Abstract
The Elispot effectively measures the frequencies of cells secreting particular molecules, especially low-frequency cells such as antigen-specific T cells. The Fluorospot assay adapted this analysis to two products per cell, and this has now been extended to three-color measurement of both mouse and human cytokine-secreting cells. Due to the increased data complexity, and particularly the need to define single-, double- and triple-producing cells, it is critical to objectively quantify spot number, size, intensity, and coincidence with other spots. An automated counting program, Exploraspot, was therefore developed to detect and quantify Fluorospots in automated fluorescence microscope images. Morphological parameters, including size, intensity, location, circularity and others are calculated for each spot, exported in FCS format, and further analyzed by gating and graphical display in popular flow cytometry analysis programs. The utility of Exploraspot is demonstrated by identification of single-, double- and triple-secreting T cells; tolerance of variable background fluorescence; and estimation of the numbers of genuine versus random multiple events.
Keywords: Exploraspot, elispot, fluorospot, cytokines, imaging, multicolor, fluorescence
Introduction
The Elispot has been invaluable for detecting the frequencies of cells secreting specific proteins, particularly antigen-specific lymphocytes that are often present at very low frequencies in normal populations, such as B cells and plasma cells secreting antibodies, and T cells secreting cytokines. The Elispot is as sensitive as competing methods such as intracellular cytokine staining and flow cytometry, and typically has a very low background. However, a significant limitation of the Elispot is its restriction to mainly single-parameter detection. Although two-color Elispots have been used [1–3], the detection of two colors by precipitating reagents has the limitation that large amounts of precipitate of one color may obscure low amounts of the other color. The utility of immunospot assays has been further extended by using fluorescent reagents in the two-color Fluorospot assay [4–6]. This opens up the potential to extend the assay to three or more parameters.
Identification of multiple products secreted by single cells is particularly important for defining antigen-specific T cells, as the cytokine patterns secreted by individual differentiated effector T cells are major determinants of their function, and it is becoming clear that there are more subsets of effector T cells than the Th1 and Th2 subsets. Among CD4 T cells, Th1, Th2, Thpp, Th0, Th17 and Treg cells all secrete different cytokine patterns [7–11], and the analysis of normal T cell populations suggests that additional types will be discovered. Thus multiparameter detection of secreted cytokines would be a valuable tool for quantifying rare antigen-specific T cell phenotypes.
Even for Elispots, automated counting is preferable to manual methods because of subjective effects, and automated image processing is essential to extract all useful information from the complex multicolor Fluorospot data. In addition to the identification and counting of spots, multicolor fluorescent assays can potentially identify double-, triple- and multiple-producer cells, and quantify the relative amounts of each product. The exact position of spots in different channels may vary slightly due to cell migration or assay effects, resulting in difficult decisions regarding which spots in different channels were derived from the same cell.
To take advantage of the potential of multicolor spot assays, we have extended the Fluorospot method to two- and three-color single-cell cytokine secretion assays for several combinations of mouse and human cytokines. These assays use multiple species-specific reagents, biotin-streptavidin and FITC-anti-FITC strategies to clearly separate the different cytokines. We have also developed software to locate and evaluate spots in two, three or more colors. Exploraspot is an automated counting program optimized for detection and quantization of Fluorospots in images captured by an automated fluorescent microscope. Several parameters, including size, intensity, locations, circularity and others are calculated for each spot, and after segmentation, coincident spots in different colors are assigned. The coincidence parameters can be adjusted by the user to optimize the distinction between double-producing cells and adjacent single-producing cells. The resulting data are exported in FCS format, which takes advantage of the sophisticated multiparameter analysis programs developed for the analysis of flow cytometry data. We present here an analysis of the accuracy and robustness of the multicolor Fluorospot and the Exploraspot program.
Methods
Reagents & Antibodies
Anti-mouse IL-4 (11B11, [12]) and anti-mouse IFN-γ (AN18) antibodies were produced in our laboratory. Purified anti-mouse IL-2 (JES6-1A12), biotinylated anti-mouse IL-2 (JES6-5H), purified anti-CD3 (145-2C11) and purified anti-CD28 (37.51) were purchased from eBioscience (San Diego, CA). Anti-mouse MIP-1α (39624.11), goat anti-mouse IL-4 (Cat. AF-404-NA), goat anti-MIP-1α (Cat. AF-450-NA) were all obtained from R&D Systems (Minneapolis, MN). Mouse anti-goat Cy3 (Cat. 205-165–108) and mouse anti-rabbit Cy2 (Cat. 211–225-109) were purchased from Jackson Immuno Research (West Grove, PA). Rabbit anti-IFN-γ (Cat. RMF-421) was purchased from Antigenix America (Huntington Station, NY).
Cells
Mouse Th1 and Th2 cells were produced as described previously [13] by stimulating sorted CD62L+ CD44− CD4+ CD8− naïve CD4 T cells derived from C57Bl/6 splenocytes with the allogeneic cell line TA3. Cells were cultured in RPMI 1640 containing 8% fetal bovine serum in the presence of 1ng/ml IL-2 and either 1ng/ml IL-12 IL-12 or 5ng/ml IL-4, respectively. After at least 5 days to allow differentiation cells were restimulated in cytokine assays. A mixed population of allospecific CD4 T cell phenotypes was derived similarly, except that the cultures were supplemented with 1 ng/ml IL-2, 5 ng/ml IL-7, 10 pg/ml IL-12, 0.2 ng/ml IL-4, and 2 µg/ml progesterone.
Human PBMC were stimulated in intracellular cytokine staining (ICS) and Fluorospot assays as described previously [5]. Briefly, PBMC were purified from whole blood by Ficoll-Hypaque separation. For ICS, cells were stimulated with influenza antigen for two hours, and then influenza-stimulated and control cultures were incubated with brefeldin and monensin to induce accumulation of intracellular cytokines. Cells were permeabilized, fixed and stained with antibodies specific for IL-2, IFNγ, CD4 and CD8. Stained cells were analyzed on a Becton Dickinson LSR-II cytometer. Fluorospot assays were carried out as below except that influenza vaccine was used as antigen.
Fluorospot
Multiscreen HTS plates (MSIPN4550, Millipore Corporation, Marlborough, MA) were coated with the specified combinations of 10 µg/ml purified anti-IL-2, anti-IL-4, and anti-MIP-1α and 2 µg/ml anti-IFN-γ antibodies as well as purified anti-CD3 at 2 µg/ml and purified anti-CD28 at 1 µg/ml for cell stimulation. After 2 hours at room temperature or overnight at 4°C, plates were washed three times with RPMI 8% FBS + 50µM β2-mercaptoethanol + Penicillin-Streptomycin-Fungizone. Cells were added along with secondary antibodies: biotinylated anti-IL-2, goat anti-IL-4, goat anti-MIP1α, and/or rabbit anti-IFNγ all at a final concentration of 1 µg/ml. After incubation for 18 h at 37°C, plates were washed in PBS containing 0.1% Tween 20 and detection reagents (Streptavidin-Cy5, mouse anti-goat IgG Cy3, mouse anti-rabbit IgG Cy2) were added to the plates at 2 µg/ml for 30 min at room temperature. Plates were then washed, allowed to soak in PBS containing 0.1% Tween 20 for 90 minutes and rinsed with cold water before being dried in an air current.
