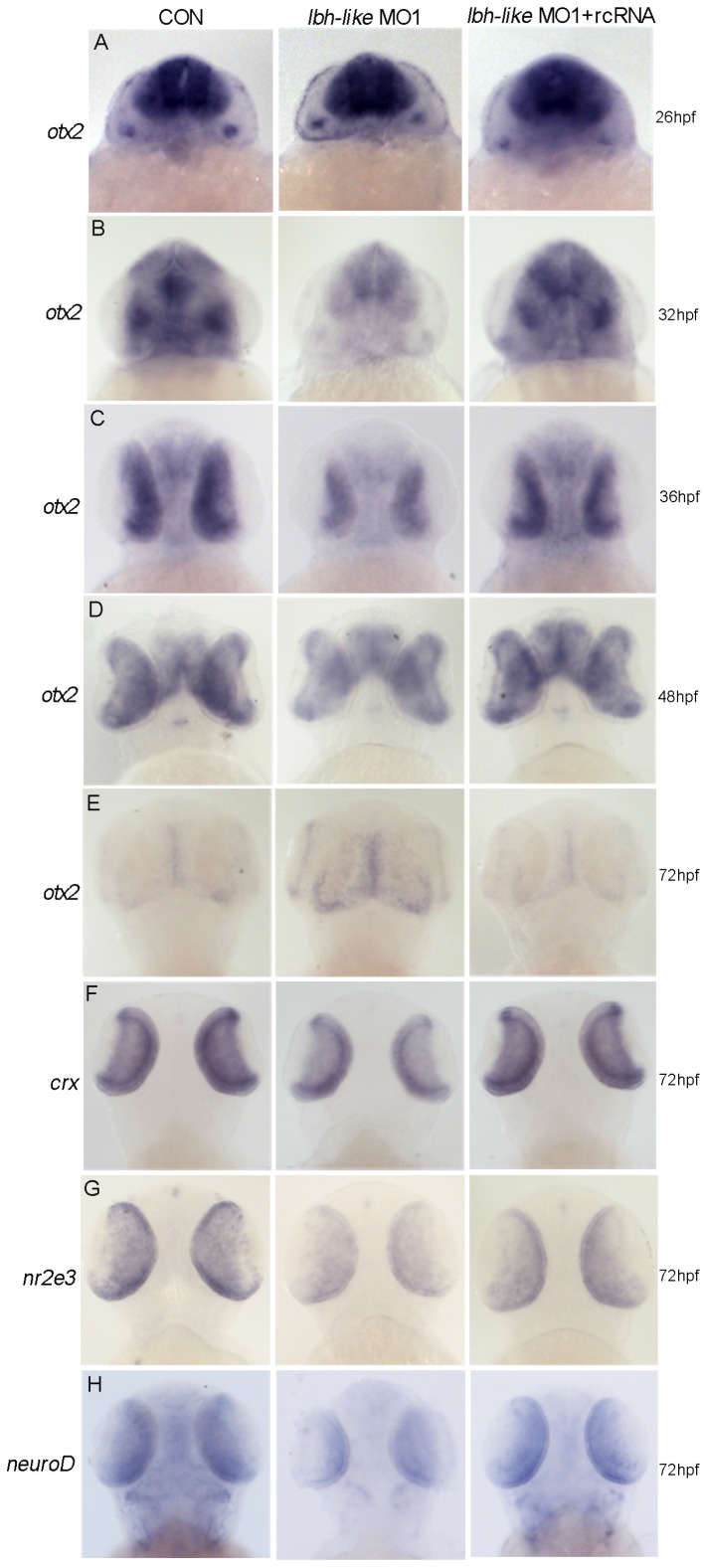
To reveal the potential mechanism of Lbh-like function during photoreceptor neurogenesis and differentiation, we screened its effects on several key genes for the development of retinal photoreceptor cells. Of these, Otx2 expressed in all photoreceptor cells 8 was revealed to play a critical role in cell fate determination of retinal photoreceptor cells. Since lbh-like message was first detected in the developing brain and eyes at 30 hpf (Fig. 1D), we widely examined the expression of otx2 at 26, 32, 36, 48 and 72 hpf. At 26 hpf, otx2 was expressed at high level throughout the midbrain and retina in lbh-like morphants, equally to that in control embryos (Fig. 7A). Interestingly, otx2 expression was significantly down-regulated in Lbh-like morphants from 32 hpf to 48 hpf, and was basically coincident with the expression pattern of lbh-like. And, co-injection with rcRNA could rescue this defect (Fig. 7B-D). At 72 hpf, when otx2 expression level rapidly decreased in control embryos, however, the otx2 transcript still kept a higher level in Lbh-like morphants (Fig. 7E). The above data implies that Lbh-like might tend to keep a certain expression level of otx2 during retina development, and the effects of Lbh-like seemed similar to a previous finding about cone-rod homeobox gene (crx) 48, a direct downstream target gene of otx2 8.
