Abstract
Hsp70 molecular chaperones are implicated in a wide variety of cellular processes, including protein biogenesis, protection of the proteome from stress, recovery of proteins from aggregates, facilitation of protein translocation across membranes, and more specialized roles such as disassembly of particular protein complexes. It is a fascinating question to ask how the mechanism of these deceptively simple molecular machines is matched to their roles in these wide-ranging processes. The key is a combination of the nature of the recognition and binding of Hsp70 substrates and the impact of Hsp70 action on their substrates. In many cases, the binding, which relies on interaction with an extended, accessible short hydrophobic sequence, favors more unfolded states of client proteins. The ATP-mediated dissociation of the substrate thus releases it in a relatively less folded state for downstream folding, membrane translocation, or hand-off to another chaperone. There are cases, such as regulation of the heat shock response or disassembly of clathrin-coats, however, where binding of a short hydrophobic sequence selects conformational states of clients to favor their productive participation in a subsequent step. This Perspective discusses current understanding of how Hsp70 molecular chaperones recognize and act on their substrates and the relationships between these fundamental processes and the functional roles played by these molecular machines.
Keywords: Hsp70 molecular chaperone, chaperone substrates, protein folding, disaggregation, translocation, complex disassembly
Graphical Abstract
Introduction
Hsp70 molecular chaperones have a deceptively simple mechanism of action: They bind via their substrate-binding domain (SBD) to short polypeptide sequences with recognition motifs that are not stringently defined but can be identified by common features. The affinity of Hsp70s for their substrates is modulated allosterically by nucleotide binding to their N-terminal actin-like nucleotide-binding domain (NBD), with ATP binding causing a reduction in affinity and ADP binding leading to high substrate affinity. The past few years have provided deep insights into the structural basis of Hsp70 allostery, much of it based on the E. coli Hsp70 family member, DnaK.1, 2 In the ADP-bound state of DnaK, the NBD and the SBD are largely independent, behaving like the separate domains connected by a flexible linker.3 Upon ATP binding, the two domains are intimately docked with a conserved hydrophobic sequence in the interdomain linker forming a key part of the packing interface between domains.4–6 Substrate binding to the ADP-bound DnaK is characterized by slow on-off kinetics. In the presence of ATP, substrate binding stimulates the ATP hydrolysis rate of the NBD, and the on and off rates for substrate binding are accelerated, leading to lower overall affinity and consequently favoring substrate release.7 Atomic structures are now available for both ADP-3 and ATP-bound DnaK,4, 5 and an activated state of DnaK with both ATP and a substrate peptide bound was recently described, in which the hydrophobic linker sequence is associated with the NBD but the domains are not fully docked on each other.8 In physiological contexts, the allosteric cycle of Hsp70s is modulated by co-chaperones including nucleotide exchange factors (NEFs), which stimulate exchange of ADP for ATP, and J proteins, which both facilitate substrate delivery to Hsp70s and catalyze ATP hydrolysis.2 The impact of J proteins in modulating Hsp70-substrate interactions is of great importance to the interplay of this chaperone system with its clients and will be discussed in greater detail in a later section. While the allosteric mechanism of Hsp70s has been elucidated to a great extent, our understanding of the impact of Hsp70 chaperone actions on their substrates remains shallow despite the fact that this is the essence of Hsp70 physiological functions.
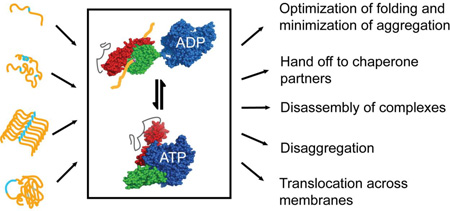
Hsp70s facilitate a stunning array of diverse functions. Their ability to do so is a consequence of their intrinsic allosteric mechanism coupled with their ability to cooperate with upstream and downstream chaperone partners. For example, all Hsp70s partner with J-protein family members, which are highly diverse, specialized, and spatially restricted.9 Hsp70 actions include optimization of folding and minimization of aggregation, both early in the biogenesis of proteins and upon stress-induced unfolding; shepherding proteins across membranes in translocation processes that require substrates to remain unfolded; promoting disassembly of large protein complexes; working with partner chaperones to mediate disaggregation reactions; and hand-off of substrates to downstream chaperones in folding and assembly reactions. When proteins fail to acquire the proper structure for downstream steps, Hsp70s work with their co-chaperones and partner chaperones to provide a quality control mechanism that includes shunting non-competent proteins to degradation.10, 11
Despite this physiological diversity, all Hsp70 functions share a reliance on their ability to bind and release short unstructured sequences in their substrates. Importantly, the extent to which this binding reaction remodels the substrate and alters its conformational ensemble is critical to the biological outcome. Questions that must be addressed to get to the heart of the action of Hsp70s on their substrates include: what is the nature of the recognition of substrates, including sequence and conformational requirements in the substrate for binding, whether and how the binding and release by the chaperone remodel the substrate, whether there is a direct mechanical action of Hsp70s on substrates, how/whether subsequent processes involving the substrate (folding, translocation, etc.) are favored because of the action of the Hsp70, and how hand-off to downstream chaperones is facilitated by Hsp70 action.
In this Perspective, we discuss what is known about the answers to these questions. This brief review makes it clear that current knowledge is limited. With a few notable exceptions, we lack adequate fundamental understanding of the nature of Hsp70 chaperone/substrate interactions to elucidate the related physiological functions. The reasons for this are not unusual: Most of our current understanding is based on model systems with purified components, in isolation from the full system responsible for the physiological function. In particular in the case of Hsp70s, the lion's share of our understanding of how they interact with substrates derives from studies of short peptides as models for protein substrates. Moreover, most experiments have been carried out on purified Hsp70s in isolation from partner chaperones. Thus, there is a pressing need to take our knowledge of Hsp70-substrate interactions to the next level of complexity. Here, we discuss examples of published work related to Hsp70/substrate interactions, and we apologize in advance to those whose work we do not cite because of the limitations of length and scope of this Perspective.
The basis of substrate recognition by Hsp70s
Structural descriptions of substrate binding: peptides
It was discovered early on that Hsp70s bind a variety of peptides seven residues in length, and that these fragments, like protein substrates, activate the ATPase activity of the chaperone.12–14 From that point on, there has been heavy reliance on peptide models to understand the basis of substrate recognition by Hsp70s. Peptide library and phage display studies with several Hsp70s showed a preference for hydrophobic residues and positive charge, but without any clear binding motif.14–18 Nonetheless, enough sequence bias has been extracted from these studies to develop predictive algorithms to identify potential Hsp70 binding sites.14–19 From the earliest phage display work, it appeared that sequence preferences may differ from one Hsp70 to another,14, 16 but the overlap in binding capacity is high and it remains an open question to what extent Hsp70s have binding sequence biases.20 Current thinking is that the J-protein partners of Hsp70s provide most of the physiological substrate specificity,9, 21 which is also consistent with the greater number of different J-proteins than Hsp70s in a given cellular compartment, and particularly the larger number in eukaryotes relative to prokaryotes.