Image Capture
Filter bottoms were removed from 96-well Fluorospot plates onto an adhesive sheet, and each of the 96 wells was captured as an individual image using a Nikon Eclipse E400 microscope and a SPOT RT SE monochrome camera, with filter cubes to resolve different fluorochromes (i.e. two or three grayscale images per well, later assigned to RGB color-space). The camera resolution and microscope magnification resulted in a resolution of 150 pixels per mm, giving an image of 1000 by 1000 pixels for each 6 mm diameter well, for a pixel size of 44.44 sq. µm. This resolution was sufficient even for small Fluorospots, e.g. those produced in four-hour assays. Images were saved as 8-bit monochrome (or 24-bit for RGB) files. Stage movement and filter cube changing were automated using IPLabs software, so that each plate could be read in 20 to 40 minutes without the presence of an operator.
Spot-Localization Process
All algorithms as well as the Graphical User Interface (GUI) were developed in MATLAB (The Mathworks, Natick, MA), which is required for their use. The image in each channel was initially processed independently from the other channels. Image contrast was enhanced using a custom Laplacian filter, and noise was reduced using a 3×3 median filter applied 1–3 times. The image was then inverted and a watershed transform applied. The watershed image was converted to binary. Non-uniform background was then approximated from the filtered image using the watershed image as a mask. The background was then subtracted from the filtered image, and segmented by a user-defined threshold. Final segmentation was achieved by multiplying the segmented image by the binary watershed image. Each segment, representing a spot, was then assigned a unique identification number. These segments were used for subsequent spot measurements.
Background Approximation
The filtered image was multiplied by the inverse watershed image, which created a map of all low intensity regions between signal peaks (all other regions became 0). The resulting image was then binned to 1/625 of its original size, with the 10th percentile value of all non-zero values assigned to each respective bin. If no non-zero values existed for a bin, the bin value became zero and was considered an empty bin. Values for empty bins were determined by interpolation using a soap film partial differential equation. The binned image was then enlarged to the original size using a 2-D interpolation by cubic spline. This resulted in a strong approximation of any non-uniform background fluorescence.
Single-channel Image Measurements
All measurements were derived from the original, unmodified image after subtraction of the background as determined above. Area equaled the number of pixels (each 44.44 sq. µm, see above) in a spot. Center of Mass was determined by measuring the average position of pixels (weighted by each pixel’s respective intensity) in a spot. Net Intensity was the summation of all intensity values within a spot minus the Local Background. Local Background equaled the mean intensity of the perimeter pixels of a spot multiplied by the area (in pixels) of the spot (note that the local background was typically very low, as most of the background was subtracted in the step described above). Circularity was determined by evaluating the radial variability of a spot by measuring the distance from the Spot Center of Mass to each of its perimeter pixels. The mean (µ) of the radii minus two standard deviations (2σ) was divided by the mean (µ), i.e. (µ-2σ)/µ, negative values were reassigned to zero, and the result multiplied by 100 to represent Circularity as a pseudo-percentage. Background Variance was defined as the statistical variance of the background approximation image. Each of these parameters was determined independently for each channel.
Image Manipulations for Random Double-Color Events
To measure the number of random double-color events in images containing more than 400 events (data for Fig. 3D), multiple fluorospot images were combined (by addition) to obtain the requisite event numbers for analysis. Image combinations used red and green channels only, with channels mismatched intentionally to simulate genuine randomness of spot locations between channels. Direct addition of N multiple images extended the maximum possible intensity value in an image to N*255. Because no more than 32 images were combined at any one time, all images for this experiment were therefore processed as 16-bit images.
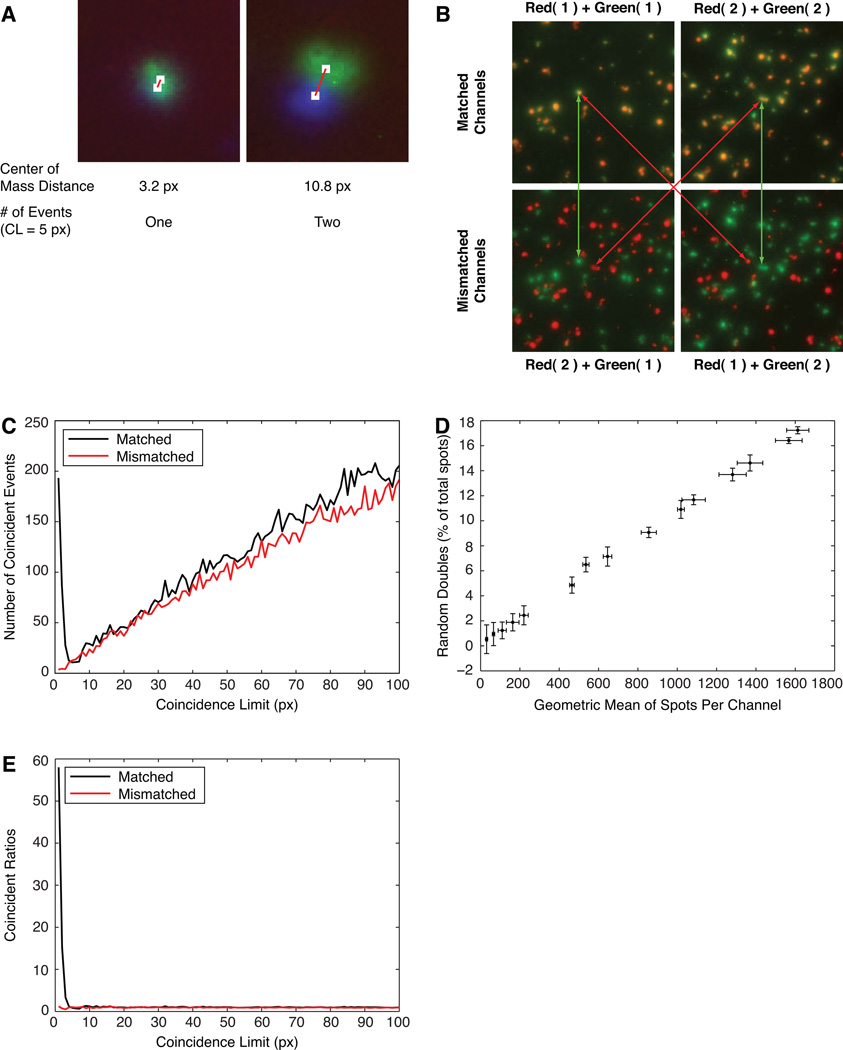
Figure 3. Assignment and analysis of coincidence limits using matched and mismatched images.
