Abstract
Extracellular superoxide dismutase (EC-SOD) is responsible for the dismutation of the superoxide radical produced in the extracellular space and known to be expressed by inflammatory cells, including macrophages and neutrophils. Here we show that EC-SOD is produced by resting macrophages and associated with the cell surface via the extracellular matrix (ECM)-binding region. Upon cellular activation induced by lipopolysaccharide, EC-SOD is relocated and detected both in the cell culture medium and in lipid raft structures. Although the secreted material presented a significantly reduced ligand-binding capacity, this could not be correlated to proteolytic removal of the ECM-binding region, because the integrity of the material recovered from the medium was comparable to that of the cell surface-associated protein. The naturally occurring R213G amino acid substitution located in the ECM-binding region of EC-SOD is known to affect the binding characteristics of the protein. However, the analysis of macrophages expressing R213G EC-SOD did not present evidence of an altered cellular distribution. Our results suggest that EC-SOD plays a dynamic role in the inflammatory response mounted by activated macrophages.
Keywords: Antioxidant, Extracellular superoxide dismutase, Inflammation, Macrophage, Free radicals
The antioxidant extracellular superoxide dismutase (EC-SOD)1 is the only extracellular protein responsible for the enzymatic dismutation of the superoxide radical (), producing molecular oxygen (O2) and hydrogen peroxide (H2O2). The protein is highly expressed in lung and vascular tissue [1,2] and is known to be associated with pulmonary and cardiovascular diseases [3,4]. The protein interacts with a number of ligands in the extracellular space, including heparan sulfate, type I collagen, and hyaluronan, and protects these structures against oxidative fragmentation induced by the superoxide radical [1,5–11].
The interaction with ligands in the extracellular matrix (ECM) is mediated by the C-terminal region of the protein (ECM-binding region), encompassing a sequence of six consecutive basic amino acid residues [7,8,12,13]. The binding capacity of this region is reduced by a naturally occurring amino acid substitution substituting glycine for arginine (R213G). Heterozygous or homozygous carriers present an increased concentration of EC-SOD in the circulation [14–16], which is likely to reflect the reduced affinity for ligands in the ECM [17,18]. The ECM-binding region can be removed immediately before secretion of the protein into the extracellular space by a two-step proteolytic event involving furin and an unknown carboxypeptidase [19–21]. Because the mature protein is a tetramer, this event supports the formation of mature EC-SOD with modulated ECM-binding capacity depending on the ratio of intact and cleaved subunits within the tetramer [5,12]. We have recently provided evidence that the proteolytic processing of the EC-SOD subunit is subject to redox control and argued that this type of regulation dictates the variable ratios of intact and cleaved subunits observed in EC-SOD extracted from different tissues [19,22].
A number of pulmonary diseases are characterized by an influx of inflammatory cells, including neutrophils and macrophages, which are known to express EC-SOD [23,24]. Using animal models of lung disease, it has been shown that overexpression of EC-SOD attenuates the inflammatory response by reducing the influx of neutrophils, whereas the absence of EC-SOD exacerbates the response [25–28]. Because fragments generated by the oxidation of ECM components, e.g., heparan sulfate, hyaluronan, and elastin, are known to attract inflammatory cells, it is likely that the attenuation of the inflammatory response by EC-SOD reflects the capacity to inhibit oxidative fragmentation [9,10]. However, recent evidence suggests that EC-SOD may also play a role in the maturation of the inflammatory response. In a mouse model using Escherichia coli-induced pneumonia, Manni et al. [29] showed that the lack of EC-SOD impaired phagocytosis of bacteria by macrophages. Using a model of intracellular bacterial infection by Listeria monocytogenes infecting primarily liver and spleen, Break et al. [30] showed that increased expression of EC-SOD was associated with reduced survival and speculated that suppression of EC-SOD activity will increase proinflammatory responses and thus aid in the clearance of bacterial infections. In addition, studies have shown that EC-SOD may also be involved in the maturation of dendritic cells and controls the migration of inflammatory cells by regulating the expression of adhesion molecules and cytokines [27,31,32]. Collectively, these studies clearly show that apart from being an antioxidant preventing molecular fragmentation, EC-SOD may also play important and diverse roles in regulating the activity of inflammatory cells and hence the orchestration of the inflammatory response.
Here we show that even though resting macrophages express EC-SOD, the protein is not secreted into the culture medium, but remains bound to the cell surface. However, upon cellular activation induced by LPS, the protein is detected in the culture medium, indicating active secretion of the protein. This event was not reflected by a concomitant decrease in surface-associated protein as evaluated by flow cytometry. Interestingly, we find that secreted EC-SOD had lost its affinity for heparin although the subunit composition was similar to the material associated to the cell surface. In addition to the secretion of EC-SOD, we find that the protein is present in lipid rafts of activated macrophages. The dynamic distribution of EC-SOD was not affected by the R213G substitution as evaluated by using cells derived from EC-SOD R213G knock-in mice. Our data suggest a dynamic role for EC-SOD in the inflammatory response mounted by macrophages.
Materials and methods
R213G EC-SOD knock-in mouse
The establishment of the R213G EC-SOD knock-in mouse is described in detail elsewhere (submitted for publication). The R213G knock-in targeting vector was created using the high-fidelity Red/ET recombination technology (Gene Bridges). Briefly, a 10.5-kb region was subcloned from a positively identified C57BL/6 BAC clone by homologous recombination. The short homology arm extended 2.25 kb 3′ of the LoxP-flanked Neo cassette and the long homology arm extended 6.13 kb to the 3′ end of the single LoxP site. The single LoxP site was inserted upstream of exon 2 in intron 1–2, and the LoxP-flanked Neo cassette was inserted downstream of exon 2 in intron 2–3. The C-to-G base mutation (amino acid substitution Arg to Gly) within exon 2 was generated by a three-step PCR mutagenesis protocol. Using conventional subcloning methods, the wild-type sequence was replaced with the PCR product harboring the point mutation. Targeted iTL BA1 (C57BL/6N × 129/SvEv) hybrid embryonic stem cells were microinjected into C57BL/6 blastocysts. Resulting chimeras with a high percentage of agouti coat color were mated to wild-type C57BL/6 mice to generate F1 heterozygous offspring. We obtained two independent C57BL/6 founder lines to generate homozygous R213G knock-in mice. DNA was sequenced periodically to verify the lack of new spontaneous mutations.
Antibodies and reagents
The following antibodies were used: goat anti-mouse EC-SOD (S-19, Santa Cruz Biotechnology, SC-32222), mouse anti-human EC-SOD (4G11G6, Santa Cruz Biotechnology, SC-101338), rabbit anti-mouse EC-SOD antiserum (in-house), horseradish peroxidase (HRP)-conjugated goat anti-rabbit Ig (DAKO, P0448), HRP-conjugated rabbit anti-mouse Ig (DAKO, P0260), Alexa Fluor 633-conjugated donkey anti-goat IgG (Invitrogen, A21082), fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse F4/80 (macrophage marker; Serotec, MCA497F), and Alexa Fluor 555-conjugated cholera toxin subunit B (Life Technologies, C-34776). GammaBind G Sepharose was obtained from GE Healthcare. LPS, octyl-β-D-glucopyranoside (β-OG), poly-L-lysine, macrophage colony-stimulating factor (m-CSF), bovine serum albumin (BSA), and Hoechst 33258 were obtained from Sigma. Complete Ultra proteinase inhibitor cocktail was obtained from Roche. Normal donkey serum was obtained from Millipore (S30-100ML) and normal goat IgG was obtained from Santa Cruz Biotechnology (sc-2028).