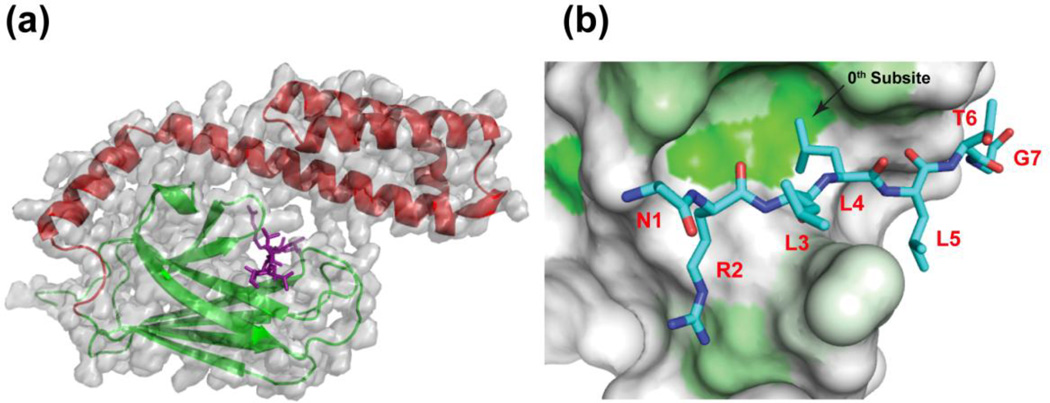
The first atomic level structural description of substrate binding by an Hsp70 revealed that the 7-residue model peptide NRLLLTG (called "NR") bound in an extended, poly-proline like conformation to a cleft in the SBD of the E. coli Hsp70, DnaK (Figure 1).22 Sub-sites for side chain interaction could be identified around a central (0) site with a deep pocket where the second Leu bound. This hydrophobic subsite explained the strong bias towards an aliphatic hydrophobic residue in DnaK binding motifs. The three subsites of interaction in DnaK on either side of the 0th subsite (i.e., −3, −2, −1, and +1, +2, +3) have the potential for quite permissive interactions, and significant hydrogen bonding to the backbone stabilizes the bound peptide. Subsequent structural studies (summarized in Table 1) have reinforced these original findings, in one case with a human Hsp7023, and added new twists: For example, the NR model peptide was observed in an alternate crystal to bind in a different register, showing that the position of the positive charge relative to the hydrophobic residue binding to the 0th subsite is not critical.24 Some peptides from antimicrobial origins reinforced this finding and showed that a peptide lacking a positive charge may bind, albeit with somewhat lower affinity.24 Polypeptides can bind in the opposite orientation to the binding mode utilized in the original crystal structure of DnaK. This mode of binding (which we call "reverse" in Table 1) was first seen in HscA, an Hsp70 that is implicated in Fe-S complex assembly in E. coli,25 and has subsequently been observed for several peptides (Table 1)24, 26, 27. The ‘reverse’ binding mode is favored when a proline residue occupies the ‘0’ position regardless of the location of the positive charge.24, 27, 28
Figure 1.
Structures illustrating the binding of peptide substrate models by Hsp70s. a) The substrate-binding domain of the E. coli Hsp70, DnaK, bound to the heptapeptide, NRLLLTG (PDB 1dkz).12 Note that the peptide (purple) is in an extended conformation, cradled by the β-subdomain (green) and fully enclosed by the α-helical lid (red). b) A view from the top of the β-subdomain (after removal of the α-helical lid) showing the hydrophobic pocket of the peptide binding site that defines the 0th subsite. Greater hydrophobicity of the SBD surface is indicated by the intensity of the green color. The second leucine of the bound NR peptide interacts with the pocket.
Table 1.
Binding modes of structurally characterized peptide/Hsp70 complexes
| Chaperonea | Peptide | Subsite Bindingc | Orientation | Origin of Peptide |
Ref |
|---|---|---|---|---|---|
| EC DnaK SBDb | NR | Forward | Phage Display | 12 | |
| EC DnaK SBD | NR | Forward | Phage Display | 14 | |
| EC DnaK SBD | LML | Forward | Phage Display Variant | 14 | |
| HS HspA1 SBD | NR | Forward | Phage Display | 13 | |
| GK DnaKd | Linker | Forward | Interdomain linker | 20 | |
| EC DnaK SBD | LYZ | Forward | Antimicrobial pyrrhocoricin | 14 | |
| EC DnaK SBD | 1-11_LYZ | Forward | Antimicrobial pyrrhocoricin | 14 | |
| EC DnaK SBD | 1-10_LYZZ | Forward | Antimicrobial pyrrhocoricin | 14 | |
| EC DnaK SBD | 1-10_LYZI | Forward | Antimicrobial pyrrhocoricin | 14 | |
| EC DnaK SBD | W/T | Forward | Antimicrobial pyrrhocoricin | 14 | |
| EC DnaK SBD | Onc72 | Forward | Antimicrobial onconin | 14 | |
| EC DnaK SBD | 1–15 | Forward | Antimicrobial PR-39 | 14 | |
| EC DnaK SBD | 1–20 | Forward | Antimicrobial, designed | 14 | |
| EC DnaK SBD | 3–11 | Forward | Antimicrobial apidaecin | 14 | |
| EC DnaK SBD | 4–11 | Forward | Antimicrobial apidaecin | 14 | |
| EC DnaK SBD | PL | Reverse | IscU, EC Fe-S assembly protein | 15 | |
| EC DnaK SBD | PP | Reverse | Variant of IscU peptide | 14 | |
| EC DnaK SBD | NR | Reverse | Phage Display Variant | 14 | |
| EC DnaK SBD | 3–11 | Reverse | Apidaecin variant, Api88 | 16 | |
| EC DnaK SBD | 12–20 | Reverse | Antimicrobial pyrrhocoricin | 14 | |
| EC DnaK SBD | 12–19 | Reverse | Antimicrobial drosocin | 14 | |
| EC DnaK SBD | 14–21 | Reverse | Antimicrobial heliocin | 14 |
EC denotes Escherichia coli; HS, Homo sapiens; GK, Geobacillus kaustophilus.
SBD constructs are from aa393 to aa603, E. coli numbering in all cases.
The residue occupying the central pocket is shown in larger font. Residues in italics were outside the binding site and generally had high B factors.
Construct is from aa1 to aa603.
Z Denotes cyclohexlyalanine.
Peptide substrates act as allosteric effectors of Hsp70s, and their action on the allosteric conformational shifts of the chaperone are governed by their affinities of binding. This relationship was elegantly demonstrated using a set of model peptides of differing hydrophobicity and E. coli DnaK variants with complementary mutations.29 The energetics of peptide binding were found to be coupled to the tendency of DnaK to shift from the domain undocked high affinity conformation to the high ATPase docked conformation: Stronger binding peptides resulted in a higher rate of ATP hydrolysis, showing the thermodynamic coupling of ligand-modulated allostery in Hsp70s and raising the question whether there are functional ramifications of the response to substrates of differing affinities.
Protein substrates: How are protein substrates recognized and bound by Hsp70s?
In their cellular roles, Hsp70s act on full-length protein substrates. Because of the difficulty of working with incompletely folded proteins, there is a relative paucity of structural data on the nature of the binding interaction between protein substrates and Hsp70s. It has been assumed by most of the field that binding of protein substrates is mediated by interaction of short highly accessible sequences with the canonical binding cleft in the SBD, analogous to the binding of a free peptide, and that therefore, the recognition sites in proteins would be those predicted based on peptide studies, and the mode of binding of Hsp70s to proteins would be comparable to that to peptides. In fact, this expectation has rarely been tested directly, and there are many questions that arise when the binding of an Hsp70 to a protein substrate is considered. Is the mode of recognition of a protein like that observed for model peptides? What happens to the rest of the substrate protein when/if an accessible heptapeptide motif is bound to the canonical binding groove? And most importantly, what is the impact of interaction with the Hsp70 on the conformational ensemble of a protein substrate?
In an intriguing x-ray crystal structure, Geobacillus kaustophilus DnaK molecules bound to their own interdomain linkers via the highly conserved hydrophobic tetrapeptide sequence (VVLL in G. kaustophilus).30 This structure not only shows how an accessible heptapeptide sequence in a protein may bind to the canonical cleft of the SBD in a manner that resembles closely the way a free peptide binds, but they also lend credence to the possibility that Hsp70s may oligomerize by binding to their own interdomain linkers.31 Oligomerization of Hsp70s has been reported by several investigators and proposed to be involved in regulation of their cellular activities.15, 32
In only a couple of cases has the expectation that proteins bind to Hsp70s using the same mode of binding as peptides been directly tested. Chen et al. used peptide scanning to identify the region of apomyoglobin that bound with highest affinity to the β-subdomain of the SBD of DnaK, and found that the NMR signals for these same residues were broadened so much as to disappear when DnaK bound to full-length apomyoglobin, again supporting the expectation that the protein substrate utilizes the same binding mode as observed for peptides.33 Similarly, Rodriquez et al. found the preferred DnaK-binding sequence within the E. coli heat-shock transcription factor σ32 by peptide scanning and also identified the sequence bound to DnaK in the context of the protein by proteolysis footprinting; their results showed that the same sequence bound in the context of the full-length σ32 as when presented as a peptide.34
Thus, it seems valid to anticipate that Hsp70s will bind linear sequences of their protein substrates via short (7-residue) stretches with at least an anchoring hydrophobic residue, and usually at least one nearby positively charged residue, as current algorithms would predict. Moreover, peptide studies raise the possibility that the binding sites for Hsp70s in proteins may bind in either of two directionalities. But how is the linear sequence presented for binding in the protein substrate? Do Hsp70s bind only unfolded substrates where linear sequences would be readily accessible? Or rather do they bind partially collapsed proteins via transiently accessible regions? Or do they take advantage of dynamic sampling of multiple conformations and select for a conformation that is suitable for binding? What happens to regions that are not bound? And are other sites in the Hsp70 apart from the canonical substrate-binding cleft in the SBD playing any role in binding?