Images were used from the experiments shown in Figure 1C (A) or Fig. 1A (B, C, D, E). A) Assignment of a coincidence limit (CL) for two pairs of spots. At CL=5 pixels, the green and blue spots would be considered as one (left) or two (right) events. B) Effect of matching and mismatching the red and green channels of duplicate wells in the two-color MIP-1α and IFNγ assay (most are genuine double-producers). C) The number of apparent double-producers found at different values of the coincidence limit, using the matched and mismatched wells shown in B. D) The frequency of random doubles with increasing numbers of spots per well, estimated from mismatched images. E) The coincidence curves comparing ratios for ‘predicted random’ to actual matched and mismatched images.
Image Manipulations for Background Tolerance
Fluorospot images (192) were divided into 3 categories containing: A: normal image: variable spot number, background variance <0.078, B: low background image: spot count <20, background variance <0.078, C: high background image: spot count <20, background variance >0.235. Intensity values for all images were then multiplied by 0.5 to allow for image addition while constraining intensities to 8 bits. Two sets of 96 RGB images were then constructed from these single-channel fluorospot images: A+B (normal + low background) and A+C (normal + high background). For example, to create an A+B image, one A and one B image were added together and assigned to the red channel, the individual A was assigned to the green channel, and the individual B assigned to blue. This multi-channel assignment strategy enabled the program to find colocalized spots, and determine which spots were conserved, gained or lost in the combined image.
Results
Demonstration of two- and three-color Fluorospot assay using defined cell types
The ability of the two-color Fluorospot assay to reliably detect double- and single-producing T cells was tested by using Th1 cells, producing both IFN-γ and MIP-1α (mostly double-producers), and a mixture of Th1 and Th2 cells producing IFN-γ and IL-4, respectively (single-producers). Visual examination of the IFN-γ and MIP-1α results for Th1 cells shows that most of the green (IFN-γ) or red (MIP-1α) spots were coincident, so that the two-channel image (Fig. 1A, bottom) contained mostly yellow and orange spots indicating that most cells produced both cytokines. In contrast, when a mixture of Th1 and Th2 cells was tested (Fig. 1B) almost all green spots (IFN-γ) were not coincident with red spots (IL-4), which confirmed that these cytokines were produced by different cells. Similar resolution of single- and double-producers has been obtained in two-color assays for mouse IL-2 + IFN-γ; mouse IL-4 + IL-5; mouse IL-2 + IL-17 (data not shown); human IL-2 + IFN-γ [5]; and human IFN-γ + IL-10 [6].
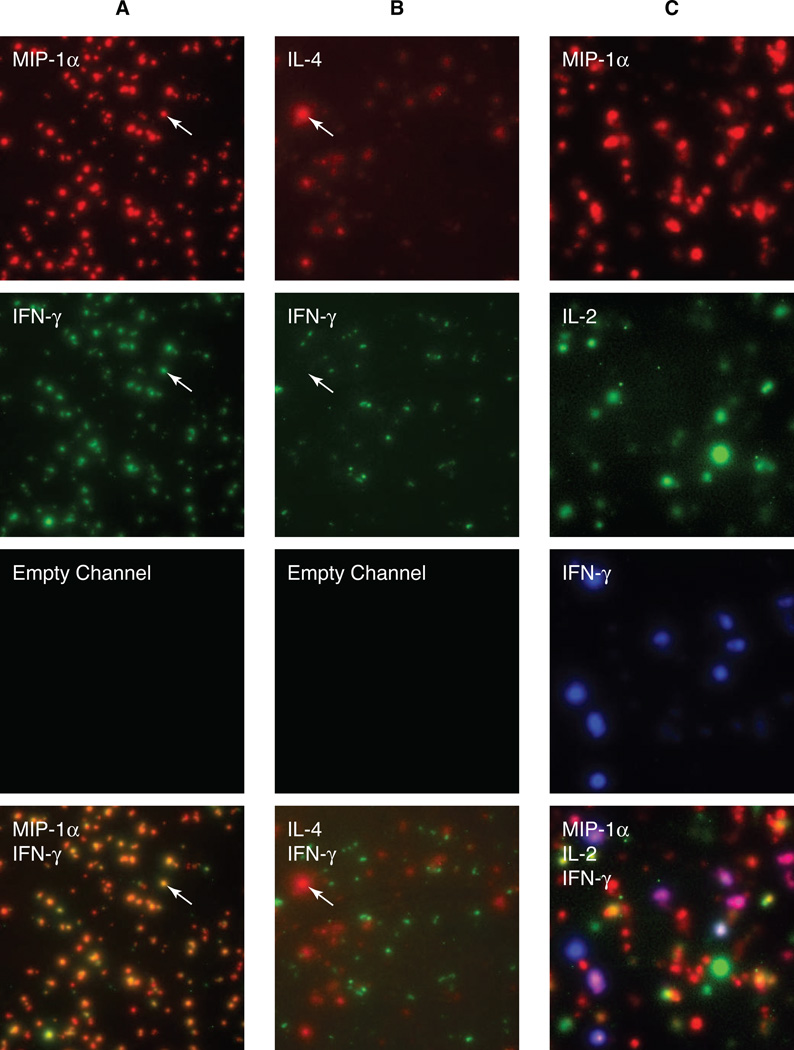
Figure 1. Two- and three-color Fluorospot assays detect various cell phenotypes.
A) 2×103 Th1 cells per well were restimulated in a Fluorospot assay for MIP-1α (red, Cy3) and IFN-γ (green, Cy2). B) 2×103 Th1 and 7×103 Th2 cells per well were restimulated in a Fluorospot assay for IL-4 (red, Cy3) and IFN-γ (green, Cy2). C) CD4 T cells expressing a range of cytokine secretion phenotypes were restimulated in a Fluorospot assay for IL-5 (red, Cy3), IL-2 (green, Cy2) and IFN-γ (blue, Cy5). The top three rows show single-channel images for red, green and blue, and the bottom rows show the composite images.
As T cell cytokine secretion phenotypes are complex [14–16], two-color assays do not fully resolve the differences between phenotypes such as Thpp (IL-2+ IFN-γ- IL-4-), Th1 (IL-2+ IFN-γ+ IL-4-), and Th0 (IL-2+ IFN-γ+ IL-4+). We have therefore established three-color Fluorospot methods for these cytokines and for mouse IL-2, IFN-γ, and MIP1-α. To demonstrate the ability of the three-color Fluorospot assay to identify complex phenotypes, naïve mouse T cells were stimulated with alloantigen in culture conditions (IL-2, IL-12, IL-4, IL-7, and progesterone) that induced differentiation into a mixture of phenotypes, then restimulated in a Fluorospot assay for IL-2, IFN-γ and MIP-1α (Fig. 1C). Most of the seven possible cytokine combinations could be identified in this population, although cells producing IFN-γ without MIP-1α were rare, as expected [13, 15].