Generation of bone marrow-derived macrophages (BMMs)
Femurs and pelvises from mice were removed, cleaned of tissue and muscle, and subsequently rinsed in ice-cold phosphate-buffered saline (PBS). Both ends of the bones were removed and bone marrow was recovered in RPMI 1640 medium supplemented with 20% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 1.5 units heparin/ml, and 10 ng/ml m-CSF (BMM medium) by flushing the bone using a 1-ml syringe and a 27-gauge needle. Cells were recovered by centrifugation for 5 min at 150 g and resuspended in fresh medium and seeded in six-well plates and placed at 37°C at 5% CO2. The cells were allowed to settle for 1 day after which medium containing nonadherent cells was transferred to a new six-well plate to allow for cells to adhere. This procedure was repeated two times. The cells were maintained in culture by changing 2/3 of the culture medium every third day and allowed to differentiate into macrophages for 7–10 days.
Analysis of EC-SOD expression by BMMs
The expression of EC-SOD by BMMs was evaluated by analyzing cell culture supernatants and lysates. Before analysis, the cells were washed in PBS and cultured for 18 h at 37 °C in serum-free medium in the absence or presence of LPS (100 ng/ml). For evaluation of EC-SOD secretion, the medium was collected and placed on ice until further analysis. Cells were washed in PBS in the absence or presence of heparin (25 units/ml) and added to ice-cold 50 mM Tris–HCl, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 20 mM N-ethylmaleimide, pH 7.4, supplemented with protease inhibitors and incubated on ice for 15 min. The lysates were subsequently cleared by centrifugation at 14,000 g and placed on ice. The material not solubilized by Triton X-100 was washed two times in PBS and subsequently resuspended in 10 mM Tris–HCl, 500 mM NaCl, 1% Triton X-100, 60 mM β-OG, pH 7.6, containing protease inhibitors and allowed to incubate on ice for 30 min. The β-OG extract was recovered by centrifugation at 14,000 g and placed on ice until further analysis. This final extract contained proteins associated with lipid raft structures.
Immunoprecipitation and Western blotting
GammaBind G Sepharose was washed in PBS, and goat anti-mouse EC-SOD antibody (S-19) was added to it and incubated 2 h at 21 °C. The resin was recovered by centrifugation at 200 g, washed three times in PBS, and cell culture supernatants or cell lysates, as indicated, were added to it. The samples were mixed end-over-end at 4 C for 16 h. As controls, we included resin incubated with PBS only or with a mouse lung homogenate prepared as previously described [33]. The resin was recovered by centrifugation and washed three times in PBS, and bound proteins were eluted by boiling the resin in the presence of SDS–PAGE sample buffer containing 0.5% SDS and 50 mM dithiothreitol. Proteins isolated by immunoprecipitation were subsequently separated by polyacrylamide gel electrophoresis using uniform 10% polyacrylamide gels and the glycine/2-amino-2-methyl-1,3-propanediol-HCl buffer system [34]. For Western blotting, separated proteins were electrophoretically transferred to a polyvinylidene difluoride membrane for 1 h at 150 mA in 10 mM 3-(cyclohexylamino)-1-propane sulfonic acid, pH 11, containing 10% ethanol [35]. The membranes were blocked with 5% (w/v) skimmed milk in 20 mM Tris–HCl, 137 mM NaCl, pH 7.4, supplemented with 0.1% Tween 20 and EC-SOD protein detected by using a rabbit anti-EC-SOD antiserum. Blots were developed by enhanced chemiluminescence using peroxidase-conjugated goat anti-rabbit Ig and data acquired using an ImageQuant LAS 4000 instrument (GE Healthcare). When indicated, the intensity of the detected bands was evaluated using the ImageJ software [36].
Heparin-binding capacity
Enzyme-linked immunosorbent assay (ELISA)
To evaluate the heparin binding capacity of EC-SOD in the presence of LPS we used an ELISA. The assay was performed essentially as described before [18]. Briefly, microtiter wells (MaxiSorb, Nunc) were coated with 50 ng heparin–BSA in 100 μl 50 mM NaHCO3, pH 9.6, overnight at 23 °C. The heparin–BSA was prepared by conjugating heparin to BSA using sodium cyanoborohydride [37]. The wells were emptied and residual binding sites blocked by the addition of 200 μl 0.1% (w/v) BSA in 20 mM Tris–HCl, 135 mM NaCl, pH 7.4 (TBS). The wells were incubated at 23 °C, washed three times using TBS containing 0.05% Tween 20 (TBST), and incubated with a twofold dilution series of purified human EC-SOD in TBST in the absence or presence of 100 ng/ml LPS as indicated. Wells were incubated overnight at 4 °C and washed three times in TBST. Bound EC-SOD was detected by the addition of MAb 4G11G6 anti-human EC-SOD diluted in TBST. Bound antibody was subsequently detected by the addition of HRP-conjugated rabbit anti-mouse antibody and wells were developed by using the O-phenylenediamine dihydrochloride system following the manufacturer's instructions. Color development was assessed by absorption at 450 nm.
Affinity chromatography
To analyze the binding capacity of EC-SOD secreted from BMMs, we used cells derived from wild-type animals cultured in the presence or absence of LPS (100 ng/ml) for 16 h as described above. The medium was recovered and diluted twofold in 20 mM Tris–HCl, pH 7.4, and passed over a small column of heparin Sepharose (~0.5 ml) previously equilibrated in 20 mM Tris–HCl, 50 mM NaCl, pH 7.4. The flowthrough was collected and stored on ice. The column was washed in 20 mM Tris–HCl, 50 mM NaCl, pH 7.4, and bound protein eluted using 20 mM Tris–HCl, 500 mM NaCl, pH 7.4. The presence of EC-SOD in the starting material, the flowthrough, and the eluate was evaluated by immunoprecipitation and Western blotting as described above.