Figure 7.
Lbh-like knockdown disrupts otx2 expression. otx2 (A-E) and its downstream target genes (F-H) were detected by WISH. Gene names are marked in the left and the stages of embryos are marked in the right. In each panel, the left embryos were injected with 4 ng control morpholino, the middle embryos with 4 ng lbh-like MO1, and the right embryos with 4 ng lbh-like MO1 and 0.1 ng rcRNA. Dorsal views.
Moreover, we analyzed the expression characterization of crx and the interacted genes nr2e3 49 and neuroD 50, 51 in Lbh-like morphants. Consistent with the expression reduction of cone and rod photoreceptor marker opsins (Fig. 4), all of the three genes displayed the reduced expression in Lbh-like morphants at 72 hpf (Fig. 7F-H and Supplementary Fig. S9), indicating that the photoreceptor differentiation is blocked. Perhaps, it is the delayed withdrawal of cone and rod progenitors from cell cycle that delays the down-regulation of otx2 accompanying with retinal differentiation, and leads to the unexpected stronger otx2 signal in Lbh-like morphants at 72 hpf (Fig. 7E). Therefore, Lbh-like is required for correct expression of otx2 during retinal development.
Lbh-like regulates photoreceptor development via Otx2
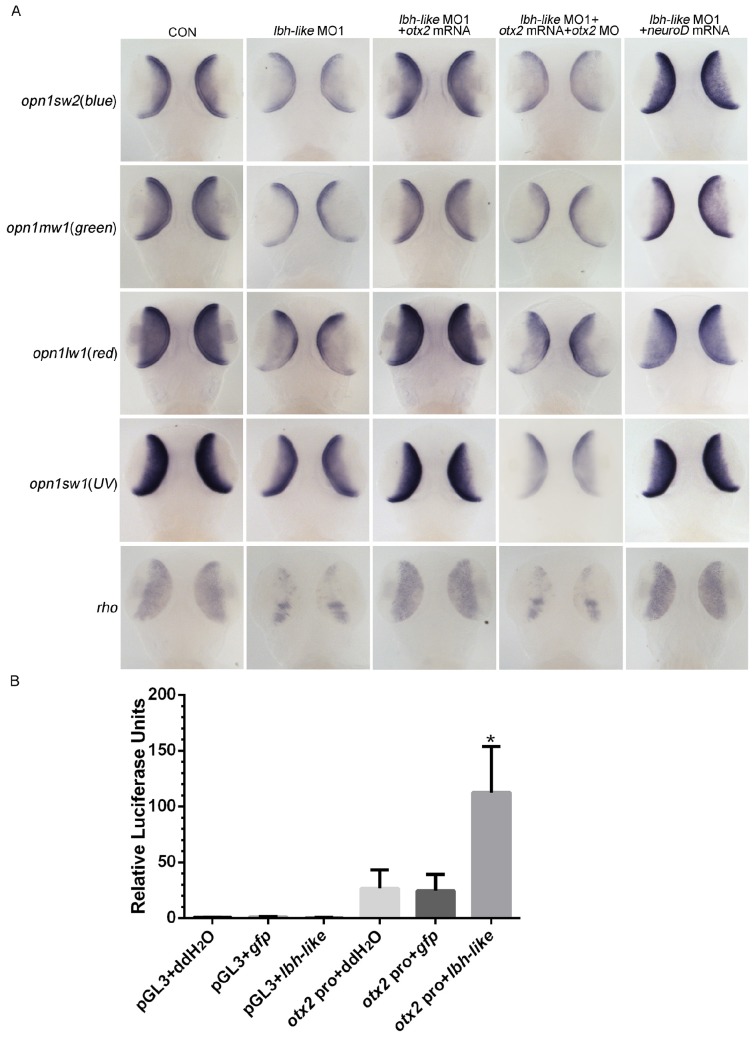
Since Otx2 is required for differentiation of photoreceptor cells, one would expect that Otx2 might be the effector of Lbh-like to regulate photoreceptor development. To address this, cone opsins and rho expression were examined in Lbh-like morphants at 3 dpf through introducing otx2-related treatments. Knockdown of Lbh-like inhibited the expressions of 4 cone opsins and rho (Fig. 8A and 4), and the inhibition could be rescued by co-injecting with otx2 mRNA or otx2 downstream transcription factor neuroD mRNA (Fig. 8A and Supplementary Fig. S10). In addition, the rescued effect of otx2 mRNA was compromised by co-injection with otx2 morpholino, which blocked the production of Otx2 protein (Fig. 8A and S10). Finally, we performed a luciferase assay to investigate the proposed mechanism by co-injection with lbh-like rcRNA and reporter vector Otx2pro-Luc. Overexpression of Lbh-like gave a high activation of Otx2pro-Luc (112.39±41.57) by up to 4.39-folds against that of control embryos co-injected with ddH2O (26.54±16.62) or gfp (24.60±14.64) mRNA (Fig. 8B). Therefore, our results indicate that Lbh-like is required for normal otx2 expression to regulate photoreceptor differentiation.