Thus two- and three-color Fluorospot assays can effectively identify a range of T cell cytokine secretion phenotypes. However, visual analysis of the results is time consuming; cannot quantify the relative intensities of different spots; and introduces subjective errors in counting and determination of coincident spots. We have therefore developed automated counting and analysis software for Fluorospot assays, using the assays above to evaluate the performance of the software.
Exploraspot Image Analysis System
Single-channel processing
Significant differences between channels can occur due to biological variations (cell movement, cytokine secretion kinetics), or technical variations (filter backgrounds, microscope alignment, convection). Therefore, the Exploraspot program initially processes each channel independently by the same basic routine, but each channel can be customized by adjusting parameters controlling background approximation, thresholding, image smoothing, and artifact exclusion. This strategy means that the program is readily scalable to three or more channels. A three-channel image will be described in the following explanation of Exploraspot processing.
Single-channel spot identification and measurement
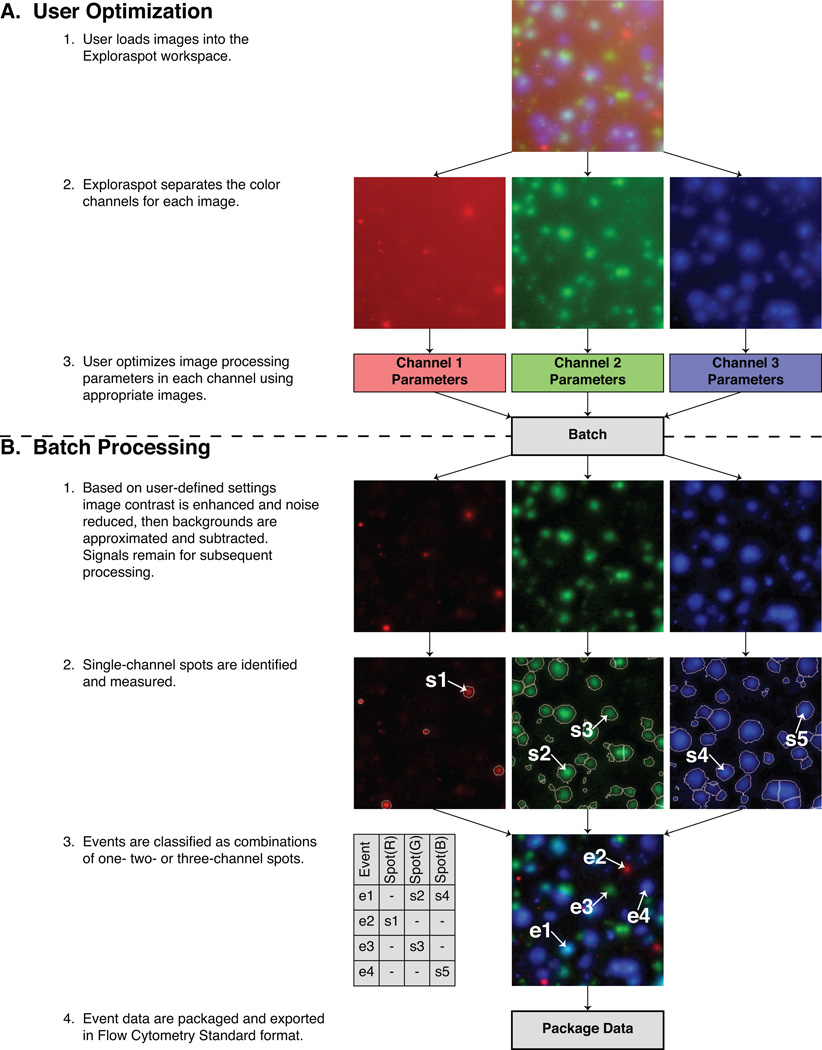
The Exploraspot quantification process begins by loading images into the Exploraspot workspace. The program separates each control image into individual channels, and provides direct visual feedback while the user optimizes the image processing parameters and artifact exclusion gates for each channel (Fig. 2A). The following algorithm (Fig. 2B) is then applied to each channel: 1. Image contrast is enhanced and noise reduced, then the background is approximated and subtracted from the original image. 2. The resulting image is segmented to identify single-channel spots. Exploraspot then measures several morphological parameters for each spot: Area, Center of Mass, Net Intensity, and Circularity. Each (single-color) spot is therefore represented computationally as a structural data element containing a value for each measurement. An example of several Spot data elements is given in Table I. These measurements are used to classify spots based on their morphological characteristics, e.g. determine whether two different-color spots were secreted by the same cell or separate cells.
Figure 2. Exploraspot processing routine for a three-color Fluorospot experiment.
Table I.
Sample Spot Data
| SPOT | CoM(x) | CoM(y) | Channel | Size (px) |
Area (sq.µm) |
Circularity | Net Intensity |
Mean Intensity |
|---|---|---|---|---|---|---|---|---|
| Null | * | * | * | 0 | 0 | 0.00 | 0 | 0.00 |
| s1 | 242.00 | 249.00 | 1 | 405 | 17982 | 87.40 | 5751 | 14.20 |
| s2 | 117.60 | 78.40 | 2 | 453 | 20113 | 83.30 | 13216 | 29.17 |
| s3 | 206.00 | 203.00 | 2 | 276 | 12254 | 82.10 | 3099 | 11.23 |
| s4 | 116.40 | 79.20 | 3 | 567 | 25175 | 85.00 | 16548 | 29.19 |
| s5 | 284.00 | 208.00 | 3 | 777 | 34499 | 82.90 | 33602 | 43.25 |
These data are inherited from Spot partners.
Classification of single- and multi-channel Events
Even though three different cytokines (three different color spots) may be generated by the same individual cell, the perimeters and centers of the spots may vary slightly in their respective channels for the reasons mentioned above. Exploraspot provides tools to both measure and correct for this variation by evaluating coincidence between spot locations in different channels.
Determination of coincidence between Spots in different channels
Coincidence is determined computationally by finding the 2-D distance between the Centers of Mass of any two spots in different channels, and applying a user-defined Coincidence Limit (CL). If the measured distance is less than or equal to the specified Coincidence Limit, then the spots are considered to be coincident (Fig. 3A).
Optimizing Coincidence Limits by Random Overlap Method
Exploraspot provides objective data for assigning the optimum CL for each experiment, and for estimating the frequency of genuine double-producing cells. This method requires replicate wells of a sample that contains a measurable proportion of genuine double-color events. Matched and mismatched images were constructed from two replicate wells, A and B, of a Fluorospot of Th1 cells that produced MIP-1α and IFN-γ (Fig. 3B). Matched images were Red(1)+Green(1) and Red(2)+Green(2); and mismatched images were Red(2)+Green(1) and Red(1)+Green(2). The matched images therefore contained some genuine double-color events, whereas any apparent double-color events in mismatched images represented random overlap rather than genuine double-secretion.