Taking the last question first: It has been proposed that the C-terminus of Hsp70s play a role in substrate binding. The Wang group reported that a truncated Hsc70 lacking the flexible C-terminal 10-kDa segment was unable to form a complex with a model unfolded protein, reduced carboxymethylated α-lactalbumin35. They postulated that the disordered C-terminus of the Hsp70 serves as an additional interaction site with substrate. In addition, two groups have reported that the C-terminal region of DnaK is required for its full function, including in vitro refolding of protein substrates. Mayer et al. reported several functional defects including inability to refold luciferase upon truncation of E. coli DnaK to 538, removing the flexible C-terminus plus the latter part of the helical lid.29 Smock et al. found that the characteristics of the extreme C-terminal ~25 amino acids of bacterial Hsp70s were conserved and explored the role of this region in E. coli DnaK. They showed that a relatively short truncation (35 residues) led to loss of viability under stringent conditions (absence of SecB) and loss of ability to efficiently refold denatured luciferase in vitro.36 In addition, the C-terminus of E. coli DnaK became more ordered when DnaK bound to a protein model substrate (a stably unfolded staphylococcal nuclease mutant) but not when bound to a peptide.36 Taken together these studies suggest that protein substrates may bind to the C-terminus of Hsp70s in addition to the canonical substrate-binding groove. This type of binding may serve multiple purposes–keeping the substrate more unfolded and also keeping the substrate in the local region of the chaperone even after ATP-mediated release, to facilitate rebinding if necessary.
In terms of the nature of the bound protein substrate, Hsp70s can bind substrates that are quite unfolded or non-native or, in some instances, fully folded or near-native, provided their binding sites are accessible and conformationally compatible with the binding of an extended region in the SBD binding cleft. Pioneering studies by Fink and coworkers showed by fluorescence and circular dichroism that a thermally unstable fragment of staphylococcal nuclease is a substrate for Hsp70 at 37 °C (when it is unfolded), and this substrate is partially unfolded and loses its α-helical structure compared with the native protein when bound to the chaperone This substrate is released from the chaperone upon lowering the temperature to 10 °C, which stabilizes the native fold.37 Similarly, N-terminal fragments from the model substrate apomyoglobin were unfolded when bound to the β-subdomain of the SBD of DnaK based on their lack of helicity and global tertiary structure.33 Note that full-length apomyoglobin is relatively well folded and did not interact with DnaK β-SBD. Studies of another model substrate, a slow folding variant of ribonuclease H, showed that the DnaK-bound client had more secondary structure than the client refolded in the absence of chaperones, suggesting that the bound substrate is maintained in a conformational state that supports productive folding.38
Protein substrates may retain some structure when bound. Schlecht et al. used disulfide cross-linking and measurement of mobility of spin labels to show that the α-helical lid of the SBD does not fully close around a protein substrate, as it does around a peptide, indicating that protein substrates may retain tertiary structure when bound.39 On the other hand, single molecule fluorescence measurements by Kellner, et al. indicated that a substrate protein (in this case denatured rhodanese) with a compact denatured state ensemble in the absence of chaperones expanded substantially in Stokes radius upon binding DnaK, indicating a more unfolded substrate while bound to the chaperone.40 Note that initial binding of DnaJ, which caused a very modest expansion of denatured rhodanese, was a prerequisite to DnaK binding. These studies also support the idea that the Hsp70 system can accommodate structured proteins around the binding site, and suggest that the SBD helical lid may interact with regions of the client in addition to the interaction between the client heptapeptide binding motif and the canonical cleft of the β-subdomain of the SBD.39, 41. The differences in binding and interaction between protein and peptide substrates were also observed to carry over into distinct influences on the kinetics of ATP hydrolysis; BiP binding to a stably unfolded antibody decelerated the hydrolysis step of the ATPase cycle while peptide binding accelerated it.42
By contrast with these examples of largely unfolded Hsp70-bound model substrates, among its natural substrates Hsc70 is proposed to bind to well-folded clathrin triskelions, but they do so via a QLMLT motif present in their C-terminal unstructured tails.43 In addition, both σ32 and the glucocorticoid receptor exemplify substrates wherein the Hsp70 binding site is transiently exposed in one conformation of a fluctuating, but not fully unfolded substrate. These last three examples illustrate the functionally critical role of Hsp70 chaperone binding: Interaction with the chaperone provides a nucleotide-modulated mechanism for selecting a conformation that is optimal for the required downstream function. The next section describes in greater detail how the simple mechanism of Hsp70s has been evolutionarily matched to functions.
The action of Hsp70s on their substrates: Relationship to physiological functions
Since the essence of the binding mechanism is the same from one Hsp70 to another, the adaptation of Hsp70s to specific functions must be a consequence of interactions with substrates and co-chaperones associated with that physiological role. For example, the nature of the chaperone substrate–viz., its conformational state when bound and how its recognition motif is presented to the Hsp70 system–can determine the action the Hsp70 exerts on its client, and the fate of the client once released from the chaperone. As described above, Hsp70s in all cases bind a segment of the protein that is either stably or transiently exposed. This binding will energetically bias the substrate towards conformations in which the binding segment is accessible; in many but not all cases this translates into a state that is less folded than the state that existed before chaperone binding. Then, upon release from binding, the substrate may re-engage in the downstream process, be it folding or interaction with a partner. If these downstream processes fail, the substrate may either re-bind to an Hsp70, or be degraded. In the paragraphs below, we relate the way Hsp70s act on their substrates to particular physiological functions of Hsp70s, thus illustrating the resourcefulness of the evolutionary process: Once a molecular machine has evolved to mediate a mechanistically simple function, specialized tasks can be facilitated by adapting the mechanism to particular needs and recruiting co-chaperones that add diversity.
The role of Hsp70s in maintaining proteins in an unfolded state: a holdase and an unfoldase
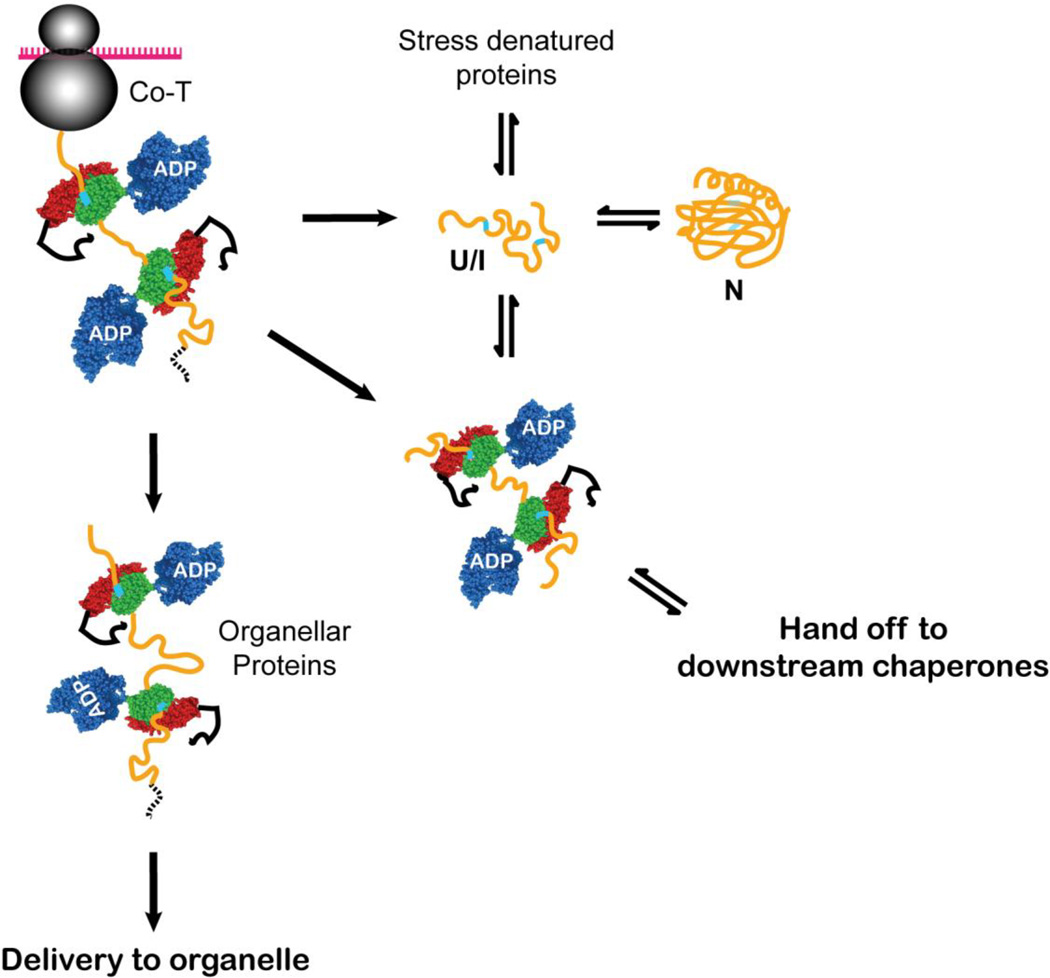
One of the major functions that has been associated with Hsp70s in vivo is maintenance of the unfolded state. In the cell, this role comes into play when a nascent chain emerges from the ribosome upon its biosynthesis, when a protein is destined to a non-cytoplasmic location and must be translocated across a membrane, in general requiring that it not fold prematurely, and when stress leads to accumulation of partially unfolded proteins that are susceptible to aggregation (Figure 2). Numerous discussions have appeared in the literature (see, for example44) debating whether Hsp70s actively unfold their substrates–to wit, an unfoldase activity, or simply bind and restrict a substrate from folding or aggregating: a holdase activity.