Confocal microscopy of primary murine macrophages derived from transgenic mice
For analysis by confocal microscopy, harvested bone marrow cells were seeded directly on poly-l-lysine-coated coverslips in BMM medium. Cells were allowed to adhere for 1 day at 37 °C at 5% CO2 after which nonadherent cells were removed. Attached cells were subsequently allowed to differentiate into macrophages for 5 days in BMM medium. For analysis of EC-SOD distribution, cells were incubated in BMM medium in the absence or presence of LPS (100 ng/ml) for 18 h. Cells were washed in PBS and incubated for 30 min in PBS containing 0.5% skimmed milk, 0.1% BSA, and 2% normal donkey serum (incubation buffer) to block nonspecific binding of antibody. Cells were washed gently in cold PBS and incubated at 4 °C for 1 h with Alexa Fluor 555-conjugated cholera toxin subunit B diluted to 0.1 μg/ml in incubation buffer. Cells were gently washed in cold PBS at 4 °C before fixation using ice-cold methanol for 5 min. This treatment ensures surface labeling of lipid rafts and renders cells permeable for intracellular staining of EC-SOD. The cells were washed in PBS and incubated for 1 h at 21 °C using goat anti-mouse EC-SOD (S-19) or the equivalent isotype control (normal goat IgG) diluted in incubation buffer. Cells were washed and incubated with Alexa Fluor 633-conjugated donkey anti-goat for 30 min at 21 °C to allow for the detection of bound antibodies. Nuclei were stained by using Hoechst 33258 (0.2 mg/ml in H2O). Cells were mounted on glass slides using fluorescence mounting medium (DAKO, S3023). Confocal microscopy was performed using a Zeiss LSM710 laser scanning microscope equipped with a × 40 C Apochromat 1.2 numerical aperture water objective. Acquired images were processed by using ImageJ software [36].
Evaluation of surface-associated EC-SOD by flow cytometry
BMMs representing all genotypes were cultured in BMM medium for 18 h in the presence or absence of 100 ng/ml LPS. Cells (106 in 100 μl PBS) were recovered by using a rubber policeman, washed twice in PBS, and incubated with FITC-conjugated rat anti-mouse F4/80 for 30 min. Cells were subsequently washed in PBS and incubated with goat anti-mouse EC-SOD (S-19) in PBS for 1 h. As a control for EC-SOD staining, cells were washed in PBS containing 25 units/ml heparin for 2 × 10 min before incubation with anti-EC-SOD antibody. Moreover, we included normal goat IgG as a negative control. Bound antibody was detected using Alexa Fluor 633-conjugated donkey anti-goat IgG. All incubations were performed in the dark. The fraction of dead cells was evaluated by using 7-amino-actinomycin D (7-AAD; BD Biosciences) according to the manufacturer's instructions. Flow cytometric analysis was performed using a LSR Fortessa cell analyzer (BD Biosciences) using a 561-nm laser to excite 7-AAD (emitted light collected in a 660/20 band pass filter), a 640-nm laser to excite Alexa Fluor 633 (emitted light collected in a 670/30 band pass filter), and a 488-nm laser to excite FITC (emitted light collected in a 530/30 band pass filter). More than 10,000 cells were counted for each sample. The obtained data were processed using the FlowJo software (version 9.6.2, TreeStar, Inc., Ashland, OR, USA).
Results
EC-SOD is present on the macrophage cell surface
To evaluate the distribution of EC-SOD in resting BMMs, we analyzed cellular compartments by immunoprecipitation. When the cells were washed with PBS containing heparin, EC-SOD could be detected in the wash fraction, whereas no EC-SOD could be detected when cells were washed with PBS only (Fig. 1). This finding shows that EC-SOD is associated with the cell surface of BMMs and that this interaction is mediated by the ECM-binding region. Analysis of the intracellular compartment represented by the Triton X-100 lysate of heparin-washed cells showed that only the intact EC-SOD subunit could be detected. This finding corroborates previous analyses showing that the EC-SOD subunit is cleaved immediately before secretion [19]. Both subunits can be detected in the Triton X-100 lysates of PBS-washed cells, representing both surface-associated and intracellular EC-SOD (Fig. 1). The residual from the Triton X-100 lysates was subsequently treated with β-OG, which is known to extract proteins associated with lipid raft structures that are insoluble in cold Triton X-100. No EC-SOD could be detected in these extracts, suggesting that the protein is not located in lipid raft structures on the cell surface of resting macrophages. Collectively, these data show that EC-SOD is produced by resting BMMs and is anchored to the cell surface via the ECM-binding region.
Fig. 1.
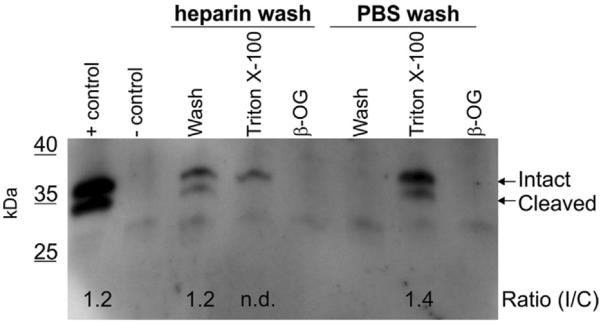
Expression of EC-SOD in wild-type BMMs. Resting macrophages (not stimulated with LPS) derived from isolated bone marrow cells were washed in PBS with or without 25 units heparin/ml as indicated. Cells were subsequently lysed in ice-cold buffer containing 1% Triton X-100 and the residual from this lysate was extracted by the addition of buffer containing 0.1% β-OG. All isolates were subjected to immunoprecipitation and subsequently analyzed by SDS–PAGE and Western blotting. A positive control representing immunoprecipitation of EC-SOD from a mouse lung homogenate was included (+control). The negative control represents an immunoprecipitate conducted in buffer only (−control). The positions of the intact and cleaved subunits of EC-SOD are indicated on the right. The intensity of the detected bands was evaluated using ImageJ analysis and used to establish the ratio between the intact and the cleaved subunit of EC-SOD as indicated.
Redistribution of EC-SOD in activated macrophages
To analyze the impact of macrophage activation on the cellular distribution of EC-SOD, we subjected isolated BMMs to LPS. Moreover, we included BMMs recovered from transgenic knock-in mice heterozygous (Arg/Gly) or homozygous (Gly/Gly) for the naturally occurring Arg213Gly amino acid substitution. This variant is known to reduce the ECM-binding capacity of EC-SOD and may thus have an impact on the cellular distribution [14,18]. When cell culture supernatants were analyzed by immunoprecipitation and Western blotting, we could not detect EC-SOD in medium recovered from resting macrophages (Fig. 2). It is interesting to note that no EC-SOD protein could be detected in culture super-natants recovered from cells representing R213G hetero- and homozygous animals, despite the expression of protein with reduced binding capacity. However, upon activation by LPS, EC-SOD protein was present in the supernatant representing all genotypes (Fig. 2). Notably, both the intact and the cleaved EC-SOD subunits were detected, suggesting that dissociation from the cell surface does not rely on proteolytic removal of the ECM-binding region. The analysis of Triton X-100 lysates from all genotypes showed that both resting and activated BMMs expressed EC-SOD, proving that the absence of protein in the culture supernatant of resting macrophages does not reflect the lack of EC-SOD expression (Fig. 2). Because lipid rafts are known to be affected by LPS activation of macrophages [38], we analyzed the presence of EC-SOD in this membrane microdomain by subsequent treatment of Triton X-100 cell lysate residuals with β-OG. The obtained data showed that EC-SOD is absent from lipid rafts in resting macrophages but could be extracted from lipid rafts from all genotypes upon cellular activation by LPS (Fig. 2). No EC-SOD could be detected when the residual of Triton X-100 was washed before extraction by β-OG, showing that the protein was indeed extracted from Triton X-100-insoluble material. Collectively, these data show that the resting macrophage produces EC-SOD and that the protein associates with the cell surface of all genotypes. However, upon cellular activation, the protein is both dissociated from the cell surface as well as relocated to lipid rafts. Notably, the reorganization of EC-SOD seems to be independent of the proteolytic removal of the ECM-binding region, as all compartments show comparable ratios between the intact and the cleaved subunits (Fig. 2).