These four images were then analyzed by calculating the frequency of single-channel spots proximal to other spots in different channels over a range of distance intervals (Fig. 3C). As expected, in mismatched images there was a very low frequency of neighbor spots at small distances, and the number increased steadily with distance. By contrast, in wells containing genuine double-color events, there was a much higher frequency of neighbor spots at small distances. At greater distances, the values for the matched images became very similar to the mismatched values. The interpretation of these results is that the mismatched images provide an estimate of the number of apparent double–color events that will appear randomly at each distance, and the excess of events in the matched images represents the number of genuine double-color events. Two values can be obtained from this analysis – first, the distance interval corresponding to the minimum of the frequency plot for matched images provides a reasonable compromise for the Coincidence Limit, ensuring that almost all genuine double-color events are identified while minimizing the number of spurious double-color events included (this value can be extracted even from a single image, although the comparison of matched and mismatched data greatly improves the precision of the measurement). Second, the comparison of matched and mismatched plots provides a sensitive measure of the presence of genuine double-color events. For example, in the experiment from which the images in Figure 1B were derived, comparison of matched (6.09±1.64%) and mismatched (2.13±0.73%) double-color events within the coincidence limit in the Th1 + Th2 mixtures confirms that few if any genuine double-producers were present. This is important because the number of apparent double-color events increases with increasing quantity of input cells (Fig. 3D), so that a substantial number of apparent double-color events may occur in crowded wells.
As the CL is user-assigned, the value can be obtained from the mismatched analysis described above, derived intuitively from inspection of the data, or a standard value can be used based on previous experience. For example, in most of our assays a CL of 5 pixels (i.e. 33.3µm) provides excellent results. The coincidence curves measuring the frequency of apparent doubles for matched and mismatched images can also be predicted from the number of spots, the area of the well, and the CL. Dividing the experimental curves by the predicted curve effectively removes noise from the frequency plot (Fig. 3E), thus simplifying the identification of an optimal Coincidence Limit.
Multichannel Events
After channel-specific image processing and coincidence determination, single-color Spot data elements are organized into higher-level structures called Events. (Fig. 2B, step 3). Coincident Spots in two or more channels are assigned to the same Event, and all remaining noncoincident Spots are also unique Events. Therefore the physical significance of an Event normally corresponds to a single cell secreting one or more cytokines. In addition to the channel-specific parameters (Table I) of the Spots belonging to an Event, each Event also has three common parameters: Event Center of Mass (mean location of constituent Spot Centers of Mass); Identity (composed of one true/false value per channel, indicating presence or absence of a Spot); and Well Location (row and column). The data structure for several sample Events is given in Table II.
Table II.
Sample Event Data
| EVENT | CoM(x) | CoM(y) | Identity Channel 1 |
Identity Channel 2 |
Identity Channel 3 |
Row | Column |
|---|---|---|---|---|---|---|---|
| e1 | 117.00 | 78.80 | Null | s2 | s4 | B | 10 |
| e2 | 242.00 | 249.00 | s1 | Null | Null | B | 10 |
| e3 | 206.00 | 203.00 | Null | s3 | Null | B | 10 |
| e4 | 284.00 | 208.00 | Null | Null | s5 | B | 10 |
Further Analysis of Fluorospot Data
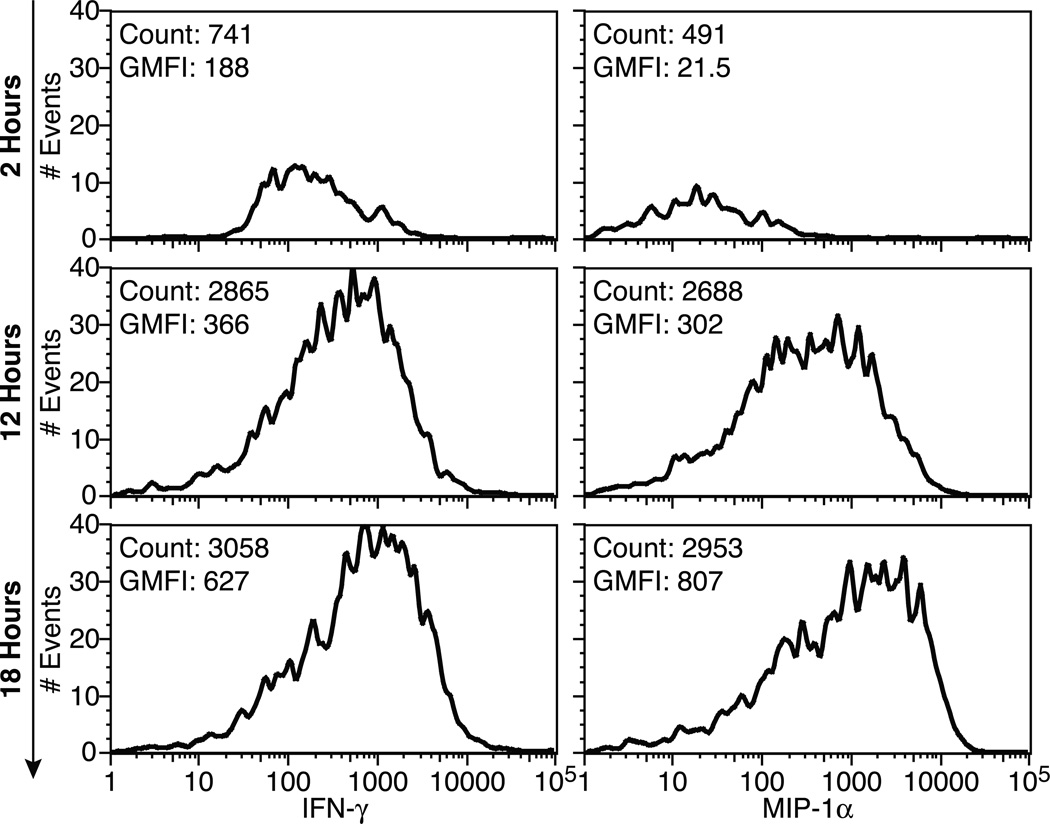
Spot and Event data are exported from Exploraspot in Flow Cytometry Standard (FCS) 3.0 format to facilitate analysis in the Flow Cytometry Analysis programs in which many researchers are proficient. This allows convenient analysis of relationships between all the parameters measured by Exploraspot, within and between samples, as well as providing excellent gating functions, batch processing and graphical presentation. For example, the differences in spot intensities after different assay times (Fig. 4) show that with increasing assay times, more cells become activated, and higher amounts of cytokine accumulate per spot. Flow Cytometry Analysis programs are particularly useful for Fluorospot assays detecting three or more secreted products, which cannot be visualized easily in a simple 2-D plot.
Figure 4. IFNγ and MIP-1α intensity time-course.
Exploraspot intensity histograms of IFN-γ (left) and MIP-1α (right) production over time (converted to FCS format and visualized using FlowJo). Data pooled from 6 replicate wells, each containing 1×103 Th1 cells per well.