Figure 8.
lbh-like affects photoreceptor development via otx2. (A) WISH analysis of four opsins and rho in the embryos at 3 dpf. Gene names are marked in the left. From left panel to right panel, embryos were injected with 4 ng control morpholino; 4 ng lbh-like MO1; 4 ng lbh-like MO1 and 35 pg otx2 mRNA; 4 ng lbh-like MO1, 35 pg otx2 mRNA and 2.5 ng otx2 MO; 4 ng lbh-like MO1 and 40 pg neuroD mRNA. Dorsal views. (B) Relative activities of luciferase driven by a 1.8kb region of the otx2 promoter (otx2 pro) in the embryos co-injected with ddH2O, lbh-like or gfp mRNA. The pGL3 vector was used as mock. The data was presented as Mean ± SD (3 independent experiments performed in quadruple,*p<0.05).
Discussion
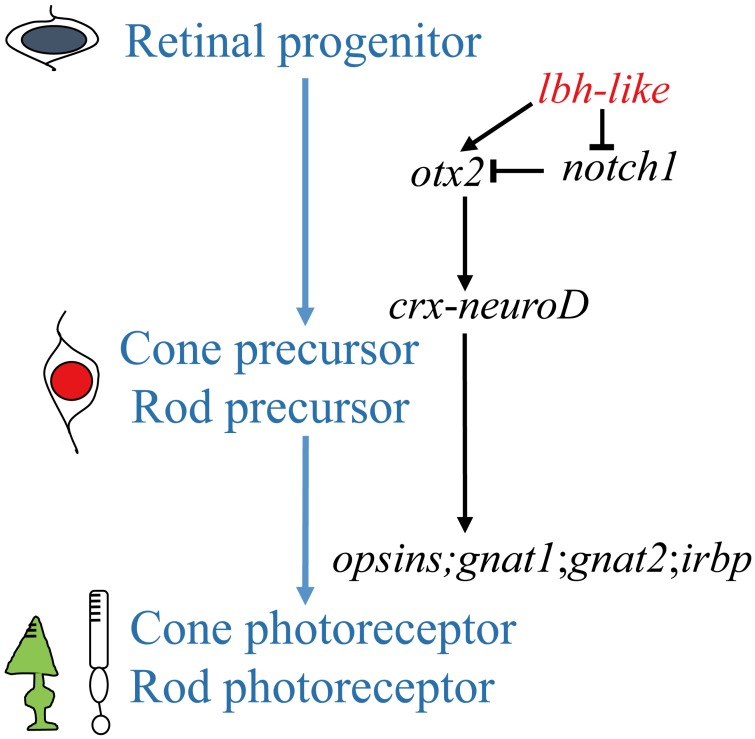
An understanding of the photoreceptor differentiation can greatly promote the development of therapies to treat inherited retina diseases because most of these diseases due to dysfunction or loss of photoreceptor cells 52. In this study, we have characterized a novel member (lbh-like) of lbh gene family distributed in PCL and OPL of embryonic retina, and have demonstrated that Lbh-like acts as a novel intrinsic factor that regulates retinal development in zebrafish. Based on these findings, we propose a functional pathway of zebrafish lbh-like in retinal development, in which Lbh-like is required for correct expression of otx2 and notch1, and under the control of otx2, crx interacts with neuroD and regulates the expression of opsins, gnat1, gnat2, and irbp. Thereby, retinal progenitor differentiates into cone and rod photoreceptor (Fig. 9).
Figure 9.
A proposed pathway of zebrafish lbh-like in retinal development. The left shows the differentiation of cone and rod photoreceptor, and the right is the regulated pathway.