Figure 2.
Hsp70s function to maintain and favor the unfolded state of their substrates for a variety of downstream outcomes, including folding to the native state (N), delivery to organelles for translocation across membranes, and hand off to downstream chaperones (or degradation machinery in quality control pathways, not shown). The Hsp70 binding may occur co-translationally on a nascent chain or post-translationally to substrates that are released from the ribosome or stress-unfolded. Upon release from the Hsp70 interaction, the substrate is in an unfolded (U) or folding-competent intermediate (I) state. In this cartoon, the polypeptide substrate is shown in yellow with Hsp70 binding sites shown in cyan; an organellar targeting sequence is shown as a dashed black line. The Hsp70 subdomains are colored blue for the NBD, green for the β-subdomain of the SBD, red for the α-helical lid, and black for the unstructured C-terminal segment. In this and subsequent figures, the Hsp70 co-chaperones (J-proteins and nucleotide exchange factors) are not shown for simplicity.
Interaction with nascent proteins as they exit the ribosome represents one of the best-established holdase functions of Hsp70s.45 The roles of the Hsp70s in this context are to delay folding until all sequence elements required for folding are accessible, to prevent the chain from acquisition of non-native interactions, to protect the nascent chain from aggregation, and to release the chain in a form that is capable of folding or interacting with a downstream chaperone. In both prokaryotes and eukaryotes Hsp70s that play these co-translational holdase functions are associated with the ribosome in complexes with J-proteins and other members of a "greeting committee" for the newborn nascent chains. For example, in yeast the Hsp70 associated with ribosome is SSB, and the SSB co-translationally binds to a subset of nascent polypeptides with its binding specificity regulated by its co-chaperone, RAC.46 Thus, the Hsp70 system has privileged access to the newly synthesized polypeptide, which presents exposed hydrophobic stretches, most likely in an extended conformation. After the chains are released from ribosome-associated Hsp70s, they are likely largely unfolded and able either to fold spontaneously, to be handed to downstream chaperones for assisted folding (including the Hsp60 chaperones and the Hsp90 chaperone system), or to be targeted to non-cytoplasmic destinations via interaction with specialized machineries using specific targeting sequences. Indeed, in vitro experiments using unfolded or partially folded model substrates indicate that substrates released from the E. coli Hsp70 are in an unfolded conformation that can either spontaneously refold to the native state or misfold and then re-bind the chaperone system.37, 47 Thus far, there is no experimental evidence that the rate of folding of the client is altered by Hsp70. For example, the staphylococcal nuclease model substrate studied by Fink and coworkers displayed the same rate of refolding after ATP-induced release from the chaperone as the free protein under the same conditions.37 Similarly, Sharma et al. found using a stably misfolded monomeric luciferase substrate that the product released from DnaK by ATP folded spontaneously by kinetics that match the unassisted refolding reaction.47
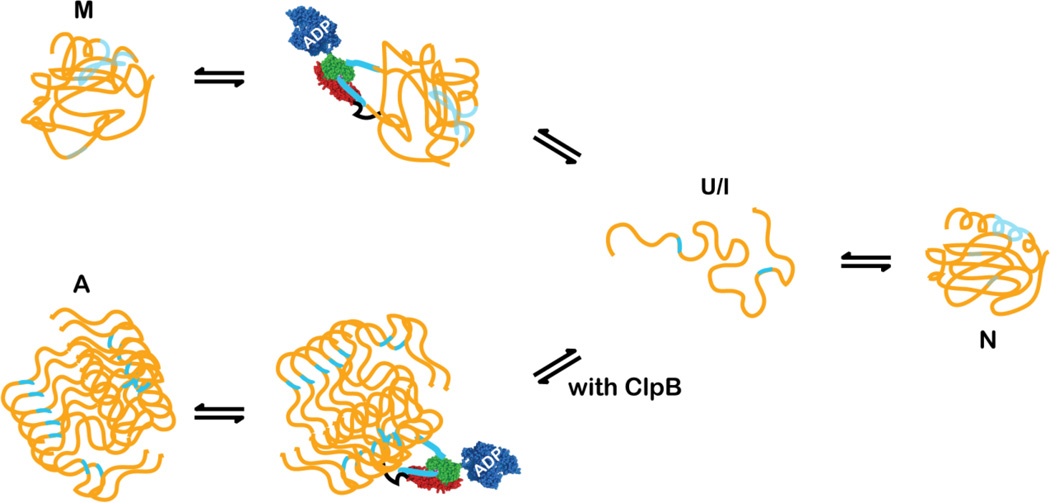
In contrast to their holdase activity, Hsp70s have been demonstrated in vitro to act as unfoldases on misfolded substrates. In the case of nascent chains emerging from the ribosome the protein substrates are more or less unfolded, and the Hsp70 system has in principle unlimited access to its binding sequences, suggesting that the conformations stabilized by Hsp70 binding are similar and biased towards unfolded states. By contrast, when a protein is stress-unfolded in the cell and needs to be protected by the Hsp70 system from aggregation, it may visit either a completely or partially unfolded conformations that have non-native interactions. Recognition motifs for Hsp70 binding are hydrophobic in character so that they may be involved in interactions favoring collapsed states. When the substrate is non-native, it will dynamically sample multiple conformations, and Hsp70 binding sites will be transiently accessible (Figure 3). Binding of these sequences will in turn favor more unfolded conformations in a conformational selection mechanism, leading upon substrate release to new opportunities for the substrate to fold productively. Thus the roles of Hsp70s in rescuing stress-unfolded proteins are a combination of holdase and unfoldase actions.
Figure 3.
Hsp70s can actively unfold misfolded states (M) and partner with other chaperones, like ClpB to undo aggregates (A). Hsp70s must bind to accessible binding sites to initiate the unfolding or disaggregation reaction. The coloring scheme is the same as in Figure 2.
Again, in vitro work supports these general functional actions of Hsp70s: For example, the stably misfolded monomeric form of luciferase described above is released from the DnaK chaperone system in an intermediate folding state, free of non-native interactions that inhibit folding because of Hsp70 action, such that the released product is capable of spontaneously refolding to the native state.47
Hsp70s as Disaggregation Machines, Helped by Partners
The unfoldase activity of Hsp70s can also be harnessed in the rescue of protein aggregates (Figure 3). Hsp70s generally partner with unfolding chaperones, like the ClpB proteins in bacteria (see below);48, 49 there are also reports that the Hsp70 system can dissagregate certain model proteins without the help of other unfoldases or chaperones.50 In either case, the Hsp70 chaperone system releases the client protein such that it may recommence proper folding, or if folding fails, be degraded.