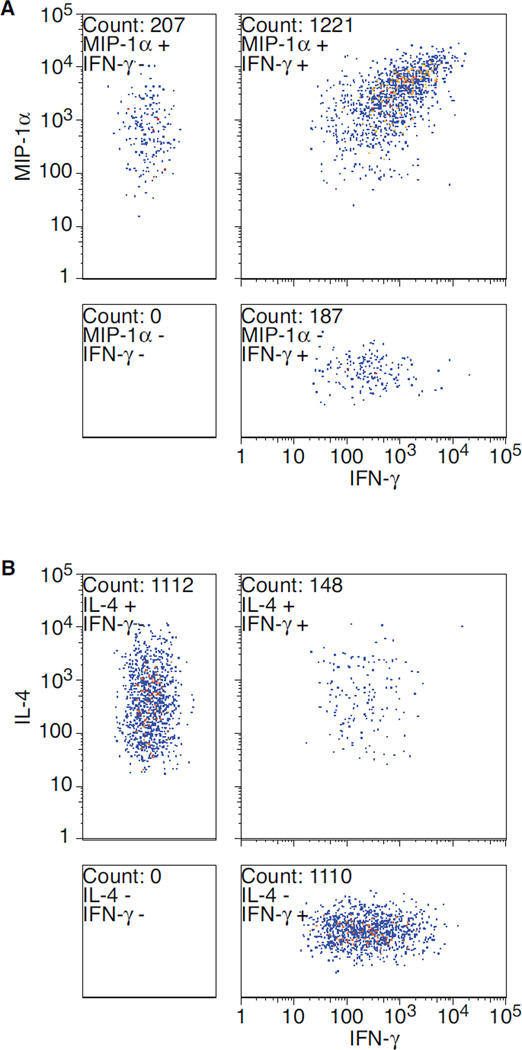
In contrast to Flow Cytometry, however, all Fluorospot Events detected by Exploraspot must contain a Spot in at least one fluorescent channel. A second difference from Flow Cytometry is that if an Event contains no Spot for a particular channel (e.g. Fig. 2B, step 3, Event: e1-red), the data point cannot be plotted numerically on the axis for that channel. Although this data could be plotted by assigning a value of 0 to all null channel parameters, a more accurate representation of the data is shown in Figure 5. The large, square plots show the conventional 2-D plot of Events with detectable spots in both channels, two separate boxes show the singleaxis values of Events that have no values in the other channels (positions on the non-value axes have been randomized to enhance visibility). The fourth box shows events that are negative in both of the channels plotted, and therefore can only contain events in assays with three or more colors.
Figure 5. Quadrant plots of two-color spot intensities for two experiments.
Exploraspot dual intensity plots of A) MIP-1α and IFN-γ production by 8×103 Th1 cells (pooled from 4 replicates), and B) IL-4 and IFN-γ production of a mixture of 8×103 Th1 and 2.8×104 Th2 cells (pooled from 4 replicates).
Robustness of Algorithm
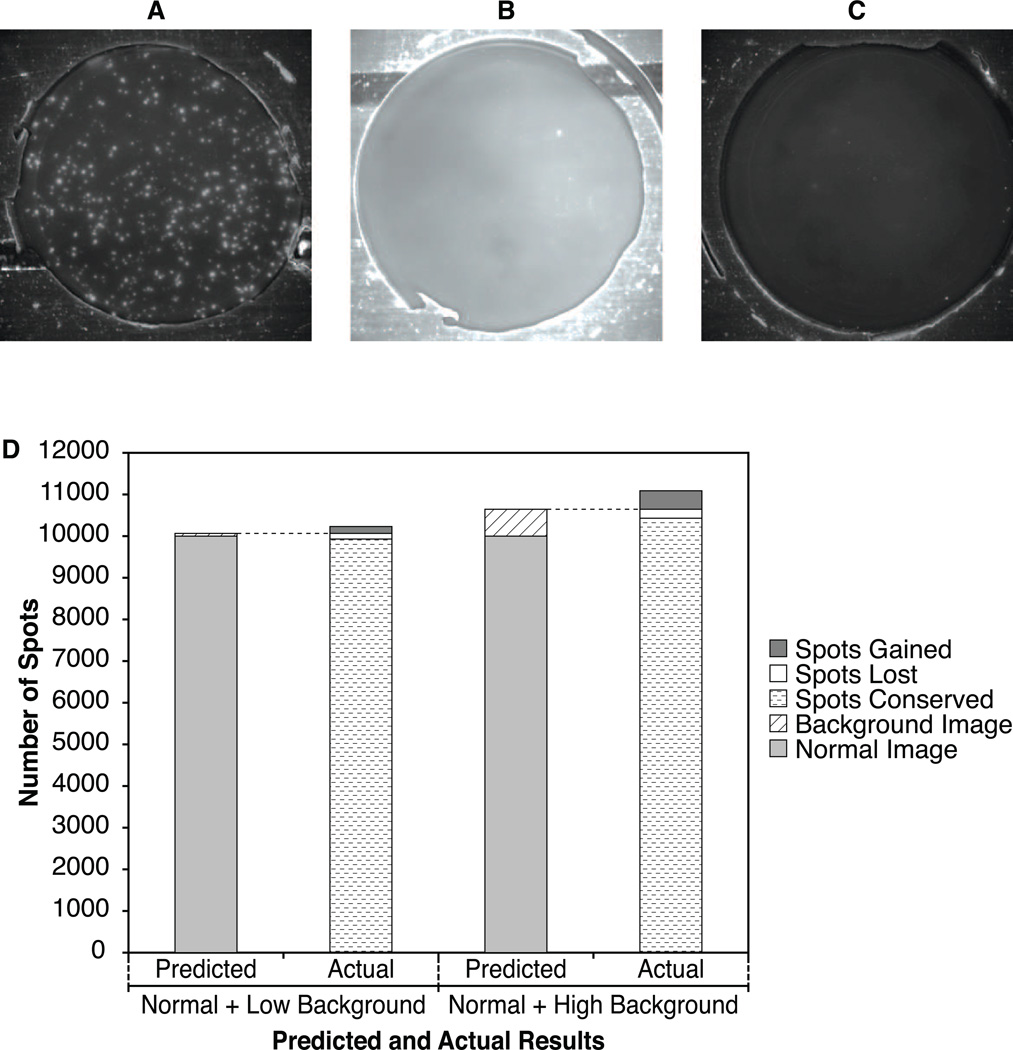
Exploraspot’s methods for approximating background allow reliable detection of spots even in the presence of the non-uniform image backgrounds that occur in some assays due to inherent properties of some cell preparations, antibodies, or operator expertise. To demonstrate this, Th1 IFN-γ Fluorospots were quantified by Exploraspot in the following normal and combined images: normal background (Fig. 6A), normal + additional low background (Fig. 6B), and normal + high background (Fig. 6C). Ideally, the Fluorospot numbers, locations and intensities in the combined image should be the sum of these values in each of the component images, i.e. spots should be neither lost nor gained as a result of the image additions. Figure 6D shows good agreement between predicted and actual Fluorospot numbers for the three sets of images. When low background was added (increasing the overall background variance 3.6-fold ± 1.2), the locations of the spots were highly conserved (98.65%) and spot intensities also correlated well (r=0.9828). Even when high backgrounds were added (increasing the overall background variance 12.3-fold ± 4.0), the locations and intensities of the spots were still well-conserved (97.99% and r=0.9755, respectively). Thus due to the spot-finding algorithm’s tolerance of background variability, there were minimal effects of the noise contributed by adding two images.