LBH family members are highly conserved small acidic nuclear proteins in vertebrates 11. As a novel member of Lbh family, Lbh-like possesses several different characters from Lbh: 1) Zebrafish Lbh contains an N-terminal hydrophobic stretch, a putative nuclear localization signal (NLS) and a C-terminal glutamate-rich acidic domain, same as its orthologous gene in human and mouse 53, 54. However, Lbh-like shares only 19% amino acid identity with Lbh and contains only a C-terminal glutamate-rich acidic domain (Fig. 1A). 2) Zebrafish lbh and lbh-like show differential expression patterns (Fig. 1B-E). 3) Zebrafish Lbh mediates migration of cranial neural crest cells 14. Our result indicates that lbh-like is also expressed in the neural crest. However, the function of lbh-like gene during neurogenesis needs more exploration.
Otx2 is expressed in retinal progenitors during final mitosis and guides the RPCs into photoreceptor precursor fate 6. In zebrafish, retinal neurogenesis is initiated around 27 hpf when ganglion cell precursors become postmitotic in a small patch of ventrally located cells 55, 56. Suzuki and his colleagues 57 traced cone genesis in transgenic zebrafish and observed mitotic division of L-cone progenitors occurred at about 30 hpf. Zebrafish otx2 is localized in dorsal diencephalon, ventral midbrain and RPE at 26 hpf 58, 59. At this stage, otx2 expression is unaffected in Lbh-like morphants (Fig. 7A). Coincidently after the expression of lbh-like, otx2 expression is significantly down-regulated in Lbh-like morphants at 32 hpf (Fig. 7B). Furthermore, the results of rescue experiments (Fig. 8A and S10) and promoter-reporter assay (Fig.8B) indicate that lbh-like regulates photoreceptor differentiation directly through affecting the expression of otx2. Interestingly, similar to the results in Crx zebrafish morphants 48 and in Crx-/- mice 60, otx2 expression level increases in Lbh-like morphants compared to controls at 72 hpf (Fig. 7E). The delayed withdrawal of cone and rod progenitors from the cell cycle in Crx or Lbh-like morphants might result in delaying the down-regulation of otx2 48. Although the underlined mechanism is unknown, lbh-like is an important mediator of otx2 concerned with photoreceptor development.
Previous studies have shown that LBH proteins regulate cell proliferation and differentiation in mammalian and avian 61, 62. In our case, smaller eye phenotype and reduced numbers of photoreceptor cells in Lbh-like morphants can be caused by changing the balance between proliferation and lineage differentiation of multipotent RPCs, or increasing the cell death. Interestingly, the increased occurrence of phospho-histone H3 positive cells and active caspase-3 positive cells were both observed in the retina of Lbh-like morphants (Fig. 6A-D). The same phenotypes were observed in zebrafish hmx1 or col 15a1 morphant retina 63, 64. Despite the increased proliferation in the retina (Fig. 6B, D), the mild smaller eye phenotype was most likely due to the high amount of retinal cell apoptosis observed in the morphant (Fig. 6A, C). The microphthalmia in the Otx2 conditional knockout mouse was also due to increased apoptosis which suggest Otx2 may play a role in supporting the survival of photoreceptor precursors 8.
Notch1 is required to maintain the progenitor state and inhibit the photoreceptor fate in the developing mammalian retina through the bHLH transcription factors Hes1 and Hes5 46, 47, 65, 66. In mice, retinal progenitors lacking Notch1 initiate the photoreceptor transcriptional program and upregulate the expression of otx2 and crx (Fig.9) 46, 47. In col 15a1 morphants, notch1a showed upregulated and broader expression in retina 64. Consistently, knockdown of zebrafish Lbh-like increased the activity of Notch pathway in eyes (Fig. 6E), which might down-regulate the expression of otx2 and perturb the balance among proliferation, lineage differentiation and survival of photoreceptor precursors.
Otx2 is also involved in survival and terminal differentiation of retinal bipolar cells. In Xenopus, Otx2 is found in bipolar cells but not in photoreceptors 67, 68. Overexpression of XOtx2 in developing retinal cells increases the number of bipolar cells 69. In human, polymorphisms in Otx2 may be a risk factor for bipolar disorder 70, 71. Koike and his colleagues 70 generated a postnatal bipolar-cell specific Otx2 conditional-knockout mouse line and observed impaired maturation of the bipolar cells. In our case, the expression of vsx1, a marker of bipolar cells, is significantly down-regulated in the retina of Lbh-like morphants (Fig. 3C), which might result from the reduction of otx2.