The main action of the Hsp70 system is to bind accessible segments at the surface of the aggregates that contain sequences with the typical motifs for chaperone interaction. Hsp70 and its co-chaperones employ ATP hydrolysis to extract the substrate from the aggregate surface51, 52 by clamping the helical lid onto the bound segment and causing local unfolding around the binding site.39 This action may also be active, in that a given substrate polypeptide can be pulled away from the aggregate by entropic pulling, as also proposed for the vectorial translocation of polypeptides across membranes (see below).53, 54 A polypeptide that cannot easily be detached from the aggregate is postulated to require more Hsp70 molecules to bind to the same polypeptide (if sites are available) and to pull in a cooperative way.53
The disaggregation machinery of E. coli comprises both the Hsp70 system and the AAA+ chaperone ClpB, working in partnership.55 In yeast, the analogous system is comprised of Hsp104 and Hsp70.56 The system that carries out this function in higher eukaryotes has not yet been definitively identified, but a disaggregase partnership has been demonstrated between mammalian Hsp70 and Hsp110,57 which is a structural homologue of Hsp70 that lacks a nucleotide-mediated allosteric conformational change and has previously been shown to act as a nucleotide exchange factor with Hsp7058. The best-characterized disaggregation machinery is the DnaK/ClpB system. DnaK initiates the disaggregation reaction and hands off the substrate to ClpB. Recent NMR and mutagenesis studies reveal that an interacting interface is formed between subdomains IB and IIB of the DnaK NBD and the coiled-coil middle (M) domain of ClpB59, 60. ClpB and GrpE bind to the same surface on the NBD of DnaK and thus compete with each other for binding. Kay and co-workers demonstrated that the ClpB-DnaK interaction does not stimulate nucleotide exchange of DnaK but instead promotes substrate release59. They propose that in the disaggregation cycle, the substrate dissociates from DnaK through the interaction between ClpB and DnaK and is brought to the substrate-processing central pore of ClpB for unfolding. Interestingly, the ClpB ATPase and threading activities are essential for substrate release from DnaK, suggesting that hand-off requires both chaperones to act on the substrate.59 Mogk and co-workers61 show that the M domains of ClpB may adopt alternative conformations, and the so-called "depressed" conformation interacts with DnaK to target aggregated substrates. This is consistent with recently solved cryo-EM structures of a ClpB variant,62 which also suggests that the hexameric ClpB ring can bind multiple Hsp70 molecules and is ideal for aggregated substrates. Although a general picture how ClpB collaborates with DnaK for disaggregation is emerging, many details still need to be elucidated. For example, experimental evidence for multiple DnaK molecules to interact with one ClpB hexameric ring is lacking. And the question why this machinery only targets aggregates as substrates remains to be answered.
Hsp70s as facilitators of protein translocation across membranes
Most proteins destined for non-cytoplasmic locations must be translocated across membranes in an unfolded state and represent another well-described set of physiological roles of Hsp70s. For post-translational translocation processes, as in mitochondrial63 and chloroplast import64, Hsp70s act within the cytoplasm to keep precursor proteins from folding prematurely, essentially a holdase function. The precursor thus retains the ability to be translocated in an extended conformation across a membrane and is protected from aggregation. Hsp70s transfer the pre-proteins to the membrane complex responsible for translocation. In the interior of organelles including the mitochondrial matrix, chloroplast stroma, or the lumen of the endoplasmic reticulum, Hsp70s interact with the translocating chain from the trans side of the membrane to ensure vectorial transport and hand-off to chaperones for further maturation and assembly of multimeric complexes. Substantial controversy has existed about how this role is carried out, whether by an active motor-like power stroke or by a Brownian ratchet mechanism. The most recent model reconciles alternative proposals by describing the action as entropic pulling53, 54, in which the binding of the Hsp70 near the translocation pore limits the available conformational space of the polypeptide, thus limiting its Brownian movements. Release of the chain on the trans side of the pore is accelerated by the entropic pulling force.
Hsp70s modulate protein substrates by conformational selection, facilitating specialized functions
Hsp70 systems act on a variety of substrates that are neither unfolded nor misfolded. Instead, in these cases the Hsp70 recognizes either a transiently unfolded and accessible binding site or an unstructured segment of polypeptide chain on an otherwise folded substrate. The action of the Hsp70 chaperone is to shift the conformational ensemble of the substrate by binding to its recognition site and favouring a conformation in which this site remains optimally presented for chaperone interaction. Two well-characterized examples of this mechanism of action–the regulation of the activity of the σ32 transcription factor in bacteria34 and disassembly of clathrin cages in coated vesicles43–illustrate the exploitation of Hsp70 action for a specialized function. In both examples, Hsp70s exert regulatory functions by binding protein segments that are accessible for chaperone binding in conformations that pre-exist on the substrate’s energy landscape (in the case of σ32, the partner J-protein acts upstream to favour exposure of the DnaK binding site). By binding and stabilizing conformations with the DnaK-binding motif exposed, Hsp70 interaction shifts the conformational equilibrium towards a functionally productive state.
The physiological consequence in bacteria of DnaJ and DnaK action on σ32 is the regulation of its proteolysis by FtsH, which turns off the heat shock response.65 When chaperones are abundant, σ32 is broken down, turning off the stress response that upregulates chaperone production. Rodriguez et al. showed that DnaJ binds segment 52 to 64 in the N-terminal domain of σ32, primarily via side chain interactions, and causes conformational changes in the client protein: first in the domain to which it binds, which becomes globally destabilized, and also near the binding site for DnaK, which is located at residues 198 to 201, causing it to become accessible. In this way, DnaJ facilitates DnaK binding.34 Subsequently, DnaK binding leads to a further conformational selection and exposure of the scissile region for FtsH cleavage. The model that fits all of the data posits that σ32 equilibrates between a closed and open conformation. DnaJ binding shifts the equilibrium towards an open state with exposure of the DnaK binding site. DnaK binding in turn triggers the proteolytic action of FtsH on σ32 and down-regulation of the heat shock response.
Hsc70, the constitutively expressed cytoplasmic Hsp70 of higher eukaryotes, is known as the uncoating ATPase based on its specialized role in disassembling clathrin triskelions from coated vesicles during endocytosis.66 This Hsc70 partners with a specialized J-protein called auxilin,67 which associates with vertices of the clathrin coats, thus bringing the Hsc70 to this geometric site. Elegant recent studies from the Kirchhausen and Harrison groups43, 68 have provided a structurally based model for the action of the Hsc70. As noted above, the recognition motif, QLMLT, resides on the mobile C-terminal tails of the clathrin molecules, which are otherwise tied up in a trimeric triskelion structure that is intimately linked to the coat architecture. The auxilin-targeted binding of Hsc70 molecules to its recognition motif on one clathrin molecule puts strain on the coat arrangement, selecting conformations that are incompatible with stable triskelions and causing weakness of triskelions two vertices away. The cooperative action of several Hsc70s in binding to locally fluctuating C-terminal tails is sufficient to tip the balance towards disassembly and uncoating of the vesicle. This example of a specialized Hsp70 function exploits the binding of an accessible sequence to effect a disassembly reaction at a much longer distance scale, through collaboration of several Hsc70s.