Figure 6. Examples of Normal, Low, and High Background.
A) Normal image: Image data from a Th1 IFNγ Fluorospot plate. B) Low Background image: Image data from a negative control well from same Th1 IFN-γ Fluorospot plate. C) High Background image: Image data from a separate Th1 IFNγ experiment with high background and low spot count. For display purposes, all three original images have been enhanced by equal brightness adjustments, but all calculations were performed on the original images. D) Fidelity of spot-finding in the presence of increased background.
Comparison to Other Methods
Although the Fluorospot, Elispot and intracellular cytokine staining (ICS) methods should enumerate all cytokine-secreting cells, there are significant differences between these methods. Spot assays normally detect cytokine secreted over a longer time period than intracellular staining, so heterogeneity in the time of cytokine synthesis may alter results obtained by intracellular staining. Also, the amount of cytokine per cell varies over a wide range, and so differences in sensitivity of different antibodies in each assay may change the detection threshold. To compare Fluorospot versus ICS methods, human PBMC were purified and stimulated with inactivated influenza X31 antigen, then production of IFNγ and IL2 was measured using both Fluorospot and ICS methods [Table III]. The overall pattern of the response was similar between the two methods, although the Fluorospot detected substantially higher numbers of cytokine-secreting cells (particularly for IL-2), and the signal:noise ratio was slightly better in the Fluorospot. The ratio of detection of IL-2 and IFNγ in the Fluorospot could be influenced by the ratio of the amounts of coating antibodies (data not shown) and similarly, the sensitivity of ICS could be altered by changes in the duration of stimulation or reagent concentrations. Note that results are expressed as cytokine-secreting cells detected per million input PBMC, to allow direct comparison of the two methods. Unavoidable cell losses during tissue culture, staining and flow cytometric analysis substantially reduce the ICS frequencies.
Table III.
Comparison of cytokine-secreting cells measured by intracellular staining (ICS) and Fluorospot.
| IL2+/IFNγ− | IL2+/IFNγ+ | IL2−/IFNγ+ | ||
|---|---|---|---|---|
| Fluorospot | No Ag | 13.3 ± 8.1 | 4.0 ± 2.0 | 4.0 ± 2.0 |
| Influenza | 133.3 ± 28.4 | 90.7 ± 14.0 | 114.7 ± 15.1 | |
| ICS | No Ag | 3.0 ± 1.3 | 0.8 ± 0.6 | 3.7 ± 0.8 |
| Influenza | 7.8 ± 5.0 | 22.2 ± 8.5 | 26.5 ± 11.0 |
Human PBMC were stimulated with influenza antigen or left unstimulated, and cytokine secretion or synthesis were measured in triplicate by Fluorospot or intracellular cytokine staining, respectively. Single and double-producing cells were measured by analysis of ICS results in FlowJo, gating on CD4+ T cells (there was no CD8 T cell response to the influenza vaccine antigen), and Fluorospot results were analyzed in Exploraspot. The table shows cytokine-secreting cell frequencies per million PBMC added to cultures. Similar results were obtained with a second subject.
Discussion
The ability of the Fluorospot assay to detect cytokine secretion is comparable to other methods, but the complexity of the data obtained from each channel in multicolor Fluorospot assays requires careful processing. As in Elispot assays, accurate and objective spot identification and counting are essential, and additional information can be obtained from spot morphology, intensity and size.
However, significant differences between channels can occur in a multicolor image for a number of reasons. Preexisting plate conditions can cause variation in background noise and intensity, or affect the inclusion and frequency of artifacts. Different antibody/cytokine combinations can influence spot morphology, and cell movement, cytokine secretion kinetics and convection can affect spot frequencies, locations and morphologies. Directional versus omnidirectional cytokine secretion [17, 18] should not significantly affect the image locations, as a lymphocyte is about the size of one pixel. While background variations should be minimized by pre-screening plates for high-quality lots, other sources of variation between channels are difficult to control, and may in fact be informative (e.g. regarding cell migration). Thus for multicolor Fluorospot analysis, it is critical to independently locate and quantify spots separately in each channel, rather than by combining channels prior to defining spots. For example, if a combined spot perimeter was used, a large spot in one channel would cause a large unnecessary background component to be added to a small spot at the same location in another channel. The Exploraspot software provides the necessary independent multichannel analysis, and also includes detailed measurement of spot morphology parameters for analysis of both Elispots and Fluorospots.
A critical additional feature of Fluorospot analysis is to decide whether spots in different colors at almost the same location are independent or coincident events. Ideally, a cell secreting three cytokines would produce similar spots in all three channels, and all three spots would have the same center of mass. In practice, spots in different channels can have different morphologies and thus different Centers of Mass due to the differences described above. To determine whether spots in two channels were produced by a single cell, or two independent cells, we considered two strategies: evaluating the total area of overlap between spots in different channels, or measuring the distance between the centers of mass of the two spots. The overlap between spots will be strongly dependent on the size of the two spots, which is in turn dependent on the amount of protein secreted by the cells, and also on some non-biologically-significant parameters such as the amount and affinity of coating antibody in the Fluorospot assay. Neither of these parameters is linked to the probability that any two spots in different channels are produced by the same cell. In contrast, the distance between the centers of mass of two spots produced by the same cell should be dependent on the distance migrated by the cell during the secretion of two cytokines that are synthesized with different kinetics. This parameter should also be affected uniformly by small errors in stage alignment between different color images. Therefore the proximity analysis in Exploraspot uses the distance between the centers of mass to determine whether spots in different channels are likely to have been produced by a single cell.
Thus Exploraspot has several strong multicolor image analysis tools, many of which are not available in other single- and multi-channel Elispot analysis programs, e.g. AID (www.elispot.com). The independent determination of spots in each channel, and subsequent proximity analysis, have several advantages as described above. Exploraspot can analyze several channels, limited only by computer processing power (e.g. ten channels on a typical desktop computer). The ability to export data as .fcs files, in addition to text and Excel files, allows convenient analysis of multiparameter data by existing powerful flow cytometry programs. Exploraspot runs on Mac or Windows computers, requiring only MATLAB for operation. The random coincidence measurement tools in Exploraspot also provide strong methods for determining the significance of two-color spots in crowded images.