Müller glia functions as radial-glial-like neural stem cells for generation or regeneration of retinal neurons in teleost fish 72. In developing fish retina, proliferating Müller glia is the source of rod progenitors 73. When retinal neurons are destroyed, Müller glia partially and transiently dedifferentiates and increases the expression of BLBP, Vsx2 and other stem cell or progenitor markers 74, 75. In Lbh-like morphants, the Müller cell maturation markers gs and cahz 32 are up-regulated at 72 hpf (Fig. 3A-B). It seems that Müller glia may respond to the injury signals (the reduction of photoreceptor and bipolar cells) and generate multipotent retinal progenitors to repair the mistakes caused by Lbh-like knockdown.
We have noticed that knockdown of Lbh-like cannot completely eliminate the formation of photoreceptors and the expression of otx2 and opsins. This suggests a complex parallel regulating network in regulating the development of retina. Insm1a has been identified as a novel factor to regulate photoreceptor differentiation in the zebrafish retina 41. Since no any expression change of insm1a (Fig. 3H) and its downstream target gene ath5 (Fig. 3G) occurs in the Lbh-like morphants at 48 hpf, lbh-like for regulating photoreceptor cell differentiation should be independent from insm1a-ath5 pathway. Therefore, our results reveal a new mediator that regulates photoreceptors development through directing the precise expression of otx2.
Supplementary Material
Supplementary Figures S1-S10. Supplementary Table S1.
Acknowledgments
This work was supported by grants from the Innovation Project of Chinese Academy of Sciences (KSCX3-EW-N-04) and the Ministry of Agriculture of China within the special Fund for Agro-scientific Research in the Public Interest (NYCYTX-49). Thanks for Wei Liu's microscope operation service.
References
- 1.Cayouette M, Barres BA, Raff M. Importance of intrinsic mechanisms in cell fate decisions in the developing rat retina. Neuron. 2003;40:897–904. doi: 10.1016/s0896-6273(03)00756-6. [DOI] [PubMed] [Google Scholar]
- 2.Livesey F, Cepko C. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–18. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 3.Marquardt T, Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 2002;25:32–8. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- 4.Takatsuka K, Hatakeyama J, Bessho Y, Kageyama R. Roles of the bHLH gene Hes1 in retinal morphogenesis. Brain Res. 2004;1004:148–55. doi: 10.1016/j.brainres.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 5.Ma W, Yan RT, Xie W, Wang SZ. A role of ath5 in inducing neuroD and the photoreceptor pathway. J Neurosci. 2004;24:7150–8. doi: 10.1523/JNEUROSCI.2266-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–76. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beby F, Lamonerie T. The homeobox gene Otx2 in development and disease. Exp Eye Res. 2013;111:9–16. doi: 10.1016/j.exer.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I. et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–63. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 9.Akagi T, Mandai M, Ooto S, Hirami Y, Osakada F, Kageyama R. et al. Otx2 homeobox gene induces photoreceptor-specific phenotypes in cells derived from adult iris and ciliary tissue. Invest Ophthalmol Vis Sci. 2004;45:4570–5. doi: 10.1167/iovs.04-0697. [DOI] [PubMed] [Google Scholar]
- 10.Rieger ME, Sims AH, Coats ER, Clarke RB, Briegel KJ. The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol. 2010;30:4267–79. doi: 10.1128/MCB.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briegel KJ, Joyner AL. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Dev Biol. 2001;233:291–304. doi: 10.1006/dbio.2001.0225. [DOI] [PubMed] [Google Scholar]
- 12.Lindley LE, Curtis KM, Sanchez-Mejias A, Rieger ME, Robbins DJ, Briegel KJ. The WNT-controlled transcriptional regulator LBH is required for mammary stem cell expansion and maintenance of the basal lineage. Development. 2015;142:893–904. doi: 10.1242/dev.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekwall AKH, Whitaker JW, Hammaker D, Bugbee W, Wang W, Firestein GS. The rheumatoid arthritis risk gene, LBH, regulates growth in fibroblast-like synoviocytes. Arthritis Rheumatol. 2015 doi: 10.1002/art.39060. DOI: 10.1002/art.39060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powder KE, Cousin H, McLinden GP, Albertson RC. A nonsynonymous mutation in the transcriptional regulator lbh is associated with cichlid craniofacial adaptation and neural crest cell development. Mol Biol Evol. 2014;31:3113–24. doi: 10.1093/molbev/msu267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei J, Li Z, Gui JF. Cooperation of Mtmr8 with PI3K regulates actin filament modeling and muscle development in zebrafish. PLoS One. 2009;4:e4979. doi: 10.1371/journal.pone.0004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dynam. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 17.Zhou L, Yao B, Xia W, Li C-J, Wang Y, Shi Y-H. et al. EST-based identification of genes expressed in the hypothalamus of male orange-spotted grouper (Epinephelus coioides) Aquaculture. 2006;256:129–39. [Google Scholar]