Hsp70s hand off substrates to downstream chaperones
The ability of Hsp70s to facilitate many physiological functions relies on their cooperation with other chaperones. The action of an Hsp70 can prepare a given substrate to be productively acted on by a downstream chaperone in a hand-off cascade. The archetypal hand-off team is the E. coli DnaK system partnership with the chaperonin system GroEL/ES.69 Abundant evidence now exists that nascent chains interact with DnaK early upon their release from the ribosome, and that a subset of particularly recalcitrant folders then is passed on from DnaK to GroEL where the ultimate folding step takes place.70 In addition, stress-unfolded proteins may be protected by DnaK and then upon their release helped to fold to their native state and avoid aggregation by interaction with the chaperonin system. Moreover, under conditions of limiting GroEL/ES, substrates reliant on this chaperonin system accumulate on DnaK.71 Despite the extensive biochemistry data available and the clear importance of this team for the folding of many proteins in E. coli, details about the actual hand-off step are poorly described. Present understanding would suggest that DnaK releases a particular substrate, which simply diffuses to a GroEL chaperonin, presumably faster than it can irreversibly misfold or aggregate, unless the chaperonin concentration is too low. Simulations based on this model have quite good success in recapitulating the behavior of the E. coli chaperone networks.72
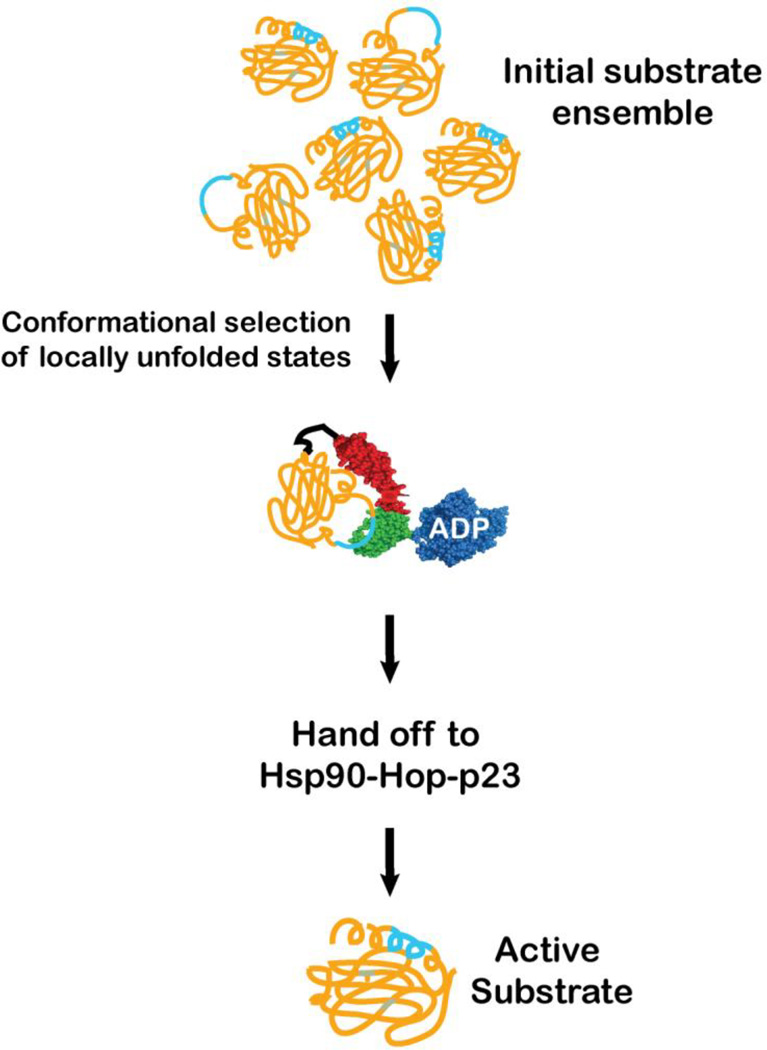
Arguably the best understood example of cooperation between the Hsp70 system and other chaperones is the hand off of substrates to Hsp90 (Figure 4). New details of the mechanism of substrate transfer between these two chaperones have been emerging in the last year. Hsp90 is known to act on substrates in the later stages of their folding.73 Agard and co-workers developed an in vitro reconstitution system for the hand off of the glucocorticoid receptor ligand-binding domain (GR-LBD) from Hsp70 to Hsp90.74 While purified GR-LBD is able to fold and bind ligand, in vivo this substrate requires Hsp70 and Hsp90 for its maturation. These authors found that interaction with Hsp70 caused a local unfolding around the ligand-binding site and consequent inactivation of GR-LBD. The Hsp70 complex with GR-LBD thus provides another example of a native protein with only local unfolding as a result of Hsp70 action. Recovery of activity from the complex of GR-LBD and Hsp70 required Hsp90 and the two co-chaperones Hop (Hsp-organizing protein) and p23, as well as ATP hydrolysis by Hsp90. Cryo-EM images of the Hsp90, Hsp70, Hop, and GR-LBD complex are consistent with Hsp70 binding between the dimeric Hsp90 molecules, and GR-LBD interaction with one monomer and Hop interaction with the other. Alvira et al. drew similar conclusions from their EM analysis of complexes of Hsp90, its co-chaperone Hop, Hsp70, and GR-LBD.75 The complex between Hsp70 and substrate (pre-formed in the presence of Hsp40) binds via the C-terminal disordered region of the Hsp70 to the first TPR domain of Hop, and the movements of this domain bring the Hsp70-GR substrate complex close to Hsp90.
Figure 4.
Hsp70s hand off substrates to downstream chaperones, like Hsp90s. The action of the Hsp70 on substrates influences them so that they interact productively with the partner chaperone. Here the example of the glucocorticoid receptor ligand-binding domain (GR-LBD) is illustrated. As described in the text, the conformational ensemble of the GR-LBD contains locally unfolded states with an exposed Hsp70 binding site (cyan). The binding of the Hsp70 selects these conformations, which are in turn delivered in a process facilitated by co-chaperones Hop and p23 to the Hsp90 for further maturation steps. The coloring scheme is as in Figure 2.
Recent mass spectrometry analysis suggests that in the dominant complex formed with Hsp90, Hop and GR-LBD, Hsp70 exists as an anti-parallel dimer, and that the Hsp70 monomers may either bind the GR-LBD substrate or the Hsp90 and Hop.(C. Robinson, personal communication) In either case, complex formation between GR-LBD and Hsp70 primes this substrate for transfer to Hsp90 by binding to dynamically accessible sequence and favoring local unfolding.
The role of the J-protein
The in-cell functions of Hsp70s always require the participation of partner J-protein co-chaperones (also called Hsp40s), which cooperate with Hsp70s in several ways including targeting them to specific cellular locations (exemplified by auxilin), stimulating the Hsp70 ATPase rate and thus substrate binding-release cycle time, delivering substrates to Hsp70s, and more.9 While many functional associations of J-proteins with Hsp70s have been described, the mechanistic roles of J-proteins in regulating Hsp70 function and how the J-protein impacts Hsp70-substrate complex formation are not well understood. The defining feature of J-proteins is the presence of a highly conserved ca. 70-residue J-domain; the NBD of Hsp70s interacts with a motif of exposed residues in this domain. Apart from their conserved J-domains, J-proteins exhibit substantial diversity of sequence and structural domains. Multiple J-proteins can collaborate with a single Hsp70, and they play important roles in driving functional and substrate diversities of Hsp70s.
Some J-proteins contain their own client-binding domains, and others do not. In this latter group, the J-proteins recruit Hsp70 molecules to a particular site of action and thereby increase the local concentration of Hsp70s near their substrates. For example, a ribosome-associated J-protein lacking a substrate-binding domain recruits soluble Hsp70s for downstream folding events.76 Similarly, the J-protein Hlj1 is tethered on the cytosolic face of the ER membrane and recruits Hsp70s for ER-associated degradation (ERAD).77 On the lumenal side of the ER membrane, the ER resident Hsp70 BiP is brought into close association with the translocating protein by interaction with a translocon-associated J-protein.78
For J-proteins with client protein-binding domains, many lines of evidence make it clear that substrates are delivered by the partner J-protein to the Hsp70s, and in many cases, the Hsp70 in such a partnership will not bind its substrate directly, but instead obligatorily requires that it be presented by the J-protein partner.79 To understand the mechanism by which J-proteins work with their Hsp70 partners in remodeling substrates, the following questions need to be addressed: Do J-proteins and Hsp70s bind to the same site or different sites of the substrate? Does the binding of J-protein alter the substrate conformational state to facilitate Hsp70 binding? How is substrate transferred from J-protein to Hsp70? The answers to these questions are only known for a few systems, and the generality of the answers cannot be assumed, given the variety of J-protein/Hsp70 partnerships and functions.
Several studies have reported that J-proteins and Hsp70s bind to different sites on their substrates, and/or they have different effects on the conformational properties of substrate, suggesting that both J-protein and Hsp70 may bind at the same time to the substrate to form a ternary complex. For example, the interaction of the DnaK and DnaJ with σ32 involves different sites, as described above.34 Importantly, binding of DnaJ to σ32 has a significant effect on the σ32 conformation, opening up the DnaK-binding site. In the case of E. coli HscA/HscB, an Hsp70/Hsp40 pair mediating transfer of iron-sulfur clusters from one protein to another during their assembly, with their substrate protein, IscU, HscB preferentially binds a relatively structured conformation of IscU, and delivers it to ATP-bound HscA. After ATP hydrolysis, and transfer of the Fe-S cluster to acceptor proteins, IscU becomes disordered and remains bound to ADP-bound HscA. Re-binding of ATP to HscA then releases disordered IscU.80, 81
While there are clearly key roles for J-protein/Hsp70 partnerships, which may include simultaneous or serial interaction with a substrate, J-proteins lack nucleotide modulation of their substrate affinity. In the partnership with Hsp70s, the player that has the most versatile activity and can be harnessed in substrate remodeling for a variety of physiological roles is the Hsp70 molecular machine. Nonetheless, Hsp70s do not perform alone!