Fig. 2.
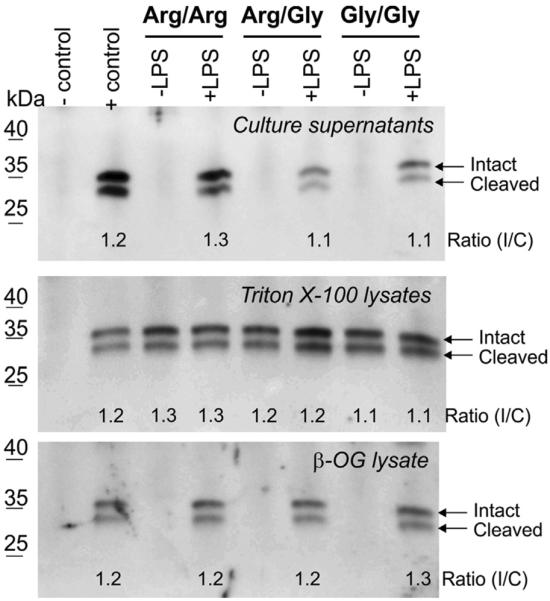
Cellular distribution of EC-SOD in resting and activated BMMs. The expression of EC-SOD in macrophages derived from wild-type (Arg/Arg), R213G heterozygous (Arg/Gly), and R213G homozygous (Gly/Gly) animals was analyzed by SDS–PAGE and Western blotting. Cells were cultured in the absence or presence of LPS (100 ng/ml) as indicated. Immunoprecipitates were prepared from cell culture medium, Triton X-100 lysates (intracellular compartment), and β-OG extracts (lipid raft structures) as described for Fig. 1. The positions of the intact and cleaved subunits of EC-SOD are indicated on the right. The intensity and ratio of the EC-SOD subunits were evaluated using ImageJ and controls were prepared as described for Fig. 1.
LPS does not affect heparin-binding capacity of EC-SOD
To determine if the secretion of EC-SOD into the medium of cells stimulated by LPS reflected a direct interaction between EC-SOD and LPS supporting dissociation of the former from the cell surface, we evaluated the heparin-binding capacity by ELISA (Fig. 3). When EC-SOD was allowed to bind to a heparin-coated surface in the absence or presence of LPS, we observed comparable titration curves, showing that heparin binding is not affected by LPS. This analysis shows that the identification of EC-SOD in the culture supernatant of cells stimulated with LPS does not reflect simple elution from the cell surface but represents an active cellular process.
Fig. 3.
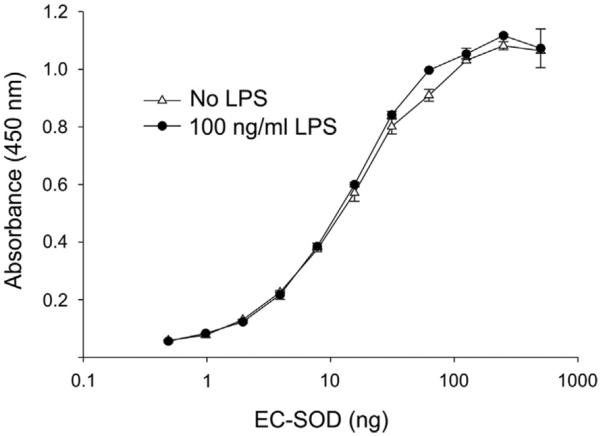
Analysis of LPS-induced dissociation of EC-SOD. The impact of LPS on the binding capacity was evaluated by ELISA using heparin-coated microtiter wells. EC-SOD was allowed to bind to the surface in the absence or presence of LPS as indicated and the level of bound protein is given as absorbance at 450 nm. The data points represent the mean of doublet estimations and error bars indicate standard deviation.
The ECM-binding capacity of secreted EC-SOD is significantly reduced
Because the ratio between the intact and the cleaved subunits of EC-SOD secreted into the medium of activated BMMs is comparable to the ratio of cell-surface-associated EC-SOD (Figs. 1 and 2), we speculated that the binding capacity of the secreted EC-SOD tetramer was reduced despite the lack of proteolytic cleavage. To investigate this, we collected the supernatant from wild-type BMMs and subjected it to heparin-Sepharose absorption. Because no EC-SOD was secreted into the cell culture medium of resting BMMs (Fig. 2), this supernatant represents a negative control. Hence, no signal could be detected in the medium, flowthrough, or eluate representing the resting macrophage (Fig. 4; −LPS). However, when medium recovered from activated macrophages was analyzed, the majority of EC-SOD in the culture supernatant was found to be present in the flow-through, whereas only a small fraction was detected in the eluate (Fig. 4; +LPS). To evaluate the binding capacity of the resin, we included a mouse lung homogenate as a positive control. The analysis of this homogenate produced relatively strong signal intensity in the eluate, proving that the binding capacity of the resin is maintained (Fig. 4; control). The ratio of the intact and cleaved subunits in the flowthrough was comparable to that of EC-SOD recovered from cell lysates or from a lung homogenate (Figs. 1 and 2). This analysis shows that the binding capacity of EC-SOD present in the cell culture supernatant of activated BMMs is significantly reduced. However, because the ratio of the intact and cleaved subunit is maintained, it is evident that the protein does not dissociate from the cell surface owing to proteolytic removal of the ECM-binding region but rather by modulating the binding capacity.
Fig. 4.
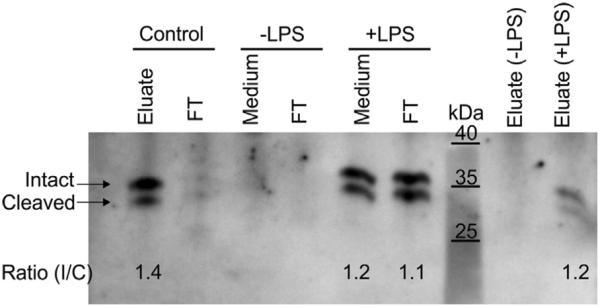
Heparin binding capacity of secreted EC-SOD. BMMs were derived from wild-type animals and incubated in the absence (−LPS) or presence (+LPS) of LPS. The cell culture medium was collected for analysis by affinity chromatography using heparin–Sepharose. The starting material (medium), the column flowthrough (FT), and the eluate were analyzed by immunoprecipitation and Western blotting. The presence of intact and cleaved EC-SOD is indicated on the left. The control samples represent absorption of EC-SOD present in a mouse lung homogenate. The intensity and ratio of the EC-SOD subunits were evaluated by using ImageJ.