- 18.Li XY, Li Z, Zhang XJ, Zhou L, Gui JF. Expression characterization of testicular DMRT1 in both Sertoli cells and spermatogenic cells of polyploid gibel carp. Gene. 2014;548:119–25. doi: 10.1016/j.gene.2014.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Xiao Q, Xia JH, Zhang XJ, Li Z, Wang Y, Zhou L. et al. Type-IV antifreeze proteins are essential for epiboly and convergence in gastrulation of zebrafish embryos. Int J Biol Sci. 2014;10:715–32. doi: 10.7150/ijbs.9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foucher I, Mione M, Simeone A, Acampora D, Bally-Cuif L, Houart C. Differentiation of cerebellar cell identities in absence of Fgf signalling in zebrafish Otx morphants. Development. 2006;133:1891–900. doi: 10.1242/dev.02352. [DOI] [PubMed] [Google Scholar]
- 21.Zhong JX, Zhou L, Li Z, Wang Y, Gui JF. Zebrafish Noxa promotes mitosis in early embryonic development and regulates apoptosis in subsequent embryogenesis. Cell Death Differ. 2014;21:1013–24. doi: 10.1038/cdd.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Wang Y, Zhou L, Li Z, Li W, Gui J. Apolipoprotein C1 regulates epiboly during gastrulation in zebrafish. Sci China Life Sci. 2013;56:975–84. doi: 10.1007/s11427-013-4563-4. [DOI] [PubMed] [Google Scholar]
- 23.Yue HM, Li Z, Wu N, Liu Z, Wang Y, Gui JF. Oocyte-specific H2A variant H2af1o is required for cell synchrony before mid-blastula transition in early zebrafish embryos. Biol Reprod. 2013;89:1–13. doi: 10.1095/biolreprod.113.108043. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Sun ZH, Zhou L, Li Z, Gui JF. Grouper tshβ promoter-driven transgenic zebrafish marks proximal kidney tubule development. PLoS One. 2014;9:e97806. doi: 10.1371/journal.pone.0097806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurokawa D, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S. Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development. 2004;131:3319–31. doi: 10.1242/dev.01220. [DOI] [PubMed] [Google Scholar]
- 26.Kurokawa D, Sakurai Y, Inoue A, Nakayama R, Takasaki N, Suda Y. et al. Evolutionary constraint on Otx2 neuroectoderm enhancers-deep conservation from skate to mouse and unique divergence in teleost. P Natl Acad Sci USA. 2006;103:19350–5. doi: 10.1073/pnas.0604686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin F, Liu Y, Lai C, Shan Y, Cheng H, Hsu P. et al. RUNX3-mediated transcriptional inhibition of Akt suppresses tumorigenesis of human gastric cancer cells. Oncogene. 2012;31:4302–16. doi: 10.1038/onc.2011.596. [DOI] [PubMed] [Google Scholar]
- 28.Alcaraz-Perez F, Mulero V, Cayuela ML. Application of the dual-luciferase reporter assay to the analysis of promoter activity in zebrafish embryos. BMC Biotechnol. 2008;8:81. doi: 10.1186/1472-6750-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekker SC, Larson JD. Morphant technology in model developmental systems. Genesis. 2001;30:89–93. doi: 10.1002/gene.1038. [DOI] [PubMed] [Google Scholar]
- 30.Huitema LF, Renn J, Logister I, Gray JK, Waltz SE, Flik G. et al. Macrophage-stimulating protein and calcium homeostasis in zebrafish. Faseb J. 2012;26:4092–101. doi: 10.1096/fj.11-202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willett CE, Zapata AG, Hopkins N, Steiner LA. Expression of zebrafish rag genes during early development identifies the thymus. Dev Biol. 1997;182:331–41. doi: 10.1006/dbio.1996.8446. [DOI] [PubMed] [Google Scholar]