Concluding Remarks: Many substrates--many functions, all exploiting the same molecular mechanism
As discussed in this Perspective, Hsp70s have an apparently simple mechanism: They bind short hydrophobic sequences in extended conformations. The kinetics of their association and dissociation with the chaperone are enhanced by ATP binding, and diminished when ADP is bound. Hsp70 co-chaperones embellish this simple mechanism in a variety of ways: substrate delivery, cellular localization, acceleration of nucleotide hydrolysis, nucleotide exchange, etc. Nonetheless, the impact of Hsp70 binding on their substrates is biophysically intriguing: including global to local unfolding, conformational selection, and altered dynamics. These actions have been exploited through evolution for an array of purposes, from the maintenance of an unfolded state to the disassembly of clathrin-coats. Hsp70s interact with a large and diverse set of substrates in vivo. For example, a quantitative proteomics study in E. coli found over seven hundred cytosolic proteins that interact with DnaK, with approximately 180 of them highly enriched on DnaK (i.e., high affinity binders).71 Connecting this vast array of substrates to specific functional roles of their Hsp70 interaction will be exciting and challenging. Understanding how Hsp70s cooperate with their co-chaperones to augment the spectrum of substrates they may interact with and to achieve greater specificity will also be fascinating. Findings such as a single residue change between the yeast Hsp70s Ssa1 and Ssa2 that determines whether they act in prion propagation or protein degradation, but via co-chaperone interactions and not substrate binding specificity,82 are tantalizingly suggestive of the uncharted territory that we must explore to fully appreciate Hsp70 physiological roles. Based on what we know currently, we suspect that Nature has harnessed this simple machine in myriad ways. Connecting the biophysics of this molecular machine to its physiological functions will be a fascinating ongoing endeavor.
Highlights.
Hsp70 molecular chaperones perform a wide array of functions using a simple mechanism.
Key to the function of Hsp70s is how they recognize and influence their substrates.
Hsp70s team with co-chaperones, which enhance functional diversity and cellular roles.
Current understanding of Hsp70-substrate interactions is limited.
Acknowledgements
We thank our colleagues Peter Chien, Charles English and Karan Hingorani for critical reading of this manuscript and Rob Vass for help with Figure 1. Work in the Gierasch laboratory on Hsp70 mechanism and chaperone networks in E. coli is supported by NIH grants GM027616 and GM101644, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Zuiderweg ER, Bertelsen EB, Rousaki A, Mayer MP, Gestwicki JE, Ahmad A. Allostery in the Hsp70 chaperone proteins. Top Curr Chem. 2013;328:99–153. doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci U S A. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell. 2012;48:863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 5.Qi R, Sarbeng EB, Liu Q, Le KQ, Xu X, Xu H, Yang J, Wong JL, Vorvis C, Hendrickson WA, Zhou L, Liu Q. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol. 2013;20:900–907. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Los Rios P, Barducci A. Hsp70 chaperones are non-equilibrium machines that achieve ultra-affinity by energy consumption. eLife. 2014;3:e02218. doi: 10.7554/eLife.02218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhuravleva A, Clerico EM, Gierasch LM. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151:1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 11.Lanneau D, Wettstein G, Bonniaud P, Garrido C. Heat shock proteins: cell protection through protein triage. ScientificWorldJournal. 2010;10:1543–1552. doi: 10.1100/tsw.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gragerov A, Gottesman ME. Different peptide binding specificities of hsp70 family members. J Mol Biol. 1994;241:133–135. doi: 10.1006/jmbi.1994.1482. [DOI] [PubMed] [Google Scholar]
- 13.Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman ME. Specificity of DnaK-peptide binding. J Mol Biol. 1994;235:848–854. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- 14.Flynn GC, Chappell TG, Rothman JE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 15.Blond-Elguindi S, Fourie AM, Sambrook JF, Gething MJ. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem. 1993;268:12730–12735. [PubMed] [Google Scholar]
- 16.Fourie AM, Sambrook JF, Gething MJ. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J Biol Chem. 1994;269:30470–30478. [PubMed] [Google Scholar]
- 17.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulosebound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Durme J, Maurer-Stroh S, Gallardo R, Wilkinson H, Rousseau F, Schymkowitz J. Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comput Biol. 2009;5:e1000475. doi: 10.1371/journal.pcbi.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivasan SR, Gillies AT, Chang L, Thompson AD, Gestwicki JE. Molecular chaperones DnaK and DnaJ share predicted binding sites on most proteins in the E. coli proteome. Mol Biosyst. 2012;8:2323–2333. doi: 10.1039/c2mb25145k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcinowski M, Rosam M, Seitz C, Elferich J, Behnke J, Bello C, Feige MJ, Becker CF, Antes I, Buchner J. Conformational selection in substrate recognition by Hsp70 chaperones. J Mol Biol. 2013;425:466–474. doi: 10.1016/j.jmb.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 21.Vembar SS, Jonikas MC, Hendershot LM, Weissman JS, Brodsky JL. J domain co-chaperone specificity defines the role of BiP during protein translocation. J Biol Chem. 2010;285:22484–22494. doi: 10.1074/jbc.M110.102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, Leu JI, Murphy ME, George DL, Marmorstein R. Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with peptide substrate. PLoS One. 2014;9:e103518. doi: 10.1371/journal.pone.0103518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahn M, Berthold N, Kieslich B, Knappe D, Hoffmann R, Strater N. Structural studies on the forward and reverse binding modes of peptides to the chaperone DnaK. J Mol Biol. 2013;425:2463–2479. doi: 10.1016/j.jmb.2013.03.041. [DOI] [PubMed] [Google Scholar]
- 25.Cupp-Vickery JR, Peterson JC, Ta DT, Vickery LE. Crystal structure of the molecular chaperone HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC. J Mol Biol. 2004;342:1265–1278. doi: 10.1016/j.jmb.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 26.Czihal P, Knappe D, Fritsche S, Zahn M, Berthold N, Piantavigna S, Muller U, Van Dorpe S, Herth N, Binas A, Kohler G, De Spiegeleer B, Martin LL, Nolte O, Strater N, Alber G, Hoffmann R. Api88 is a novel antibacterial designer peptide to treat systemic infections with multidrug-resistant Gram-negative pathogens. ACS Chem Biol. 2012;7:1281–1291. doi: 10.1021/cb300063v. [DOI] [PubMed] [Google Scholar]
- 27.Tapley TL, Cupp-Vickery JR, Vickery LE. Sequence-dependent peptide binding orientation by the molecular chaperone DnaK. Biochemistry. 2005;44:12307–12315. doi: 10.1021/bi051145r. [DOI] [PubMed] [Google Scholar]
- 28.Tapley TL, Cupp-Vickery JR, Vickery LE. Structural determinants of HscA peptide-binding specificity. Biochemistry. 2006;45:8058–8066. doi: 10.1021/bi0606187. [DOI] [PubMed] [Google Scholar]
- 29.Mayer MP, Schroder H, Rudiger S, Paal K, Laufen T, Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struct Biol. 2000;7:586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- 30.Chang YW, Sun YJ, Wang C, Hsiao CD. Crystal structures of the 70-kDa heat shock proteins in domain disjoining conformation. J Biol Chem. 2008;283:15502–15511. doi: 10.1074/jbc.M708992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aprile FA, Dhulesia A, Stengel F, Roodveldt C, Benesch JL, Tortora P, Robinson CV, Salvatella X, Dobson CM, Cremades N. Hsp70 oligomerization is mediated by an interaction between the interdomain linker and the substrate-binding domain. PLoS One. 2013;8:e67961. doi: 10.1371/journal.pone.0067961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson AD, Bernard SM, Skiniotis G, Gestwicki JE. Visualization and functional analysis of the oligomeric states of Escherichia coli heat shock protein 70 (Hsp70/DnaK) Cell Stress Chaperones. 2012;17:313–327. doi: 10.1007/s12192-011-0307-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z, Kurt N, Rajagopalan S, Cavagnero S. Secondary structure mapping of DnaK-bound protein fragments: chain helicity and local helix unwinding at the binding site. Biochemistry. 2006;45:12325–12333. doi: 10.1021/bi0612263. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez F, Arsène-Ploetze F, Rist W, Rüdiger S, Schneider-Mergener J, Mayer MP, Bukau B. Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones. Molecular cell. 2008;32:347–358. doi: 10.1016/j.molcel.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 35.Hu SM, Wang C. Involvement of the 10-kDa C-terminal fragment of hsc70 in complexing with unfolded protein. Arch Biochem Biophys. 1996;332:163–169. doi: 10.1006/abbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 36.Smock RG, Blackburn ME, Gierasch LM. Conserved, disordered C terminus of DnaK enhances cellular survival upon stress and DnaK in vitro chaperone activity. J Biol Chem. 2011;286:31821–31829. doi: 10.1074/jbc.M111.265835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palleros DR, Shi L, Reid KL, Fink AL. hsp70-protein complexes. Complex stability and conformation of bound substrate protein. J Biol Chem. 1994;269:13107–13114. [PubMed] [Google Scholar]