Cellular distribution of EC-SOD in BMMs
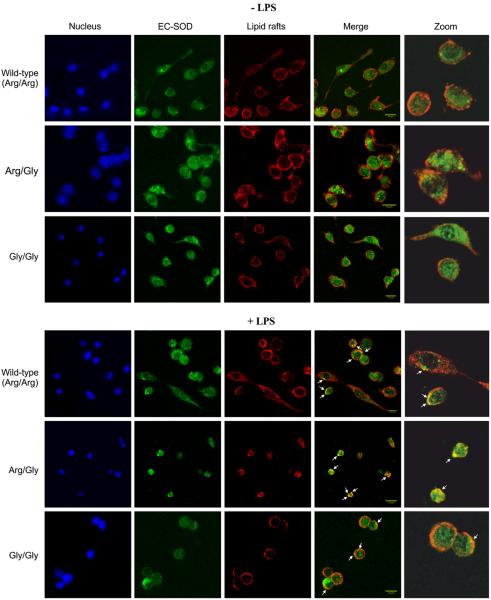
To further evaluate the cellular distribution of EC-SOD, we analyzed BMMs by using confocal microscopy. The analysis was performed by costaining using the β subunit of cholera toxin to allow for the detection of lipid rafts. In the absence of LPS, the staining for wild-type EC-SOD showed a diffuse distribution of the protein, probably reflecting the anchoring of the protein on the cell surface and the absence of compartmentalization (Fig. 5). The same pattern was obtained for BMMs isolated from R213G ECSOD heterozygous or homozygous animals. Costaining for lipid raft structures did not reveal any discernible colocalization in any of the analyzed cell cultures, suggesting that EC-SOD is not present in lipid raft structures in resting macrophages, corroborating the previous observations (Fig. 2). When the cells were activated by LPS, the staining pattern of EC-SOD did not change for any of the genotypes, indicating that there is no major reorganization of the protein. However, we were able to detect areas of colocalization between the β subunit of cholera toxin and EC-SOD (Fig. 5), suggesting the localization of EC-SOD in lipid rafts. However, the major fraction of the proteins seems to be associated with nonraft structures. In conclusion, confocal microscopy shows that EC-SOD is diffusely distributed in the cell and that no significant difference exists between the three genotypes.
Fig. 5.
Analysis of cellular distribution by confocal microscopy. Differentiated cells were maintained in normal medium (−LPS) or exposed to medium containing LPS (+LPS) for 16 h. Before fixation, cells were incubated with cholera toxin subunit B (red) to identify lipid rafts located in the cell membrane. Cells were subsequently fixed in methanol and stained for EC-SOD (green). Cell nuclei were stained using Hoechst 33258 (blue). The channels for lipid rafts and EC-SOD were merged to visualize colocalization (yellow) as indicated by white arrows. Sections of colocalization are enlarged for clarity. Scale bar, 14 μm. The images were acquired using an LSM 710 laser scanning microscope and processed using ImageJ software.
FACS analysis of cell-surface-associated EC-SOD
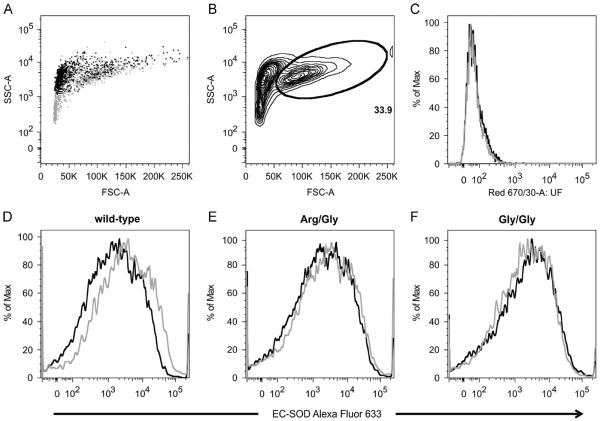
To further evaluate the presence of cell-surface-associated EC-SOD, we analyzed BMMs by flow cytometry. The gating strategy was tuned to select viable cells by the back-gating of cells identified by 7-AAD staining, defining approximately 34% of the population as live cells (Fig. 6A and B). The specificity of the protocol was confirmed by the staining of cells using an isotype control antibody, which showed the absence of any signal above the background of unstained cells (Fig. 6C). When resting macrophages derived from wild-type animals were stained by an anti-EC-SOD antibody, we could detect a broad distribution of signal intensity, showing that EC-SOD is indeed associated with the cell surface (Fig. 6D). A comparable signal distribution was obtained from wild-type LPS-stimulated macrophages, indicating that ECSOD remains associated with the cell surface upon cellular activation. The analysis of cells isolated from both heterozygous (Fig. 6E) and homozygous (Fig. 6F) animals produced comparable results showing that the protein remains associated with the cell surface despite the reduced binding capacity of the ECM-binding region. These data corroborate the analyses performed by Western blotting and confocal microscopy (Figs. 2 and 5). Importantly, the data show that EC-SOD detected in the cell culture supernatant of LPS-stimulated BMMs (Fig. 2) does not reflect a significant decrease in cell surface-associated EC-SOD.
Fig. 6.
Histograms representing analysis of BMMs stained for surface-associated EC-SOD. (A) Back-gating of dead cells (all cells, gray dots; 7-AAD positive, black dots). (B) Forward- and side-scatter plot with a gate defining live cells (33.9%). (C) Histogram of WT cells unstained (black line) or stained with isotype control (gray line). BMMs isolated from (D) wild-type, (E) R213G heterozygous (Arg/Gly), or (F) homozygous (Gly/Gly) animals were stained for EC-SOD using Alexa Fluor 633. Cells were analyzed in the resting state (black line) or after overnight activation by LPS (100 ng/ml) (gray line). Pooled data from two independent experiments are represented.
Discussion
This study shows that the spatial expression of EC-SOD in BMMs is affected by the stimulation of cells by LPS. We observe that macrophages secrete EC-SOD into the medium only when they are stimulated by LPS. However, we do not detect any difference in the ratio between intact and cleaved subunits when comparing cell-surface-associated and secreted EC-SOD, but observe that the affinity for heparin of the secreted material is significantly reduced (Fig. 4). It thus appears that secretion relies on modifications other than proteolytic removal of the ECM-binding region. Previous studies of interstitial lung disease induced by bleomycin or asbestos have shown that EC-SOD relocates from the lung parenchyma to the bronchoalveolar lavage fluid (BALF) [39,40]. Likewise, the content of EC-SOD in BALF was found to increase when using a model of bacterial pneumonia characterized by the absence of interstitial inflammation [41]. In this model, the presence of EC-SOD in BALF was not associated with a decrease in EC-SOD in the lung parenchyma, which is likely to reflect the finding that inflammatory cells are the source of the protein [41]. However, in both models the recovery of EC-SOD from BALF was associated with an increased level of the cleaved EC-SOD subunit, indicating that relocation is associated with proteolytic removal of the ECM-binding region and hence the generation of EC-SOD tetramers with reduced binding capacity. Although our data corroborate the observation that secretion of EC-SOD is induced by inflammation, this was not associated with increased proteolysis (Fig. 2). As the affinity of EC-SOD for heparin has previously been shown to be modified by posttranslational modifications other than proteolytic cleavage [42], we speculate that the secretion might reflect this type of event. This modification may subsequently render the protein more susceptible to cleavage by proteases present in the inflamed tissue as previously suggested [39,40].