- 32.Peterson RE, Fadool JM, McClintock J, Linser PJ. Müller cell differentiation in the zebrafish neural retina: evidence of distinct early and late stages in cell maturation. J Comp Neurol. 2001;429:530–40. doi: 10.1002/1096-9861(20010122)429:4<530::aid-cne2>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 33.Durr K, Holzschuh J, Filippi A, Ettl AK, Ryu S, Shepherd IT. et al. Differential roles of transcriptional mediator complex subunits Crsp34/Med27, Crsp150/Med14 and Trap100/Med24 during zebrafish retinal development. Genetics. 2006;174:693–705. doi: 10.1534/genetics.105.055152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohtoshi A, Wang SW, Maeda H, Saszik SM, Frishman LJ, Klein WH. et al. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr Biol. 2004;14:530–6. doi: 10.1016/j.cub.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 35.Vitorino M, Jusuf PR, Maurus D, Kimura Y, Higashijima S, Harris WA. Vsx2 in the zebrafish retina: restricted lineages through derepression. Neural Dev. 2009;4:14. doi: 10.1186/1749-8104-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diekmann H, Stuermer CA. Zebrafish Neurolin-a and -b, orthologs of ALCAM, are involved in retinal ganglion cell differentiation and retinal axon pathfinding. J Comp Neurol. 2009;513:38–50. doi: 10.1002/cne.21928. [DOI] [PubMed] [Google Scholar]
- 37.Godinho L, Williams PR, Claassen Y, Provost E, Leach SD, Kamermans M. et al. Nonapical symmetric divisions underlie horizontal cell layer formation in the developing retina in vivo. Neuron. 2007;56:597–603. doi: 10.1016/j.neuron.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 38.Yazulla S, Studholme KM. Neurochemical anatomy of the zebrafish retina as determined by immunocytochemistry. J Neurocytol. 2001;30:551–92. doi: 10.1023/a:1016512617484. [DOI] [PubMed] [Google Scholar]
- 39.Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H. Retinal ganglion cell genesis requires lakritz, a Zebrafish atonal Homolog. Neuron. 2001;30:725–36. doi: 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 40.Masai I, Stemple DL, Okamoto H, Wilson SW. Midline signals regulate retinal neurogenesis in zebrafish. Neuron. 2000;27:251–63. doi: 10.1016/s0896-6273(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 41.Forbes-Osborne MA, Wilson SG, Morris AC. Insulinoma-associated 1a (Insm1a) is required for photoreceptor differentiation in the zebrafish retina. Dev Biol. 2013;380:157–71. doi: 10.1016/j.ydbio.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brockerhoff SE, Rieke F, Matthews HR, Taylor MR, Kennedy B, Ankoudinova I. et al. Light stimulates a transducin-independent increase of cytoplasmic Ca2+ and suppression of current in cones from the zebrafish mutant nof. J Neurosci. 2003;23:470–80. doi: 10.1523/JNEUROSCI.23-02-00470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stenkamp DL, Cunningham LL, Raymond PA, Gonzalez-Fernandez F. Novel expression pattern of interphotoreceptor retinoid-binding protein (IRBP) in the adult and developing zebrafish retina and RPE. Mol Vis. 1998;4:26. [PubMed] [Google Scholar]
- 44.Iida A, Shinoe T, Baba Y, Mano H, Watanabe S. Dicer plays essential roles for retinal development by regulation of survival and differentiation. Invest Ophthalmol Vis Sci. 2011;52:3008–17. doi: 10.1167/iovs.10-6428. [DOI] [PubMed] [Google Scholar]
- 45.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA. et al. p53 activation by knockdown technologies. PLoS Genetics. 2005;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006;133:1367–78. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- 47.Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006;133:913–23. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- 48.Shen YC, Raymond PA. Zebrafish cone-rod (crx) homeobox gene promotes retinogenesis. Dev Biol. 2004;269:237–51. doi: 10.1016/j.ydbio.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 49.Peng G-H, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14:747–64. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- 50.Seko Y, Azuma N, Ishii T, Komuta Y, Miyamoto K, Miyagawa Y. et al. Derivation of human differential photoreceptor cells from adult human dermal fibroblasts by defined combinations of CRX, RAX, OTX2 and NEUROD. Genes Cells. 2014;19:198–208. doi: 10.1111/gtc.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seko Y, Azuma N, Kaneda M, Nakatani K, Miyagawa Y, Noshiro Y. et al. Derivation of human differential photoreceptor-like cells from the iris by defined combinations of CRX, RX and NEUROD. PLoS One. 2012;7:e35611. doi: 10.1371/journal.pone.0035611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boucherie C, Sowden JC, Ali RR. Induced pluripotent stem cell technology for generating photoreceptors. Regen Med. 2011;6:469–79. doi: 10.2217/rme.11.37. [DOI] [PubMed] [Google Scholar]