- 38.Sekhar A, Santiago M, Lam HN, Lee JH, Cavagnero S. Transient interactions of a slow-folding protein with the Hsp70 chaperone machinery. Protein Sci. 2012;21:1042–1055. doi: 10.1002/pro.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlecht R, Erbse AH, Bukau B, Mayer MP. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat Struct Mol Biol. 2011;18:345–351. doi: 10.1038/nsmb.2006. [DOI] [PubMed] [Google Scholar]
- 40.Kellner R, Hofmann H, Barducci A, Wunderlich B, Nettels D, Schuler B. Single-molecule spectroscopy reveals chaperone-mediated expansion of substrate protein. Proc Natl Acad Sci U S A. 2014;111:13355–13360. doi: 10.1073/pnas.1407086111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcinowski M, Holler M, Feige MJ, Baerend D, Lamb DC, Buchner J. Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat Struct Mol Biol. 2011;18:150–158. doi: 10.1038/nsmb.1970. [DOI] [PubMed] [Google Scholar]
- 42.Mayer M, Reinstein J, Buchner J. Modulation of the ATPase cycle of BiP by peptides and proteins. J Mol Biol. 2003;330:137–144. doi: 10.1016/s0022-2836(03)00556-4. [DOI] [PubMed] [Google Scholar]
- 43.Bocking T, Aguet F, Harrison SC, Kirchhausen T. Single-molecule analysis of a molecular disassemblase reveals the mechanism of Hsc70-driven clathrin uncoating. Nat Struct Mol Biol. 2011;18:295–301. doi: 10.1038/nsmb.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattoo RU, Goloubinoff P. Molecular chaperones are nanomachines that catalytically unfold misfolded and alternatively folded proteins. Cell Mol Life Sci. 2014;71:3311–3325. doi: 10.1007/s00018-014-1627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 46.Willmund F, del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma SK, De los Rios P, Christen P, Lustig A, Goloubinoff P. The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat Chem Biol. 2010;6:914–920. doi: 10.1038/nchembio.455. [DOI] [PubMed] [Google Scholar]
- 48.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 49.Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci U S A. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diamant S, Ben-Zvi AP, Bukau B, Goloubinoff P. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J Biol Chem. 2000;275:21107–21113. doi: 10.1074/jbc.M001293200. [DOI] [PubMed] [Google Scholar]
- 51.Hubbard TJ, Sander C. The role of heat-shock and chaperone proteins in protein folding: possible molecular mechanisms. Protein Eng. 1991;4:711–717. doi: 10.1093/protein/4.7.711. [DOI] [PubMed] [Google Scholar]
- 52.Rothman JE. Polypeptide chain binding proteins: catalysts of protein folding and related processes in cells. Cell. 1989;59:591–601. doi: 10.1016/0092-8674(89)90005-6. [DOI] [PubMed] [Google Scholar]
- 53.De Los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc Natl Acad Sci U S A. 2006;103:6166–6171. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goloubinoff P, De Los Rios P. The mechanism of Hsp70 chaperones: (entropic) pulling the models together. Trends Biochem Sci. 2007;32:372–380. doi: 10.1016/j.tibs.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 55.Weibezahn J, Schlieker C, Tessarz P, Mogk A, Bukau B. Novel insights into the mechanism of chaperone-assisted protein disaggregation. Biol Chem. 2005;386:739–744. doi: 10.1515/BC.2005.086. [DOI] [PubMed] [Google Scholar]
- 56.Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 57.Mattoo RU, Sharma SK, Priya S, Finka A, Goloubinoff P. Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates. J Biol Chem. 2013;288:21399–21411. doi: 10.1074/jbc.M113.479253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover J, Kay L. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science. 2013;339:1080–1083. doi: 10.1126/science.1233066. [DOI] [PubMed] [Google Scholar]
- 60.Doyle SM, Shastry S, Kravats AN, Shih YH, Miot M, Hoskins JR, Stan G, Wickner S. Interplay between E. coli DnaK, ClpB and GrpE during Protein Disaggregation. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.10.013. E-pub ahead of print, Oct. 29, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seyffer F, Kummer E, Oguchi Y, Winkler J, Kumar M, Zahn R, Sourjik V, Bukau B, Mogk A. Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat Struct Mol Biol. 2012;19:1347–1355. doi: 10.1038/nsmb.2442. [DOI] [PubMed] [Google Scholar]
- 62.Carroni M, Kummer E, Oguchi Y, Wendler P, Clare D, Sinning I, Kopp J, Mogk A, Bukau B, Saibil H. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. eLife. 2014;3:02481. doi: 10.7554/eLife.02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paila YD, Richardson LG, Schnell DJ. New Insights into the Mechanism of Chloroplast Protein Import and Its Integration with Protein Quality Control, Organelle Biogenesis and Development. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.08.016. E-pub ahead of print, Aug. 28, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guisbert E, Herman C, Lu CZ, Gross CA. A chaperone network controls the heat shock response in E. coli. Genes Dev. 2004;18:2812–2821. doi: 10.1101/gad.1219204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chappell TG, Konforti BB, Schmid SL, Rothman JE. The ATPase core of a clathrin uncoating protein. J Biol Chem. 1987;262:746–751. [PubMed] [Google Scholar]
- 67.Ungewickell E, Ungewickell H, Holstein SE, Lindner R, Prasad K, Barouch W, Martin B, Greene LE, Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- 68.Xing Y, Bocking T, Wolf M, Grigorieff N, Kirchhausen T, Harrison SC. Structure of clathrin coat with bound Hsc70 and auxilin: mechanism of Hsc70-facilitated disassembly. EMBO J. 2010;29:655–665. doi: 10.1038/emboj.2009.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 70.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 71.Calloni G, Chen T, Schermann S, Chang H-C, Genevaux P, Agostini F, Tartaglia G, Hayer-Hartl M, Hartl F. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 72.Powers ET, Powers DL, Gierasch LM. FoldEco: a model for proteostasis in E. coli. Cell Rep. 2012;1:265–276. doi: 10.1016/j.celrep.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157:1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alvira S, Cuellar J, Rohl A, Yamamoto S, Itoh H, Alfonso C, Rivas G, Buchner J, Valpuesta JM. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat Commun. 2014;5:5484. doi: 10.1038/ncomms6484. [DOI] [PubMed] [Google Scholar]
- 76.Hundley HA, Walter W, Bairstow S, Craig EA. Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science. 2005;308:1032–1034. doi: 10.1126/science.1109247. [DOI] [PubMed] [Google Scholar]
- 77.Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 78.Johnson AE, van Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 79.Misselwitz B, Staeck O, Rapoport T. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 80.Kim JH, Tonelli M, Frederick RO, Chow DC, Markley JL. Specialized Hsp70 chaperone (HscA) binds preferentially to the disordered form, whereas J-protein (HscB) binds preferentially to the structured form of the iron-sulfur cluster scaffold protein (IscU) J Biol Chem. 2012;287:31406–31413. doi: 10.1074/jbc.M112.352617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim JH, Alderson TR, Frederick RO, Markley JL. Nucleotide-dependent interactions within a specialized Hsp70/Hsp40 complex involved in Fe-S cluster biogenesis. J Am Chem Soc. 2014;136:11586–11589. doi: 10.1021/ja5055252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sharma D, Masison DC. Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p. Proc Natl Acad Sci U S A. 2011;108:13665–13670. doi: 10.1073/pnas.1107421108. [DOI] [PMC free article] [PubMed] [Google Scholar]