Although EC-SOD protein was secreted from LPS-stimulated macrophages, we could not detect a concomitant reduction in the amount of surface staining, as evaluated by flow cytometry (Fig. 6). This finding correlates with previous data showing no significant change in EC-SOD staining in macrophages recovered from rat lungs exposed to LPS [23]. Recently, it was shown, using cultured astrocytes, that LPS stimulation produced a transient reduction in surface-associated EC-SOD [43]. The apparent steady-state level of surface-associated EC-SOD was established after approximately 36 h. The recovery of EC-SOD from the culture supernatant of BMMs exposed to LPS observed in this study may thus reflect a transient dissociation of protein from the cell surface followed by the establishment of the steady-state level on the cell surface. This is currently under investigation.
In addition to secretion of EC-SOD by LPS stimulation, we also find that a minor part of the protein associates with lipid raft structures in the membrane, as evaluated by β-OG extraction (Fig. 2). Lipid rafts are known to be domains on the cell surface capable of mediating protein-protein interaction upon both intraand extracellular stimuli [44]. Using adenovirus-transduced human umbilical vein endothelial cells expressing EC-SOD and mouse lung tissue homogenates, Oshikawa et al. [45] likewise showed that a minor part of the protein was associated with lipid rafts. These authors argued that the localization of EC-SOD to this functional domain of the cell membrane supports local production of hydrogen peroxide, which subsequently stimulates angiogenesis by mediating the oxidation of proteins involved in the regulation of endothelial cell migration. It is interesting to speculate that ECSOD functions in lipid rafts of macrophages to support the development of redox-regulated events involved in the inflamma-tory response, e.g., by reducing the oxidative activation of NF-κB [31,46]
EC-SOD has previously been detected in intracellular membrane-bound vesicles of inflammatory cells as evaluated by immunohistochemistry of tissue sections representing challenged and inflamed tissue [23,29,41,47]. Owing to the lack of resolution of confocal microscopy, we did not detect any significant intracellular staining in BMMs stimulated by LPS. Nor did we detect any significant redistribution of the protein, even though areas of costaining for EC-SOD/lipid rafts could be detected in LPS-stimulated cells (Fig. 5). It is interesting to speculate that the presence of EC-SOD in intracellular membrane-bound vesicles reflects endocytosis mediated by lipid rafts encompassing ECSOD in activated macrophages.
R213G EC-SOD has been associated with cardiopulmonary disease in population studies showing that heterozygous carriers have a decreased susceptibility to COPD, whereas the incidence of ischemic heart disease is increased [16,48]. The basis of these associations has recently been addressed by the analysis of an R213G EC-SOD knock-in mouse (submitted for publication). These authors show that the immunohistochemical staining of R213G EC-SOD in aorta and lung tissues is significantly reduced relative to wild-type animals and that the concentration of EC-SOD in both plasma and BALF is increased. In the present study, we did not observe any significant dissociation of the protein from resting BMMs as evaluated by the absence of protein in cell culture supernatants representing cells derived from R213G heterozygous or homozygous animals (Fig. 2). It is not clear why R213G EC-SOD is lost in tissue, whereas it remains bound to the macrophage cell surface. In vascular and pulmonary tissue, EC-SOD is expressed primarily by vascular smooth muscle cells [49] and by epithelial and alveolar type II cells [50], respectively, and thought to diffuse into the interstitial space. Indeed, in contrast to isolated resting macrophages, cultured human smooth muscle cells were shown to produce and secrete EC-SOD into the culture medium [49]. This suggests that the secretion of EC-SOD from smooth muscle cells differs from isolated macrophages. In concert, these findings may explain the apparent discrepancy between resting macrophages and tissue recovered from R213G EC-SOD animals.
This paper shows that EC-SOD expression in macrophages is dynamic and spatially affected by the stimulation of cells by LPS. The translocation of EC-SOD from the cell surface to lipid rafts may represent an important function, as lipid rafts are platforms for cellular signaling. Moreover, the section of EC-SOD from activated cells suggests that the macrophage is a key player in the global defense against superoxide in tissues. Although the response of macrophages in inflammation is complex and may not adequately be reflected by the use of cultured BMMs, we believe that the present paper positions EC-SOD as an important player in the cellular response to infection.
Acknowledgments
This work was supported by the Lundbeck Foundation and Aase og Ejnar Danielsens Fond (S.V.P.) and by the Flight Attendants Medical Research Institute (FAMRI 09_2050) and National Institutes of Health (NIH HL 11-1288) (R.P.B.). R.H.G. is the recipient of a Ph.D. stipend from Aarhus University. Dr. Charlotte Christie Pedersen from the FACS core facility at Aarhus University is gratefully acknowledged for help and guidance in experimental setup of flow cytometric analyses. Dr. Morten S. Nielsen, Aarhus University, is greatly acknowledged for his kind input on performing confocal microscopy.