- 53.Ai J, Wang Y, Tan K, Deng Y, Luo N, Yuan W. et al. A human homolog of mouse Lbh gene, hLBH, expresses in heart and activates SRE and AP-1 mediated MAPK signaling pathway. Mol Biol Rep. 2008;35:179–87. doi: 10.1007/s11033-007-9068-4. [DOI] [PubMed] [Google Scholar]
- 54.Al-Ali H, Rieger ME, Seldeen KL, Harris TK, Farooq A, Briegel KJ. Biophysical characterization reveals structural disorder in the developmental transcriptional regulator LBH. Biochem Bioph Res Co. 2010;391:1104–9. doi: 10.1016/j.bbrc.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Avanesov A, Malicki J. Approaches to study neurogenesis in the zebrafish retina. Method Cell Biol. 2004;76:333–84. doi: 10.1016/s0091-679x(04)76016-1. [DOI] [PubMed] [Google Scholar]
- 56.Malicki J, Jo H, Wei X, Hsiung M, Pujic Z. Analysis of gene function in the zebrafish retina. Methods. 2002;28:427–38. doi: 10.1016/s1046-2023(02)00262-1. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki SC, Bleckert A, Williams PR, Takechi M, Kawamura S, Wong RO. Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. P Natl Acad Sci USA. 2013;110:15109–14. doi: 10.1073/pnas.1303551110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Allende ML, Finkelstein R, Weinberg ES. Expression of two zebrafish orthodenticle-related genes in the embryonic brain. Mech Dev. 1994;48:229–44. doi: 10.1016/0925-4773(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 59.Mori H, Miyazaki Y, Morita T, Nitta H, Mishina M. Different spatio-temporal expressions of three otx homeoprotein transcripts during zebrafish embryogenesis. Mol Brain Res. 1994;27:221–31. doi: 10.1016/0169-328x(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 60.Livesey F, Furukawa T, Steffen M, Church G, Cepko C. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol. 2000;10:301–10. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- 61.Briegel KJ, Baldwin HS, Epstein JA, Joyner AL. Congenital heart disease reminiscent of partial trisomy 2p syndrome in mice transgenic for the transcription factor Lbh. Development. 2005;132:3305–16. doi: 10.1242/dev.01887. [DOI] [PubMed] [Google Scholar]
- 62.Conen K, Nishimori S, Provot S, Kronenberg H. The transcriptional cofactor Lbh regulates angiogenesis and endochondral bone formation during fetal bone development. Dev Biol. 2009;333:348–58. doi: 10.1016/j.ydbio.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boisset G, Schorderet DF. Zebrafish hmx1 promotes retinogenesis. Exp Eye Res. 2012;105:34–42. doi: 10.1016/j.exer.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 64.Gonzalez-Nunez V, Nocco V, Budd A. Characterization of drCol 15a1b: a novel component of the stem cell niche in the zebrafish retina. Stem Cells. 2010;28:1399–411. doi: 10.1002/stem.461. [DOI] [PubMed] [Google Scholar]
- 65.Wakeham A, Correia K, Samper E, Brown S, Aguilera R, Nakano T. et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–48. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 66.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kablar B, Vignali R, Menotti L, Pannese M, Andreazzoli M, Polo C. et al. Xotx genes in the developing brain of Xenopus laevis. Mech Dev. 1996;55:145–58. doi: 10.1016/0925-4773(96)00497-2. [DOI] [PubMed] [Google Scholar]
- 68.Perron M, Kanekar S, Vetter ML, Harris WA. The genetic sequence of retinal development in the ciliary margin of the Xenopus eye. Dev Biol. 1998;199:185–200. doi: 10.1006/dbio.1998.8939. [DOI] [PubMed] [Google Scholar]
- 69.Viczian AS. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130:1281–94. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- 70.Koike C, Nishida A, Ueno S, Saito H, Sanuki R, Sato S. et al. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol. 2007;27:8318–29. doi: 10.1128/MCB.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sabunciyan S, Yolken R, Ragan CM, Potash JB, Nimgaonkar VL, Dickerson F. et al. Polymorphisms in the homeobox gene OTX2 may be a risk factor for bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:1083–6. doi: 10.1002/ajmg.b.30523. [DOI] [PubMed] [Google Scholar]
- 72.Lenkowski JR, Raymond PA. Müller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish. Prog Retin Eye Res. 2014;40:94–123. doi: 10.1016/j.preteyeres.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–40. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagashima M, Barthel LK, Raymond PA. A self-renewing division of zebrafish Müller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013;140:4510–21. doi: 10.1242/dev.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S10. Supplementary Table S1.