As the number of events in a well increases, the measured number in a single channel may be underestimated because of overlap between spots (in the same channel) due to random juxtaposition of cells. Larger numbers of spots, larger sizes of individual spots, or irregular-shaped spots will all contribute to underestimation of events in a single channel. By combining images from replicate wells in a single channel, the counting function of Exploraspot provides a practical way to estimate the magnitude of the loss due to overcrowding. If the number of spots detected in the combined image is similar to the sum of the spots detected in the two component images, then it is likely that the spots in the original images are accurately counted with little loss due to overcrowding. This is a stringent test because the number of spots in the combined image is doubled, thus overestimating potential overlap. If significant numbers of spots are lost due to random overlap, the results need to be further analyzed, or preferably a lower dilution of cells should be used in the assay.
For the images processed so far, our present resolution (1500 pixels/cm, 8 bits per channel) is an appropriate compromise between finding small spots, and keeping files small enough for rapid processing and efficient data storage. It is also possible to magnify small regions within a well, and reconstruct a high resolution image when necessary.
The ability of Exploraspot to identify objects in multicolor fluorescent Elispot images also provides a method to quantify objects in other images. For example, Exploraspot has been used to quantify fluorescent cells in histological sections, and to determine the relative staining by multiple fluorescent antibodies [19]. Further applications may emerge, as Exploraspot can potentially quantify the numbers and properties of multicolor objects in any image, provided the objects can be defined as discrete events.
Acknowledgments
Supported by NIH grant AI54593 and NIH contract N01-AI-50020
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Quast S, Zhang W, Shive C, Kovalovski D, Ott PA, Herzog BA, Boehm BO, Tary-Lehmann M, Karulin AY, Lehmann PV. IL-2 absorption affects IFN-gamma and IL-5, but not IL-4 producing memory T cells in double color cytokine ELISPOT assays. Cell. Immunol. 2005;237(1):28–36. doi: 10.1016/j.cellimm.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Snyder JE, Bowers WJ, Livingstone AM, Lee FE, Federoff HJ, Mosmann TR. Measuring the frequency of mouse and human cytotoxic T cells by the Lysispot assay: independent regulation of cytokine secretion and short-term killing. Nat. Med. 2003;9(2):231–235. doi: 10.1038/nm821. [DOI] [PubMed] [Google Scholar]
- 3.Karulin AY, Hesse MD, Tary-Lehmann M, Lehmann PV. Single-cytokine-producing CD4 memory cells predominate in type 1 and type 2 immunity. J. Immunol. 2000;164(4):1862–1872. doi: 10.4049/jimmunol.164.4.1862. [DOI] [PubMed] [Google Scholar]
- 4.Gazagne A, Claret E, Wijdenes J, Yssel H, Bousquet F, Levy E, Vielh P, Scotte F, Goupil TL, Fridman WH, Tartour E. A Fluorospot assay to detect single T lymphocytes simultaneously producing multiple cytokines. J. Immunol. Meth. 2003;283(1–2):91–98. doi: 10.1016/j.jim.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Divekar AA, Zaiss DM, Lee FE, Liu D, Topham DJ, Sijts AJ, Mosmann TR. Protein vaccines induce uncommitted IL-2-secreting human and mouse CD4 T cells, whereas infections induce more IFN-gamma-secreting cells. J. Immunol. 2006;176(3):1465–1473. doi: 10.4049/jimmunol.176.3.1465. [DOI] [PubMed] [Google Scholar]
- 6.Lee FE, Walsh EE, Falsey AR, Liu N, Liu D, Divekar A, Snyder-Cappione JE, Mosmann TR. The balance between influenza- and RSV-specific CD4 T cells secreting IL-10 or IFNgamma in young and healthy-elderly subjects. Mech. Ageing Dev. 2005;126(11):1223–1229. doi: 10.1016/j.mad.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986;136(7):2348–2357. [PubMed] [Google Scholar]
- 8.Sad S, Mosmann TR. Single IL-2-secreting precursor CD4 T cell can develop into either Th1 or Th2 cytokine secretion phenotype. J. Immunol. 1994;153(8):3514–3522. [PubMed] [Google Scholar]
- 9.Firestein GS, Roeder WD, Laxer JA, Townsend KS, Weaver CT, Hom JT, Linton J, Torbett BE, Glasebrook AL. A new murine CD4+ T cell subset with an unrestricted cytokine profile. J. Immunol. 1989;143(2):518–525. [PubMed] [Google Scholar]
- 10.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 2005;201(2):233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groux H, O'Garra A, Bigler M, Rouleau M, Antonenko S, de Vries JE, Roncarolo MG. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389(6652):737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 12.Ohara J, Paul WE. Production of a monoclonal antibody to and molecular characterization of B-cell stimulatory factor-1. Nature. 1985;315(6017):333–336. doi: 10.1038/315333a0. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, Mosmann T. Synthesis of several chemokines but few cytokines by primed uncommitted precursor CD4 T cells suggests that these cells recruit other immune cells without exerting direct effector functions. Eur. J. Immunol. 2004;34(6):1617–1626. doi: 10.1002/eji.200424939. [DOI] [PubMed] [Google Scholar]
- 14.Mosmann TR, Sad S. The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol. Today. 1996;17(3):138–146. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 15.De Rosa SC, Lu FX, Yu J, Perfetto SP, Falloon J, Moser S, Evans TG, Koup R, Miller CJ, Roederer M. Vaccination in humans generates broad T cell cytokine responses. J. Immunol. 2004;173(9):5372–5380. doi: 10.4049/jimmunol.173.9.5372. [DOI] [PubMed] [Google Scholar]
- 16.Langrish CL, McKenzie BS, Wilson NJ, de Waal Malefyt R, Kastelein RA, Cua DJ. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol. Rev. 2004;202:96–105. doi: 10.1111/j.0105-2896.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 17.Kupfer A, Mosmann TR, Kupfer H. Polarized expression of cytokines in cell conjugates of helper T cells and splenic B cells. Proc. Natl. Acad. Sci. USA. 1991;88(3):775–779. doi: 10.1073/pnas.88.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huse M, Lillemeier BF, Kuhns MS, Chen DS, Davis MM. T cells use two directionally distinct pathways for cytokine secretion. Nat. Immunol. 2006;7(3):247–255. doi: 10.1038/ni1304. [DOI] [PubMed] [Google Scholar]
- 19.Richter M, Ray SJ, Chapman TJ, Austin SJ, Rebhahn J, Mosmann TR, Gardner H, Kotelianski V, deFougerolles AR, Topham DJ. Collagen distribution and expression of collagen-binding alpha1beta1 (VLA-1) and alpha2beta1 (VLA-2) integrins on CD4 and CD8 T cells during influenza infection. J. Immunol. 2007;178(7):4506–4516. doi: 10.4049/jimmunol.178.7.4506. [DOI] [PubMed] [Google Scholar]