Abbreviations
- BMM
bone marrow-derived macrophage
- β-OG
β-octylglucoside
- BALF
bronchoalveolar lavage fluid
- ECM
extracellular matrix
- EC-SOD
extracellular superoxide dismutase
- LPS
lipopolysaccharide
References
- [1].Oury TD, Chang LY, Marklund SL, Day BJ, Crapo JD. Immunocyto-chemical localization of extracellular superoxide dismutase in human lung. Lab. Invest. 1994;70:889–898. [PubMed] [Google Scholar]
- [2].Oury TD, Day BJ, Crapo JD. Extracellular superoxide dismutase in vessels and airways of humans and baboons. Free Radic. Biol. Med. 1996;20:957–965. doi: 10.1016/0891-5849(95)02222-8. [DOI] [PubMed] [Google Scholar]
- [3].Fukai T, Folz RJ, Landmesser U, Harrison DG. Extracellular superoxide dismutase and cardiovascular disease. Cardiovasc. Res. 2002;55:239–249. doi: 10.1016/s0008-6363(02)00328-0. [DOI] [PubMed] [Google Scholar]
- [4].Fattman CL, Schaefer LM, Oury TD. Extracellular superoxide dismutase in biology and medicine. Free Radic. Biol. Med. 2003;35:236–256. doi: 10.1016/s0891-5849(03)00275-2. [DOI] [PubMed] [Google Scholar]
- [5].Marklund SL. Human copper-containing superoxide dismutase of high molecular weight. Proc. Natl. Acad. Sci. USA. 1982;79:7634–7638. doi: 10.1073/pnas.79.24.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Karlsson K, Lindahl U, Marklund SL. Binding of human extracellular superoxide dismutase C to sulphated glycosaminoglycans. Biochem. J. 1988;256:29–33. doi: 10.1042/bj2560029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Petersen SV, Oury T, Oestergaard L, Valnickova Z, Wegrzyn J, Thogersen IB, Jacobsen C, Bowler RP, Fattman CL, Crapo JD, Enghild JJ. Extracellular superoxide dismutase (EC-SOD) binds to type I collagen and protects against oxidative fragmentation. J. Biol. Chem. 2004;279:13705–13710. doi: 10.1074/jbc.M310217200. [DOI] [PubMed] [Google Scholar]
- [8].Gao F, Koenitzer JR, Tobolewski JM, Jiang D, Liang J, Noble PW, Oury TD. Extracellular superoxide dismutase inhibits inflammation by preventing oxidative fragmentation of hyaluronan. J. Biol. Chem. 2008;283:6058–6066. doi: 10.1074/jbc.M709273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kliment CR, Tobolewski JM, Manni ML, Tan RJ, Enghild J, Oury TD. Extracellular superoxide dismutase protects against matrix degradation of heparan sulfate in the lung. Antioxid. Redox Signaling. 2008;10:261–268. doi: 10.1089/ars.2007.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yao H, Arunachalam G, Hwang JW, Chung S, Sundar IK, Kinnula VL, Crapo JD, Rahman I. Extracellular superoxide dismutase protects against pulmonary emphysema by attenuating oxidative fragmentation of ECM. Proc. Natl. Acad. Sci. USA. 2010;107:15571–15576. doi: 10.1073/pnas.1007625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kliment CR, Oury TD. Extracellular superoxide dismutase protects cardiovascular syndecan-1 from oxidative shedding. Free Radic. Biol. Med. 2011;50:1075–1080. doi: 10.1016/j.freeradbiomed.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sandstrom J, Carlsson L, Marklund SL, Edlund T. The heparin-binding domain of extracellular superoxide dismutase C and formation of variants with reduced heparin affinity. J. Biol. Chem. 1992;267:18205–18209. [PubMed] [Google Scholar]
- [13].Adachi T, Kodera T, Ohta H, Hayashi K, Hirano K. The heparin binding site of human extracellular-superoxide dismutase. Arch. Biochem. Biophys. 1992;297:155–161. doi: 10.1016/0003-9861(92)90654-f. [DOI] [PubMed] [Google Scholar]
- [14].Sandstrom J, Nilsson P, Karlsson K, Marklund SL. 10-fold increase in human plasma extracellular superoxide dismutase content caused by a mutation in heparin-binding domain. J. Biol. Chem. 1994;269:19163–19166. [PubMed] [Google Scholar]
- [15].Folz RJ, Peno-Green L, Crapo JD. Identification of a homozygous missense mutation (Arg to Gly) in the critical binding region of the human EC-SOD gene (SOD3) and its association with dramatically increased serum enzyme levels. Hum. Mol. Genet. 1994;3:2251–2254. doi: 10.1093/hmg/3.12.2251. [DOI] [PubMed] [Google Scholar]
- [16].Juul K, Tybjaerg-Hansen A, Marklund S, Heegaard NH, Steffensen R, Sillesen H, Jensen G, Nordestgaard BG. Genetically reduced antioxidative protection and increased ischemic heart disease risk: the Copenhagen City Heart Study. Circulation. 2004;109:59–65. doi: 10.1161/01.CIR.0000105720.28086.6C. [DOI] [PubMed] [Google Scholar]
- [17].Petersen SV, Thogersen IB, Valnickova Z, Nielsen MS, Petersen JS, Poulsen ET, Jacobsen C, Oury TD, Moestrup SK, Crapo JD, Nielsen NC, Kristensen T, Enghild JJ. The concentration of extracellular superoxide dismutase in plasma is maintained by LRP-mediated endocytosis. Free Radic. Biol. Med. 2010;49:894–899. doi: 10.1016/j.freeradbiomed.2010.06.019. [DOI] [PubMed] [Google Scholar]
- [18].Petersen SV, Olsen DA, Kenney JM, Oury TD, Valnickova Z, Thogersen IB, Crapo JD, Enghild JJ. The high concentration of Arg213→Gly extracellular superoxide dismutase (EC-SOD) in plasma is caused by a reduction of both heparin and collagen affinities. Biochem. J. 2005;385:427–432. doi: 10.1042/BJ20041218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Enghild JJ, Thogersen IB, Oury TD, Valnickova Z, Hojrup P, Crapo JD. The heparin-binding domain of extracellular superoxide dismutase is proteolytically processed intracellularly during biosynthesis. J. Biol. Chem. 1999;274:14818–14822. doi: 10.1074/jbc.274.21.14818. [DOI] [PubMed] [Google Scholar]
- [20].Bowler RP, Nicks M, Olsen DA, Thogersen IB, Valnickova Z, Hojrup P, Franzusoff A, Enghild JJ, Crapo JD. Furin proteolytically processes the heparin-binding region of extracellular superoxide dismutase. J. Biol. Chem. 2002;277:16505–16511. doi: 10.1074/jbc.M105409200. [DOI] [PubMed] [Google Scholar]
- [21].Olsen DA, Petersen SV, Oury TD, Valnickova Z, Thogersen IB, Kristensen T, Bowler RP, Crapo JD, Enghild JJ. The intracellular proteolytic processing of extracellular superoxide dismutase (EC-SOD) is a two step event. J. Biol. Chem. 2004;279:22152–22157. doi: 10.1074/jbc.M401180200. [DOI] [PubMed] [Google Scholar]
- [22].Gottfredsen RH, Tran SM, Larsen UG, Madsen P, Nielsen MS, Enghild JJ, Petersen SV. The C-terminal proteolytic processing of extracellular superoxide dismutase is redox regulated. Free Radic. Biol. Med. 2012;52:191–197. doi: 10.1016/j.freeradbiomed.2011.10.443. [DOI] [PubMed] [Google Scholar]
- [23].Loenders B, Van Mechelen E, Nicolai S, Buyssens N, Van Osselaer N, Jorens PG, Willems J, Herman AG, Slegers H. Localization of extracellular superoxide dismutase in rat lung: neutrophils and macrophages as carriers of the enzyme. Free Radic. Biol. Med. 1998;24:1097–1106. doi: 10.1016/s0891-5849(97)00434-6. [DOI] [PubMed] [Google Scholar]
- [24].Luoma JS, Stralin P, Marklund SL, Hiltunen TP, Sarkioja T, Yla-Herttuala S. Expression of extracellular SOD and iNOS in macrophages and smooth muscle cells in human and rabbit atherosclerotic lesions: colocalization with epitopes characteristic of oxidized LDL and peroxynitrite-modified proteins. Arterioscler. Thromb. Vasc. Biol. 1998;18:157–167. doi: 10.1161/01.atv.18.2.157. [DOI] [PubMed] [Google Scholar]
- [25].Folz RJ, Abushamaa AM, Suliman HB. Extracellular superoxide dismutase in the airways of transgenic mice reduces inflammation and attenuates lung toxicity following hyperoxia. J. Clin. Invest. 1999;103:1055–1066. doi: 10.1172/JCI3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ghio AJ, Suliman HB, Carter JD, Abushamaa AM, Folz RJ. Overexpression of extracellular superoxide dismutase decreases lung injury after exposure to oil fly ash. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;283:L211–L218. doi: 10.1152/ajplung.00409.2001. [DOI] [PubMed] [Google Scholar]
- [27].Bowler RP, Nicks M, Tran K, Tanner G, Chang LY, Young SK, Worthen GS. Extracellular superoxide dismutase attenuates lipopolysaccharide-induced neutrophilic inflammation. Am. J. Respir. Cell Mol. Biol. 2004;31:432–439. doi: 10.1165/rcmb.2004-0057OC. [DOI] [PubMed] [Google Scholar]
- [28].Fattman CL, Tan RJ, Tobolewski JM, Oury TD. Increased sensitivity to asbestos-induced lung injury in mice lacking extracellular superoxide dismutase. Free Radic. Biol. Med. 2006;40:601–607. doi: 10.1016/j.freeradbiomed.2005.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Manni ML, Tomai LP, Norris CA, Thomas LM, Kelley EE, Salter RD, Crapo JD, Chang LY, Watkins SC, Piganelli JD, Oury TD. Extracellular superoxide dismutase in macrophages augments bacterial killing by promoting phagocytosis. Am. J. Pathol. 2011;178:2752–2759. doi: 10.1016/j.ajpath.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Break TJ, Jun S, Indramohan M, Carr KD, Sieve AN, Dory L, Berg RE. Extracellular superoxide dismutase inhibits innate immune responses and clearance of an intracellular bacterial infection. J. Immunol. 2012;188:3342–3350. doi: 10.4049/jimmunol.1102341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Laurila JP, Laatikainen LE, Castellone MD, Laukkanen MO. SOD3 reduces inflammatory cell migration by regulating adhesion molecule and cytokine expression. PLoS One. 2009;4:e5786. doi: 10.1371/journal.pone.0005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kwon MJ, Han J, Kim BH, Lee YS, Kim TY. Superoxide dismutase 3 suppresses hyaluronic acid fragments mediated skin inflammation by inhibition of toll-like receptor 4 signaling pathway: superoxide dismutase 3 inhibits reactive oxygen species-induced trafficking of toll-like receptor 4 to lipid rafts. Antioxid. Redox Signaling. 2012;16:297–313. doi: 10.1089/ars.2011.4066. [DOI] [PubMed] [Google Scholar]
- [33].Fattman CL, Enghild JJ, Crapo JD, Schaefer LM, Valnickova Z, Oury TD. Purification and characterization of extracellular superoxide dismutase in mouse lung. Biochem. Biophys. Res. Commun. 2000;275:542–548. doi: 10.1006/bbrc.2000.3327. [DOI] [PubMed] [Google Scholar]
- [34].Bury AF. Analysis of protein and peptide mixtures: evaluation of three sodium dodecyl sulfate-polyacrylamide gel electrophoresis buffer systems. J. Chromatogr. 1981;213:491–500. [Google Scholar]
- [35].Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- [36].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Najjam S, Gibbs RV, Gordon MY, Rider CC. Characterization of human recombinant interleukin 2 binding to heparin and heparan sulfate using an ELISA approach. Cytokine. 1997;9:1013–1022. doi: 10.1006/cyto.1997.0246. [DOI] [PubMed] [Google Scholar]
- [38].Triantafilou M, Triantafilou K. Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. 2002;23:301–304. doi: 10.1016/s1471-4906(02)02233-0. [DOI] [PubMed] [Google Scholar]
- [39].Fattman CL, Chu CT, Kulich SM, Enghild JJ, Oury TD. Altered expression of extracellular superoxide dismutase in mouse lung after bleomycin treatment. Free Radic. Biol. Med. 2001;31:1198–1207. doi: 10.1016/s0891-5849(01)00699-2. [DOI] [PubMed] [Google Scholar]
- [40].Tan RJ, Fattman CL, Watkins SC, Oury TD. Redistribution of pulmonary EC-SOD after exposure to asbestos. J. Appl. Physiol. 2004;97:2006–2013. doi: 10.1152/japplphysiol.00480.2004. [DOI] [PubMed] [Google Scholar]
- [41].Tan RJ, Lee JS, Manni ML, Fattman CL, Tobolewski JM, Zheng M, Kolls JK, Martin TR, Oury TD. Inflammatory cells as a source of airspace extracellular superoxide dismutase after pulmonary injury. Am. J. Respir. Cell Mol. Biol. 2006;34:226–232. doi: 10.1165/rcmb.2005-0212OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Adachi T, Ohta H, Hayashi K, Hirano K, Marklund SL. The site of nonenzymic glycation of human extracellular-superoxide dismutase in vitro. Free Radic. Biol. Med. 1992;13:205–210. doi: 10.1016/0891-5849(92)90016-a. [DOI] [PubMed] [Google Scholar]
- [43].Iitsuka I, Motoyoshi-Yamashiro A, Moriyama M, Kannan-Hayashi Y, Fujimoto Y, Takano K, Murakami K, Yoneda Y, Nakamura Y. Extracellular superoxide dismutase in cultured astrocytes: decrease in cell-surface activity and increase in medium activity by lipopolysaccharide-stimulation. Neurochem. Res. 2012;37:2108–2116. doi: 10.1007/s11064-012-0832-z. [DOI] [PubMed] [Google Scholar]
- [44].Simons K, Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- [45].Oshikawa J, Urao N, Kim HW, Kaplan N, Razvi M, McKinney R, Poole LB, Fukai T, Ushio-Fukai M. Extracellular SOD-derived H2O2 promotes VEGF signaling in caveolae/lipid rafts and post-ischemic angiogenesis in mice. PLoS One. 2010;5:e10189. doi: 10.1371/journal.pone.0010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kim Y, Kim BH, Lee H, Jeon B, Lee YS, Kwon MJ, Kim TY. Regulation of skin inflammation and angiogenesis by EC-SOD via HIF-1alpha and NF-kappaB pathways. Free Radic. Biol. Med. 2011;51:1985–1995. doi: 10.1016/j.freeradbiomed.2011.08.027. [DOI] [PubMed] [Google Scholar]
- [47].Fattman CL, Chang LY, Termin TA, Petersen L, Enghild JJ, Oury TD. Enhanced bleomycin-induced pulmonary damage in mice lacking extracellular superoxide dismutase. Free Radic. Biol. Med. 2003;35:763–771. doi: 10.1016/s0891-5849(03)00402-7. [DOI] [PubMed] [Google Scholar]
- [48].Juul K, Tybjaerg-Hansen A, Marklund S, Lange P, Nordestgaard BG. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2006;173:858–864. doi: 10.1164/rccm.200509-1387OC. [DOI] [PubMed] [Google Scholar]
- [49].Stralin P, Karlsson K, Johansson BO, Marklund SL. The interstitium of the human arterial wall contains very large amounts of extracellular superoxide dismutase. Arterioscler. Thromb. Vasc. Biol. 1995;15:2032–2036. doi: 10.1161/01.atv.15.11.2032. [DOI] [PubMed] [Google Scholar]
- [50].Su WY, Folz R, Chen JS, Crapo JD, Chang LY. Extracellular superoxide dismutase mRNA expressions in the human lung by in situ hybridization. Am. J. Respir. Cell Mol. Biol. 1997;16:162–170. doi: 10.1165/ajrcmb.16.2.9032123. [DOI] [PubMed] [Google Scholar]